Abstract
The complete positive-sense single-stranded RNA genome of Cassava brown streak virus (CBSV; genus Ipomovirus; Potyviridae) was found to consist of 9,069 nucleotides and predicted to produce a polyprotein of 2,902 amino acids. It was lacking helper-component proteinase but contained a single P1 serine proteinase that strongly suppressed RNA silencing. Besides the exceptional structure of the 5′-proximal part of the genome, CBSV also contained a Maf/HAM1-like sequence (678 nucleotides, 226 amino acids) recombined between the replicase and coat protein domains in the 3′-proximal part of the genome, which is highly conserved in Potyviridae. HAM1 was flanked by consensus proteolytic cleavage sites for ipomovirus NIaPro cysteine proteinase. Homology of CBSV HAM1 with cellular Maf/HAM1 pyrophosphatases suggests that it may intercept noncanonical nucleoside triphosphates to reduce mutagenesis of viral RNA.
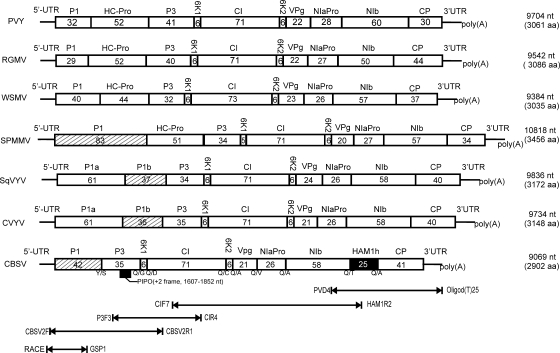
Cassava (Manihot esculenta Crantz; Euphorbiaceae) is an important tropical subsistence crop that is affected by cassava brown streak disease (CBSD) in the Indian Ocean coastal lowlands of East Africa (16, 35). There was a recent outbreak of CBSD at higher altitudes around Lake Victoria in Uganda and Tanzania (2, 26). The disease is caused by Cassava brown streak virus (CBSV), a whitefly-transmitted member of genus Ipomovirus that belongs to the family Potyviridae, which contains the largest number (ca. 200) of positive single-stranded RNA viruses infecting plants (13, 25, 27, 28). This virus family is divided into the genus Bymovirus with bipartite genomes and the genera Ipomovirus, Macluravirus, Potyvirus, Rymovirus, and Tritimovirus, containing monopartite viruses that encode a large polyprotein autoproteolytically cleaved into 10 mature proteins (Fig. 1) (13, 40). Additionally, a small open reading frame (ORF) created by frameshifting was recently detected in the P3 protein encoding region (10). Among members of the Potyviridae, ipomoviruses are exceptional in variability of protein-encoding sequences at the 5′ end of the genome (Fig. 1). Sweet potato mild mottle virus (SPMMV) (11) contains a single P1 serine proteinase at the polyprotein N terminus, whereas Cucumber vein yellowing virus (CVYV) (17, 22, 41) and Squash vein yellowing virus (SqVYV) (23) contain two P1 proteinases (P1a and P1b) that are evolutionary diversified (40). In addition, SqVYV and CVYV lack the multifunctional helper component proteinase (HC-Pro) (17, 23, 40), which is located second in the polyprotein (Fig. 1) in other monopartite Potyviridae members (1, 13) and which acts as a suppressor of RNA silencing (3, 7, 19, 46). This function has been adopted by P1b in CVYV (39, 41).
FIG. 1.
Comparison of the viral genomes in four genera of Potyviridae. The complete sequences of the type members of the genera Potyvirus (Potato virus Y, PVY; accession no. NC_001616), Rymovirus (Ryegrass mosaic virus, RGMV; AF035818), Tritimovirus (Wheat streak mosaic virus, WSMV; AF057533) and Ipomovirus (SPMMV; Z73124) are shown. Furthermore, genome structures of three additional ipomoviruses, SqVYV (EU259611), CVYV (NC_006941), and CBSV (FJ039520), whose genome sequence was determined in this study, are shown. Genera Macluravirus and Bymovirus were excluded because no complete viral genome sequence was available and because the genomes are bipartite, respectively. The box represents viral polyprotein translated from a large ORF. A small ORF created by +2 frameshifting is found in all the depicted viruses and shown in the drawing for CBSV below P3 as a black box (PIPO). The 5′UTR and 3′UTR are shown as bold lines. The numbers indicated after the 3′UTR refer to the genome size (nucleotides) and the size of the polyprotein (amino acids). The amino acids at the predicted proteolytic cleavage sites of the polyprotein are shown below the polyprotein. The estimated molecular weights of the mature proteins (in kilodaltons) are indicated in the box for each protein. The names of proteins above the polyprotein are as follows: P1, a serine proteinase, the first protein; P1a and P1b, two diversified serine proteinases; HC-Pro, helper component cysteine proteinase; P3, the third protein; 6K1 and 6K2, 6-kDa proteins; CI, cylindrical inclusion protein; VPg, viral genome-linked protein; NIa-Pro, the main viral proteinase; NIb, replicase. The primers used to amplify and sequence the CBSV genome in segments are shown at the bottom. Primers given at the bottom of the figure, from right to left, respectively (sequence and genomic position of the primers are in parentheses), are oligo(T)25 and PVD4 (5′-CTCAATGTTCCTGATGATGA-3′, 6606 to 6625), HAM1R2 (5′-GGACTATTAGTCATCTTCACT-3′, 7292 to 7312) and CIF7 (5′-TGAACTACAAGGAACATCTG-3′, 2930 to 2949), CIR4 (5′-TGTCAACCAAATGAGCTGTG-3′, 3512 to 3531) and P3F3 (5′-ATGGAGTATGTAGGCAAACAT-3′, 1410 to 1430), and CBSV2R1 (5′-CATATGTCATGATTGTAATAG-3′, 2665 to 2685) and CBSV2F (5′-AAATAAACATGACATAAGAATAC-3′, 3 to 25). The primer GSP1 (5′-TGTTGCCCTCTCCGTCAAGC-3′, 191 to 210) was used for 5′ RACE.
The genome structure of CBSV has hitherto not been known. Only partial coat protein (CP)-encoding sequences of coastal lowland isolates (27, 28) and complete CP sequences of highland isolates from East Africa are available, revealing that these isolates belong to two phylogenetically different strains (26). The complete sequence of the emergent highland strain of CBSV was determined in this study. Isolate MLB3 (26) from the Kagera region, Northwestern Tanzania, was mechanically transmitted to Nicotiana rustica, particles were purified from systemically infected leaves, and RNA was extracted from virions and used for cDNA synthesis as described previously (26). Overlapping fragments of the viral genome were amplified by PCR initially using degenerate primers designed according to the most-conserved regions of the SPMMV, CVYV, and SqVYV genomes and sequenced. Larger genomic segments were subsequently amplified using CBSV-specific primers (Fig. 1) and the high-fidelity Phusion DNA polymerase (Finnzymes, Espoo, Finland). At least two independently amplified fragments corresponding to each genomic segment (Fig. 1) were sequenced in both directions or sequenced directly without cloning as described previously (26). The 5′ end of the viral genome was determined using rapid amplification of cDNA ends (RACE) as described previously (32), by using SuperScript III reverse transcriptase (Invitrogen, Life Science Technologies, United Kingdom) for cDNA synthesis according to the manufacturer's instructions.
Nucleotide sequences were assembled by SeqMan (version 5.03; DNAStar, Madison, WI). Sequences of other viruses of Potyviridae were retrieved from the validated DPVweb database (http://www.dpvweb.net./seqs/plantviruses.php) and NCBI database (http://www.ncbi.nlm.nih.gov/). Polyprotein cleavage sites were predicted according to the previous comprehensive studies (1, 23, 40). Multiple sequence alignments of deduced amino acid sequences were done with ClustalX (version 1.83) using default settings. The sequences were exported to GeneDoc (version 2.6.002) for manual adjustment. The percent identities of nucleotides and deduced amino acid sequences were determined using the sequence distances option in the MegAlign program (version 5.03) from DNAStar, with default settings. Phylogenetic analyses were carried out according to the neighbor-joining method in MEGA4 (38). Vector NTI Advance software from Invitrogen (version 10.3.0) was used to estimate the molecular weights of polypeptides.
The genome of CBSV highland isolate MLB3 was 9,069 nucleotides (nt) long, excluding the poly(A) tail, and hence shorter than the genomes of ipomoviruses SPMMV (10,818 nt), SqVYV (9,836 nt), and CYVV (9,734 nt) (Fig. 1). The 5′ untranslated region (5′UTR; 134 nt) was followed by a single ORF (start codon AUG) terminated by the stop codon UAA at positions 8841 to 8843. The 3′UTR was 226 nt. Prediction of the proteolytic cleavage sites and motifs conserved in the polyproteins of Potyviridae members (1, 10, 17, 23, 33) revealed that CBSV encodes only 9 of the 10 expected proteins of Potyviridae, namely, P1, P3, 6K1, CI, 6K2, VPg, NIaPro, NIb, and CP (Fig. 1). No sequence homologous to HC-Pro was detected. PCR amplification of five additional highland isolates (BSA4, IGA8, LWR2, MLB9, and NTG10; see reference 26) of CBSV with primers CBSV2F1 and CBSV2R1 (Fig. 1) resulted in a 2.7-kb PCR product also in these isolates, consistent with a lack of the HC-Pro encoding sequence (data not shown). CBSV is hence the first member of Potyviridae that encodes a single P1 serine proteinase but lacks HC-Pro. Two other ipomoviruses, CVYV and SqVYV, lack HC-Pro but have two P1 proteinases (Fig. 1).
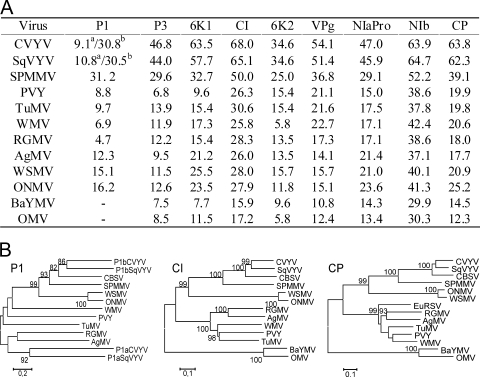
The P1 of CBSV (362 amino acids [aa]) is most closely related to the P1 of SPMMV and P1b of CVYV and SqVYV (Fig. 2A), which all are related to the P1 of tritimoviruses (Fig. 2B) (40). The conserved histidine, aspartic acid, and serine of the catalytic triad HDS (H-7X-D-34X-S) of P1 (44, 45) were observed at positions 265, 273, and 308 in CBSV. It matches with the spacing in P1b of CVYV and SqVYV, although the amino acid sequences of the proteins per se are only 31% and 30% identical, respectively (Fig. 2A). High divergence of the P1 proteins is characteristic of Potyviridae (40). The identity between P1a of CVYV and SqVYV and P1 of CBSV was very low (Fig. 2A).
FIG. 2.
Identities and phylogenetic relationships between mature proteins of CBSV, other ipomoviruses, and the type members of other genera in Potyviridae. (A) Percentage amino acid sequence identities between the mature proteins of CBSV and other viruses. In CVYV and SqVYV, P1a and P1b were compared to CBSV P1 (superscripts a and b, respectively). −, bymoviruses do not contain P1. (B) Phylogenetic analysis by the neighbor-joining method of three mature proteins of CBSV and other representative members of Potyviridae. AgMV, Agropyron mosaic virus, genus Rymovirus (sequence accession no. AY623626); ONMV, Oat necrotic mottle virus, genus Tritimovirus (AY377938); WMV, Watermelon mosaic virus, genus Potyvirus (EU660588); TuMV, Turnip mosaic virus, genus Potyvirus (D83184); BaYMV, Barley yellow mosaic virus, genus Bymovirus (NC_002990); OMV, Oat mosaic virus, genus Bymovirus (AJ306718); PVY, Potato virus Y; RGMV, Ryegrass mosaic virus; WSMV, Wheat streak mosaic virus; and EuRSV in the CP tree, genus Potyvirus (AY697300). Other acronyms are explained in the legend to Fig. 1. Only bootstrap values higher than 70% (of 1,000 replicates) are shown. Scale indicates Kimura units (38).
The deduced P3 sequence (294 aa) of CBSV was more identical to CVYV, SqVYV, and SPMMV P3 than were the P3 proteins of other Potyviridae members (Fig. 2A). The small ORF PiPo created by a +2 frameshift (10) was identified in the P3 encoding region of CBSV (positions 1607 to 1852) and consisted of 82 codons, compared to 99 and 79 codons in SPMMV and CVYV, respectively (10). P3 and the other mature proteins, 6K1 and 6K2 (52 aa each), CI (628 aa), VPg (185 aa), NIaPro (234 aa), NIb (502 aa), and CP (367 aa), showed the highest sequence identities with SqVYV and CVYV, followed by SPMMV (Fig. 2A), and much lower amino acid sequence identities and more distant phylogenetic relatedness with other members of the Potyviridae (Fig. 2A and B).
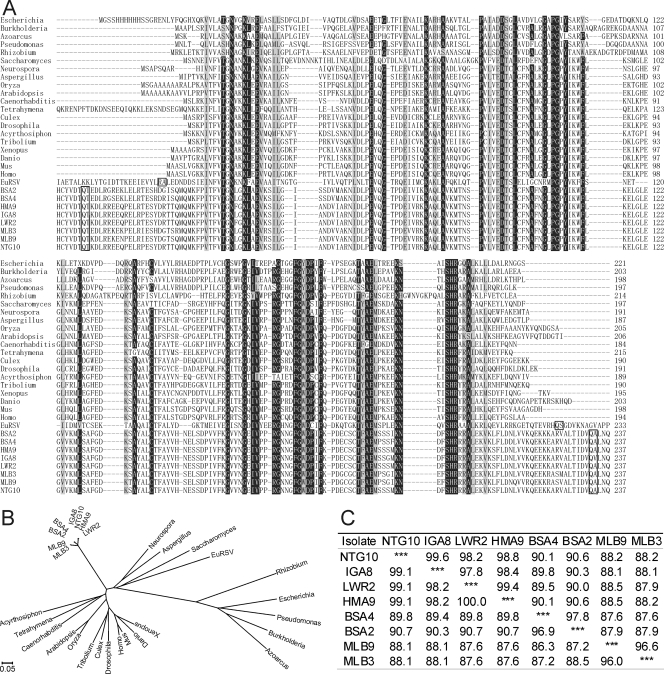
Surprisingly, alignment of the complete nucleotide and polyprotein amino acid sequences of CBSV, CVYV, SPMMV, and SqVYV revealed that CBSV contains an insertion (678 nt) resulting in a novel polypeptide of 226 aa between the replicase (NIb) and CP flanked by the predicted proteolytic cleavage sites VDTQ2309/T and IDVQ2535/A for the main viral proteinase, NIaPro (Fig. 1) (17, 23). BLASTN and BLASTP searches indicated that the novel sequence is homologous and shares the conserved amino acid motifs with the Maf/HAM1 superfamily of proteins known for prokaryotic and eukaryotic organisms, including bacteria, fungi, plants, insects, frogs, fishes, warm-blooded animals, and humans (Fig. 3A and B). The Maf/HAM1 proteins are nucleoside triphosphate (NTP) pyrophosphatases that reduce mutagenesis by intercepting noncanonical NTPs and preventing their incorporation into DNA or RNA (14). HAM1 of yeast (Saccharomyces cerevisiae) reduces sensitivity to 6-N-hydroxylaminopurine (HAP), which causes a hypermutable phenotype in phages, bacteria, yeast, and other eukaryotic organisms (29). HAM1 also has purine nucleoside triphosphatase activity on ITP and HAP triphosphate (8) and an ability to decompose and detoxify abnormal pyrimidine and purine nucleotides (37). The HAM1 homolog (HAM1h) was detected for all eight highland isolates of CBSV tested from Uganda and Tanzania (Fig. 3A) and shared 87.6 to 99.6% and 86.3 to 100% nucleotide and amino acid identity, respectively, among isolates (Fig. 3C). That HAM1h is an integral part of the CBSV genome was verified by mechanical transmission of the CBSV isolates to Nicotiana benthamiana, Nicotiana tabacum, and Nicotiana occidentalis and an analysis of virus progeny from the systemically infected leaves by reverse transcription-PCR using primer pairs in which one primer targeted HAM1h and the other primer the NIb- or CP-encoding region (data not shown). HAM1-like sequence was also detected for Euphorbia ringspot virus (EuRSV; 00.057.0.81.036 in the ICTVdB database, version 4; NCBI database accession number AY397600), belonging to the genus Potyvirus (13). Also in EuRSV, HAM1h was situated between NIb and CP and flanked by the predicted proteolytic cleavage sites of Potyvirus NIaPro (1) (Fig. 3A). No other viruses carrying HAM1-like sequences were found in sequence databases.
FIG. 3.
Comparison of the amino acid sequences of HAM1h proteins in eight CBSV isolates and one isolate of EuRSV with the HAM1 proteins of prokaryotic and eukaryotic organisms, including genetic model species. (A) Alignment of the amino acid sequences. The highly conserved motifs are shown against a black background, whereas the somewhat less-conserved motifs are highlighted in gray. The predicted proteolytic cleavage sites flanking HAM1h in CBSV and EuRSV are boxed, and the last C-terminal and first N-terminal amino acids of NIb and CP, respectively, are shown. Prokaryotes, Escherichia coli (sequence accession 1K7K_A), Burkholderia multivorans (YP_001945326), plant endophytes Pseudomonas fluorescens (YP_262903) and an Azoarcus sp. (CAL96580), and the root nodule-inducing and -inhabiting Rhizobium leguminosarum (CAK05869). Fungi, unicellular fungus (yeast) S. cerevisiae (CAA89597) and filamentous fungi Neurospora crassa (XP_955963) and Aspergillus fumigatus (XP_754075). Plants, rice (Oryza sativa; NP_001064763) and Arabidopsis thaliana (NP_567410). Animals, nematode Caenorhabditis elegans (AAG00041), cilial protozoan Tetrahymena thermophila (XP_977249), aphid Acyrthosiphon pisum (XP_001952278), fly Drosophila ananassae (EDV32196), mosquito Culex pipiens (XP_001843604), flour beetle Tribolium castaneum (XP_974197), frog Xenopus laevis (AAI10772), zebrafish Danio rerio (NP_001093456), and mouse Mus musculus (CAM16449), and human (Homo sapiens; AAK21848). The isolates of CBSV (origin and accession number in parentheses) are NTG10 (Ntungamo, FJ026002), IGA8 (Iganga, FJ026000), LWR2 (Luwero, FJ025999), HAM9 (Hoima, FJ039521), BAS4 (Busia, FJ026001), and BAS2 (Busia, FJ039522) from Uganda and MLB9 (Muleba, FJ025998) and MLB3 (Muleba, FJ039520) from Tanzania. The sequence of EuRSV belongs to an isolate from the United States (AY697300). (B) A neighbor-joining tree of the deduced amino acid sequences of HAM1h proteins and the aforementioned related proteins. The N and C termini upstream and downstream from the first and last semiconserved (gray shading) amino acid residue, respectively (positions 25 and 212 in HAM1 of E. coli) (see panel A) were excluded from analysis. Scale indicates Kimura units (38). (C) Percent nucleotide (values above and to the right of the asterisks in the table) and amino acid (values below and to the left of asterisks in the table) identities of HAM1h-encoding sequences among isolates of CBSV.
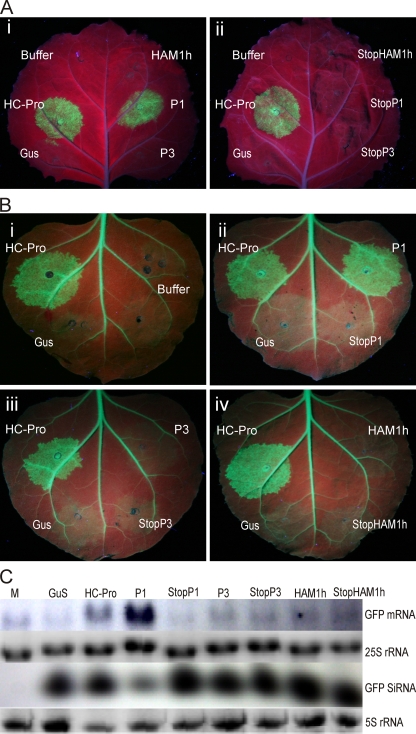
P1, P3, and HAM1h of CBSV were tested for their ability to suppress RNA silencing using the previously described agroinfiltration assay (7, 18, 21). Agrobacterium tumefaciens (strain C58c1; pGV3850) was transformed with binary vectors (pA35Shp200) each expressing one of the proteins under Cauliflower mosaic virus 35S promoter, and the 5′UTR of Potato virus A (PVA) was used for translation enhancement (21). Beta-glucuronidase (GUS) and PVA HC-Pro were included as negative and positive controls, respectively (21). Leaves of N. benthamiana were coinfiltrated with Agrobacterium strains for expression of the gfp gene for green fluorescent protein (GFP), gfp-specific double-stranded (hairpin) RNA to induce “strong gfp silencing” (18), and a third construct to express the putative silencing suppressor, as described previously (21). Silencing was suppressed by CBSV P1 and PVA HC-Pro, allowing continued GFP expression, whereas GFP fluorescence faded out by 5 days postinfiltration following infiltration with other constructs (Fig. 4A), including mutated P1-, P3-, and HAM1h-encoding sequences containing a stop codon and frameshift in the beginning of the ORF to prevent protein expression. Hence, the P1 protein was required for silencing suppression. It contains the conserved LXKA (aa 109 to 120) and zinc finger motif (cysteine residues 128, 131, 145, 148) required for efficient silencing suppression by CVYV P1b (39). Second, gfp transcripts were overexpressed by agroinfiltration in leaves of transgenic N. benthamiana constitutively expressing gfp (line 16c, courtesy of D. C. Baulcombe) (7) to induce gene cosuppression or “weak gfp silencing” (18). CBSV P1 and PVA HC-Pro suppressed gfp silencing, in contrast to the other constructs (Fig. 4B). Visual observation of GFP fluorescence correlated with the accumulation of gfp mRNA, whereas a lack of fluorescence correlated with a high accumulation of gfp-specific small interfering RNA (siRNA) detected by Northern blot analysis (Fig. 4C) using an [α-32P]rUTP-labeled gfp RNA probe (21) as described previously (30). CBSV P1 exhibited highly efficient suppression of gfp silencing and siRNA accumulation, whereas HC-Pro only partially prevented siRNA accumulation as reported previously (21, 46). Results of the three experiments were similar.
FIG. 4.
Results of an agroinfiltration assay to test suppression of RNA silencing by the P1, P3, and HAM1h proteins of CBSV. (A) “Silencing on the spot” to induce “strong silencing” of the gfp gene for GFP (18) was achieved by coexpressing gfp from one A. tumefaciens strain and double-stranded (hairpin) RNA homologous to gfp from another strain in coinfiltrated leaf tissue of N. benthamiana, and (i) coinfiltration of a third strain expressing CBSV P1, P3, or HAM1h to suppress gfp silencing. Agrobacterium strains expressing HC-Pro of PVA (genus Potyvirus) or GUS were included as the positive or negative control, respectively. “Buffer” indicates that buffer instead of the third strain was used. In panel ii, the third infiltrated strain at the right side of the leaf expressed constructs in which translation of the CBSV protein was prevented by the introduced mutations (StopP1, StopP3, and StopHAM1h). Leaves were illuminated with UV light and photographed from the underside with a digital camera 5 days postinfiltration. (B) Cosuppression of gfp in transgenic N. benthamiana plants (line 16c) constitutively expressing gfp (note the green fluorescence in veins) (7). The spots were agroinfiltrated with a mixture of two Agrobacterium strains, one expressing gfp to achieve cosuppression (silencing) of gfp and another expressing P1, P3, or HAM1h or constructs in which translation of the CBSV protein was prevented (StopP1, StopP3, and StopHAM1h). Leaves were illuminated with UV light, photographed from the underside with a digital camera, and analyzed for gfp mRNA and siRNA accumulation 5 days postinfiltration. (C) Detection of gfp mRNA and siRNA in the leaf tissues illustrated in panel B by Northern blot analysis using a radiolabeled probe. M, leaf tissue of the 16c line mock infiltrated with buffer. Upper panel, accumulation of gfp mRNA in the respective infiltrated regions. Lower panel, accumulation of gfp-derived siRNA. Ethidium bromide-stained gels of 5S rRNA were used as loading controls.
Conclusions.
Valli et al. (40) have proposed scenarios on how the hypothesized ancestor polyprotein structure P1a-P1b-HCPro-P3 has evolved to the P1a-P1b-P3 structure of CVYV and SqVYV, which has involved the adoption of silencing suppression functions by P1b and the loss of HC-Pro. Evolution in this direction is observed for SPMMV, in which the large P1 protein (82 kDa) suppresses silencing, while HC-Pro contributes only to the durability of silencing suppression (15). Furthermore, in the tritimovirus Wheat streak mosaic virus, HC-Pro does not suppress silencing and is redundant for infectivity and symptom induction (34). Our data suggest that CBSV represents the latest step of evolution which shapes the polyprotein N terminus in ipomo- and tritimo-like viruses, because CBSV lacks both P1a and HC-Pro and uses the P1b-like P1 protein for suppression of silencing.
Intriguingly, CBSV has recombined a Maf/HAM1-like sequence to the 3′-proximal part of the viral genome, which is highly conserved in Potyviridae (13). The NIb/CP junction can accommodate heterologous genes in engineered potyvirus clones without compromising infectivity (see references 4, 20, and 43 and the references therein), but natural integration of foreign sequences has not been reported. While HAMh1 could not suppress RNA silencing, homology of the protein with cellular Maf/HAM1 NTP pyrophosphatases suggests that HAMh1 might intercept noncanonical NTPs to reduce mutation rates of viral RNA. When mutation rates exceed a critical threshold, the virus may experience an “error catastrophe,” i.e., decreased infectivity and extinction of the virus population (9). For example, the presence of ribavirin, an IMP dehydrogenase inhibitor, causes depletion of the canonical GTP pool and increases the mutation rate 10-fold in poliovirus (12), which is related to viruses of the family Potyviridae (13). Suppression of the mutation rates might provide a particular advantage to the virus under conditions that are stressful to the host plant. For example, oxidative stress increases mutation rates (5) and causes early senescence (31), which is observed with older leaves of CBSD-affected cassava plants (16, 26, 35). EuRSV also infects plants of the Euphorbiaceae family (ICTVdB database, version 4) and may encode HAM1h for similar reasons. Another strategy to counteract deleterious mutations is exhibited by replicases of many viruses of the Flexiviridae and some viruses of the Closteroviridae that contain an AlkB domain similar to the AlkB proteins in plant-infecting bacteria (6). However, Blackberry virus Y which represents a putative new genus of Potyviridae contains the AlkB domain inside the P1 proteinase (36). This virus is the only Potyviridae member besides CBSV and EuRSV that is known to carry an insertion of cellular origin. AlkB domains in the aforementioned viral proteins are functional in repairing methylation damage of nucleic acids by oxidative demethylation, as also shown for the homologous proteins in cellular organisms (42). The AlkB proteins of viruses have not been found to suppress RNA silencing (42), which was also the case with CBSV HAMh1 in this study. Since a large proportion of the AlkB-containing viruses infect perennial, often woody, host plants, it is anticipated that RNA repair by AlkB proteins may be advantageous for the stability of viruses in hosts infected for long periods of time under various environmental conditions (24). It has also been proposed that conditions enhancing the methylation of nucleic acids might promote the incorporation and maintenance of AlkB sequences by viruses (6). The benefits for viral fitness from incorporation of HAM1h in CBSV and EuRSV deserve to be investigated thoroughly.
Nucleotide sequence accession number.
The CBSV genome sequence determined in this study was submitted to GenBank under accession number FJ039520.
Acknowledgments
We thank Minna-Liisa Rajamäki and Wilmer Cuellar for technical advice and Alois Kullaya, Fred Tairo, and Samuel Kyamanywa for supporting this study.
This work was part of the BIO-EARN program funded by Sida, Sweden.
Footnotes
Published ahead of print on 22 April 2009.
REFERENCES
- 1.Adams, M. J., J. F. Antoniw, and F. C. Beaudoin. 2005. Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Mol. Plant Pathol. 6471-487. [DOI] [PubMed] [Google Scholar]
- 2.Alicai, T., C. A. Omongo, M. N. Maruthi, R. J. Hillocks, Y. Baguma, R. Kawuki, A. Bua, G. W. Otim-Nape, and J. Colvin. 2007. Re-emergence of cassava brown streak disease in Uganda. Plant Dis. 9124-29. [DOI] [PubMed] [Google Scholar]
- 3.Anandalakshmi, R., G. J. Pruss, X. Ge, R. Marathe, A. C. Mallory, T. H. Smith, and V. B. Vance. 1998. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 9513079-13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arazi, T., S. H. Slutsky, Y. M. Shiboleth, Y. Wang, M. Rubinstein, S. Barak, J. Yang, and A. Gal-On. 2001. Engineering zucchini yellow mosaic potyvirus as a non-pathogenic vector for expression of heterologous proteins in cucurbits. J. Biotechnol. 8767-82. [DOI] [PubMed] [Google Scholar]
- 5.Boyko, A., F. Zemp, J. Filkowski, and I. Kovalchuk. 2006. Double-strand break repair in plants is developmentally regulated. Plant Physiol. 141488-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bratlie, M. S., and F. Drabløs. 2005. Bioinformatic mapping of AlkB homology domains in viruses. BMC Genomics 61. doi: 10.1186/1471-2164-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 176739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Burgis, N. E., and R. P. Cunnigham. 2007. Substrate specificity of the RdgB protein, a deoxyribonucleoside triphosphate pyrophosphohydrolase. J. Biol. Chem. 2823531-3538. [DOI] [PubMed] [Google Scholar]
- 9.Cases-González, C., M. Arribas, E. Domingo, and E. Lázaro. 2008. Beneficial effects of population bottlenecks in an RNA virus evolving at increased error rate. J. Mol. Biol. 3841120-1129. [DOI] [PubMed] [Google Scholar]
- 10.Chung, B. Y. W., W. A. Miller, J. F. Atkins, and A. E. Firth. 2008. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 1055897-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colinet, D., J. Kummert, and P. Lepoivre. 1998. The nucleotide sequence and genome organization of the whitefly transmitted sweet potato mild mottle virus: a close relationship with members of the family Potyviridae. Virus Res. 53187-196. [DOI] [PubMed] [Google Scholar]
- 12.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 986895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). 2005. Virus taxonomy: the eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 14.Galperin, M. Y., O. V. Moroz, K. S. Wilson, and A. G. Murzin. 2006. House cleaning, a part of good housekeeping. Mol. Microbiol. 595-19. [DOI] [PubMed] [Google Scholar]
- 15.Giner, A., M. García-Chapa, L. Lakatos, J. Burgyan, and J. J. López-Moya. 2008. Involvement of P1 and HCPro proteins of sweet potato mild mottle ipomovirus (SPMMV) in suppression of gene silencing, abstr. P3-4, p. 90. International Conference. Genetic Control of Plant Pathogenic Viruses and Their Vectors: Towards New Resistance Strategies. ResistVir, Puerto de Santa Maria, Spain. http://www.resistvir-db.org/conference_2008.htm.
- 16.Hillocks, R. J., and D. K. Jennings. 2003. Cassava brown streak disease: a review of present knowledge and research needs. Int. J. Pest Manag. 49225-234. [Google Scholar]
- 17.Janssen, D., G. Martín, L. Velasco, P. Gómez, E. Segundo, L. Ruiz, and I. M. Cuadrado. 2005. Absence of a coding region for the helper component-proteinase in the genome of Cucumber vein yellowing virus, a whitefly-transmitted member of the family Potyviridae. Arch. Virol. 1501439-1447. [DOI] [PubMed] [Google Scholar]
- 18.Johansen, L. K., and J. C. Carrington. 2001. Silencing on the spot: induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasschau, K. D., and J. C. Carrington. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95461-470. [DOI] [PubMed] [Google Scholar]
- 20.Kelloniemi, J., K. Mäkinen, and J. P. T. Valkonen. 2008. Three heterologous proteins simultaneously expressed from a chimeric potyvirus: infectivity, stability and the correlation of genome and virion lengths. Virus Res. 135282-291. [DOI] [PubMed] [Google Scholar]
- 21.Kreuze, J. F., E. I. Savenkov, W. Cuellar, X. Li, and J. P. T. Valkonen. 2005. Viral class 1 RNase III involved in suppression of RNA silencing. J. Virol. 797227-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecoq, H., C. Desbiez, B. Delécolle, S. Cohen, and A. Mansour. 2000. Cytological and molecular evidence that the whitefly-transmitted Cucumber vein yellowing virus is a tentative member of the family Potyviridae. J. Gen. Virol. 812289-2293. [DOI] [PubMed] [Google Scholar]
- 23.Li, W., M. E. Hilf, S. E. Webb, C. A. Baker, and S. Adkins. 2008. Presence of P1b and absence of HC-Pro in Squash vein yellowing virus suggests a general feature of the genus Ipomovirus in the family Potyviridae. Virus Res. 135213-219. [DOI] [PubMed] [Google Scholar]
- 24.Martelli, G. P., M. J. Adams, J. F. Kreuze, and V. V. Dolja. 2007. Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 4573-100. [DOI] [PubMed] [Google Scholar]
- 25.Maruthi, M. N., R. J. Hillocks, K. Mtunda, M. D. Raya, M. Muhanna, H. Kiozia, A. R. Rekha, J. Colvin, and J. M. Thresh. 2005. Transmission of cassava brown streak virus by Bemisia tabaci (Gennadius). J. Phytopathol. 153307-312. [Google Scholar]
- 26.Mbanzibwa, D. R., Y. P. Tian, A. K. Tugume, S. B. Mukasa, F. Tairo, S. Kyamanywa, A. Kullaya, and J. P. T. Valkonen. 2009. Genetically distinct strains of Cassava brown streak virus in the Lake Victoria basin and the Indian Ocean coastal area of East Africa. Arch. Virol. 154353-359. [DOI] [PubMed] [Google Scholar]
- 27.Monger, W. A., S. Seal, S. Cotton, and G. D. Foster. 2001. Identification of different isolates of cassava brown streak virus and development of a diagnostic test. Plant Pathol. 50768-775. [Google Scholar]
- 28.Monger, W. A., S. Seal, A. M. Isaac, and G. D. Foster. 2001. Molecular characterization of the cassava brown streak virus coat protein. Plant Pathol. 50527-534. [Google Scholar]
- 29.Noskov, V. N., K. Staak, P. V. Shcherbakova, S. G. Kozmin, K. Negishi, B. C. Ono, H. Hayatsu, and Y. I. Pavlov. 1996. HAM1, the gene controlling 6-N-hydroxylaminopurine sensitivity and mutagenesis in the yeast Saccharomyces cerevisiae. Yeast 1217-29. [DOI] [PubMed] [Google Scholar]
- 30.Pall, G. S., and A. J. Hamilton. 2008. Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 31077-1084. [DOI] [PubMed] [Google Scholar]
- 31.Schippers, J. H. M., A. Nunes-Nesi, R. Apetrei, J. Hille, A. R. Fernie, and P. P. Dijkwel. 2008. The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 202909-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scotto-Lavino, E., G. Du, and M. A. Frohman. 2006. 5′ end cDNA amplification using classic RACE. Nat. Protoc. 12555-2562. [DOI] [PubMed] [Google Scholar]
- 33.Shukla, D. D., C. W. Ward, and A. A. Brunt. 1994. The Potyviridae. CAB International, Wallingford, United Kingdom.
- 34.Stenger, D. C., B. A. Young, F. Qu, T. J. Morris, and R. French. 2007. Wheat streak mosaic virus lacking helper component-proteinase is competent to produce disease synergism in double infections with Maize chlorotic mottle virus. Phytopathology 971213-1221. [DOI] [PubMed] [Google Scholar]
- 35.Storey, H. H. 1936. Virus diseases of East African plants. VI. A progress report on studies of the disease of cassava. East Afr. Agric. J. 234-39. [Google Scholar]
- 36.Susaimuthu, J., I. E. Tzanetakis, R. C. Gergerich, and R. R. Martin. 2008. A member of a new genus in the Potyviridae infects Rubus. Virus Res. 131145-151. [DOI] [PubMed] [Google Scholar]
- 37.Takayama, S., M. Fujii, A. Kurosawa, N. Adachi, and D. Ayusawa. 2007. Overexpression of HAM1 gene detoxifies 5-bromodeoxyuridine in the yeast Saccharomyces cerevisiae. Curr. Genet. 52203-211. [DOI] [PubMed] [Google Scholar]
- 38.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 39.Valli, A., G. Dujovny, and J. A. García. 2008. Proteinase activity, self interaction, and small interfering RNA binding of the silencing suppressor P1b from cucumber vein yellowing ipomovirus. J. Virol. 82974-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valli, A., J. J. López-Moya, and J. A. García. 2007. Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J. Gen. Virol. 881016-1028. [DOI] [PubMed] [Google Scholar]
- 41.Valli, A., A. M. Martín-Hernández, J. J. López-Moya, and J. A. García. 2006. RNA silencing suppression by a second copy of the P1 serine proteinase of Cucumber vein yellowing ipomovirus, a member of the family Potyviridae that lacks the cysteine proteinase HCPro. J. Virol. 8010055-10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van den Born, E., M. V. Omelchenko, A. Bekkelund, V. Leihne, E. V. Koonin, V. V. Dolja, and P. Ø. Falnes. 2008. Viral AlkB proteins repair RNA damage by oxidative demethylation. Nucleic Acids Res. 365451-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varrelmann, M., and E. Maiss. 2000. Mutations in the coat protein gene of Plum pox virus suppress particle assembly, heterologous encapsidation and complementation in transgenic plants of Nicotiana benthamiana. J. Gen. Virol. 81567-576. [DOI] [PubMed] [Google Scholar]
- 44.Verchot, J., K. L. Herndon, and J. C. Carrington. 1992. Mutational analysis of the tobacco etch potyviral 35-kDa proteinase: identification of essential residues and requirements for autoproteolysis. Virology 190298-306. [DOI] [PubMed] [Google Scholar]
- 45.Verchot, J., E. V. Koonin, and J. C. Carrington. 1991. The 35-kDa protein from the N-terminus of the potyviral polyprotein functions as a third virus-encoded proteinase. Virology 185527-535. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, X., P. Du, L. Lu, Q. Xiao, W. Wang, X. Cao, B. Ren, C. Wei, and Y. Li. 2008. Contrasting effects of HC-Pro and 2b viral suppressors from Sugarcane mosaic virus and Tomato aspermy cucumovirus on the accumulation of siRNAs. Virology 374351-360. [DOI] [PubMed] [Google Scholar]