Abstract
Replication and infectivity of hepatitis C virus (HCV) with a defective genome is ambiguous. We molecularly cloned 38 HCV isolates with defective genomes from 18 patient sera. The structural regions were widely deleted, with the 5′ untranslated, core, and NS3-NS5B regions preserved. All of the deletions were in frame, indicating that they are translatable to the authentic terminus. Phylogenetic analyses showed self-replication of the defective genomes independent of full genomes. We generated a defective genome of chimeric HCV to mimic the defective isolate in the serum. By using this, we demonstrated for the first time that the defective genome, as it is circulating in the blood, can be encapsidated as an infectious particle by trans complementation of the structural proteins.
Viruses with a deletion mutation in their genome have been identified as defective interfering (DI) particles for many virus species (1, 3, 9, 16). Part of the DI virus genome is deleted, but regions indispensable for replication and packaging are preserved. Most DI viruses occur spontaneously in the course of cell culture infected with a high titer of wild-type viruses. Hepatitis C virus (HCV) with a defective genome has been found in liver and serum specimens of some HCV patients (4, 8, 15). HCV has a plus-strand RNA genome that encodes the viral core, E1, E2, and p7 structural proteins and NS2, NS3, NS4A, NS4B, NS5A, and NS5B nonstructural proteins (10). According to the reports, the deletions have been found mainly in the structural region and most of the deletions are in frame, but some deletions are out of frame (4), raising questions about whether the defective HCV genome is merely a by-product of a full genome or a self-replicating genome and whether it can be encapsidated into an infectious virus particle.
In the present study, we molecularly cloned 38 HCV isolates with defective genomes from HCV patient sera to address these questions by genetic analyses and infection experiments. As long as we explored, all of the deletions were in frame, indicating the potential to support translation from the authentic initiation codon to the termination codon, although the structural region was widely deleted, as reported previously. Phylogenetic analyses evidenced self-replication of the defective genomes independent of full genomes. We demonstrated for the first time, by trans complementation experiments, that the defective genome, as it is circulating in the blood, can be encapsidated as an infectious particle, designated HCVCCD.
First, to amplify HCV cDNAs in 21 serum specimens from 18 HCV patients (genotype 1b), we performed three sets of long-distance reverse transcription (RT)-PCRs flanking (i) the 5′ untranslated region (UTR) to the 5′ part of the NS3 region, (ii) the remaining part of the NS3 region to the end of NS5B, and (iii) the 5′ UTR to the end of the NS5B region (Fig. 1A). The specimens were collected with informed consent. cDNA was synthesized with RNase H-deficient reverse transcriptase Superscript III (Invitrogen, Carlsbad, CA) at a higher temperature (55°C) to reduce template switching and mispriming. PCRs were performed in a (hemi)nested manner with high-fidelity polymerase KOD plus or KOD FX (Toyobo, Osaka, Japan) as described previously (5). For some target nucleotide positions, a mixture of two or three primers was used to reduce mismatches due to sequence heterogeneity (Table 1). Of the 21 specimens examined, representative results are shown in Fig. 1. An amplicon of the 5′ UTR-NS3 region of the predicted size (ca. 3.7 kb) was detected in all specimens (18/18), and representative results are shown in Fig. 1B. In addition, a shorter amplicon suggestive of a defective HCV genome was simultaneously present in four specimens from 1 (R4) of 12 cases of clinically mild hepatitis and from 3 (T5, K3, and K4-pre) of 6 cases of active hepatitis (clinical data not shown). Defective genomes were found in the patients with relatively higher copy numbers of HCV RNA (>8.1 × 105 copies/ml in the 5′ UTR, Table 2), suggesting that the coexistence of a defective genome is related to hepatitis severity. The authentic-size amplicon was poorly detected when coexisting with a defective HCV genome shorter than 2 kb (T5 and K3), presumably because of preferential amplification of the shorter amplicon. A shorter amplicon was not detected for the NS3-NS5B region, while the amplicon of the predicted size (ca. 5.6 kb) was detected in all of the specimens, albeit with various efficiencies (Fig. 1C). The 5′ UTR-NS5B region, which covers almost the whole genome, was then amplified, and an amplicon of the predicted size (9.4 kb) was detected in 10 specimens from nine patients (5 specimens in Fig. 1D). Of the successfully amplified specimens, two (R4 and K4-pre) also contained a shorter amplicon, in accordance with the results of the 5′ UTR-NS3 PCR. The NS3-NS5B region is essential for autonomous replication of HCV as an RNA replicon in vitro (5-7). It has been shown that NS5A is the only nonstructural protein that can trans complement HCV replication (13). They used a nonadaptive mutation of NS5A as a replication-incompetent NS5A protein instead of a deletion mutant protein. Thus, we speculate that deletion of the NS3-NS5B region cannot be complemented in trans. Intriguingly, the shorter amplicon was not detected after full-term interferon treatment in patient K4 (K4-af24), although it was detected prior to treatment (K4-pre) (Fig. 1B and D). The possible reasons for this are that (i) the defective genome disappeared naturally, (ii) packaging of the defective genome by the helper virus was impeded by an unknown mechanism of interferon, or (iii) replication of the defective genome is preferentially inhibited by the interferon pathway. Further studies are needed to reveal the effect of a defective HCV genome on the pathogenesis and treatment of HCV.
FIG. 1.
Representative results of long-distance RT-PCRs for serum HCV. (A) The three sets of long-distance RT-PCR used: 5′ UTR-NS3 (5′ UTR to the 5′ part of the NS3 region), NS3-NS5B (the remaining part of the NS3 region to the end of NS5B), and 5′ UTR-NS5B (5′ UTR to the end of the NS5B region). The nucleotide positions of the 5′ and 3′ ends of each amplicon are indicated in parentheses. PCR products were electrophoresed and stained with ethidium bromide. Results of the representative 11 specimens (eight patients) are shown for 5′ UTR-NS3 (B), NS3-NS5B (C), and 5′ UTR-NS5B (D). Serum specimens were collected from patients K4 and K6 before interferon treatment (pre), at the end of the full 48-week treatment period (tx48), and 24 weeks after the full treatment period (af24). DNA molecular size markers are at both sides of panels B to D.
TABLE 1.
Primers used for long-distance and quantitative RT-PCRs in this study
| Test and region(s) | Direction | Primer(s)a | Sequenceb | Positionc |
|---|---|---|---|---|
| Long-distance PCR | ||||
| 5′ UTR-NS3 | RT | 606R/712R | GTTTCCATAGACTC(A/G)ACGGG | 3930-3949 |
| 5′ UTR-NS3, 5′ UTR-NS5B | 1st forward | 420 | GGCGACACTCCACCATAGATCACTC | 1-42 |
| 5′ UTR-NS3 | 1st reverse | 605R/713R | ACCGGAATGACATCAGCATG(T/C)CTCGT | 3741-3766 |
| 5′ UTR-NS3, 5′ UTR-NS5B | 2nd forward | AscT7-420 | ATCGTAGGCGCGCCTCTAATACGACTCACTATAGC | 1-42 |
| CAGCCCCCGATTGGGGGCGACACTCCACCATAGATCACTC | ||||
| 5′ UTR-NS3 | 2nd reverse | 604R/714R | CGAGGTCCTGGTCTACATT(G/A)GTGTACAT | 3639-3666 |
| NS3-NS5B, 5′ UTR-NS5B | RT | 386R | AATGGCCTATTGGCCTGGAG | 9390-9392 |
| NS3-NS5B | 1st forward | 602/723 | CCACCGCAACACAATCTTTCCT(G/A)GCGAC | 3529-3556 |
| NS3-NS5B, 5′ UTR-NS5B | 1st reverse | 719R/720R/721R | GAGTGTTTAGCTCCCCGTTCA(T/C/G)CGGTTGGG | 9363-9392 |
| NS3-NS5B | 2nd forward | 603/724 | CAAAGGGTCCAATCACCCA(A/G)ATGTACAC | 3619-3646 |
| NS3-NS5B, 5′ UTR-NS5B | 2nd reverse | 607R/654R/722R | CGGTTGGGGAGCAGGTA(G/A/G)A(T/T/C)GCCTAC | 9345-9370 |
| Quantitative PCR | ||||
| 5′ UTR | RT | 738RH | ACTCGCAAGCACCCTATCAGGC | 291-312 |
| 5′ UTR | Forward | 736 | AAGCGTCTAGCCATGGCGTTAGTA | 73-96 |
| 5′ UTR | Reverse | 737R | GGCAGTACCACAAGGCCTTTCG | 272-293 |
| 5′ UTR | Probe | 733FB | FAM-TCTGCGGAACCGGTGAGTACAC-BHQ1 | 147-168 |
| E2 | RT | 743RH/744RH/753RH/753RH | CAACGCTCTCCTCG(A/A/G/G)GTCCA(A/G/A/G)TTGCA | 2271-2296 |
| E2 | Forwardd | 751/752 | GGCCTCCACATGGCAA(C/T)TGGTTCGG | 1972-1993 |
| E2 | Forwardd | 739/740 | CCGCCGCAAGGCAACTGGTT(C/T)GG | 1974-1993 |
| E2 | Reverse | 741R/742R | GCCTCGGGGTGCTTCCGGAAGCA(G/A)TCCGT | 2088-2116 |
| E2 | Probe | 734FB/735FB | FAM-TGGATGAA(T/C)AGCACTGGGTTCACCAAGAC-BHQ1 | 2001-2029 |
Primers separated by slashes harbor a nucleotide substitution(s) (in parentheses) in the sequence in the same order.
An underline and a double underline indicate recognition sequences for AscI and BsrGI, respectively, with which the PCR products were subcloned into plasmid vector pASGT5. Italics denote the T7 promoter, which was used to synthesize RNA in vitro from the T5S2 isolate (Fig. 4A).
Nucleotide positions correspond to the HCV-JS sequence (12).
Forward primers for E2 were mixed in the reaction mixture.
TABLE 2.
Quantitative PCRs for the 5′ UTR and E2 regions of HCVa
| Region for quantification | No. of copies/ml
|
5′ UTR/E2 ratio | |
|---|---|---|---|
| 5′ UTR | E2 | ||
| R2 | 2.0 × 106 | 1.7 × 106 | 1.17 |
| R4 | 5.3 × 106 | 2.2 × 106 | 2.44 |
| T5 | NDb | NDb | |
| K1 | 8.3 × 105 | 8.4 × 105 | 0.99 |
| K2 | 3.6 × 105 | 3.5 × 105 | 1.01 |
| K3 | 8.6 × 105 | 5.1 × 105 | 1.7 |
| K4pre | 8.1 × 105 | 3.3 × 105 | 2.45 |
| K4af24 | 4.5 × 105 | 4.4 × 105 | 1.01 |
For quantification of the 5′ UTR and E2 regions, the TaqMan Fast PCR Universal mixture and the 7500 Fast Real-Time PCR system (Applied Biosystems) were used in a two-step method with the primers and probes shown in Table 1 according to the manufacturer's protocol. The copy number of HCV was determined by the standard-curve method with serial dilutions of the synthesized full-length HCV RNA.
ND, not determined due to sample shortage.
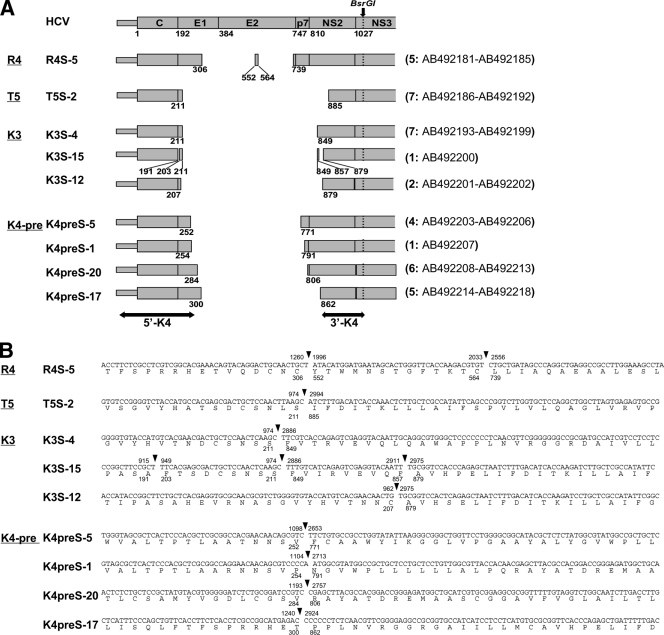
A total of 38 isolates with defective HCV genomes were molecularly cloned into plasmid vector pASGT (unpublished data) from the shorter amplicons of the 5′ UTR-NS3 PCR from four serum specimens (R4, T5, K3, and K4-pre in Fig. 1B) at the AscI and BsrGI restriction sites. The nucleotide sequences were determined with an autosequencer (3730 DNA analyzer; Applied Biosystems, Foster City, CA). Sequence analyses revealed that the structural region was widely deleted in all of the defective isolates and that the deletion ranges were quite diverse among the isolates (extending up to the NS2 region) (Fig. 2A). In contrast, the 5′ UTR and core regions were constantly preserved, suggesting that these regions, as well as the NS3-NS5B region, are indispensable for the production of HCV with a defective genome. Intriguingly, defective genomes with different deletion patterns coexisted in single specimens from two patients (three patterns in patient K3 and four patterns in patient K4-pre). Moreover, two deletions in a single genome were observed in five isolates from patient R4 (isolate R4S-5). As many as three deletions in a single genome were observed in the isolate from patient K3 (e.g., isolate K3S-15), in which two small deletions resulted in two tiny residual fragments. Such diversity in deletion ranges indicates flexibility of the remaining structural region for the replication of defective HCV genomes. Nevertheless, all of the deletions identified in the 38 isolates were in frame (Fig. 2B), implying that these defective HCV genomes have the potential for translation from the core to the authentic end of NS5B without a frameshift.
FIG. 2.
Sequence analysis of the defective HCV genomes. A total of 38 isolates were molecularly cloned into a plasmid vector and sequenced. Data from representative isolates are presented. Nucleotide positions and deduced amino acid positions correspond to those of genotype 1b strain HCV-JS (12). (A) Defects located in the structural region were compared. The remaining regions are illustrated as shaded boxes. Below the boxes are numbers indicating amino acid positions at the end of each remaining region. At the top of the panel is the HCV genome with the amino acid position at the N terminus of each HCV protein below. The BsrGI restriction site that was used to clone the PCR products is shown as a dotted line. Each value in parentheses at the right is the number of isolates showing the same deletion pattern, followed by the GenBank accession number(s). The two-headed arrows indicate the 5′ and 3′ maximum overlapping regions among the defective HCV isolates in the K4-pre specimen that are compared in the following phylogenetic analyses (5′-K4 and 3′-K4; see Fig. 3). (B) Deletion breakpoints and their adjacent nucleotides and deduced amino acid sequences are indicated. Solid triangles denote breakpoints, and numbers indicate the nucleotide positions (above) and amino acid positions (below) at the junctions.
To determine the ratio of defective to full genomes, we performed quantitative PCRs targeting a relatively conserved E2 sequence, which is commonly deleted in the defective genomes, with primers listed in Table 1. Calculation of the 5′ UTR/E2 ratio, which must theoretically be 1 without the existence of the defective genome, showed higher values (1.7 to 2.45) in specimens containing the defective genomes (R4, K3, and K4pre in Table 2), indicating that the defective genome level in serum is 0.7 to 1.45 times the full genome level. However, to clarify the impact of defective genomes on pathogenesis and their effect on the treatment of HCV, accumulation of more data is needed.
The nucleotide sequence comparison of 38 defective HCV isolates showed sequence diversity. Such diversity was observed even among isolates obtained from the same specimen. Perhaps such diversity is a result of self-replication and the subsequent evolution of the defective HCV genome. To explore this possibility, phylogenetic analyses were performed on the nucleotide sequence data from patient K4. Sequences at the 5′ and 3′ maximum overlapping regions located outside the deletions were separately compared (Fig. 2A), and phylogenetic trees were created by the neighbor-joining method with GENETYX software (Genetyx Inc., Tokyo, Japan). As a result, isolates with the same deletion pattern formed genetic clusters that were distinct from each other, as well as from those of nondefective HCV isolates (Fig. 3A and B). Similar results were obtained for the other patients with defective HCV genomes (data not shown). These results suggest that a defective HCV genome is capable of replication to accumulate mutations and to evolve independently of the nondefective HCV genome.
FIG. 3.
Phylogenetic analyses of defective HCV genomes. Nucleotide sequence data from 33 isolates from patient K4 were used for phylogenetic analyses. The defective HCV genome (16 isolates) and the nondefective HCV genome coexisting before interferon treatment (9 isolates; GenBank accession no. AB492219 to AB492227) and those after treatment (8 isolates; GenBank accession no. AB492228 to AB492235) were compared in the 5′ and 3′ maximum overlapping regions separately (5′-K4 and 3′-K4 in Fig. 2A). Phylogenetic trees were created for the respective regions (A and B). In the isolate designations, pre and af24 stand for before and after interferon treatment and S and L stand for defective and nondefective HCV genomes, respectively. Isolates with the same deletion pattern (according to K4-pre in Fig. 2) are shaded in the same color. Asterisks denote the representative isolates illustrated in Fig. 2.
Next, the ability of the defective HCV genome to be encapsidated and released from cells as HCVCCD was examined. A genotype 1b replicon RNA lacking the structural region was synthesized by using defective isolate T5S-2 from patient T5 (Fig. 2 and 4A) as the template in an in vitro transcription system (MEGAscript T7 kit; Ambion, Inc., Austin, TX) under the control of the T7 promoter. Also, capped mRNA encoding the genotype 1b structural proteins from the same patient (designated C-NS2 in Fig. 4A) was synthesized in vitro with the mMessage mMachine T7 kit (Ambion). Both synthesized RNAs were cotransfected into Huh7.5 hepatoma cells. However, HCVCCD was not obtained, presumably because of low replication or virus productivity of genotype 1b HCV per se. In fact, we transfected the defective RNA alone and observed the replication and protein expression of HCV, but with low efficiency (data not shown). Thus, to augment virus productivity, a JFH1-based chimeric HCV genome (genotype 1b/2a) and its deletion mutant were generated to mimic isolate T5S-2 (designated TNS2J1 and TNS2J1ΔS, respectively, Fig. 4A). JFH1 is genotype 2a HCV isolate that can produce high levels of infectious virus (14). To verify the virus productivity of TNS2J1, Huh7.5 cells (10-cm plate) were transfected with 10 μg of in vitro-synthesized RNA from TNS2J1 or JFH1 by lipofection with TransMessenger transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's protocol. Two days later, the culture medium was concentrated 10-fold and inoculated into naïve Huh7.5 cells (four-well chamber slide). Cells inoculated with the culture medium from TNS2J1 RNA-transfected cells markedly expressed HCV protein, as shown by immunofluorescent staining (Fig. 4B). The percentage of HCV-positive cells in chimera-infected cells, 40% (565/1,240), was greater than that of JFH1, 3% (37/1,210), demonstrating that the chimeric genome TNS2J1 can produce infectious HCV more robustly than JFH1 can (P < 0.0001).
FIG. 4.
In vitro infectivity of deletion mutant of chimeric HCV conferred by trans complementation of structural proteins. (A) Schematics of the following HCV genomic constructs: the defective HCV isolate (T5S-2), JFH1, chimeric virus of genotypes 1b and 2a (TNS2J1), and its deletion mutant (TNS2J1ΔS). C-NS2 and C-NS3 are fragments encoding the region from the core to the C terminus of the NS2 region and to the C terminus of the serine protease moiety in NS3, respectively. For the trans complementation experiments, the latter two constructs were inserted into pcDNA3.1 (Invitrogen) to synthesize capped mRNAs or into retroviral vector pCX4bsr (GenBank accession no. AB086384) to establish packaging cell lines stably expressing the proteins. Shaded and open boxes represent genotypes 1b (isolate from patient T5) and 2a (JFH1), respectively. The numbers below the boxes are amino acid positions at deletion breakpoints or at PCR-based recombination junctions. Naïve Huh7.5 cells were inoculated with the culture medium from cells transfected with JFH1 or TNS2J1 RNA (B), from cells cotransfected with TNS2J1ΔS RNA together with the structural region mRNA (C-NS2 or C-NS3P) or TNS2J1ΔS RNA alone (C), and from the packaging cell line (C-NS2/Huh7.5 or C-NS3P/Huh7.5) transfected with TNS2J1ΔS RNA and parental Huh7.5 cells transfected with TNS2J1ΔS RNA (D). HCV protein was detected by human HCV serum (1:500) by the indirect immunofluorescent method with Alexa Fluor 568 goat anti-human immunoglobulin G (1:200; red; Invitrogen). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; blue).
Taking advantage of this chimeric genome, we conducted trans complementation experiments. To mimic the T5S-2 isolate, the region corresponding to the defect found in T5S-2 was identically deleted from the TNS2J1 genome (designated TN2J1ΔS, Fig. 4A). Ten micrograms of synthesized RNA of TN2J1ΔS was cotransfected into Huh7.5 cells (10-cm plate) together with 10 μg of synthesized capped mRNA encoding the structural region, including part of the nonstructural region of TNS2J1, designated C-NS2 or C-NS3P (Fig. 4A). Two days later, the culture medium was concentrated and inoculated into naïve Huh7.5 cells as previously described. HCV protein was expressed when cells were inoculated with the medium of cells cotransfected with TN2J1ΔS RNA and C-NS2 or C-NS3P mRNA, whereas no expression was observed in the case of TN2J1ΔS RNA alone (Fig. 4C). To stably provide the structural proteins in trans, packaging cell lines were established by retroviral transduction (2) of Huh7.5 cells with genes encoding the C-NS2 or C-NS3P region (Fig. 4A). These packaging cell lines were transfected with TN2J1ΔS RNA, and HCV protein was expressed in cells inoculated with the culture medium from the RNA-transfected packaging cells (Fig. 4D). Notably, the construct C-NS2 helped to produce HCVCCD more efficiently than C-NS3P did (Fig. 4C). We observed less expression of the structural proteins with the C-NS3 construct than with the C-NS2 construct in a transient expression experiment (data not shown). One possible reason for this is that the C-NS3 construct needs one additional process, i.e., cleavage between NS2 and NS3, to produce NS2 and may affect the other proteins. Otherwise, it is simply because of the difference in the lengths of the constructs. These results indicate that a defective HCV genome lacking the structural region can be encapsidated by trans complementation of the structural proteins, thus conferring infectivity in vivo. Recently, a trans-packaging system consisting of an HCV subgenomic replicon and a reporter gene was also reported in which an intragenotypic chimera (2a/2a) was used as the most efficient packaging construct (11). Our packaging system used an efficient intergenotypic chimera (1b/2a) to encapsidate a genome mimicking a naturally occurring deletion (1b). Thus, although its efficiency may be different, our system could be a useful tool for the study of HCVCCD of chimeric genome 1b/2a or genotype 1b.
Taken together, genetic analyses of the defective HCV genome showed the potential of its translation and self-replication. These defective genomes can be encapsidated into infectious virus-like particles by trans complementation of the structural proteins in vitro. The 5′ UTR and core regions, which are preserved in defective HCV genomes, are targets for the clinical quantification of HCV. Therefore, measured values may represent additive values for defective and nondefective HCVs and the method used for HCV quantification should be reevaluated.
Acknowledgments
We thank H. Kato, R. Shiina, and H. Yamamoto for technical assistance; T. Wakita for the gift of JFH1; C. Rice for the gift of Huh7.5 cells; and T. Akagi for the gift of retroviral vector pCX4bsr.
This work was supported by a grant-in-aid for scientific research (C) from the Japan Society for the Promotion of Science (KAKENHI18590454) and a grant-in-aid for research on hepatitis from the Ministry of Health, Labor, and Welfare.
Footnotes
Published ahead of print on 15 April 2009.
REFERENCES
- 1.Brinton, M. A. 1983. Analysis of extracellular West Nile virus particles produced by cell cultures from genetically resistant and susceptible mice indicates enhanced amplification of defective interfering particles by resistant cultures. J. Virol. 46860-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, C. J., K. Sugiyama, H. Kubo, C. Huang, and S. Makino. 2004. Murine coronavirus nonstructural protein p28 arrests cell cycle in G0/G1 phase. J. Virol. 7810410-10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang, A. S., and D. Baltimore. 1970. Defective viral particles and viral disease processes. Nature 226325-327. [DOI] [PubMed] [Google Scholar]
- 4.Iwai, A., H. Marusawa, Y. Takada, H. Egawa, K. Ikeda, M. Nabeshima, S. Uemoto, and T. Chiba. 2006. Identification of novel defective HCV clones in liver transplant recipients with recurrent HCV infection. J. Viral Hepat. 13523-531. [DOI] [PubMed] [Google Scholar]
- 5.Kato, N., K. Sugiyama, K. Namba, H. Dansako, T. Nakamura, M. Takami, K. Naka, A. Nozaki, and K. Shimotohno. 2003. Establishment of a hepatitis C virus subgenomic replicon derived from human hepatocytes infected in vitro. Biochem. Biophys. Res. Commun. 306756-766. [DOI] [PubMed] [Google Scholar]
- 6.Kishine, H., K. Sugiyama, M. Hijikata, N. Kato, H. Takahashi, T. Noshi, Y. Nio, M. Hosaka, Y. Miyanari, and K. Shimotohno. 2002. Subgenomic replicon derived from a cell line infected with the hepatitis C virus. Biochem. Biophys. Res. Commun. 293993-999. [DOI] [PubMed] [Google Scholar]
- 7.Lohmann, V., F. Körner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 8.Noppornpanth, S., S. L. Smits, T. X. Lien, Y. Poovorawan, A. D. Osterhaus, and B. L. Haagmans. 2007. Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients. J. Virol. 8112496-12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poidinger, M., R. J. Coelen, and J. S. Mackenzie. 1991. Persistent infection of Vero cells by the flavivirus Murray Valley encephalitis virus. J. Gen. Virol. 72(Pt. 3)573-578. [DOI] [PubMed] [Google Scholar]
- 10.Shimotohno, K. 1995. Hepatitis C virus as a causative agent of hepatocellular carcinoma. Intervirology 38162-169. [DOI] [PubMed] [Google Scholar]
- 11.Steinmann, E., C. Brohm, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2008. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J. Virol. 827034-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugiyama, K., N. Kato, T. Mizutani, M. Ikeda, T. Tanaka, and K. Shimotohno. 1997. Genetic analysis of the hepatitis C virus (HCV) genome from HCV-infected human T cells. J. Gen. Virol. 78(Pt. 2)329-336. [DOI] [PubMed] [Google Scholar]
- 13.Tong, X., and B. A. Malcolm. 2006. Trans-complementation of HCV replication by non-structural protein 5A. Virus Res. 115122-130. [DOI] [PubMed] [Google Scholar]
- 14.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yagi, S., K. Mori, E. Tanaka, A. Matsumoto, F. Sunaga, K. Kiyosawa, and K. Yamaguchi. 2005. Identification of novel HCV subgenome replicating persistently in chronic active hepatitis C patients. J. Med. Virol. 77399-413. [DOI] [PubMed] [Google Scholar]
- 16.Yoon, S. W., S. Y. Lee, S. Y. Won, S. H. Park, S. Y. Park, and Y. S. Jeong. 2006. Characterization of homologous defective interfering RNA during persistent infection of Vero cells with Japanese encephalitis virus. Mol. Cells 21112-120. [PubMed] [Google Scholar]