Abstract
Papillomavirus-like particles (VLPs) based on L1 capsid protein represent a promising prophylactic vaccine against human papillomavirus (HPV) infections. However, cell-mediated immune responses against this antigen are believed to be of limited therapeutic value in established HPV-infected cervical lesions and, for this reason, have not been intensively investigated in cervical cancer patients. In this study we analyzed and quantified by real-time PCR (RT-PCR) the RNA expression levels of E6, E7, and L1 genes in flash-frozen HPV-16 cervical carcinomas. In addition, the kinetics of expression of E6, E7, and L1 in HPV-16-infected primary cell lines established as long-term cultures in vitro was also evaluated at RNA and protein levels. Finally, in order to evaluate the therapeutic potential of L1-specific CD4+ and CD8+ T lymphocytes responses in cervical cancer patients, L1 VLP-loaded dendritic cells (DCs) were used to stimulate peripheral blood lymphocytes from cervical cancer patients and such responses were compared to those elicited by the E7 oncoprotein. We show that 22 of 22 (100%) flash-frozen cervical biopsy samples collected from HPV-16-positive cervical cancer patients harbor L1, in addition to E6 and E7 RNA, as detected by RT-PCR. E7 RNA copy number (mean, 176.2) was significantly higher in HPV-16-positive cervical cancers compared to the E6 RNA copy number (mean, 47.3) and the L1 copy number (mean, 58.3) (P < 0.0001 and P < 0.001, respectively). However, no significant differences in expression levels between E6 and L1 were found. Kinetic studies of E6, E7, and L1 RNA and protein expression levels in primary tumors showed a sharp reduction in L1 expression after multiple in vitro passages compared to E6 and E7. Autologous DCs pulsed with HPV-16 VLPs or recombinant full-length E7 elicited strong type 1 L1- and E7-specific responses in CD4+ and CD8+ T cells from cervical cancer patients. Importantly, L1 VLP-specific CD8+ T lymphocytes expressed strong cytolytic activity against autologous tumor cells and were as effective as E7-specific cytotoxic T lymphocytes in lysing naturally HPV-16-infected autologous tumor cells. Taken together, these data demonstrate a consistent expression of L1 in primary cervical tumors and the possibility of inducing effective L1/tumor-specific CD4+ and CD8+ T-lymphocyte responses in patients harboring HPV-infected cervical cancer. These results may have important implications for the treatment of patients harboring established HPV-infected lesions with L1 VLPs or combined E7/L1 DC-based vaccinations.
Human papillomavirus (HPV) infection represents the most important risk factor for the development of cervical cancer. Although more than 100 distinct HPV genotypes have been described, and at least 20 are associated with cervical cancer, HPV type 16 (HPV-16) is by far the most frequently detected in cervical neoplasia regardless of the geographical origin of the patients (4). In the last few years significant advances have been made in the development of candidate prophylactic vaccine against cervical cancer and HPV-related infections. In several large prospective randomized studies, virus-like particles consisting of the HPV-16 and HPV-18 major capsid protein L1 (L1-VLPs) have shown promise in protecting young healthy females against persistent infection with HPV-16 and HPV-18 and their associated cervical intraepithelial neoplasia (reviewed in reference 12). These data strongly suggest that the implementation of large-scale L1-VLP-based prophylactic vaccinations have the potential to dramatically reduce worldwide cervical cancer rates in the years to come.
Unfortunately, because HPV infection is endemic in humans and there is a long latency from HPV infection to the development of invasive cervical cancer in women, even if prophylactic L1-based vaccinations are implemented on a worldwide scale today it would take decades to perceive any significant benefit. Consistent with this view, an estimated 5 million cervical cancer deaths will occur in the next 20 years due to existing HPV infections (4, 12). Thus, the current development of therapeutic vaccines for protection against persistent HPV infections, cervical cancer, and its precursor lesions remains an area of great interest.
Although the interactions between the host immune system and HPV-infected cells are still not completely understood, several lines of evidence suggest that protection against HPV-related infections by L1-VLP-based vaccines is likely conferred by the generation of high levels of neutralizing antibodies (12, 38). Nevertheless, a potential crucial role of L1-specific T-cell responses and the involvement of T cells in mediating the production of neutralizing antibodies and antiviral effect in infected hosts has been previously hypothesized (8, 24). This point may be particularly noteworthy in patients harboring HPV-infected cervical lesions because several studies have demonstrated the critical importance of both cytotoxic (CD8+) and helper (CD4+) T cells in achieving clinical responses (1, 5, 16-18, 20, 23). However, limited information is currently available to evaluate whether cell-mediated immune responses to L1-VLP may have any significant therapeutic effect in cervical cancer patients harboring HPV-16 positive tumors. Furthermore, to our knowledge, no direct comparison of the therapeutic efficacy of L1 and E7-specific immune responses against naturally HPV-16-infected cervical cancer have been yet reported in human patients.
In the present study we have analyzed and quantified by highly sensitive real-time PCR (RT-PCR) the RNA levels of E6, E7, and L1 in flash-frozen biopsy specimens obtained from HPV-16-infected cervical carcinomas and in short- and long-term primary cultures of HPV-16-positive cervical tumors. In addition, we have studied the kinetics of expression of these genes and proteins during the establishment of HPV-16-positive primary tumors in vitro. Finally, using completely autologous systems of naturally infected HPV-16-positive human tumors, we have carefully studied the phenotype and function of L1-specific CD4+ and CD8+ T-lymphocyte responses generated by VLP-loaded dendritic cells (DCs) and compared their therapeutic potential to those elicited by DC loaded with the E7 oncoprotein.
MATERIALS AND METHODS
Flash-frozen tumor biopsy samples and establishment of primary HPV-16-positive cervical cancer cell lines.
HPV-16-positive flash-frozen biopsy specimens from a total of 22 patients diagnosed with frankly invasive stage IB and IIIB cervical cancer (staged according to the F.I.G.O. clinical staging system) were obtained at the time of surgery and/or staging through the Gynecologic Oncology Division and the Pathology Department, University of Arkansas for Medical Sciences, and the University of Brescia School of Medicine, under the approval of the institutional review boards. Each sample was histologically analyzed by a staff pathologist, and only tumor samples containing at least 90% tumor epithelial cells were retained for RNA extraction. All biopsy samples were sharp dissected and snap-frozen in liquid nitrogen within 30 min from collection. Patient characteristics are presented in Table 1. Normal keratinocyte control samples were obtained from cervical biopsy samples of hysterectomy specimens from women diagnosed with benign disease and a previous report of a normal cervical cytological evaluation. Establishment of primary cell lines from HPV-16-positive cervical tumor samples was performed as previously described (31). Briefly, single cell suspensions were obtained by processing tumor samples under sterile conditions at room temperature. Viable tumor tissue was mechanically minced in RPMI 1640 to portions no larger than 1 to 2 mm3 and washed twice with RPMI 1640. The portions of minced tumor were then placed into 250-ml flasks containing 30 ml of enzyme solution (0.14% collagenase type I [Sigma] and 0.01% DNase [Sigma] at 2,000 Kunitz units (KU)/mg) in RPMI 1640 and incubated on a magnetic stirring apparatus either for 2 h at 37°C or overnight at 4°C. Enzymatically dissociated tumor was then filtered through 150-μm-pore-size nylon mesh to generate a single cell suspension. The resultant cell suspension was then washed twice in RPMI 1640 plus 10% human AB serum (Gemini Bioproducts, Calabasas, CA) before being seeded in tissue culture flasks in serum-free keratinocyte medium, supplemented with 5 ng of epidermal growth factor/ml and 35 to 50 μg of bovine pituitary extract/ml (Invitrogen, Grand Island, NY) at 37°C. The purity and homogeneity of fresh tumor cultures was tested by morphology, immunohistochemistry staining, and/or flow cytometry with antibodies against cytokeratins and p16 (CDKN2). In this regard, only primary cultures that had at least 95% viability and contained >99% epithelial cells expressing p16 were used for total RNA extraction in kinetic studies and cytotoxicity assays (see below).
TABLE 1.
Characteristics of cervical cancer patients and expression of E6, E7, and L1 mRNA copy numbers by RT-PCR
| Patient | Age (yr) | Stage | Grade | Histology | Mean no. of copies
|
||
|---|---|---|---|---|---|---|---|
| E6 | E7 | L1 | |||||
| Kcx1 | 46 | IB | G2 | Squamous | 8 | 42.25 | 0.62 |
| Kcx2 | 41 | IIB | G2 | Squamous | 12.13 | 194 | 0.38 |
| Kcx3 | 45 | IB | G1 | AdenoCa | 45.25 | 239 | 7.46 |
| Kcx4 | 32 | IB | G1 | AdenoCa | 168.9 | 238.8 | 388 |
| Kcx5 | 51 | IIB | G2 | Squamous | 4.29 | 13.93 | 0.44 |
| Kcx6 | 59 | IB | G2 | Squamous | 59.7 | 315.17 | 194 |
| Kcx7 | 39 | IB | G3 | Squamous | 22.63 | 222.86 | 0.12 |
| Kcx8 | 42 | IIB | G3 | Squamous | 64 | 256 | 27.86 |
| Kcx9 | 47 | IB | G2 | Squamous | 73.52 | 315.17 | 78.79 |
| Kcx10 | 39 | IIB | G2 | Squamous | 13.93 | 97 | 3.73 |
| Kcx11 | 45 | IIIB | G3 | Squamous | 26 | 137.2 | 8 |
| Kcx12 | 43 | IB | G3 | Squamous | 59.7 | 337.8 | 0.1 |
| Kcx13 | 51 | IB | G2 | Squamous | 12.1 | 59.7 | 1.15 |
| Kcx14 | 52 | IIB | G2 | Squamous | 50.6 | 292 | 29.2 |
| Kcx15 | 23 | IB | G2 | Squamous | 54.2 | 127.1 | 18 |
| Kcx16 | 42 | IB | G3 | Squamous | 27.1 | 59.3 | 0.1 |
| Kcx17 | 52 | IIIB | G2 | Squamous | 176 | 254.2 | 288 |
| Kcx18 | 48 | IB | G2 | Squamous | 2.1 | 4.6 | 1.6 |
| Kcx19 | 22 | IB | G2 | Squamous | 8.8 | 6.6 | 2 |
| Kcx20 | 38 | IB | G2 | Squamous | 18.1 | 18.5 | 42.5 |
| Kcx21 | 29 | IB | G2 | Squamous | 20 | 11.4 | 0.1 |
| Kcx22 | 71 | IB | G2 | Squamous | 114.5 | 634.7 | 190 |
HPV genotyping.
Sequence-specific primers for the L1 and E7 proteins were used to confirm or exclude by PCR the presence of the HPV-16 genotype in a total of 41 fresh-frozen tumor biopsy samples, three primary (Kcx19, Kcx21, and Kcx22) and four established cell lines (i.e., CasKi, SiHa, HT-3, and C33A, obtained from American Type Culture Collection, Rockville, MD), as well as four normal cervical keratinocyte controls. Briefly, for HPV-16 E7, 0.1 to 1 μg of each genomic DNA sample was amplified in a 50-μl reaction containing 0.3 μM concentrations of each of the individual primers (5′-ATG GAG ATA CAC CTA CAT TGC-3′ [forward] and 5′-GGT TTC TGA GAA CAG ATG GGG C-3′ [reverse]) in the presence of 1× PCR buffer, 2.5 μM MgCl2, 0.8 μM deoxynucleoside triphosphates, and 0.025 U of AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA)/μl. Amplifications were performed in the Applied Biosystems GeneAmp PCR System 2700 (Applied Biosystems) at 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min, with a final extension of 72°C for 7 min. For HPV-16 L1, 0.1 to 1 μg of each genomic DNA sample was amplified in a 50-μl reaction containing 0.3 μM concentrations of each of the individual primers (5′-TCT ACT TGC CTC CTG TCC CAG TAT-3′ [forward] and 5′-CCA GCC GCT GTG TAT CTG GAT TAT-3′ [reverse]) in the presence of 1× PCR buffer, 2.5 μM MgCl2, 0.8 μM deoxynucleoside triphosphates, and 0.025 U of AmpliTaq DNA polymerase (Applied Biosystems)/μl. Amplifications were performed in the Applied Biosystems GeneAmp PCR System 2700 (Applied Biosystems) at 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min, with a final extension of 72°C for 7 min. The PCR products were stored at 4°C before electrophoresis on a 2% agarose gel. β-Tubulin gene amplification performed with the primers 5′-CGC ATC AAC GTG TAC TAC AA-3′ (forward) and 5′-TAC GAG CTG GTG GAC TGA GA-3′ (reverse) (each at concentrations of 0.25 μM) was used as a positive internal control. SiHa and Caski cervical carcinoma cell lines DNA (HPV-16 positive) were used as a positive external control. C33a and HT3 (HPV-negative) carcinoma cell lines DNA and a water template were used as negative controls.
L1, E6, and E7 expression by qRT-PCR.
The amount of L1, E6, and E7 HPV-16 RNA copy number was quantified by real time-PCR analysis in all 22 flash-frozen HPV-16-positive cervical cancer biopsy specimens. Control samples included in all of the assays were RNA extracted from four HPV-16-negative flash-frozen biopsy specimens derived from normal cervical keratinocytes, two established HPV-16-negative cancer cell lines (i.e., C33a and HT-3), and two HPV-16-positive cell lines (i.e., SiHa and CaSki) which harbor 1 to 2 and >500 copies of HPV-16 viral genome, respectively (6). In addition, to evaluate whether the expression of viral genes in HPV-16-infected cervical tumors may vary due to in vitro manipulation and serial passages, the kinetics of expression of L1, E6, and E7 HPV-16 RNA were evaluated in two primary naturally HPV-16-infected cell lines established in our laboratory at different time points of their in vitro growth (i.e., passage 0 [P0] up to passage P23). Quantitative RT-PCR (qRT-PCR) was performed with an ABI Prism 7500 sequence analyzer using the manufacturer's recommended protocol (Applied Biosystems). Each reaction was run in duplicate. The comparative threshold cycle (CT) method was used for the calculation of amplification fold as specified by the manufacturer. Briefly, 5 μg of total RNA from each sample was reverse transcribed by using SuperScript III first-strand cDNA synthesis (Invitrogen). Portions (10 μl) of reverse-transcribed RNA samples (from 500 μl of the total volume) were amplified by using the TaqMan Universal PCR master mix (Applied Biosystems) to produce PCR products specific for L1, E6, and E7 HPV-16. Primers specific for GAPDH (assays ID Hs99999905_m1) and 18S rRNA and empirically determined ratios of 18S competimers (Applied Biosystems) were used as control for the amounts of cDNA generated from each sample. Primers for L1, E6, and E7 HPV-16 were obtained from Applied Biosystems. The L1 primer sequences were 5′-CCC AAA CGC ACC GAA TAG TTA C-3′ (forward) and 5′-ATT CCA ATT CCC CTG CAA ACT-3′ (reverse). E6 primer sequences were 5′-CCC AAA CGC ACC GAA TAG TTA C-3′ (forward) and 5′-ATT CCA ATT CCC CTG CAA ACT-3′ (reverse). E7 primer sequences were 5′-CCC AAA CGC ACC GAA TAG TTA C-3′ (forward) and 5′-ATT CCA ATT CCC CTG CAA ACT-3′ (reverse). The number of copies of HPV-16 RNA per sample was estimated by subtracting the CT number at which HPV-16 was detected from that at which GAPDH was detected, to give the δCT for the sample. The δCT derived for the HPV-16-positive cervical carcinoma cell line SiHa, which has a known HPV-16 load of one to two copies/cell (6), was then subtracted from the sample δCT to give the sample δδCT. The formula 2δδCT gives the approximate number of RNA copies of HPV-16 per sample relative to SiHa cells.
Western blot analysis.
To validate the qRT-PCR kinetic data at the protein level, E7 and L1 protein expression were evaluated in Kcx22 (i.e., one of the primary HPV-16-positive cell lines established as short- and long-term culture and used in the kinetic studies and cytotoxicity experiments). Briefly, primary Kcx22 cells grown to confluence in T150 cm flasks were harvested at early passages (i.e., P3) and late passage (i.e., P23) and compared for E7 and L1 expression by Western blot analysis before being tested in cytotoxicity experiments (see below). Briefly, cells were harvested and washed once with phosphate-buffered saline (PBS). Supernatants were removed, and cell pellets were transferred to ice and immediately resuspended in 0.5 ml of lysis buffer (10 mM Tris-HCl [pH 7.4], 1% sodium dodecyl sulfate [SDS], 1 mM sodium metavanadate) with 1 mM phenylmethylsulfonyl fluoride and 10 μg of aprotinin, leupeptin, and pepstatin/ml. The lysates were passed 10 times through a 20-gauge needle fitted to a 3-ml syringe and then sonicated five times for 5 s and 5 s pause on an ice bed. After 30-min incubation on ice the samples were centrifuged at 14,000 rpm for 5 min to eventually eliminate the pellet of debris. Total protein concentration was then determined by using Bio-Rad protein assay dye (catalog no. 500-0006; Bio-Rad, Hercules CA) according to manufacturing instructions. Aliquots of the total cell lysate were then prepared and stored at −20°C. Electrophoresis of the samples in SDS-10% polyacrylamide gel for L1 (20 μg of total lysate loaded) and SDS-15% polyacrylamide gel for E7-16 (20 μg of total cell lysate loaded) were then performed. Recombinant L1-VLP (1 ng) and E7-16 (0.1 ng) were used as positive controls and C33a cell line (20 μg of total cell lysate loaded) was used as a negative control. Blotting to nitrocellulose membrane (Hybond ECL, catalog no. RPN203D; Amersham Biosciences, Piscataway, NJ) was performed for 45 min at 4°C. The membranes were then incubated for 30 min at room temperature with 0.5% glutaraldehyde and washed twice with 0.5% PBS-Tween 20. After blocking with 5% milk for 90 min at room temperature, the membranes were washed at room temperature with 0.5% PBS-Tween 20 and probed with anti-L1 (CAMVIR-1, catalog no. 554171; BD Pharmingen San Diego CA) and anti-E7-16 (ED17, catalog no. sc-6981; Santa Cruz Biotechnology, Santa Cruz, CA). Primary antibodies dilution were 1:5,000 for L1 and 1:500 for E7-16, and incubations were done overnight at 4°C. Secondary antibody incubations were done at room temperature for 1 h using goat anti-mouse antibody at 1:1,000 dilution for both L1 and E7-16. Detection of signal was obtained by using ECL Western blotting detection reagents from Amersham Biosciences.
Production of E7 proteins and L1-VLPs.
Purified E7 oncoprotein of HPV-16 was generated using a previously characterized plasmid encoding glutathione S-transferase (GST) E7 fusion protein as previously described (31). Briefly, GST and derivative fusion protein was maintained in Escherichia coli BL21, and protein expression was induced in cultures at an optical density at 600 nm of 0.6 by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration, 0.1 mM). Cultures were grown for 6 h for large-scale preparations (2,000 to 4,000 ml). Cells were pelleted and washed once in ice-cold PBS (pH 7.4) and then resuspended for disruption using sonication on ice in short bursts. We then added 20% Triton X-100 in PBS to a final concentration of 1%, followed by incubation with gentle mixing for 30 min to aid solubilization of the fusion proteins. Bacterial lysate was centrifuged at 12,000 × g for 10 min at 4°C, and the supernatant was purified by using a glutathione-Sepharose 4B RediPack column in accordance with the procedures suggested by the manufacturer (Pharmacia Biotech., Inc., Piscataway, NJ). Cleavage of E7 oncoprotein from GST was then achieved by using a site-specific protease (i.e., human thrombin; Sigma, St. Louis, MO) at 100 U in 2 ml of PBS at room temperature for 16 h. E7 protein was eluted from the column by spinning (1,000 × g for 5 min), and the collected material was filter sterilized. Purity of the protein was analyzed by SDS-polyacrylamide gel electrophoresis and Coomassie blue staining, while quantification was obtained spectrophotometrically using the Bio-Rad protein assay. Preparations were typically >98% pure E7 oncoprotein with a level of endotoxin not exceeding 1 endotoxin U/ml (31). Baculovirus produced and previously characterized HPV-16 L1-VLPs were kindly provided by J. T. Schiller (National Institutes of Health, Bethesda, MD).
Isolation of peripheral blood mononuclear cells (PBMC) and generation of DCs.
PBMC were separated from heparinized venous blood by Ficoll-Hypaque (Sigma) density gradient centrifugation and either cryopreserved in RPMI 1640 (Gibco-BRL, Grand Island, NY) plus 10% dimethyl sulfoxide and 30% autologous plasma or immediately used for DC generation. Briefly, PBMC obtained from 42 ml of peripheral blood were placed into six-well culture plates (Costar, Cambridge, MA) in AIM-V (Gibco-BRL, Grand Island, NY) at 0.5 × 107 to 1 × 107/3 ml per well. After 2 h at 37°C, nonadherent cells were removed, and the adherent cells were cultured at 37°C in a humidified 5% CO2-95% air incubator in medium supplemented with recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF; 800 U/ml; Immunex, Seattle, WA) and interleukin-4 (IL-4; 1,000 U/ml; Genzyme, Cambridge, MA). Every 2 days, 1 ml of spent medium was replaced by 1.5 ml of fresh medium containing 1,600 U of GM-CSF/ml and 1,000 U of IL-4/ml to yield final concentrations of 800 and 500 U/ml, respectively. Final maturation of monocyte-derived DCs was induced by exposure during the last 48 h of culture (i.e., days 5 to 7) to tumor necrosis factor alpha (1,000 U/ml), IL-1β (500 U/ml; R&D Systems), and PGE2a (0.5 μM/ml; Sigma) (15). The DC purity (i.e., cells strongly expressing HLA-DR+, CD86+, CD83+, CD80+, CD40+, and CD14−) by flow cytometry ranged from 62 to 90% of the total cell population (26). After final maturation (see below), DCs were harvested to be used for priming experiments in vitro.
DC pulsing.
Two methods of DC pulsing were used in these studies with comparable results in multiple experiments. In the first (i.e., overnight pulsing), immature DCs (i.e., after 4 days of culture) were loaded overnight with 50 μg of HPV-16 L1-VLPs or HPV-16 E7/ml before inducing their final maturation (i.e., days 5 to 7) with the cocktail of the inflammatory cytokines (26). In the second, fully mature DCs (i.e., day 7 of culture) were harvested and washed twice in AIM-V and added to 50-ml polypropylene tubes (Falcon, Oxnard, CA). The cationic lipid DOTAP (Boehringer Mannheim, Indianapolis, IN) was used to deliver the HPV-16 L1-VLPs or HPV-16 E7 proteins into cells (28). Briefly, 50 μg of L1 VLPs or E7 oncoprotein and DOTAP (125 mg in 500 ml of AIM-V)/ml were mixed in 12-by-75-mm polystyrene tubes at room temperature for 20 min. The complex was added to the DCs in a total volume of 2 to 5 ml of AIM-V, followed by incubation at 37°C in an incubator with occasional agitation for 3 h. The cells were washed twice with PBS and resuspended in AIM-V as described below.
In vitro generation of HPV L1- and E7-specific CTLs.
Fresh or cryopreserved responder PBMC were washed and resuspended in AIM-V at 10 × 106 to 20 × 106 cells/well in six-well culture plates (Costar) with HPV-16 L1-VLPs or HPV-16 E7-pulsed autologous DC (ratios from 20:1 to 30:1 responders [PBMC/DC]). The cultures were supplemented with recombinant human GM-CSF (500 U/ml) and incubated at 37°C. Human recombinant IL-2 (10 U/ml, Aldesleukin; Chiron Therapeutics, Emeryville, CA) was added to the cultures after 7 days. In the majority of the experiments, a second stimulation with HPV-16 L1-VLPs or HPV-16 E7 pulsed autologous DCs was done at 14 to 21 days before CD8+ cells were separated from the bulk cultures by positive selection with CD8 Dynabeads (Dynal, Inc., Lake Success, NY). CD8+ T cells were further expanded in number for 5 to 7 days using autologous or allogeneic irradiated peripheral blood lymphocytes (5,000 cGy) at 106 cells/well and anti-CD3 monoclonal antibody (MAb; Ortho Pharmaceutical Corp., Raritan, NJ) at 0.2 mg/ml plus 5% autologous plasma and 100 U of IL-2/ml in 24-well plates (Costar), before being assayed for cytotoxic-T-lymphocyte (CTL) activity. Epstein-Barr virus (EBV)-transformed autologous lymphoblastoid B-cell lines (LCL) were established by coculture of PBMC with EBV-containing supernatant from the B95.8 cell line in the presence of 1 mg of cyclosporine (Sandoz, Camberley, United Kingdom)/ml and were maintained in AIM-V supplemented with 10% human AB serum (Gemini Bioproducts, Calabasas, CA).
Cytotoxic activity.
A 5-h chromium (51Cr) release assay was performed as previously described (31) to measure the cytotoxic reactivity of DC HPV-16 L1-VLPs or HPV-16 E7-stimulated CD8+ T lymphocytes. In addition to autologous cervical tumor cells, EBV-transformed autologous LCL were used as HLA identical HPV-negative control targets. To determine the structures on the target cells involved in lysis, MAbs were used to block cytotoxicity. 51Cr-labeled tumor targets were preincubated with MAbs specific for monomorphic HLA class I W6/32 (50 μg/ml) or isotype control (immunoglobulin G1K [IgG1K] MAb isotype standard anti-TNP, 10 μg/ml; Pharmingen, San Diego, CA). The effector cells and 51Cr-labeled targets were then incubated in a final volume of 200 ml/microwell at 37°C with 6% CO2.
T-cell proliferation assay.
CD4+ T cells, derived from CD8+-depleted T-cell populations were restimulated once with L1- and E7-pulsed DCs at a 20:1 ratio, and 2 weeks later were enriched by positive selection with CD4-Dynabeads (Dynal, Inc., Lake Success, NY) to obtain a population that was more than 99% pure. Specific lymphoproliferative responses against E7 and L1 were tested using autologous DCs or LCL pulsed with HPV-16 L1-VLPs or HPV-16 E7 in the presence of DOTAP as described above for DC pulsing. Irradiated (5,000 cGy) E7- or L1-pulsed or unpulsed autologous DCs or LCL were seeded in a 96-well/plates (5 × 104 to 20 × 104 cells/well) with different numbers of CD4+ T cells and tested for specific proliferation after 72 h. Cultures were pulsed with 1 mCi of [3H]thymidine/well for the last 16 h, and the incorporated radioactivity was measured as described previously (31). All assays were carried out in triplicate wells.
Phenotypic analysis of T cells.
Enriched cultures of CD8+ T cells from E7- and L1-stimulated DCs were phenotyped at the time of first cytotoxicity and thereafter in order to correlate cytolytic specificity with a particular lymphoid subset. In addition, because recent reports have suggested an association of cervical cancer with the presence of CD4+ regulatory T cells (Tregs) specific for HPV antigens (37), enriched cultures of E7- and L1-stimulated CD4+ T cells were phenotyped at the time of the first proliferation assays to evaluate a potential difference in the number of CD4+ CD25+ FOXP3+ Tregs and CD4+ CD25+ FOXP3− effector T cells. Flow cytometry was performed using MAbs directly conjugated against the following human leukocyte antigens: Leu-4 (CD3, pan-T cells), Leu-3 (CD4, T helper/inducer), Leu-2a (CD8, T cytotoxic/suppressor), Leu-19 (CD56, NK/K cells), anti-CD25 fluorescein isothiocyanate (FITC), and anti-TcR-α/β or TcR-γ/δ (Becton Dickinson, San Jose, CA). FOXP3 expression by CD25+ CD4+ T cells was evaluated by using an Alexa Fluor 647 anti-human FOXP3 staining kit (BD Biosciences, San Jose, CA) and analyzed on a FACScan (Becton Dickinson).
Flow cytometric analysis of intracellular cytokines in E7 and L1-specific CTLs.
HPV-16 L1- and E7-specific cytotoxic CD8+ T cells were tested at about 7 to 10 weeks after priming. Cells were rested for 14 days after last antigen stimulation prior to activation by solid-phase OKT-3. Unstimulated and PMA and ionomycin stimulated CD8+ T cells were used as negative and positive controls, respectively. Briefly, T cells (7.5 × 105/ml) were incubated overnight at 37°C on solid-phase OKT-3 in AIM-V 5% autologous plasma plus 10 μg of brefeldin A/ml. Similar numbers of CD8+ T-cell populations were incubated at 37°C for 6 h in AIM-V 5% autologous plasma plus 50 ng of phorbol myristate acetate/ml and 500 ng of ionomycin/ml. Then, 10 μg of brefeldin A/ml was added for the final 3 h of incubation. Controls (nonactivated cultures) were incubated overnight in the presence of brefeldin A only. The cells were harvested, washed, and fixed with 2% paraformaldehyde in PBS for 20 min at room temperature, after which they were washed and stored overnight in PBS at 4°C. Cells were then washed and permeabilized by incubation in PBS plus 1% bovine serum albumin (BSA) and 0.5% saponin (S-7900; Sigma) for 10 min at room temperature for intracellular staining. Activated and control cells were stained with FITC-conjugated anti-IFN-γ and phycoerythrin (PE)-conjugated anti-IL-4 and isotype-matched controls (FITC-anti-IgG2a and PE-anti-IgG1) from Becton Dickinson. After staining, cells were washed twice with PBS plus 1% BSA and 0.5% saponin, once with PBS plus 0.5% BSA, and fixed a second time with 2% paraformaldehyde in PBS. All analyses were conducted with a FACScan, utilizing CellQuest software (Becton Dickinson).
Statistics.
Significance analysis was performed by using the Student t test for paired data. Only P values of <0.05 were considered significant.
RESULTS
Determination of HPV-16 L1, E6, and E7 RNA expression levels by qRT-PCR in flash-frozen cervical cancer biopsy samples.
Fresh-frozen biopsy samples from 22 cervical cancer patients out of 41 tested (53%) were found to be positive for the presence of HPV-16 infection by PCR using HPV-16 E7- and L1-specific primers and further evaluated by qRT-PCR. Patient characteristics are presented in Table 1. With no exception, tumors found to be positive for HPV-16 by conventional PCR using E7 specific primers were also positive when HPV-16 L1-specific primers were used (data not shown). Mirror sections of the tumor biopsy specimens used for DNA genotyping were used for total RNA extraction and real-time PCR evaluation. HPV-16 L1, E6, and E7 RNA expression levels in HPV-16-positive flash-frozen tumor biopsy samples were normalized to the expression levels found in SiHa cell line as described in Materials and Methods. Although L1, E6, and E7 expression was not detectable in any of the HPV-16-negative cervical keratinocyte control biopsy samples or in the two HPV-16-negative cell lines (i.e., C33a and HT3) tested (data not shown), expression of L1, E6, and E7 was found in 100% (i.e., 22 of 22, Table 1) of the HPV-16-positive cervical cancer biopsy specimens, as well as in both of the HPV-16-positive control cell lines (i.e., SiHa and Caski) tested. The E7 RNA copy numbers (mean, 176.2) were found to be significantly higher than the copy numbers detected for E6 (mean, 47.3) and L1 (mean, 58.3) (P < 0.0001 and P < 0.001, respectively) in the same tumors. Surprisingly, no significant differences in expression levels between E6 and L1 were found in the fresh-frozen cervical tumor tested (P = 0.449). Indeed, the level of expression of L1 was higher than the expression levels of E6 in 6 of 22 evaluated samples (Table 1).
Kinetics of expression of L1, E6, and E7 mRNA as determined by qRT-PCR in primary HPV-16-positive cervical cancer cell lines.
To evaluate whether the expression of HPV viral genes in naturally HPV-16-infected cervical tumors is significantly affected by their growth and manipulation in vitro, the kinetics of expression of L1, E6, and E7 HPV-16 RNA were evaluated at different time points of in vitro culture (i.e., different in vitro passages) in two primary naturally HPV-16-infected cell lines recently established in our laboratory (i.e., Kcx21 and Kcx22). As representatively shown in Fig. 1 for the Kcx22 primary tumor cell line, the expression levels of E6, E7, and L1 were found to consistently decrease during the in vitro culture of both primary tumors tested (Table 2). Importantly, however, while the expression levels of E6 and E7 remained well above the expression levels found in SiHa (i.e., the calibrator HPV-16-positive cell line) even after 20 in vitro passages, L1 expression levels were found to drastically decrease after P5 in both primary tumors tested with minimal L1 expression detectable after P10 (Fig. 1 and Table 2).
FIG. 1.
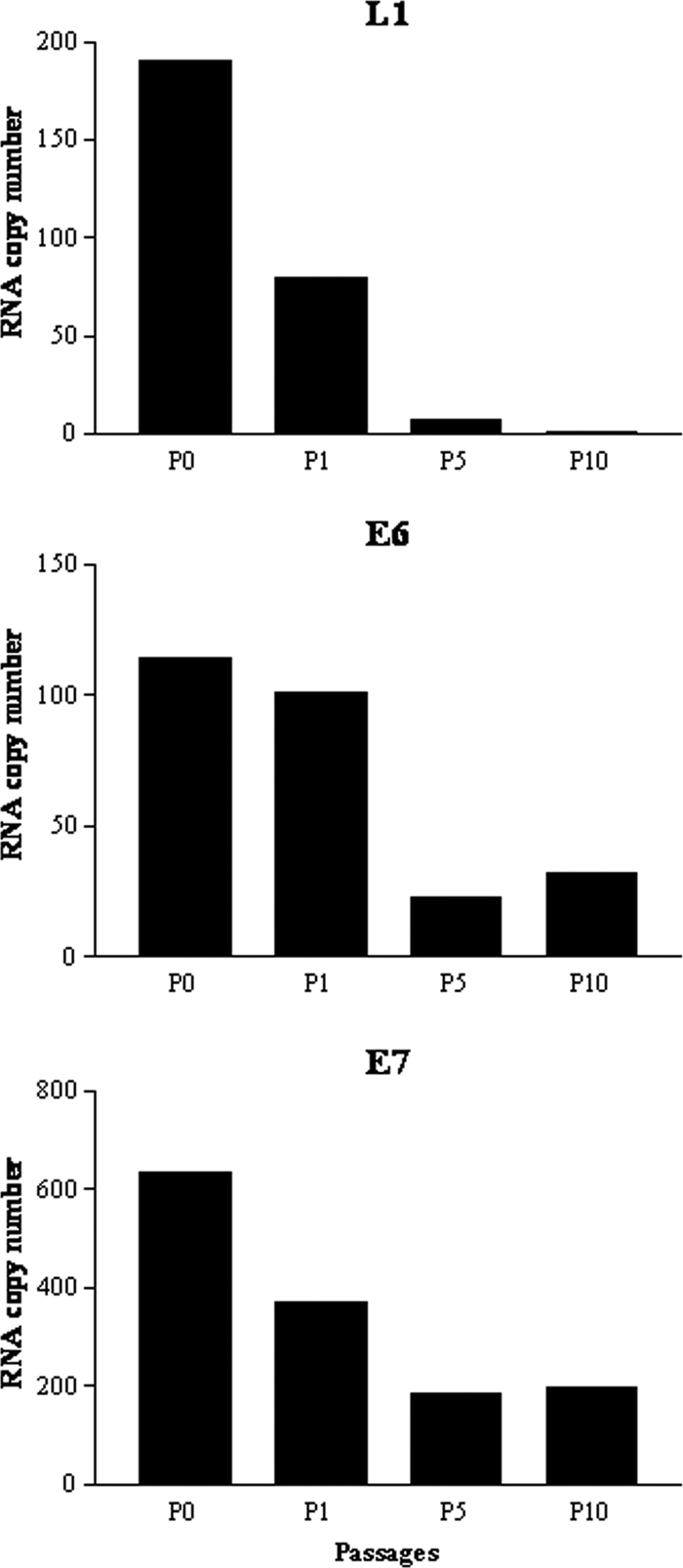
Representative qRT-PCR analysis of L1, E6, and E7 expression in a Kcx22 primary HPV-16-positive cell line at different time points of in vitro culture. y axis, fold induction relative to SiHa expression; x axis, each sample from Kcx22 tested for L1, E6, and E7 expression at different time points (i.e., tumors never passed in culture to a new flask = P0, as well as P1, P5, and P10). A clear downward trend of L1 expression to almost undetectable levels over passage number is evident.
TABLE 2.
Expression of E6, E7, and L1 mRNA in primary tumors at different time points of in vitro culture
| Patient | Time point (passage [P]) | mRNA expression
|
||
|---|---|---|---|---|
| E6 | E7 | L1 | ||
| Kcx21 | P0 | 20 | 11.4 | 0.14 |
| Kcx21 | P2 | 18 | 6.3 | 0.19 |
| Kcx21 | P3 | 9.8 | 4.2 | 0.09 |
| Kcx21 | P7 | 7.9 | 3 | 0.01 |
| Kcx21 | P10 | 7 | 3 | 0.01 |
| Kcx22 | P0 | 114.56 | 634.73 | 190 |
| Kcx22 | P1 | 101.13 | 369.65 | 79.34 |
| Kcx22 | P5 | 22.63 | 184.82 | 7.31 |
| Kcx22 | P6 | 8.11 | 87.43 | 0.12 |
| Kcx22 | P20 | 2.03 | 14.72 | 0.06 |
Kinetics of expression of HPV-16 L1 and E7 proteins as determined by Western blot analysis in the HPV-16-positive Kcx22 cervical cancer cell line.
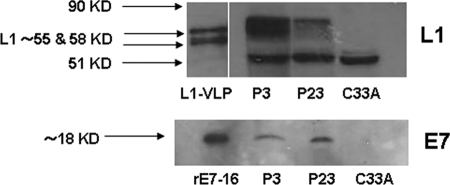
To validate the experimental kinetic results obtained by RT-PCR at protein level, L1 and E7 HPV-16 protein expression were analyzed by Western blotting in the Kcx22 primary cell line (i.e., one of the primary autologous tumor used in the cytotoxicity experiments, see below) at P3 and P23 of in vitro culture. As representatively shown in Fig. 2, the protein expression level of E7 was similar when P3 and P23 were compared by Western blot analysis (lower panel). In contrast, a decrease in the L1 protein expression level was consistently noted by comparing early (P3) versus late (P23) in vitro passages in multiple experiments (Fig. 2, upper panel).
FIG. 2.
Representative Western blot analysis of L1 (upper panel) and E7 (lower panel) expression in the Kcx22 primary HPV-16-positive cell line at P3 and P23 of in vitro culture. Unlike E7 protein, a visible reduction in L1 protein expression level was consistently noted by comparing early (P3) versus late (P23) in vitro passages in multiple experiments. L1-VLP, recombinant L1 protein; rE7, recombinant E7 protein; C33A, HPV-16-negative control cell line. The double band at approximately ca. 55 to 58 kDa corresponds to posttranslational modification of L1, while the band at ∼51 kDa represents a cellular protein cross-reacting with MAb Camvir-1 (33).
L1-specific DC-stimulated CD8+ CTLs are able to kill autologous cervical cancer cells in vitro.
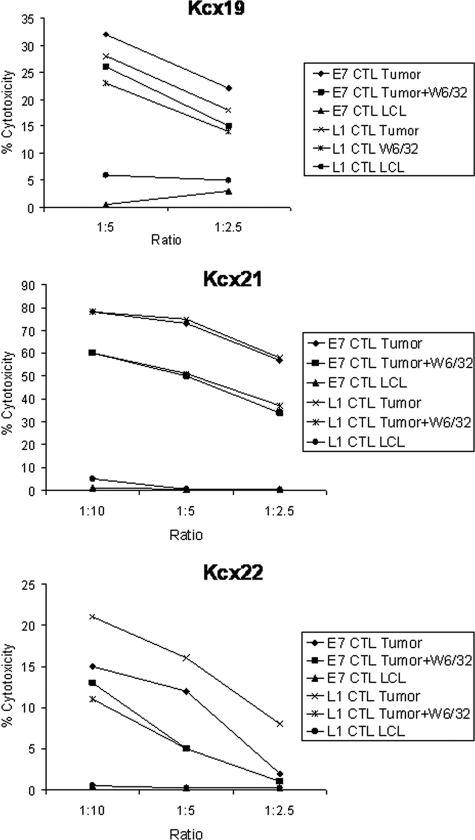
To evaluate whether L1 viral protein may be used as a therapeutic target to recognize and kill HPV-16-positive cervical tumor cells, L1-VLP-pulsed DCs from three different cervical cancer patients were used to elicit tumor-specific CTLs in vitro. Because real-time PCR and Western blot kinetics studies showed a consistent reduction in L1 RNA copy number and L1 protein expression in the HPV-16-positive primary cell lines after a few in vitro passages, we initially evaluated the ability of L1-specific CTLs to kill early in vitro passages of the primary autologous cell lines (i.e., P1 to P3). As representatively shown in Fig. 3 for all cervical cancer patients evaluated, variable levels of HLA class I-restricted lysis of autologous tumor cells were consistently detected when autologous CD8+ T-cell populations stimulated with L1 VLP were challenged against the autologous tumor cells at different effector/target cell ratios. Negligible cytotoxicity was observed against autologous HLA-identical EBV-transformed LCL, while blocking studies indicated that tumor-specific lysis by L1-specific CD8+ T cells was significantly inhibited by MAbs specific for HLA class I (P < 0.05) in all of the evaluated patients (Fig. 3).
FIG. 3.
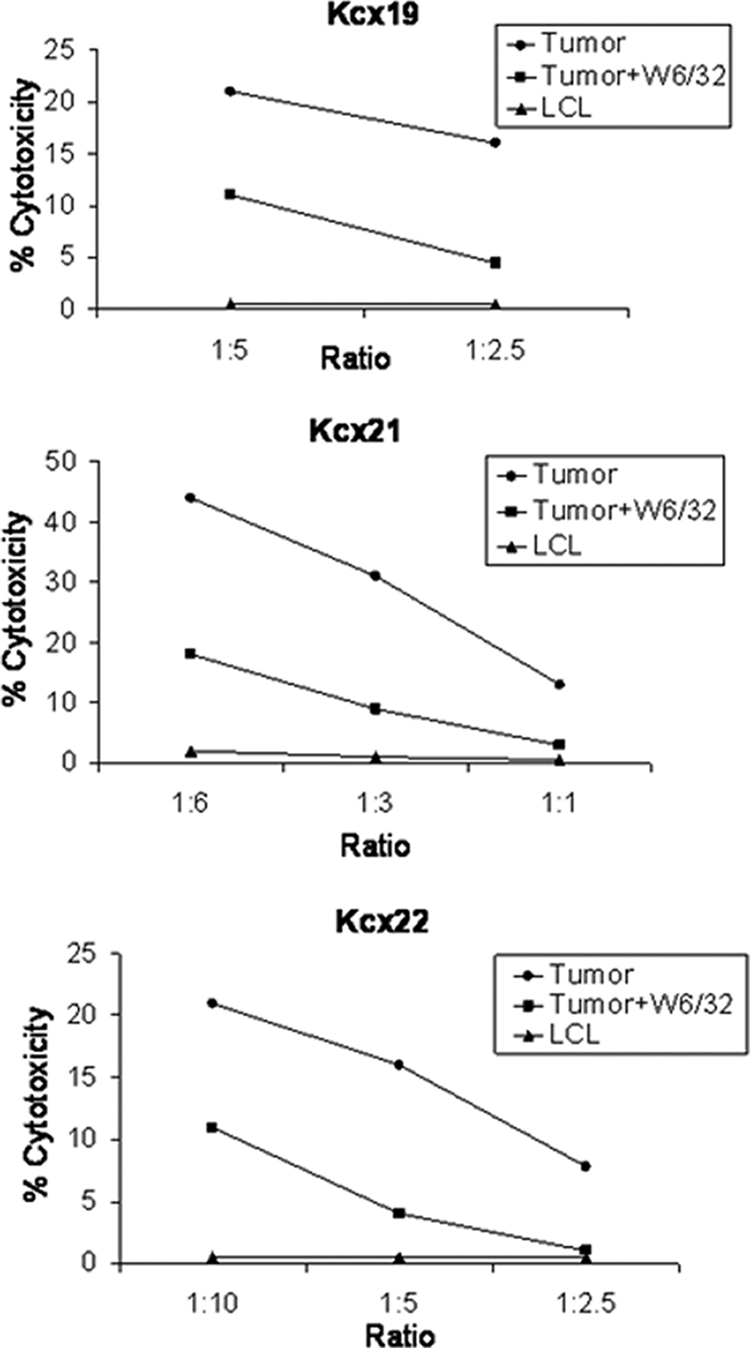
Tumor-specific HPV-16 L1-specific CD8+ CTL responses induced by L1-pulsed DCs in Kcx19, Kcx21, and Kcx22 patients, measured in a 5-h 51Cr-release assay. Percentage lysis against autologous tumor cells and HLA-identical HPV-16-negative control LCL at different effector/target cell ratio is shown. Anti-HLA class I blocking antibody (W6/32) was used at 50 μg/ml. A representative experiment for each patient is shown.
L1-specific DC-stimulated CD8+ CTLs are as effective as E7-specific CD8+ CTLs in their ability to kill autologous cervical cancer cells in vitro.
In order to compare the therapeutic efficacy of L1-specific CTLs to that of E7-specific CTLs in matched experiments, CD8+ CTLs from two patients were compared in their ability to induce killing of autologous HPV-16-infected tumor cells (i.e., P1 to P3). Cytotoxicity assays were conducted after a minimum of 7 weeks after stimulation of T cells with L1-pused DCs or E7-pused DCs. As representatively shown in Fig. 4, both the L1-specific and the E7-specific CTLs were highly cytotoxic against the primary autologous tumor cells, with no significant difference noted in their ability to kill the tumor target in multiple experiments. Blocking studies indicated that tumor-specific lysis by L1- and E7-specific CD8+ T cells were significantly inhibited by MAbs specific for HLA class I (P < 0.05), while limited cytotoxicity was observed against autologous HLA-identical EBV-transformed LCL (Fig. 4).
FIG. 4.
Tumor-specific HPV-16 L1-specific CD8+ CTL responses induced by L1-pulsed DCs and tumor-specific HPV-16 E7-specific CD8+ CTL responses induced by E7-pulsed DCs in Kcx19, Kcx21, and Kcx22 patients, as measured in a 5-h 51Cr-release assay. The percent lysis against autologous tumor cells and HLA-identical HPV-16-negative control LCL at different effector/target cell ratio is shown. Anti-HLA class I blocking antibody (W6/32) was used at 50 μg/ml. A representative experiment for each patient is shown.
L1-specific DC-stimulated CD8+ CTL killing of autologous cervical cancer cells expressing low levels of L1 VLPs in vitro.
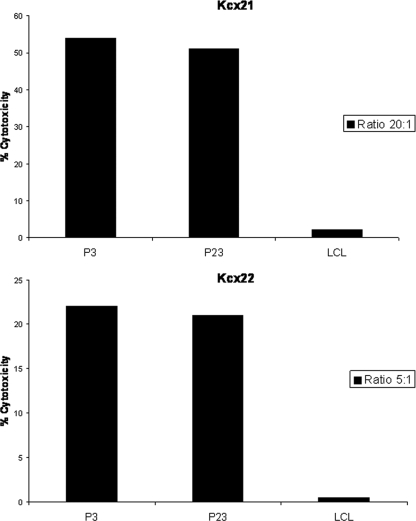
Because kinetics studies by real-time PCR and Western blot analysis showed a significant decrease in L1 RNA copy number and L1 protein expression in the most advanced in vitro passages (i.e., greater than P20) of both primary tumors tested, we investigated the ability of L1-specific CTLs to recognize and kill early (i.e., P3) versus late (i.e., P23) autologous tumor cell lines from patient Kcx21 and Kcx22. As representatively shown in Fig. 5, no significant differences were found when L1-specific DC-stimulated CD8+ CTLs from both patients were tested against autologous tumor cells expressing higher (i.e., P3) versus lower (P23) levels of L1 in multiple experiments. In this regard, in patient Kcx21, high cytotoxicity was detected against both P3 and P23 tumor cells, while negligible cytotoxicity was found against HLA-identical autologous LCL (Fig. 5). Similarly, in separate cytotoxicity assays from two different DC primings with patient Kcx22 peripheral blood lymphocytes, the killing of P3 primary tumor cells ranged from 17 to 22%, while that of P23 primary tumor cells ranged from 14 to 18% (P = not significant, Fig. 5). Confirming the specificity of the immune response against HPV-16 antigens, low to negligible cytotoxic activity was observed against autologous EBV-transformed LCL of patient Kcx22 (Fig. 5).
FIG. 5.
Tumor-specific HPV-16 L1-specific CD8+ CTL responses induced by L1-pulsed DCs in Kcx21 and Kcx22 patients, measured in a 5-h 51Cr-release assay. The percent lysis against autologous tumor cells at early passages (i.e., P3) and late passages (i.e., P23) and HLA-identical HPV-16-negative control LCL at different effector/target cell ratios is shown.
L1-specific DC-stimulated CD4+ T lymphocytes are as effective as E7-specific CD4+ T lymphocytes in their ability to induce strong proliferative responses against HPV-16 antigens.
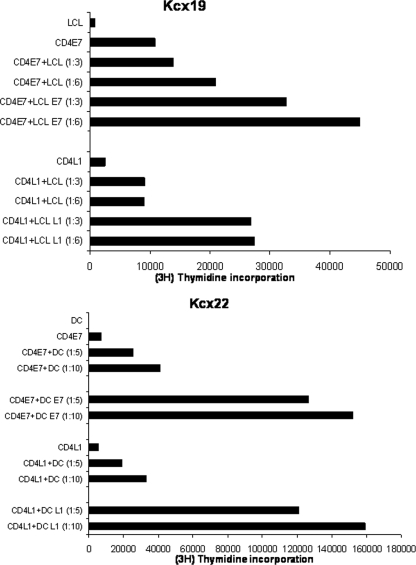
L1- and E7 DC-stimulated CD4+ T cells (purity > 99%) were tested and compared in their ability to induce specific proliferation against either L1- or E7-pulsed autologous DCs and/or LCL at different effector/target cell ratios. As controls, unpulsed autologous DCs or LCL or autologous DOTAP-pulsed DCs or LCL were used. As representatively shown in Fig. 6, strong and specific proliferations were detected in L1-specific and E7-specific CD4+ T lymphocytes when challenged with L1- and E7-pulsed autologous antigen-presenting cells as target cells, respectively. The proliferation of both E7- and L1-specific CD4+ T-cell populations were significantly higher (P < 0.01) than those induced by DC/LCL alone or DOTAP-pulsed DC/LCL (Fig. 6). However, no significant differences in proliferation capability were detectable when L1-specific and E7-specific CD4+ T lymphocytes were compared.
FIG. 6.
(Upper panel) CD4+ T-cell proliferation in response to stimulation with HPV E7- or L1-pulsed autologous irradiated LCL (LCL/E7 and LCL/L1) at different effector/target cell ratios in patient Kcx19. (Lower panel) CD4+ T-cell proliferation in response to stimulation with HPV E7- or L1-pulsed autologous DCs (DC/E7 and DC/L1) at different effector/target cell ratio in patient Kcx22. [3H]thymidine (1 mCi/well) was added to each well for at least 16 h of the assay, and proliferation was determined by measuring the [3H]thymidine incorporation. The difference in mean proliferations determined by measuring the [3H]thymidine incorporation in the presence of LCL/E7-DC/E7 or LCL/L1-DC/L1 compared to that in the presence of LCL or DC controls is significant at P < 0.01 as determined by the Student t test for both patients. In crossing experiments, no proliferation was detected in L1 VLP or E7-specific CD4+ T cells when autologous LCL or DCs were loaded with full-length E7 (instead of L1) or L1 (instead of E7) were used as stimulators.
Phenotypic analysis.
Flow cytometric analysis was used to determine the phenotypes of the populations of DC-L1- and DC-E7-stimulated CD4+ and CD8+ T cells before proliferation and cytotoxicity assays, respectively. In this regard, to characterize in more detail the nature and phenotype of CD4+ T-cell responses induced after DC immunization with L1 and E7 antigens in vitro, the percentages of CD4+ CD25+ FOXP3+ (Treg) cells and CD4+ CD25+ FOXP3− (effectors) cells were evaluated at the time of the first proliferation assay after at least 10 days from the last antigen stimulation for the expression of CD25 and CD4 surface antigens and intracellular FOXP3 by flow cytometry. As representatively shown in Table 3, results from multiple independent matched priming experiments with L1 and E7 in Kcx21 and Kcx22 cervical cancer patients demonstrated consistent numbers of CD4+ CD25+ FOXP3+ (i.e., Treg) and CD4+ CD25+ FOXP3− (i.e., effectors) T cells in both patients after in vitro DC stimulation. Importantly, although no significant differences were detected in the percentages of CD4+ CD25+ FOXP3+ Treg cells or in the percentages of L1- or E7-specific CD4+ effector T cells between the L1- and E7-specific CD4+ T-cell populations (Table 3), the numbers of CD4+ CD25+ FOXP3− effector cells (i.e., L1- or E7-specific effector cells) were found to significantly outnumber those of the Treg counterparts (i.e., CD4+ CD25+ FOXP3+) in all matched priming experiments (P = 0.01).
TABLE 3.
Expression of FOXP3 in E7- and L1-specific DC-stimulated CD4+ CD25+ T cells
| Patient | Expression (%) of:
|
|||
|---|---|---|---|---|
| CD4+ CD25+ FOXP3+
|
CD4+ CD25+ FOXP3−
|
|||
| E7 | L1 | E7 | L1 | |
| Kcx19 | 24 | 22 | 37 | 36 |
| Kcx21 | 9 | 9 | 15 | 18 |
a CD4+ T cells were tested as described in Materials and Methods after resting at least 10 days from last the restimulation. Differences in the no. of L1 CD4+ CD25+ FOXP3+ versus E7 CD4+ CD25+ FOXP3+ for both patients were not significant, while differences in the number of L1 CD4+ CD25+ FOXP3+ versus L1 CD4+ CD25+ FOXP3- and E7 CD4+ CD25+ FOXP3+ versus E7 CD4+ CD25+ FOXP3− for both patients were found to be significant at P > 0.01.
When CTLs were phenotyped by fluorescence-activated cell sorting before cytotoxicity assays, we found these populations to be comprised of CD3+ CD8+ and CD4− T cells, with a low proportion of CD56 antigen-positive cells. Further analysis revealed the populations to be TCR-α/β+ (97 to 99%), TCR-γ/δ+ (1 to 2%), with a large percentage of the cells expressing CD25+ and HLA-DR+ but not CD16− (data not shown). The expression of CD56 on T lymphocytes was further analyzed by two-color immunofluorescence. By this technique, CD8+ T cells were compared for coexpression of CD56. Although the data are not shown, similar percentages of CD8+ T lymphocytes (1 to 5%) coexpressed the CD56 surface antigen during culture with no significant differences between L1- and E7-specific CTLs.
Intracellular cytokine expression by HPV-16 L1- and E7-specific T cells.
To evaluate whether cytokine expression in L1 and E7-stimulated CD8+ T cells segregated in discrete IFN-γ+ IL-4− and IFN-γ− IL-4+ subsets and may significantly differ in the E7- and L1-specific populations of CTLs, we used flow cytometry for the detection of intracellular cytokine expression at the single cell level. Two-color flow cytometric analyses of intracellular IFN-γ and IL-4 expression by CTLs were performed after 7 to 8 weeks of culture and thereafter as described in Materials and Methods. For Kcx19 and Kcx22, the majority of CD8+ T cells specific to either L1 (mean ± the standard deviation = 45% ± 12%; range, 35 to 51%) or E7 (43% ± 14%; range, 32 to 49%) contained intracellular IFN-γ, while a second minor subset contained both intracellular IFN-γ and IL-4 and a third subset contained only IL-4 (L1: 15% ± 5%; range, 13 to 19%) (E7: 13% ± 4%; range, 11 to 18%). Although the percentage of L1- and E7-specific CTLs containing intracellular IFN-γ and IL-4 was found to vary from experiment to experiment in several repetitive analyses from different primings in both patients, no significant differences in the percentages of IFN-γ- and IL-4-positive T cells in L1-specific versus E7-specific T-cell populations were observed. Unactivated (i.e., resting) CD8+ T cells failed to stain for IFN-γ or IL-4 and, similarly, FITC-conjugated anti-IgG2a and PE-conjugated anti-IgG1 isotype controls did not stain either activated or unactivated CD8+ T cells (data not shown).
DISCUSSION
It has long been speculated that cell-mediated responses to late HPV proteins are unlikely to have any significant therapeutic role against established HPV lesions. This “common knowledge” stems from previous observations showing low to undetectable expression of HPV L1 proteins in established cervical lesions (11, 34), as well as restriction of late gene expression to terminally differentiated epithelial cells by transcriptional and posttranscriptional mechanisms (11, 34). Consistent with this view, cell-mediated immune responses to VLPs have been almost exclusively investigated in prophylactic vaccination studies of healthy individuals (14, 22). However, T cells are known to be exquisitely sensitive to antigenic stimulation since they can kill, proliferate, and produce cytokines in response to antigen presenting cells displaying as few as one to a few hundred specific peptide-major histocompatibility complexes (9, 13, 35, 36). Thus, it is likely that even a limited amount of viral protein expression in established HPV-infected lesions may be able to trigger therapeutic responses in cervical cancer patients. In agreement with this hypothesis, evidence has been previously provided to show that cells expressing levels of L1 protein that are below detection by most present-day assays can still be targeted by CTLs induced by vaccination with VLP (8, 9, 10, 19, 22, 24). Indeed, in animal models, L1 HPV-16 VLPs may efficiently induce protective antitumor responses after a single low-dose injection in the absence of any adjuvant, even if L1 protein is undetectable because of a very low steady-state level in the proliferating tumor cells (8). Furthermore, in at least one human trial, there is evidence to suggest that cell-mediated immunity induced by vaccination with VLP, might have therapeutic effects against HPV-infected established genital lesions (41). Taken together, these data strongly suggest that low-level expression of HPV-16 L1 protein potentially occurring in proliferating cervical cancer cells may serve as a tumor vaccine target in therapeutic human clinical trials.
In the present study, in order to carefully evaluate this hypothesis, we first investigated and compared L1, E6, and E7 RNA copy numbers in multiple HPV-16-positive flash-frozen biopsy samples and primary cervical cancer cell lines by real-time RT-PCR. We confirmed the expression of L1, E6, and E7 in all HPV-16-infected cervical biopsy specimens and primary tumor cell lines studied, as well as in both the established HPV-16-positive cervical cancer cell lines (i.e., SiHa and CaSki) used as positive controls. A significantly higher level of E7 RNA transcript, compared to E6 and L1 transcripts, was consistently found in HPV-16-positive cervical tumors. These data are thus consistent with the results of recent studies evaluating E6 and E7 RNA copy numbers in HPV-16-infected cervical tumors and ThinPrep pap tests. These reports also demonstrated real-time PCR methods as sensitive and reliable indicators of the direct involvement of E6 and E7 viral oncogenes in cervical cancer carcinogenesis (7, 39). Much to our surprise, however, when E6 RNA copy numbers were compared to L1 RNA copy numbers in HPV-16-infected flash-frozen cervical cancer biopsy samples, no significant differences in mRNA expression levels were found. Moreover, in 6 of 22 HPV-16-infected tumors we detected higher L1 mRNA copy numbers compared to E6. Taken together, these data demonstrate that the L1 transcript is consistently detected in HPV-16-infected invasive cervical cancer lesions and that this late viral protein, in analogy to E6 and E7, may represent an excellent target antigen for T-cell-mediated immunotherapy of HPV-infected cervical cancer patients.
Next, we evaluated the kinetics of expression of L1, E6, and E7 RNA copy numbers in two primary HPV-16-positive squamous cervical cancer cell lines recently established in our laboratory. Kinetic studies showed a significant progressive reduction of L1 mRNA levels compared to E6 and E7 expression levels after multiple in vitro passages in both cell lines. Although it remains poorly understood why L1 transcripts downregulate quickly during in vitro culture or why E6 and E7 transcripts, after an initial decrease, persist at relatively high levels compared to the SiHa calibrator cell line, we speculated that the in vitro culture conditions necessary to establish and maintain primary tumor cell lines viability, including culturing cell lines in low concentrations of calcium and magnesium in order to prevent keratinocyte maturation/differentiation (42), may have played a stronger role in the downregulation of L1 transcripts than E6 and E7. This hypothesis is indeed consistent with the previous reported evidence showing that during HPV infection, late gene expression, unlike early gene expression, is restricted to terminally differentiated epithelial cells by transcriptional and posttranscriptional mechanisms (11, 34). Western blot analysis of primary cervical cancer cell lines confirmed the kinetics of expression of L1 transcripts detected by RT-PCR at protein level and showed less and less detectable L1 proteins with increasing in vitro passages of the primary tumors.
DCs are the most potent antigen-presenting cells known in humans and play a crucial role during the priming and reactivation of antigen-specific immune responses (2, 32). This unique function, as well as the recent standardization of DC culture conditions in vitro, has provided the opportunity to evaluate their potential for the immunological treatment of cancer patients. Consistent with this view, several human phase I and II trials have been initiated using tumor antigen pulsed DCs, and promising clinical results have been reported in at least some patients, in the absence of significant toxicity (reviewed in reference 21). In cervical cancer patients, after successful induction of E7-specific T-cell responses using autologous monocyte-derived DCs in vitro (29-31), our research group has recently carried out multiple clinical trials targeting advanced stage and, more recently, early stage patients harboring HPV-16 and HPV-18 cervical cancer with a full-length E7-DC-based therapeutic vaccine (25-28).
In the present study, we provide evidence that L1-loaded monocyte-derived DCs may induce effective L1-specific cellular immune responses in human patients harboring HPV-16-positive tumors. Thus, these results differ from the currently licensed prophylactic vaccine formulations that primarily induce antibody responses with minimal cellular responses and that have been reported to have no therapeutic effect on prevalent nonmalignant infection by HPV (15). More importantly, L1-specific HLA class I-restricted CD8+ CTL responses were found to be as effective as those induced by E7 protein in killing HPV-16 naturally infected autologous cervical tumors. Furthermore, when primary HPV-16-infected tumors at different time points of in vitro culture were used as targets in cytotoxicity assays, no difference in the ability of L1-specific CTLs to kill primary tumors harboring high versus low copy numbers of L1 transcripts were found. These data therefore support the exquisite sensitivity of human L1-specific CTLs in their ability to kill autologous targets expressing low levels of L1, as previously demonstrated in animal models (8). Taken all together, these results suggest that L1 protein may represent another important target to add to E6 and E7 for the therapeutic DC-based vaccination of patients harboring established HPV-infected cervical lesions.
Recent reports have suggested a potential increase in the number of CD4+ FOXP3+ Tregs in the peripheral blood lymphocytes of myeloma patients vaccinated with DCs (3). In addition, evidence of an association of cervical cancer with the presence of CD4+ Tregs specific for HPV antigens has been recently provided (37, 40). In the present study, no detectable differences were found in the number of CD4+ CD25+ FOXP3+ (regulatory) T cells after stimulation with DC loaded with L1 versus E7. More importantly, phenotypic characterization of L1 and E7 responsive CD4+ T cells demonstrated a high number of CD4+ CD25+ FOXP3− (effector) T cells and the numbers of these cells were found to be significantly higher than those of the FOXP3+ subset of CD4+ T cells. Taken together, these data suggest that in HPV-16-infected cervical cancer patients, both E7 and L1 viral antigens may significantly increase the number of antigen specific DC-activated CD4+ CD25+ FOXP3− T cells and that the expansion of these cells is superior to that of potentially harmful Tregs.
Finally, using two-color flow cytometric analysis of intracellular cytokine expression at the single cell level, we found that a strongly polarized type pattern of cytokine secretion is inducible in both E7- and L1-primed CD8+ CTL populations. These results demonstrated no significant differences in type I versus type II immune responses generated against L1 versus E7 in cervical cancer patients harboring HPV-16-positive tumors. Taken together, our results support the view that targeting both L1 and E7 antigens may represent a more efficient approach to increase cellular immunity against viral antigens in cervical cancer patients by DC-based vaccination. The design and implementation of clinical trials with L1/E7-pulsed autologous DCs will ultimately determine the validity of this combined approach.
Acknowledgments
We thank John T. Schiller (National Institutes of Health, Bethesda, MD) for the generous gifts of HPV-16 L1 VLPs, Lisa Patriub for editing the manuscript, and Daniel DiMaio (Yale Cancer Center, New Haven, CT) for helpful suggestions and the critical review of the manuscript.
This study was supported in part by grants from the Angelo Nocivelli, the Berlucchi, and the Camillo Golgi Foundations, Brescia, Italy; NIH R01 CA122728-01A2 to A.S., and grants 501/A3/3 and 0027557 from the Italian Institute of Health to A.S. This investigation was also supported by NIH research grant CA-16359 from the National Cancer Institute and a TARE award from the Yale Cancer Center.
Footnotes
Published ahead of print on 22 April 2009.
REFERENCES
- 1.Aiba, S., M. Rokugo, and H. Tagami. 1986. Immunohistologic analysis of the phenomenon of spontaneous regression of numerous flat warts. Cancer 581246-1251. [DOI] [PubMed] [Google Scholar]
- 2.Banchereau, J., and R. M. Steinman. 1998. Dendritic cells and the control of immunity. Nature 392245-252. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, D. K., M. V. Dhodapkar, E. Matayeva, R. M. Steinman, and K. M. Dhodapkar. 2006. Expansion of FOXP3high regulatory T cells by human dendritic cells (DCs) in vitro and after injection of cytokine-matured DCs in myeloma patients. Blood 1082655-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosch, F. X., A. Lorincz, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman, N., H. D. Birley, A. M. Renton, N. F. Hanna, B. K. Ryait, M. Byrne, D. Taylor-Robinson, and M. A. Stanley. 1994. Immunological events in regressing genital warts. Am. J. Clin. Pathol. 102768-774. [DOI] [PubMed] [Google Scholar]
- 6.Cubie, H. A., A. L. Seagar, E. McGoogan, J. Whitehead, A. Brass, M. J. Arends, and M. W. Whitley. 2001. Rapid real-time PCR to distinguish between high risk human papillomavirus types 16 and 18. Mol. Pathol. 5424-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Boer, M. A., E. S. Jordanova, G. G. Kenter, A. A. Peters, W. E. Corver, J. B. Trimbos, and G. J. Fleuren. 2007. High human papillomavirus oncogene mRNA expression and not viral DNA load is associated with poor prognosis in cervical cancer patients. Clin. Cancer Res. 13132-138. [DOI] [PubMed] [Google Scholar]
- 8.De Bruijn, M. L., H. L. Greenstone, H. Vermeulen, C. J. Melief, D. R. Lowy, J. T. Schiller, and W. M. Kast. 1998. L1-specific protection from tumor challenge elicited by HPV16 virus-like particles. Virology 250371-376. [DOI] [PubMed] [Google Scholar]
- 9.De la Rosa, G. P., A. Monroy-García, M. Mora-García, C. G. Peña, J. Hernández-Montes, B. Weiss-Steider, and M. A. Gómez Lim. 2009. An HPV-16 L1-based chimeric human papillomavirus-like particles containing a string of epitopes produced in plants is able to elicit humoral and cytotoxic T-cell activity in mice. Virol. J. 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demotz, S., H. M. Grey, and A. Sette. 1990. The minimal number of class II MHC-antigen complexes needed for T-cell activation. Science 2491028-1030. [DOI] [PubMed] [Google Scholar]
- 11.Frattini, M. G., H. B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. USA 933062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frazer, I. H., J. T. Cox, E. J. Mayeaux, Jr., E. L. Franco, A. B. Moscicki, J. M. Palefsky, D. G. Ferris, A. S. Ferenczy, and L. Villa. 2006. Advances in prevention of cervical cancer and other human papillomavirus-related diseases. Pediatr. Infect. Dis. J. 25(Suppl. 2)S65-S81. [DOI] [PubMed] [Google Scholar]
- 13.Harding, C. V., and E. R. Unanue. 1990. Quantitation of antigen-presenting cell MHC class II/peptide complexes necessary for T-cell stimulation. Nature 346574-576. [DOI] [PubMed] [Google Scholar]
- 14.Harro, C. D., Y. Y. Pang, R. B. Roden, A. Hildesheim, Z. Wang, M. J. Reynolds, T. C. Mast, R. Robinson, B. R. Murphy, R. A. Karron, J. Dillner, J. T. Schiller, and D. R. Lowy. 2001. Safety and immunogenicity trial in adult volunteers of a human papillomavirus 16 L1 virus-like particle vaccine. J. Natl. Cancer Inst. 93284-292. [DOI] [PubMed] [Google Scholar]
- 15.Hildesheim, A., R. Herrero, S. Wacholder, A. C. Rodriguez, D. Solomon, M. C. Bratti, J. T. Schiller, P. Gonzalez, G. Dubin, C. Porras, S. E. Jimenez, and D. R. Lowy. 2007. Effect of human papillomavirus 16/18 L1 viruslike particle vaccine among young women with preexisting infection: a randomized trial. JAMA 298743-753. [DOI] [PubMed] [Google Scholar]
- 16.Hopfl, R., K. Heim, N. Christensen, K. Zumbach, U. Wieland, B. Volgger, A. Widschwendter, S. Haimbuchner, E. Muller-Holzner, M. Pawlita, H. Pfister, and P. Fritsch. 2000. Spontaneous regression of CIN and delayed-type hypersensitivity to HPV-16 oncoprotein E7. Lancet 3561985-1986. [DOI] [PubMed] [Google Scholar]
- 17.Kadish, A. S., G. Y. Ho, R. D. Burk, Y. Wang, S. L. Romney, R. Ledwidge, and R. H. Angeletti. 1997. Lymphoproliferative responses to human papillomavirus (HPV) type 16 proteins E6 and E7: outcome of HPV infection and associated neoplasia. J. Natl. Cancer Inst. 891285-1293. [DOI] [PubMed] [Google Scholar]
- 18.Kadish, A. S., P. Timmins, Y. Wang, G. Y. Ho, R. D. Burk, J. Ketz, W. He, S. L. Romney, A. Johnson, R. Angeletti, M. Abadi, et al. 2002. Regression of cervical intraepithelial neoplasia and loss of human papillomavirus (HPV) infection is associated with cell-mediated immune responses to an HPV type 16 E7 peptide. Cancer Epidemiol. Biomarkers Prev. 11483-488. [PubMed] [Google Scholar]
- 19.Monroy-Garcia, A., B. Weiss-Steider, J. Hernandez-Montes, V. F. Ortiz-Navarrete, A. Banuelos-Panuco, A. Acosta-Araujo, A. Diaz-Quinonez, C. M. Lopez-Graniel, G. Herbert, J. Granados, C. de Leo, R. M. Silva-Lopez, and M. L. Mora-Garcia. 2002. Identification of two homologous antigenic peptides derived from L1 HPV-16 and -18 proteins specific for the HLA-B*3901 allele. Arch. Virol. 1471933-1942. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa, M., D. P. Stites, S. Farhat, J. R. Sisler, B. Moss, F. Kong, A. B. Moscicki, and J. M. Palefsky. 1997. Cytotoxic T lymphocyte responses to E6 and E7 proteins of human papillomavirus type 16: relationship to cervical intraepithelial neoplasia. J. Infect. Dis. 175927-931. [DOI] [PubMed] [Google Scholar]
- 21.Osada, T., T. M. Clay, C. Y. Woo, M. A. Morse, and H. K. Lyerly. 2006. Dendritic cell-based immunotherapy. Int. Rev. Immunol. 25377-413. [DOI] [PubMed] [Google Scholar]
- 22.Pinto, L. A., J. Edwards, P. E. Castle, C. D. Harro, D. R. Lowy, J. T. Schiller, D. Wallace, W. Kopp, J. W. Adelsberger, M. W. Baseler, J. A. Berzofsky, and A. Hildesheim. 2003. Cellular immune responses to human papillomavirus (HPV)-16 L1 in healthy volunteers immunized with recombinant HPV-16 L1 virus-like particles. J. Infect. Dis. 188327-338. [DOI] [PubMed] [Google Scholar]
- 23.Rogozinski, T. T., S. Jablonska, and M. Jarzabek-Chorzelska. 1988. Role of cell-mediated immunity in spontaneous regression of plane warts. Int. J. Dermatol. 27322-326. [DOI] [PubMed] [Google Scholar]
- 24.Rudolf, M. P., J. D. Nieland, D. M. DaSilva, M. P. Velders, M. Muller, H. L. Greenstone, J. T. Schiller, and W. M. Kast. 1999. Induction of HPV16 capsid protein-specific human T-cell responses by virus-like particles. Biol. Chem. 380335-340. [DOI] [PubMed] [Google Scholar]
- 25.Santin, A. D., S. Bellone, M. Gokden, M. J. Cannon, and G. P. Parham. 2002. Vaccination with HPV-18 E7-pulsed dendritic cells in a patient with metastatic cervical cancer. N. Engl. J. Med. 3461752-1753. [DOI] [PubMed] [Google Scholar]
- 26.Santin, A. D., S. Bellone, M. Palmieri, A. Ravaggi, C. Romani, R. Tassi, J. J. Roman, A. Burnett, S. Pecorelli, and M. J. Cannon. 2006. HPV16/18 E7-pulsed dendritic cell vaccination in cervical cancer patients with recurrent disease refractory to standard treatment modalities. Gynecol. Oncol. 100469-478. [DOI] [PubMed] [Google Scholar]
- 27.Santin, A. D., S. Bellone, M. Palmieri, A. Zanolini, A. Ravaggi, E. E. Siegel, J. J. Roman, S. Pecorelli, and M. J. Cannon. 2008. Human papillomavirus type 16 and 18 E7-pulsed dendritic cell vaccination in stage IB or IIA cervical cancer patients: a phase I escalating dose trial. J. Virol. 821968-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santin, A. D., P. L. Hermonat, A. Ravaggi, S. Bellone, C. Cowan, S. Korourian, S. Pecorelli, M. J. Cannon, and G. P. Parham. 2000. Development, characterization, and distribution of adoptively transferred peripheral blood lymphocytes primed by human papillomavirus 18 E7-pulsed autologous dendritic cells in a patient with metastatic adenocarcinoma of the uterine cervix. Eur. J. Gynaecol. Oncol. 2117-23. [PubMed] [Google Scholar]
- 29.Santin, A. D., P. L. Hermonat, A. Ravaggi, S. Bellone, S. Pecorelli, J. J. Roman, M. J. Cannon, and G. P. Parham. 2000. Interleukin-10 increases Th1 cytokine production and cytotoxic potential in human papillomavirus-specific CD8+ cytotoxic T lymphocytes. J. Virol. 744729-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santin, A. D., P. L. Hermonat, A. Ravaggi, S. Bellone, J. J. Roman, S. Jayaprabhu, S. Pecorelli, G. P. Parham, and M. J. Cannon. 2001. Expression of CD56 by human papillomavirus E7-specific CD8+ cytotoxic T lymphocytes correlates with increased intracellular perforin expression and enhanced cytotoxicity against HLA-A2-matched cervical tumor cells. Clin. Cancer Res. 7804s-810s. [PubMed] [Google Scholar]
- 31.Santin, A. D., P. L. Hermonat, A. Ravaggi, M. Chiriva-Internati, D. Zhan, S. Pecorelli, G. P. Parham, and M. J. Cannon. 1999. Induction of human papillomavirus-specific CD4+ and CD8+ lymphocytes by E7-pulsed autologous dendritic cells in patients with human papillomavirus type 16- and 18-positive cervical cancer. J. Virol. 735402-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuler, G., and R. M. Steinman. 1997. Dendritic cells as adjuvants for immune-mediated resistance to tumors. J. Exp. Med. 1861183-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shafti-Keramat, S., A. Handisurya, E. Kriehuber, G. Meneguzzi, K. Slupetzky, and R. Kirnbauer. 2003. Different heparan sulfate proteoglycans serve as cellular receptors for human papillomaviruses. J. Virol. 7713125-13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stoler, M. H., C. R. Rhodes, A. Whitbeck, S. M. Wolinsky, L. T. Chow, and T. R. Broker. 1992. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum. Pathol. 23117-128. [DOI] [PubMed] [Google Scholar]
- 35.Sykulev, Y., M. Joo, I. Vturina, T. J. Tsomides, and H. N. Eisen. 1996. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T-cell response. Immunity 4565-571. [DOI] [PubMed] [Google Scholar]
- 36.Valitutti, S., S. Muller, M. Cella, E. Padovan, and A. Lanzavecchia. 1995. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature 375148-152. [DOI] [PubMed] [Google Scholar]
- 37.van der Burg, S. H., S. J. Piersma, A. de Jong, J. M. van der Hulst, K. M. Kwappenberg, M. van den Hende, M. J. Welters, J. J. Van Rood, G. J. Fleuren, C. J. Melief, G. G. Kenter, and R. Offringa. 2007. Association of cervical cancer with the presence of CD4+ regulatory T cells specific for human papillomavirus antigens. Proc. Natl. Acad. Sci. USA 10412087-12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villa, L. L., R. L. Costa, C. A. Petta, R. P. Andrade, K. A. Ault, A. R. Giuliano, C. M. Wheeler, L. A. Koutsky, C. Malm, M. Lehtinen, F. E. Skjeldestad, S. E. Olsson, M. Steinwall, D. R. Brown, R. J. Kurman, B. M. Ronnett, M. H. Stoler, A. Ferenczy, D. M. Harper, G. M. Tamms, J. Yu, L. Lupinacci, R. Railkar, F. J. Taddeo, K. U. Jansen, M. T. Esser, H. L. Sings, A. J. Saah, and E. Barr. 2005. Prophylactic quadrivalent human papillomavirus (types 6, 11, 16, and 18) L1 virus-like particle vaccine in young women: a randomised double-blind placebo-controlled multicentre phase II efficacy trial. Lancet Oncol. 6271-278. [DOI] [PubMed] [Google Scholar]
- 39.Wang-Johanning, F., D. W. Lu, Y. Wang, M. R. Johnson, and G. L. Johanning. 2002. Quantitation of human papillomavirus 16 E6 and E7 DNA and RNA in residual material from ThinPrep Papanicolaou tests using real-time polymerase chain reaction analysis. Cancer 942199-2210. [DOI] [PubMed] [Google Scholar]
- 40.Welters, M. J., G. G. Kenter, S. J. Piersma, A. P. Vloon, M. J. Lowik, D. M. Berends-van der Meer, J. W. Drijfhout, A. R. Valentijn, A. R. Wafelman, J. Oostendorp, G. J. Fleuren, R. Offringa, C. J. Melief, and S. H. van der Burg. 2008. Induction of tumor-specific CD4+ and CD8+ T-cell immunity in cervical cancer patients by a human papillomavirus type 16 E6 and E7 long peptides vaccine. Clin. Cancer Res. 14178-187. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, L. F., J. Zhou, S. Chen, L. L. Cai, Q. Y. Bao, F. Y. Zheng, J. Q. Lu, J. Padmanabha, K. Hengst, K. Malcolm, and I. H. Frazer. 2000. HPV6b virus-like particles are potent immunogens without adjuvant in man. Vaccine 181051-1058. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, W., S. K. Tyring, M. Brysk, and T. S. Chan. 1996. Immortalization of differentiated human keratinocytes by human papillomavirus (HPV) 16 DNA. J. Dermatol. Sci. 13140-152. [DOI] [PubMed] [Google Scholar]