Abstract
Replication of hepatitis C virus (HCV) RNA occurs on intracellular membranes, and the replication complex (RC) contains viral RNA, nonstructural proteins, and cellular cofactors. We previously demonstrated that cyclophilin A (CyPA) is an essential cofactor for HCV infection and the intracellular target of cyclosporine's anti-HCV effect. Here we investigate the mechanism by which CyPA facilitates HCV replication. Cyclosporine treatment specifically blocked the incorporation of NS5B into the RC without affecting either the total protein level or the membrane association of the protein. Other nonstructural proteins or viral RNAs in the RC were not affected. NS5B from the cyclosporine-resistant replicon was resistant to this disruption of RC incorporation. We also isolated membrane fractions from both naïve and HCV-positive cells and found that CyPA is recruited into membrane fractions in HCV-replicating cells via an interaction with RC-associated NS5B, which is sensitive to cyclosporine treatment. Finally, we introduced point mutations in the prolyl-peptidyl isomerase (PPIase) motif of CyPA and demonstrated a critical role of this motif in HCV replication in cDNA rescue experiments. We propose a model in which the incorporation of the HCV polymerase into the RC depends on its interaction with a cellular chaperone protein and in which cyclosporine inhibits HCV replication by blocking this critical interaction and the PPIase activity of CyPA. Our results provide a mechanism of action for the cyclosporine-mediated inhibition of HCV and identify a critical role of CyPA's PPIase activity in the proper assembly and function of the HCV RC.
Hepatitis C virus (HCV), of the family Flaviviridae, is an enveloped, positive-stranded RNA virus. Spread mostly by blood-borne transmission, HCV infects more than 170 million people worldwide. The viral genome is composed of a single open reading frame (ORF) plus 5′- and 3′-nontranslated regions. The ORF encodes a large polyprotein that is cleaved by cellular and viral proteases into 10 viral proteins. The structural proteins, including the capsid protein (core), two glycoproteins (E1 and E2), and a small ion channel protein (p7), reside in the N-terminal half of the polyprotein. The rest of the ORF encodes six nonstructural (NS) proteins: NS2, NS3, NS4A, NS4B, NS5A, and NS5B. NS3 through NS5B assemble into a replication complex (RC) and are necessary and sufficient for HCV RNA replication in cell culture (8, 42). NS3 is a multifunctional protein with both a serine protease and an RNA helicase activity. The protease activity is responsible for cleavage at the NS3-NS4A, NS4A-NS4B, NS4B-NS5A, and NS5A-NS5B junctions (5), and the helicase activity is probably required to unwind the double-stranded RNA intermediates formed during replication (38). NS4A serves as an essential cofactor for the NS3 protease and anchors the NS3 protein to intracellular membranes (25, 36, 39). NS4B induces the formation of a “membranous web” that is probably the site of HCV replication (16). It also contains a GTP-binding motif that is required for replication (17). The web is derived from the endoplasmic reticulum (ER) compartment, although proteins of early-endosome origin have also been found to locate to the web (62). NS5A is a phosphoprotein and an integral component of the viral RC. The precise function of NS5A in replication is still unknown but appears to be regulated by phosphorylation and its interaction with several cellular proteins (19, 22, 24, 51, 52, 59, 63, 67). In addition, it may be involved in the transition from replication and particle formation (4, 45, 64). NS5B is the RNA-dependent RNA polymerase that is responsible for copying the RNA genome of the virus during replication. Several cellular cofactors interact with NS5B and modulate its activity in the context of the viral RC (22, 24, 35, 69, 71).
Positive-stranded RNA viruses alter the intracellular membranes of host cells to form an RC in which RNA replication occurs. Modifications include the proliferation and reorganization of certain cellular membranes (1). HCV forms an RC associated with altered cellular membranes (16, 23), and crude RCs (CRCs) that maintain the replicase activity in vitro can be isolated by membrane sedimentation or flotation techniques (2, 3, 18, 27, 37).
Cyclosporine is a widely used immunosuppressive and anti-inflammatory drug for organ transplant patients. It functions by forming an inhibitory complex with cyclophilins (CyPs) that inhibits the phosphatase activity of calcineurin, which is important for T-cell activation. In recent years, cyclosporine and its derivatives have been shown to be highly effective in suppressing HCV replication in vitro (44, 49, 53, 68) and in vivo (30). The mechanism of this inhibition is independent of its immunosuppressive function and distinct from that of interferon (IFN) (44, 53, 56, 68).
We recently showed that HCV infection in vitro is inhibited when CyPA, a major intracellular target of cyclosporine, is downregulated by RNA interference, and mutations in NS5B that confer cyclosporine-resistant binding to CyPA contribute to the cyclosporine resistance of the replicons harboring these mutations (56, 71). Here we report that CyPA is recruited into the HCV RC together with NS5B in HCV replicon or in HCV-infected cells. Cyclosporine disrupts the association between RC-incorporated NS5B and CyPA and results in an exclusion of the polymerase from the viral RC. We also show that the prolyl-peptidyl isomerase (PPIase) motif of CyPA is essential for HCV replication.
MATERIALS AND METHODS
Cell lines and antibodies.
The genotype 1b-derived GS5 and RS1-2 replicon cell lines were described previously (56). Huh-7.5 cells were provided by Charles Rice and Apath, LLC. Anti-calnexin polyclonal and anti-Grp94 monoclonal antibodies were purchased from Stressgen Bioreagents (Ann Arbor, MI), anti-CyPA antibody was purchased from Biomol International (Plymouth, PA), anti-NS5A and anti-NS5B antibodies were purchased from Virogen (Boston, MA), and anti-NS3 polyclonal antibody was a gift from Guangxiang Luo (University of Kentucky). Monoclonal antibodies specific for isolate JFH-1 were produced in-house.
PK and S7 nuclease digestion.
Replicon, infected cells, or NS5B-transfected cells were permeabilized with 50 μg/ml of digitonin in buffer B (20 mM HEPES-KOH [pH 7.7], 110 mM KOAc, 2 mM MgOAc2, 1 mM EGTA) for 5 min at 25°C and incubated with various concentrations of proteinase K (PK) (Qiagen, Valencia, CA) in buffer B for 5 min at 37°C. The PK-treated cells were covered with immunoprecipitation buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5% NP-40) and incubated on ice for 30 min, after which the resulting lysate was centrifuged at 16,000 × g for 15 min. The supernatant was collected. S7 nuclease digestions of cell lysate and CRC were performed as previously described, with 0.8 U/μl of nuclease (55).
Trypsin digestion of cell lysates.
GS5 cells (2 × 106 cells) were seeded into T-25 flasks and treated with 4 μg/ml cyclosporine for 22 h before being lysed in 1 ml of immunoprecipitation buffer on ice for 30 min. Cell lysates were subjected to centrifugation at 13,500 rpm for 15 min at 4°C. Forty microliters of the supernatant was diluted fivefold in trypsin digestion buffer (10 mM Tris-HCl, 10 mM potassium acetate, 10 mM magnesium acetate, and 1 mM dithiothreitol [pH 7.6]) and digested with trypsin at room temperature for 30 min. The digestion was stopped by adding 1 μl of 200 μM of PMSF and 40 μl of 2× sodium dodecyl sulfate (SDS) loading buffer to the mixture, followed by boiling. The digested samples were subjected to 8% SDS-polyacrylamide gel electrophoresis (PAGE) and detected by anti-NS5B and anti-CyPA antibodies on Western blots.
Isolation and PK digestion of the CRCs.
Approximately 1 × 108 GS5 cells were scraped into 2 ml of phosphate-buffered saline (PBS) and collected by centrifugation. The cell pellet was then resuspended in 4 ml of hypotonic buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM PMSF, and 2 μg of leupeptin) and lysed using a Dounce homogenizer. The nuclei and unbroken cells were removed by centrifugation at 1,000 × g for 10 min at 4°C. The supernatant was then centrifuged at 68,500 × g for 1 h at 4°C. The pellet, which is the membrane fraction (MF) and contains the CRC, was resuspended in 100 μl of incomplete replication buffer (100 mM HEPES [pH 7.4], 50 mM NH4Cl, 7 mM KCl) containing freshly added spermidine (1 mM). To determine the PK sensitivity of NS3, NS5A, and NS5B in the CRC, we diluted 10 μl of the MF preparation with 30 μl of incomplete replication buffer containing 350 μg/ml of PK and incubated the mixture at 37°C for 10 min.
In vitro PK sensitivity assay.
A recombinant form of NS5B lacking the 21 C-terminal amino acids (NS5B Δ21) was kindly provided by Volker Lohmann of the University of Heidelberg. Digestion of NS5B Δ21 (1.8 to 3 μg) was carried out in PK digestion buffer (1 mM CaCl2, 50 mM Tris-HCl [pH 8.0]) for 5 min at 37°C using various concentrations of PK (50, 100, 200, and 250 ng/μl). The assay was performed in the presence of a negative control fusion protein, glutathione S-transferase (GST)-protocadherin β5 ectodomain 2 (EC2), or a GST-CyPA fusion protein. The expression and purification of these proteins via glutathione-Sepharose 4B beads were carried out according to the manufacturer's protocol (GE Healthcare, Piscataway, NJ), and 600 to 1,800 ng of purified fusion proteins was mixed with NS5B Δ21 prior to PK treatment. After digestion, the recombinant proteins were detected either by Coomassie staining or by Western blotting.
In vitro transcription, electroporation, and JFH-1-infected cells.
Full-length JFH-1 cDNA was a gift from Takaji Wakita (National Institute of Infectious Diseases, Japan). In vitro transcription of JFH-1 RNA and electroporation of Huh-7.5 cells were performed as described previously (72). For PK sensitivity analysis and isolation of MFs, cells collected 19 days after electroporation were used.
Quantification of Western blot bands.
The band intensity on the Western blots was quantified using Quantity One software (Bio-Rad, Hercules, CA) with a Bio-Rad gel documentation system. We loaded increasing amounts of cell lysate alongside known amounts of recombinant GST-CyPA onto SDS gels and then probed the membrane after immunoblotting with anti-CyPA to determine the total of amount of CyPA in the Huh-7.5 cells.
Rescue of HCV replication by CyPA cDNA.
The cotransfection of replicon RNA and a short interfering RNA-resistant CyPA cDNA was described previously (71). The F60A and F113A mutations were engineered into the CyPA cDNA using the QuikChange kit (Stratagene, La Jolla, CA).
Recombinant proteins and binding assays.
Wild-type and mutant cDNAs of CyPA were cloned into vector pGEX-2T, and recombinant proteins were purified according to the manufacturer's protocol (GE Healthcare, Piscataway, NJ). Binding reactions were performed as previously described (71) except that replicon lysates were digested with PK before being used in the binding reaction mixture.
Immunofluorescence microscopy.
Con1 replicon Rep1b cells were seeded onto coverslips and treated with 4 μg/ml of cyclosporine for 22 h. The cells were then permeabilized with 50 μg/ml of digitonin, followed by treatment with PK (0.5 μg/ml) for 5 min. The cells were then fixed and stained with anti-NS5B antibody and fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (IgG). Images were taken using a Zeiss LSM 510 confocal microscope.
BrUTP labeling of nascent HCV RNA in PK-digested cells.
Rep1b cells (3 × 105 cells) were seeded onto a coverslip in a 12-well plate and treated with 4 μg/ml of cyclosporine where indicated. At 18.5 h posttreatment, cells were treated with 5 μg/ml of actinomycin D (Sigma, St. Louis, MO) for 30 min. A total of 10 mM BrUTP (Sigma) was then transfected into the cells using Lipofectamine 2000 (Qiagen) according to the manufacturer's instruction. Cells were washed with 1× PBS and permeabilized with 50 μg/ml of digitonin, followed by PK (0.5 μg/ml) treatment at 37°C for 5 min. Cells were then fixed with 4% paraformaldehyde for 15 min and stained with anti-bromodeoxyuridine monoclonal (Exalpha Biologicals, Inc., Watertown, MA) and anti-NS5B polyclonal (Virogen) antibodies for 2 h at room temperature. Cells were washed with PBS-0.2% Triton X-100 three times for 15 min each and stained with fluorescein isothiocyanate-conjugated goat anti-mouse IgG and tetramethyl rhodamine isocyanate-conjugated goat anti-rabbit IgG (Sigma) for 2 h at room temperature. After three more washes with PBS-0.2% Triton X-100, cells were mounted with Vectashield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole) (Vector Laboratories, Inc., Burlingame, CA). Images were captured with an LSM 510 confocal laser scanning microscope (Carl Zeiss) and analyzed with Photoshop CS2 software.
RESULTS
Cyclosporine treatment increased the sensitivity of HCV NS5B to PK.
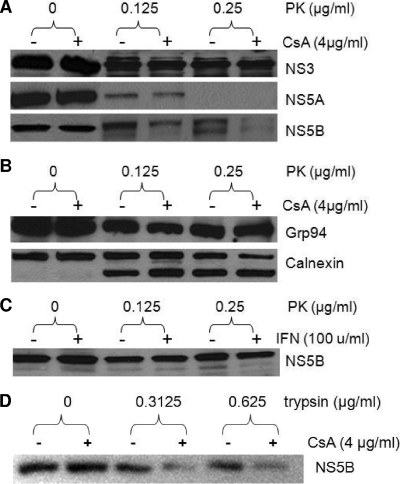
HCV NS proteins are located on the cytosolic side of the intracellular membranes, probably of ER origin. In the replicon cells, only a small fraction of each NS protein is associated with the RC; the majority is not (46, 55). These two fractions of NS proteins could be distinguished by their sensitivity to PK digestion after cell permeabilization by digitonin. The NS proteins incorporated into the RC are protected from PK digestion, much as ER luminal proteins are protected in the same assay. The mechanism of this selective incorporation is not clear but may involve interactions between the HCV replicase and cellular chaperones. We examined the effect of cyclosporine treatment on the PK sensitivities of several cellular and HCV proteins from replicon cells. Although the treatment of GS5 cells with 4 μg/ml cyclosporine for up to 22 h did not affect the total level of proteins, it significantly reduced the amount of PK-resistant NS5B (Fig. 1A). In contrast, the PK sensitivities of NS3 and NS5A, two other NS proteins that are critical for the initiation of HCV RNA synthesis (7), were not affected by the cyclosporine treatment. Overall, NS5A is less resistant to PK digestion in this assay regardless of cyclosporine treatment. The heightened PK sensitivity of NS5A compared to those of other NS proteins was observed previously (46, 55). In addition, the PK sensitivities of two ER chaperone proteins, Grp94 and calnexin, also remained largely unchanged by the treatment (Fig. 1B). A smaller calnexin band appeared with PK treatment and presumably represents the N-terminal portion of the protein, which resides in the ER lumen and is protected from PK digestion (46, 55).
FIG. 1.
Effect of cyclosporine on the PK sensitivity of HCV and cellular proteins. (A) Cyclosporine (CsA) reduces the PK resistance of NS5B but not that of NS3 or NS5A. GS5 cells were treated with cyclosporine and PK where indicated. The cell lysate was then subjected to Western blotting for the detection of HCV proteins. PK treatment sometimes generates a doublet band of Con1 NS5B by SDS-PAGE; both were recognized by the antibody and were affected by cyclosporine treatment. (B) PK treatment or cyclosporine treatment did not compromise the integrity of ER membranes. The samples in A were subjected to the detection of two ER-associated chaperone proteins, Grp94 and calnexin. The antibody against calnexin recognizes the N terminus of the protein, which resides in the ER lumen. (C) IFN-α treatment did not alter the PK sensitivity of the NS5B protein. Results shown are representative of results from three to six independent experiments. (D) Cyclosporine increases the sensitivity of the NS5B protein to trypsin in replicon cells. GS5 cells were treated with cyclosporine for 22 h and lysed. The cell lysate was then digested with trypsin at the indicated concentrations before being subjected to Western blotting and detection with anti-NS5B antibodies. The protein loaded into each lane was obtained from 1 × 105 cells.
To determine whether cyclosporine's effect on the PK sensitivity of NS5B is an indirect result of the general inhibition of viral RNA replication, we treated GS5 cells with IFN-α for 22 h and then performed the PK sensitivity experiment. Neither total nor PK-resistant NS5B was affected by this short-term IFN treatment (Fig. 1C). The activity of IFN was confirmed, as it completely inhibited the replication of the GS5 replicon in a standard 3-day treatment (data not shown).
We reasoned that if the effect of cyclosporine on NS5B's PK sensitivity was indeed the result of a blockage of RC incorporation, then this effect would not be limited to PK. Therefore, we also tested NS5B's sensitivity to another protease, trypsin, in a similar experiment. Again, cyclosporine treatment increased the sensitivity of NS5B to trypsin, suggesting that the increased protease sensitivity by cyclosporine was not specific to a particular protease (Fig. 1D).
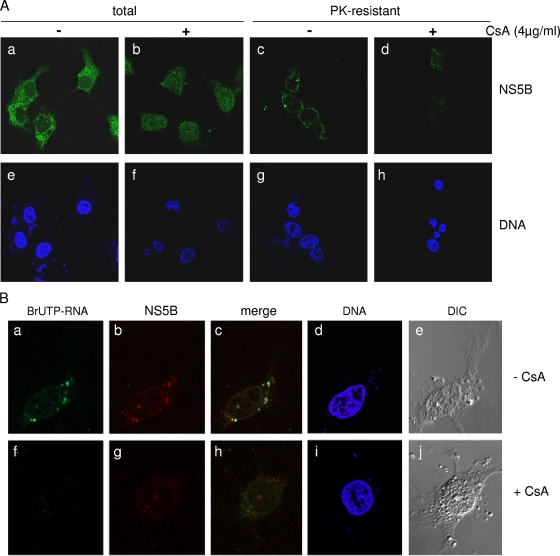
We also carried out immunofluorescence staining experiments to determine if cyclosporine had any effect on the subcellular localization of PK-resistant NS5B. Digestion with PK prior to fixing and staining removed the majority of the cytoplasmic NS5B, leaving a distinct perinuclear and reticular pattern of staining that presumably represents the location of the RC on the ER membrane (Fig. 2Ac). Cyclosporine treatment abolished this concentration in the perinuclear region and significantly reduced the staining intensity of PK-resistant NS5B (Fig. 2Ad), further suggesting that cyclosporine inhibits the incorporation of NS5B into the RC. Somewhat surprisingly, in the absence of PK digestion, cyclosporine also had a significant effect on the subcellular localization of NS5B (diffused throughout the cell versus exclusive cytoplasmic staining) (Fig. 2Aa and b), although the total amount of NS5B did not seem to be significantly affected, as shown in Fig. 1A. To further confirm that the NS5B staining after PK digestion represents RC-associated polymerase, we determined the subcellular location of HCV nascent RNA in PK-digested replicon cells by metabolic labeling with BrUTP in the presence of 5 μg/ml actinomycin D, followed by immunostaining with anti-bromodeoxyuridine monoclonal antibody and confocal microscopy. In the absence of cyclosporine treatment, the BrU signal (Fig. 2Ba) and the PK-resistant NS5B signal (Fig. 2Bb) colocalized to dot-like structures around the nucleus, consistent with data from a previous study of the HCV RC (47). Cyclosporine treatment effectively blocked the synthesis of nascent HCV RNA, eliminating the BrUTP signal; it also dispersed perinuclear punctuate staining of NS5B, consistent with a block in the RC incorporation of NS5B. Differential interference contrast images showed normal morphologies for most of the PK-treated cells, suggesting that the perinuclear staining of NS5B was not a result of cells rounding up.
FIG. 2.
Cyclosporine alters the subcellular location of NS5B and reduces levels of PK-resistant NS5B within replication complexes. (Aa and e) Cytoplasmic and punctuate localization of NS5B in untreated replicon cells. (b and f) Cyclosporine (CsA) treatment results in a diffused staining pattern for total NS5B. (c and g) PK digestion removes nonprotected NS5B, revealing perinuclear and reticular staining. (d and h) Cyclosporine reduces the accumulation of NS5B in the perinuclear area as well as the staining intensity of PK-resistant NS5B. (Ba to e) Colocalization of nascent HCV RNA and NS5B in PK-resistant, dot-like structures around the nucleus. (f to j) Cyclosporine inhibits the synthesis of nascent RNA and the incorporation of NS5B into PK-resistant replication complexes. DNA was stained with DAPI.
Cyclosporine blocked the incorporation of NS5B into the CRC without affecting its membrane association.
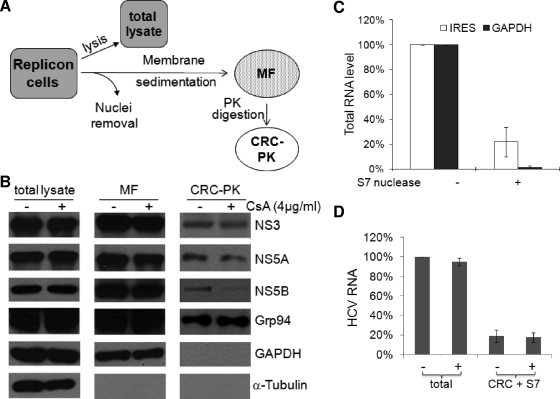
The reduction of NS5B's resistance to PK by cyclosporine treatment suggested that the treatment inhibits the incorporation of NS5B into the HCV RC. At least two possibilities could account for this effect: (i) cyclosporine blocks the proper membrane insertion of NS5B, and (ii) cyclosporine suppresses the inclusion of NS5B into the RC without affecting the membrane association of the protein. We therefore isolated the MF from GS5 cells and further treated it with PK to derive a preparation that contained PK-resistant membrane proteins, which should include the HCV CRC as well as ER luminal proteins. We designated this preparation CRC-PK (Fig. 3A). The levels of NS3, NS5A, NS5B, and several cellular proteins in the MF, CRC-PK, and total lysate were then examined. Cyclosporine treatment did not reduce the NS5B levels in the total lysate or the MF, but it specifically decreased the levels of NS5B in the CRC-PK sample. Levels of NS3, NS5A, and Grp94 were not affected in any of the three samples (Fig. 3B). We also monitored the distributions of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and α-tubulin to examine the quality of these fractions. The former, which has been shown to associate with intercellular membranes (13), copurified with MF but not CRC-PK. On the other hand, α-tubulin was found only in the cytosolic fraction but not in the MF or CRC-PK.
FIG. 3.
Cyclosporine blocks the incorporation of NS5B into the CRC of HCV. (A) Schematic representation of the different subcellular fractions analyzed for cyclosporine sensitivity. (B) CRC incorporation, but not membrane association, of NS5B is reduced by cyclosporine (CsA) treatment. The amounts of NS3 and NS5A in CRC-PK were not affected by cyclosporine treatment. The loading amounts of the different fractions were obtained from 1 × 105 cells for total lysate, from 4 × 106 cells for the MF, and from 1 × 107 cells for CRC-PK. The detection of the different proteins was achieved by the reprobing of either the same membrane or membranes that resulted from gels loaded with identical samples. (C) A fraction of the HCV RNA exhibits resistance to S7 nuclease treatment. RNA samples were extracted from replicon lysates incubated with or without S7 nuclease and subjected to real-time RT-PCR with HCV IRES- or GAPDH-specific primers; the untreated samples were normalized to 100% for HCV IRES and GAPDH, respectively. (D) Cyclosporine treatment did not influence the level of CRC-associated, nuclease-resistant HCV RNA. GS5 cells were treated with cyclosporine for 22 h before being lysed for RNA extraction (total) or CRC isolation and nuclease treatment (CRC + S7). The HCV RNA level was determined by quantitative RT-PCR with HCV IRES primers, and the value for the untreated total sample was normalized to 100%.
Like RC-incorporated proteins, the HCV RNA encapsulated in the RC is protected and exhibits resistance to nuclease treatment (46, 55). When a denucleated replicon lysate was treated with S7 nuclease, 5 to 25% of the HCV RNA, as detected by internal ribosome entry site (IRES)-specific probes, remained resistant to nuclease treatment (Fig. 3C). In contrast, the level of GAPDH RNA was reduced to less than 1% of that in the untreated sample. These results suggest that S7 nuclease treatment effectively removes cytoplasmic mRNA and that a fraction of the HCV RNA, presumably in the RC, is protected. We next determined whether the same conditions (concentration and duration of treatment) of cyclosporine treatment would inhibit the incorporation of HCV RNA into the CRC. We first isolated CRCs from GS5 cells treated or untreated with cyclosporine and digested the CRCs with S7 nuclease before extracting RNA for the detection of HCV RNA with reverse transcription (RT)-PCR. Cyclosporine treatment did not significantly affect either the total HCV RNA or the CRC-associated HCV RNA level (Fig. 3D). Note that in this assay, we detected all HCV RNAs and not just the nascent HCV RNA, as in Fig. 2B. Taken together, these results indicate that cyclosporine specifically inhibits the RC incorporation of NS5B without affecting its membrane association or compromising the integrity of the whole RC.
CyPA was associated with CRC-incorporated HCV replicase in a cyclosporine-sensitive manner.
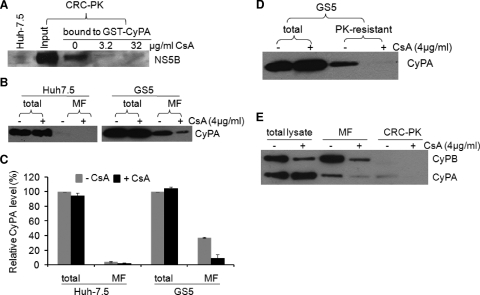
The intracellular target of cyclosporine is the CyP family of proteins. Several CyPs have been implicated in HCV replication (50, 69, 71). We have recently shown that CyPA, the most abundant isoform, is also the main mediator of cyclosporine's inhibitory effect on HCV (71). NS5B from total replicon lysates interacts with GST-CyPA in vitro and in vivo (71). Given the effect of cyclosporine on the RC incorporation of NS5B, we next determined whether CRC-associated NS5B could also interact with CyPA. CRC-PK was isolated from GS5 cells that were lysed before being mixed with GST-CyPA; the mixtures were then incubated with glutathione beads for isolation of interacting proteins. As shown in Fig. 4A, RC-associated NS5B bound to GST-CyPA in this assay. The addition of cyclosporine to the binding reaction mixtures significantly suppressed this interaction. These results indicate that NS5B within the CRC retain its ability to interact with CyPA, although whether the in vivo association between CyPA and NS5B is direct or mediated by another protein is currently unknown.
FIG. 4.
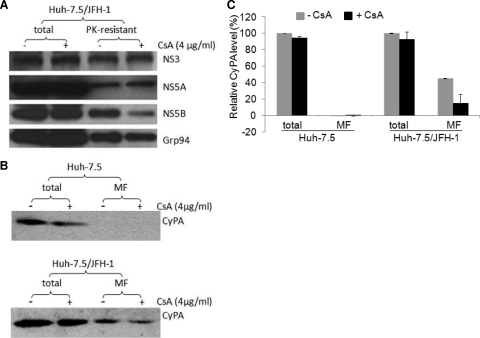
CyPA interacts with CRC-associated NS5B and is recruited to the MF and CRC. (A) CRC-associated NS5B interacts with CyPA in a cyclosporine (CsA)-sensitive manner. The PK-digested CRC was resuspended in lysis buffer and then incubated with GST proteins in a pulldown assay in the absence or in the presence of increasing amounts of cyclosporine. (B) CyPA is recruited to MFs in replicon cells. Huh-7.5 or GS5 cells were treated with 4 μg/ml of cyclosporine for 22 h before being lysed. MFs were then isolated and analyzed for the presence of CyPA by Western blotting. Proteins isolated from 4 × 105 cells were loaded into each lane. (C) The protein bands were quantified using Quantity One software. The total level of CyPA without cyclosporine treatment was set at 100%. Data represent results from three independent experiments. (D) Cyclosporine reduces the PK resistance of CyPA in replicon cells. GS5 cells were treated with cyclosporine and PK where indicated. The resulting cell lysate was then subjected to SDS-PAGE and Western blotting for the detection of CyPA. The PK concentration used was 0.5 μg/ml. (E) CyPA, but not CyPB, is incorporated into the CRC in a cyclosporine-sensitive manner. The fractions depicted in Fig. 3A were analyzed for the presence of both CyPA and CyPB. For total lysate and the MF, proteins isolated from 4 × 105 cells were loaded into each lane, and for the CRC-PK fraction, proteins isolated from 1 × 106 cells were loaded.
CyPA was recruited to intracellular membranes and the viral CRC in cells replicating HCV RNA.
An association of CyPA with PK-resistant NS5B could result in the recruitment of CyPA, presumed to be a cytosolic protein, to intracellular membranes by the HCV replicase. We isolated MFs from equal numbers of naïve Huh-7.5 and GS5 cells and then determined the level of CyPA associated with membranes by Western blotting (Fig. 4B). Approximately 3% of the total CyPA protein was found in the MF isolated from Huh-7.5 cells, whereas close to 40% was found in the MF isolated from GS5 cells (Fig. 4C). More importantly, the level of membrane-associated CyPA was reduced to ∼3% in GS5 cells when the cells were pretreated with cyclosporine for 22 h (Fig. 4C). The low level of CyPA in the MF of Huh-7.5 cells did not respond to cyclosporine treatment, indicating either a constitutive level of membrane association of CyPA or the incomplete removal of this abundant cytosolic protein from the MF. Nevertheless, the level of membrane association of CyPA in replicon cells was more than 10 times higher than that in naïve cells and also sensitive to cyclosporine. This result, together with the disruption of the interaction between CyPA and RC-associated NS5B by cyclosporine, suggested that CyPA is recruited to membranes in replicon cells through its interaction with NS5B.
We next determined whether CyPA is part of the RC. PK-resistant CyPA could be detected in GS5 cells but was eliminated by cyclosporine treatment (Fig. 4D). Furthermore, a fraction of the total CyPA could be detected in the CRC-PK preparation in a cyclosporine-sensitive manner (Fig. 4E, right). The block of the RC incorporation of CyPA appeared to be due to the disruption of its membrane association (Fig. 4E, middle). Strikingly different results were obtained for CyPB, a related CyP protein that constitutively associates with ER membranes. First, cyclosporine similarly reduced CyPB levels in both the total lysate and MF, consistent with data from a previous report showing that cyclosporine treatment releases CyPB from the intracellular membrane and into the extracellular medium (54). Second, CyPB was not detected in the CRC fraction in any significant quantity even in the absence of cyclosporine treatment despite a high level of membrane association (Fig. 4E). These results suggest that CyPA accompanies NS5B into the RC but dissociates from both the RC and the intracellular membranes when its interaction with NS5B is disrupted by cyclosporine.
Cell culture-derived infectious HCV virions (HCVcc) produced from a genotype 2a isolate, JFH-1, efficiently infect Huh-7.5 cells in vitro (12, 40, 66, 72). To determine whether the effect of cyclosporine on the RC incorporation of NS5B applies to HCV-infected cells, we performed PK sensitivity assays using the HCVcc system. Cyclosporine treatment again reduced the level of PK-resistant NS5B without affecting levels of NS3, NS5A, or Grp94 (Fig. 5A). Moreover, CyPA was also recruited to the MF in JFH-1-infected cells in a cyclosporine-sensitive manner (Fig. 5B and C). The effect of cyclosporine on either NS5B or CyPA was less drastic in this system, a result consistent with data from a previous study showing that the replication of HCV genotype 2a was less sensitive to cyclosporine treatment (31).
FIG. 5.
Effect of cyclosporine on PK resistance of NS5B and membrane enrichment of CyPA in HCV-infected cells. (A) PK-resistant NS5B in JFH-1-infected cells exhibits cyclosporine (CsA) sensitivity. Huh-7.5 cells that were infected with HCVcc were treated with or without cyclosporine for 22 h and then processed as described in the legend of Fig. 1. Results for treatment with 0.25 μg/ml PK are shown. (B) The level of membrane association of CyPA is increased in the infected cells in a cyclosporine-sensitive manner. Proteins isolated from 4 × 105 cells were loaded into each lane. (C) Quantitative results for of three independent experiments similar to the one shown above (B). The Western blotting bands were quantified using Quantity One software with a Bio-Rad gel documentation system (see Materials and Methods).
CyPA did not confer PK resistance to NS5B in the absence of RC formation.
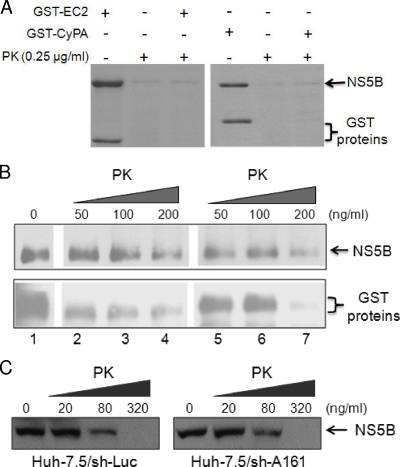
The interaction between CyPA and NS5B raised the possibility that CyPA binding alters the conformation of NS5B, rendering it resistant to PK digestion independent of any RC formation. To address this, we performed an in vitro PK sensitivity assay with NS5B in the presence of either GST-CyPA or a control GST fusion protein. We first used a PK concentration (250 ng/ml) at which most of the recombinant NS5B would be digested. As shown in Fig. 6A, GST-CyPA had no effect on NS5B's PK sensitivity compared with that of a control GST fusion protein, which contained an extracellular domain of the protocadherin β5 protein. We also performed this assay with limiting concentrations of PK. At a low concentration of PK (50 ng/ml), little digestion of either NS5B or the GST recombinant proteins could be seen (Fig. 6B, lanes 2 and 5). In contrast, the addition of 200 ng/ml of PK resulted in a significant removal of both NS5B and the GST proteins (Fig. 6B, lanes 4 and 7). Again, GST-CyPA did not specifically protect NS5B from PK digestion compared to the control protein under the limiting PK concentrations (Fig. 6B). To further examine the effect of CyPA on NS5B's PK sensitivity in vivo without an active RC, we compared the PK sensitivities of transfected NS5B in control and CyPA knockdown Huh-7.5 cells. The knockdown of CyPA had no significant effect on the PK sensitivity of NS5B, again under limiting PK concentrations ranging from 20 to 320 ng/ml (Fig. 6C). Together, these data argue against a role of CyPA in the protection of NS5B against PK digestion in the absence of RC formation.
FIG. 6.
Effect of CyPA on the PK sensitivity of NS5B in the absence of RC formation. (A) Recombinant NS5B Δ21 was readily digested by 250 ng/ml of PK in the presence of the GST-CyPA protein. Approximately 1,800 ng of NS5B and 600 ng of the GST proteins were used. Proteins were detected by Coomassie staining. (B) GST-CyPA does not protect NS5B from PK digestion under limiting PK concentrations. NS5B Δ21 was mixed with both GST fusion proteins (lane 1) or GST-EC2 (lanes 2 to 4) or GST-CyPA (lanes 5 to 7) individually prior to incubation with increasing concentrations of PK (lanes 2 to 4 and 5 to 7). Approximately 3,000 ng of NS5B and 1,800 ng of the GST proteins were used. Proteins were detected with Western blotting. (C) The PK sensitivity of NS5B expressed in the absence of RC formation is independent of CyPA short hairpin (sh) RNA-mediated knockdown. An NS5B-expressing plasmid was transfected into the indicated Huh-7.5 derivative cell lines. Forty-eight hours posttransfection, the cells were treated with digitonin and increasing concentrations of PK as described in Materials and Methods. The CyPA knockdown in the sh-A161 cell line was shown previously (71).
NS5B from a cyclosporine-resistant replicon retained PK resistance and RC incorporation with cyclosporine treatment.
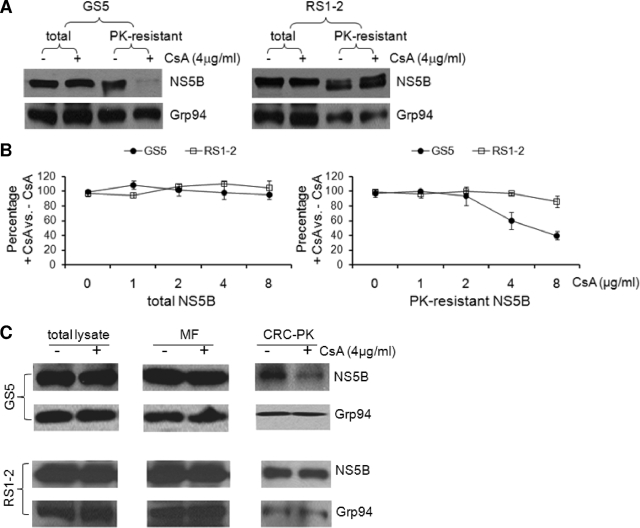
We previously isolated cyclosporine-resistant replicons, and the most resistant clone, RS1-2, conferred an eightfold resistance to cyclosporine in replication assays (56). We performed the PK sensitivity assay with the GS5 (cyclosporine-sensitive) and RS1-2 (cyclosporine-resistant) replicons. In contrast to that of GS5 NS5B, the level of PK-resistant NS5B from the RS1-2 replicon cells was not reduced by treatment with 4 μg/ml of cyclosporine (Fig. 7A, top). The detection of Grp94 confirmed that this difference was not a result of more-efficient digestion by the PK on proteins from the GS5 cells (Fig. 7A, bottom). To examine the difference more quantitatively, we performed the experiments with multiple doses of cyclosporine and quantified the levels of NS5B proteins, both total and the PK-resistant form, by normalizing to the loading control, Grp94. Increasing concentrations of cyclosporine had minimal effects on the levels of total NS5B proteins from either GS5 or RS1-2 cells (Fig. 7B, left), whereas the effect of cyclosporine on the PK sensitivity of GS5 NS5B was dosage dependent. Moreover, RS1-2 NS5B was resistant to cyclosporine at up to 8 μg/ml of the compound (Fig. 7B, right). Consistent with the PK digestion results, NS5B of RS1-2 cells exhibited cyclosporine-resistant RC incorporation (Fig. 7C).
FIG. 7.
RC incorporation of NS5B from a cyclosporine-resistant replicon was resistant to cyclosporine. (A) GS5 and RS1-2 replicon cells were treated in parallel with cyclosporine (CsA) and PK (0.25 μg/ml) as indicated. The cell lysates were then subjected to the detection of NS5B and Grp94. The anti-Grp94 antibody also recognized an additional band of higher-than-expected molecular weight that was completely removed by PK treatment (not shown). Proteins isolated from 4 × 105 cells were loaded into each lane. (B) Dosage-dependent effect of cyclosporine on the PK sensitivity of the GS5, but not the RS1-2, NS5B protein. Experiments described above (A) were performed with 0 to 8 μg/ml of cyclosporine. The protein bands detected by Western blotting were quantified, normalized to Grp94 for loading and PK digestion, and then plotted as the ratio of the protein level in the cyclosporine-treated sample to that in the untreated sample. (C) The CRC incorporation of NS5B in RS1-2 cells was resistant to cyclosporine. Isolation of the MF and digestion of the CRC were performed with RS1-2 cell lysates as described in the legend of Fig. 3. The loading amounts of the different fractions were obtained from 1 × 106 cells for total lysate and the MF and from 2.5 × 106 cells for CRC-PK.
Critical role of the PPIase motif of CyPA for HCV replication.
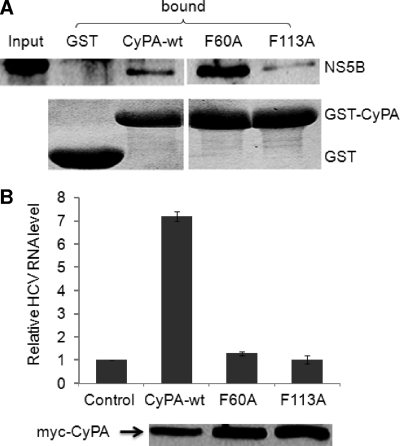
The PPIase activity of CyPA catalyzes the cis-trans isomerization of peptide bonds connecting proline residues. This isomerization process is often the rate-limiting step in protein folding and could be important for the proper RC incorporation of NS5B. To probe the role of PPIase activity in HCV replication, we generated point mutations in the active site of CyPA that abolished its PPIase activity (41, 73). The mutant CyPA proteins were then tested for their abilities to bind to NS5B and to rescue HCV replication in a CyPA knockdown cell line. In GST pulldown assays, the mutant CyPA protein interacted with NS5B to various degrees. For example, the F60A mutant pulled down more NS5B from the lysate than did the wild type, whereas the F113A mutant was slightly impaired in NS5B binding (Fig. 8A). In contrast, mutant cDNAs all behaved essentially like the negative control in the rescue experiment where a cDNA of CyPA was cotransfected into CyPA knockdown cells with a replicon RNA. Without any CyPA cDNA, HCV replication in the CyPA knockdown cells was reduced the 2 to 3% of that in the short hairpin RNA control cells (71). When a wild-type cDNA of CyPA was cotransfected with replicon RNA, however, the level of replication was increased more than sevenfold to result in a partial rescue of replication (71) (Fig. 8B). Neither mutant was capable of rescuing HCV replication to any appreciable degree (Fig. 8B). Western blotting showed comparable expression levels for all the mutants. These data indicate that the PPIase activity of CyPA is essential for HCV replication.
FIG. 8.
The PPIase motif of CyPA is essential for HCV replication. (A) CyPA protein that bears mutations in the active site of PPIase retained binding to NS5B. GST or GST fusion proteins were incubated with GS5 lysate in a GST pulldown assay. Bound NS5B was detected by Western blotting (top), and the recombinant proteins were detected by Coomassie staining (bottom). (B) The PPIase mutants failed to rescue HCV replication in CyPA knockdown cells. In vitro-transcribed Rep1b RNA was coelectroporated into Huh-7.5/sh-A161 cells with myc-tagged CyPA expression plasmids (71). RNA was extracted at 7 h and 4 days after electroporation for analysis of HCV IRES and GAPDH RNA levels. After normalization to GAPDH levels, the ratio of the day 4 HCV RNA levels to the 7-h HCV RNA levels was calculated and plotted. The RNA replication level of the vector control sample was set at 1. wt, wild type.
DISCUSSION
HCV NS proteins copurify with intracellular membranes. Although membrane association is important for the function of NS proteins in replication (48), it is not sufficient for their incorporation into the RC. Less than 5% of each NS protein is incorporated into the RC and exhibits resistance to PK digestion (46, 55). On the basis of these results, a model of the relationship between the intracellular membrane and the HCV RC has been proposed (55). In this model, invagination of the ER membrane forms vesicular membrane structures that house the HCV NS proteins, viral RNA, and host cofactors, all active components of the functional RCs. The interior of the RC is accessible to nucleotides but not to macromolecules such as PK or nucleases. Similar vesicular membrane-bound structures (spherules) have been observed in cells harboring the RC of another positive-strand RNA virus, brome mosaic virus (57). Despite this attractive model, little is known about the process by which the HCV NS proteins are selected and assembled into the RC, and specific inhibitors of RC assembly have not been identified. Here we report that cyclosporine interferes with the formation of a functional RC by inhibiting the incorporation of NS5B into the HCV RC in human hepatoma cells harboring either a subgenomic replicon or a full-length infectious clone. Cyclosporine treatment specifically reduced the level of PK-resistant NS5B but not that of NS5A, NS3, or RC-associated HCV RNA. These results suggest that the exclusion of the polymerase from the RC does not disrupt the overall structure of the RC, consistent with data from a previous report showing that the expression of NS4B alone was sufficient to induce the formation of the “membranous web,” the presumed structure of the RC (16).
CyPA is the most abundant member of the CyP subfamily of immunophilins and the intracellular target of cyclosporine for its regulation of immune function and human immunodeficiency virus (HIV) infection (10, 26). Here we show that CyPA interacts with the HCV replicase and is recruited to MFs and into the CRC in a cyclosporine-sensitive manner. These data are most consistent with a model in which CyPA is recruited to the CRC through the CyPA-NS5B interaction, which is also important for the proper inclusion of the viral polymerase in the RC. Cyclosporine, on the other hand, inhibits the membrane recruitment of CyPA and RC incorporation of NS5B by blocking the CyPA-NS5B interaction. Because cyclosporine does not affect the membrane association of NS5B, our results suggest that CyPA is required for a step in the RC assembly pathway downstream of the membrane association of NS5B, which occurs posttranslationally through a C-terminal transmembrane anchor (32). We also report here that a point mutation in the active site of CyPA abolishes the cofactor function of CyPA without reducing NS5B binding, suggesting a critical role of the PPIase activity of CyPA in a replication step after NS5B binding. This novel mode of action is distinct from the mechanism of either calcineurin inhibition or HIV regulation. In calcineurin signaling, cyclosporine inhibits T-cell activation by forming a cyclosporine-CyPA complex that in turn binds to and suppresses the activity of calcineurin (41). Cyclosporine binding, but not the PPIase activity, is required for calcineurin inhibition (73). In HIV-infected cells, cyclosporine regulates viral replication by competitively disrupting an essential interaction between CyPA and the HIV Gag (CA) protein (43, 65), and the F60A and F113A mutations in CyPA significantly decreased this interaction (9). Interestingly, although CyPA was originally identified as being an HIV virion-associated protein (21, 65), more-recent data suggest that it is the CyPA in the target cells, not that in the virion, that is important for HIV infection (28, 60).
Proving definitively that the CyPA-mediated catalysis of the isomerization of a specific peptide bond is functionally relevant for a biological process has been difficult, partly because of the overlap of the active site and the hydrophobic, protein-binding pocket of this single-domain protein (note that cyclosporine can block protein binding, the PPIase function, or both). Our study provides strong evidence for an involvement of the PPIase activity, and not just protein-protein interactions, in HCV replication, because the F60A and F113A mutants maintained NS5B binding. Although the proline substrate of CyPA that is pertinent for HCV replication remains unidentified, our data nevertheless uncover a clever strategy by which a human virus hijacks a cellular enzyme to facilitate the assembly of a macromolecular complex that is essential for viral replication.
Our data suggest that up to 40% of the total CyPA protein could be recruited onto the MFs in HCV replicon cells at least partly by interactions with NS5B. We estimated the total amount of CyPA in an Huh-7.5 cell using semiquantitative Western blotting (with recombinant CyPA used as a standard) to be ∼1.21 × 107 molecules (data not shown). The abundance of the NS5B protein in replicon cells was estimated to be ∼2.2 ×106 copies (200 fg) per cell (55). Therefore, a 1:1 stoichiometry would not make sense for NS5B to be able to recruit 40% of the CyPA to the membrane. Several reasons could account for this apparent discrepancy. First, multiple copies of CyPA could be binding to one NS5B molecule, as the latter contains many proline residues; second, other HCV proteins such as NS5A may contribute to the recruitment when they complex with NS5B, forming an even larger surface for CyPA to bind.
Cyclosporine-resistant HCV replicons have been described, and mutations associated with resistance map to NS5B and NS5A (20, 56). A correlation between enhanced CyPA and RNA binding by mutant NS5B and cyclosporine resistance of the replicon was observed previously (71). We now report that the resistant replicons are less sensitive to cyclosporine's inhibitory effect on NS5B incorporation into the RC as well, lending further support to the notion that the interaction with CyPA is important for the proper function of NS5B. Interestingly, mutations in NS5A alone could also confer resistance to cyclosporine (20), even though we could not detect any effect of cyclosporine on NS5A's incorporation into the RC or any interaction between NS5A and CyPA. Because NS5A interacts with NS5B and serves as functional cofactor for the replicase (15, 58), it is possible that mutations in NS5A modulate its interaction with NS5B in the context of RC formation, thus indirectly conferring resistance to cyclosporine. This hypothesis remains to be tested.
Cyclosporine treatment of HeLa cells has been shown to inhibit the membrane association of CyPB and to result in the secretion of the CyPB-cyclosporine complex (54). We indeed consistently detected a decrease in the intracellular level of CyPB upon cyclosporine treatment in both Huh-7.5 and replicon cells (Fig. 4E and data not shown), but unlike the situation with the membrane association of CyPA, this inhibitory effect of cyclosporine on the CyPB level was not specific to cells harboring an HCV genome. In addition, reducing the CyPB level alone by RNA interference had no significant effect on HCV replication or infection (31, 56, 71). These data argue against a reduction of the intracellular CyPB level as a significant contributor to the effect of cyclosporine on NS5B inclusion in the membrane-associated viral RC.
The requirement of cellular chaperones for viral polymerases to function was reported previously for a variety of other viruses, including both bacteriophages and human viruses (11, 14, 29, 34, 61, 70). The cellular mechanism underlying this requirement differs from one virus to another but is usually related to intracellular trafficking, RC assembly, and RNA binding. For example, the nuclear import of the herpes simplex virus type 1 polymerase depends on the Hsp90 chaperone system, whereas hepatitis B virus reverse transcriptase exhibits a strict dependence on cellular chaperones for ɛ RNA binding and protein-mediated priming (11, 61). Interestingly, Flock House virus, a positive-stranded RNA virus of the family Nodaviridae, requires the heat shock proteins Hsp70 and Hsp90 for RC assembly and efficient polymerase synthesis, respectively (14, 70). For HCV, an inhibitory effect of cyclosporine on template binding was also observed (56, 69). Whether or not RNA binding is a prerequisite for the incorporation of NS5B into the HCV RC is currently not known. Overall, current data suggest that CyPA, a cellular chaperone with peptidylprolyl-cis-trans-isomerase activity, is recruited to MFs by the HCV polymerase to ensure that the membrane-anchored viral RNA-dependent RNA polymerase maintains the optimal conformation for RC incorporation and template binding. A similar recruitment of cellular cofactors to the intracellular membrane sites where viral RCs assemble was reported previously for other positive-stranded RNA viruses such as poliovirus (6, 33). Such a recruiting mechanism would give the RNA viruses that replicate on specialized membrane structures access to cytosolic proteins that are important for RNA replication.
Acknowledgments
This work was supported by the James and Esther King Biomedical Research Program and the American Heart Association.
We thank Takaji Wakita, Charlie Rice, Timothy Tellinghuisen, Volker Lohmann, Guangxiang Luo, and Fanxiu Zhu for reagents; Andre Irsigler for real-time RT-PCR; and Anne B. Thistle for proofreading.
Footnotes
Published ahead of print on 22 April 2009.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 778181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aizaki, H., K. J. Lee, V. M. Sung, H. Ishiko, and M. M. Lai. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324450-461. [DOI] [PubMed] [Google Scholar]
- 3.Ali, N., K. D. Tardif, and A. Siddiqui. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 7612001-12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appel, N., M. Zayas, S. Miller, J. Krijnse-Locker, T. Schaller, P. Friebe, S. Kallis, U. Engel, and R. Bartenschlager. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager, R. 1999. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J. Viral Hepat. 6165-181. [DOI] [PubMed] [Google Scholar]
- 6.Belov, G. A., M. H. Fogg, and E. Ehrenfeld. 2005. Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor. J. Virol. 797207-7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binder, M., D. Quinkert, O. Bochkarova, R. Klein, N. Kezmic, R. Bartenschlager, and V. Lohmann. 2007. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J. Virol. 815270-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 2901972-1974. [DOI] [PubMed] [Google Scholar]
- 9.Braaten, D., H. Ansari, and J. Luban. 1997. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 712107-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 201300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burch, A. D., and S. K. Weller. 2005. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J. Virol. 7910740-10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai, Z., C. Zhang, K. S. Chang, J. Jiang, B. C. Ahn, T. Wakita, T. J. Liang, and G. Luo. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 7913963-13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campanella, M. E., H. Chu, and P. S. Low. 2005. Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane. Proc. Natl. Acad. Sci. USA 1022402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castorena, K. M., S. A. Weeks, K. A. Stapleford, A. M. Cadwallader, and D. J. Miller. 2007. A functional heat shock protein 90 chaperone is essential for efficient flock house virus RNA polymerase synthesis in Drosophila cells. J. Virol. 818412-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dimitrova, M., I. Imbert, M. P. Kieny, and C. Schuster. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 775401-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 765974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Einav, S., M. Elazar, T. Danieli, and J. S. Glenn. 2004. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J. Virol. 7811288-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Hage, N., and G. Luo. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 842761-2769. [DOI] [PubMed] [Google Scholar]
- 19.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. USA 10113038-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandes, F., D. S. Poole, S. Hoover, R. Middleton, A. C. Andrei, J. Gerstner, and R. Striker. 2007. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology 461026-1033. [DOI] [PubMed] [Google Scholar]
- 21.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372359-362. [DOI] [PubMed] [Google Scholar]
- 22.Gao, L., H. Aizaki, J.-W. He, and M. M. C. Lai. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 783480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 775487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamamoto, I., Y. Nishimura, T. Okamoto, H. Aizaki, M. Liu, Y. Mori, T. Abe, T. Suzuki, M. M. Lai, T. Miyamura, K. Moriishi, and Y. Matsuura. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 7913473-13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamill, P., and F. Jean. 2005. Enzymatic characterization of membrane-associated hepatitis C virus NS3-4A heterocomplex serine protease activity expressed in human cells. Biochemistry 446586-6596. [DOI] [PubMed] [Google Scholar]
- 26.Handschumacher, R. E., M. W. Harding, J. Rice, R. J. Drugge, and D. W. Speicher. 1984. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226544-547. [DOI] [PubMed] [Google Scholar]
- 27.Hardy, R. W., J. Marcotrigiano, K. J. Blight, J. E. Majors, and C. M. Rice. 2003. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J. Virol. 772029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatziioannou, T., D. Perez-Caballero, S. Cowan, and P. D. Bieniasz. 2005. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 79176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu, J., and C. Seeger. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 931060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue, K., T. Umehara, U. T. Ruegg, F. Yasui, T. Watanabe, H. Yasuda, J. M. Dumont, P. Scalfaro, M. Yoshiba, and M. Kohara. 2007. Evaluation of a cyclophilin inhibitor in hepatitis C virus-infected chimeric mice in vivo. Hepatology 45921-928. [DOI] [PubMed] [Google Scholar]
- 31.Ishii, N., K. Watashi, T. Hishiki, K. Goto, D. Inoue, M. Hijikata, T. Wakita, N. Kato, and K. Shimotohno. 2006. Diverse effects of cyclosporine on hepatitis C virus strain replication. J. Virol. 804510-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivashkina, N., B. Wolk, V. Lohmann, R. Bartenschlager, H. E. Blum, F. Penin, and D. Moradpour. 2002. The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J. Virol. 7613088-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaboord, B. F., and S. J. Benkovic. 1996. Dual role of the 44/62 protein as a matchmaker protein and DNA polymerase chaperone during assembly of the bacteriophage T4 holoenzyme complex. Biochemistry 351084-1092. [DOI] [PubMed] [Google Scholar]
- 35.Kim, C. S., S. K. Seol, O. K. Song, J. H. Park, and S. K. Jang. 2007. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis C virus replication. J. Virol. 813852-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, J. L., K. A. Morgenstern, C. Lin, T. Fox, M. D. Dwyer, J. A. Landro, S. P. Chambers, W. Markland, C. A. Lepre, E. T. O'Malley, S. L. Harbeson, C. M. Rice, M. A. Murcko, P. R. Caron, and J. A. Thomson. 1996. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87343-355. [DOI] [PubMed] [Google Scholar]
- 37.Lai, V. C., S. Dempsey, J. Y. Lau, Z. Hong, and W. Zhong. 2003. In vitro RNA replication directed by replicase complexes isolated from the subgenomic replicon cells of hepatitis C virus. J. Virol. 772295-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam, A. M., and D. N. Frick. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 80404-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, C., B. M. Pragai, A. Grakoui, J. Xu, and C. M. Rice. 1994. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J. Virol. 688147-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 41.Liu, J., J. D. Farmer, Jr., W. S. Lane, J. Friedman, I. Weissman, and S. L. Schreiber. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66807-815. [DOI] [PubMed] [Google Scholar]
- 42.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 43.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 731067-1078. [DOI] [PubMed] [Google Scholar]
- 44.Ma, S., J. E. Boerner, C. TiongYip, B. Weidmann, N. S. Ryder, M. P. Cooreman, and K. Lin. 2006. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob. Agents Chemother. 502976-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyanari, Y., K. Atsuzawa, N. Usuda, K. Watashi, T. Hishiki, M. Zayas, R. Bartenschlager, T. Wakita, M. Hijikata, and K. Shimotohno. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 91089-1097. [DOI] [PubMed] [Google Scholar]
- 46.Miyanari, Y., M. Hijikata, M. Yamaji, M. Hosaka, H. Takahashi, and K. Shimotohno. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J. Biol. Chem. 27850301-50308. [DOI] [PubMed] [Google Scholar]
- 47.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 787400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moradpour, D., F. Penin, and C. M. Rice. 2007. Replication of hepatitis C virus. Nat. Rev. Microbiol. 5453-463. [DOI] [PubMed] [Google Scholar]
- 49.Nakagawa, M., N. Sakamoto, N. Enomoto, Y. Tanabe, N. Kanazawa, T. Koyama, M. Kurosaki, S. Maekawa, T. Yamashiro, C. H. Chen, Y. Itsui, S. Kakinuma, and M. Watanabe. 2004. Specific inhibition of hepatitis C virus replication by cyclosporin A. Biochem. Biophys. Res. Commun. 31342-47. [DOI] [PubMed] [Google Scholar]
- 50.Nakagawa, M., N. Sakamoto, Y. Tanabe, T. Koyama, Y. Itsui, Y. Takeda, C. H. Chen, S. Kakinuma, S. Oooka, S. Maekawa, N. Enomoto, and M. Watanabe. 2005. Suppression of hepatitis C virus replication by cyclosporin A is mediated by blockade of cyclophilins. Gastroenterology 1291031-1041. [DOI] [PubMed] [Google Scholar]
- 51.Neddermann, P., M. Quintavalle, C. Di Pietro, A. Clementi, M. Cerretani, S. Altamura, L. Bartholomew, and R. De Francesco. 2004. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 7813306-13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okamoto, T., Y. Nishimura, T. Ichimura, K. Suzuki, T. Miyamura, T. Suzuki, K. Moriishi, and Y. Matsuura. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 255015-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paeshuyse, J., A. Kaul, E. De Clercq, B. Rosenwirth, J. M. Dumont, P. Scalfaro, R. Bartenschlager, and J. Neyts. 2006. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 43761-770. [DOI] [PubMed] [Google Scholar]
- 54.Price, E. R., M. Jin, D. Lim, S. Pati, C. T. Walsh, and F. D. McKeon. 1994. Cyclophilin B trafficking through the secretory pathway is altered by binding of cyclosporin A. Proc. Natl. Acad. Sci. USA 913931-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinkert, D., R. Bartenschlager, and V. Lohmann. 2005. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 7913594-13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robida, J. M., H. B. Nelson, Z. Liu, and H. Tang. 2007. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J. Virol. 815829-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9505-514. [DOI] [PubMed] [Google Scholar]
- 58.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 27711149-11155. [DOI] [PubMed] [Google Scholar]
- 59.Sklan, E. H., K. Staschke, T. M. Oakes, M. Elazar, M. Winters, B. Aroeti, T. Danieli, and J. S. Glenn. 2007. A Rab-GAP TBC domain protein binds hepatitis C virus NS5A and mediates viral replication. J. Virol. 8111096-11105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sokolskaja, E., D. M. Sayah, and J. Luban. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 7812800-12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stahl, M., J. Beck, and M. Nassal. 2007. Chaperones activate hepadnavirus reverse transcriptase by transiently exposing a C-proximal region in the terminal protein domain that contributes to ɛ RNA binding. J. Virol. 8113354-13364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stone, M., S. Jia, W. D. Heo, T. Meyer, and K. V. Konan. 2007. Participation of Rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J. Virol. 814551-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taguwa, S., T. Okamoto, T. Abe, Y. Mori, T. Suzuki, K. Moriishi, and Y. Matsuura. 2008. Human butyrate-induced transcript 1 interacts with hepatitis C virus NS5A and regulates viral replication. J. Virol. 822631-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372363-365. [DOI] [PubMed] [Google Scholar]
- 66.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, C., M. Gale, Jr., B. C. Keller, H. Huang, M. S. Brown, J. L. Goldstein, and J. Ye. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18425-434. [DOI] [PubMed] [Google Scholar]
- 68.Watashi, K., M. Hijikata, M. Hosaka, M. Yamaji, and K. Shimotohno. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 381282-1288. [DOI] [PubMed] [Google Scholar]
- 69.Watashi, K., N. Ishii, M. Hijikata, D. Inoue, T. Murata, Y. Miyanari, and K. Shimotohno. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19111-122. [DOI] [PubMed] [Google Scholar]
- 70.Weeks, S. A., and D. J. Miller. 2008. The heat shock protein 70 cochaperone YDJ1 is required for efficient membrane-specific flock house virus RNA replication complex assembly and function in Saccharomyces cerevisiae. J. Virol. 822004-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, F., J. M. Robotham, H. B. Nelson, A. Irsigler, R. Kenworthy, and H. Tang. 2008. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 825269-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 1029294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zydowsky, L. D., F. A. Etzkorn, H. Y. Chang, S. B. Ferguson, L. A. Stolz, S. I. Ho, and C. T. Walsh. 1992. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 11092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]