Abstract
Coronaviruses encode large replicase polyproteins which are proteolytically processed by viral proteases to generate mature nonstructural proteins (nsps) that form the viral replication complex. Mouse hepatitis virus (MHV) replicase products nsp3, nsp4, and nsp6 are predicted to act as membrane anchors during assembly of the viral replication complexes. We report the first antibody-mediated Western blot detection of nsp6 from MHV-infected cells. The nsp6-specific peptide antiserum detected the replicase intermediate p150 (nsp4 to nsp11) and two nsp6 products of approximately 23 and 25 kDa. Analysis of nsp6 transmembrane topology revealed six membrane-spanning segments and a conserved hydrophobic domain in the C-terminal cytosolic tail.
Coronaviruses are enveloped, positive-stranded RNA viruses that sequester host cell membranes to assemble viral replication complexes in the cytoplasm of infected cells (2, 21). In the case of murine coronavirus mouse hepatitis virus (MHV), three viral proteases process the replicase polyproteins (3, 4, 5, 9, 12, 13, 14, 16, 18, 19, 24, 26) into intermediates and 16 mature nonstructural protein (nsp) products (Fig. 1A). It is unclear whether the intermediate forms or the mature nsps are responsible for assembly of the viral replication complex. The replicase proteins nsp3, nsp4, and nsp6 contain transmembrane (TM)-spanning sequences that are proposed to be important for sequestering endoplasmic reticulum (ER) membranes to form the double-membrane vesicles which are the site of viral RNA synthesis (11, 17). However, the mechanism used by the nsps to generate double-membrane vesicles is not yet understood. Recent reports (8, 15, 22, 23, 28) and the study presented here have unraveled the membrane topology of these nsps. nsp4 is a glycoprotein with four TM domains (8, 22, 23, 28). nsp3 anchors its 213-kDa multidomain protein to ER membranes, likely using two TM domains (15, 22). Recently, nsp6 was shown to contain six TM domains (22); however, the authors were unable to resolve which of two C-terminal hydrophobic domains can act as the final membrane-spanning region.
FIG. 1.
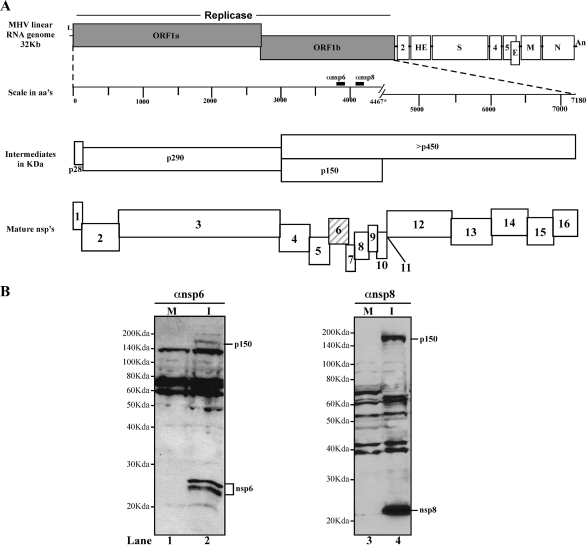
Schematic diagram of MHV RNA genome, indicating the proteolytic processing scheme of the replicase polyprotein and Western blot detection of MHV nsp6. (A) MHV-A59 linear RNA genome with the canonical representation of replicase, structural, and accessory genes. The replicase polyprotein intermediates and mature nsps generated during processing are depicted. The mature nsp6 replicase protein (hatched box) and the antibodies used to detect nsp6 and nsp8 (solid black boxes) are indicated. aa's, amino acids. (B) Western blot analysis of nsp6. Whole-cell lysates were prepared from mock-infected (M) and MHV-infected (I) HeLa-MHVR cells, and the lysates were separated by 12.5% SDS-PAGE. Products were detected by probing with nsp6- or nsp8-specific antibodies.
In this report, we show the first antibody-mediated detection of MHV-A59 nsp6 in virally infected cells. We also report the TM topology of nsp6, as determined by glycosylation tagging and N-linked glycosylation sequence insertion mutagenesis approaches, providing evidence that nsp6 contains six membrane-spanning segments with a large C-terminal tail exposed to the cytosol. Multiple alignment of the nsp6 amino acid sequences from each coronavirus group revealed a high level of conservation at the C-terminal end, suggesting an evolutionarily conserved function.
To detect nsp6 replicase protein in MHV-A59-infected cells, we used a polyclonal rabbit antiserum directed against a peptide (PLGVYNYKISVQEL) from the C-terminal region of nsp6. We detected the replicase intermediate p150 (nsp4 to nsp11) and two nsp6-specific products of 23 and 25 kDa (Fig. 1B, lane 2) in MHV-infected HeLa-MHVR (25) cells by Western blot analysis. We found similar mature products of nsp6 in MHV-infected murine cell lines 17Cl-1 and DBT (data not shown). The same MHV-infected cell lysate was used to detect nsp8 replicase protein with a specific antibody that also recognizes p150 (Fig. 1B, lane 4). The reason for the existence of multiple forms of nsp6 is currently unknown, although posttranslational modification or alternative processing of nsp6 cannot be ruled out at this point. Future experiments will be directed at purification and analysis of the two forms of nsp6 detected here.
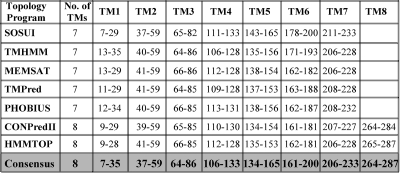
To develop a framework for understanding the membrane topology of nsp6, we first performed nsp6 bioinformatics analysis. Five out of the seven bioinformatics tools predicted that nsp6 would encode seven TM domains, whereas two programs predicted that it would encode eight TM domains (Fig. 2). However, because both the N and C termini of nsp6 must be processed in the cytosol by the viral 3C-like protease (3CLpro), we expected nsp6 to encode an even number of TM domains and established a consensus TM domain prediction for nsp6 (Fig. 2, bottom row). The consensus provided a working model for generating plasmid DNA constructs for evaluating the membrane topology of MHV nsp6. First, we employed enhanced green fluorescent protein (EGFP) glycosylation tagging (EGFPglyc) experiments as previously used for determining the membrane topology of other viral replicase TM proteins (20, 22). This approach allowed us to determine the localization of the tagged protein based on the differences in the mobility of the endoglycosidase H (endo H)-treated protein versus that of the untreated protein by the use of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. Based on the consensus topology model (Fig. 3A) suggesting a maximum of eight TM domains, we generated a series of plasmid DNA constructs starting with the N-terminal putative TM1 domain, and successively larger constructs were fused at their carboxyl terminus in frame with EGFPglycV5. The plasmid DNAs were individually transfected into BsrT7 cells (6), and the newly synthesized fusion proteins were radiolabeled with 100 μCi of [35S]methionine-cysteine per ml from 20 to 22 h posttransfection. Chimeric proteins expressed from the cell lysates were immunoprecipitated with V5 antibody, either endo H treated or mock treated, separated using 12.5% SDS-PAGE, and analyzed by autoradiography as described previously (15).
FIG. 2.
Summary of TM predictions for MHV nsp6 obtained from membrane topology bioinformatics tools. The nsp6 amino acid sequence (amino acids 3637 to 3923 in the MHV A59 genome are numbered 1 to 287 for simplicity) was analyzed for TM-spanning domains by the use of various bioinformatics tools, and the residue numbers with predicted TM domains are displayed. The consensus TM topology of MHV nsp6 used as a basis for the topology experiments is depicted at the bottom row (shaded in gray).
FIG. 3.
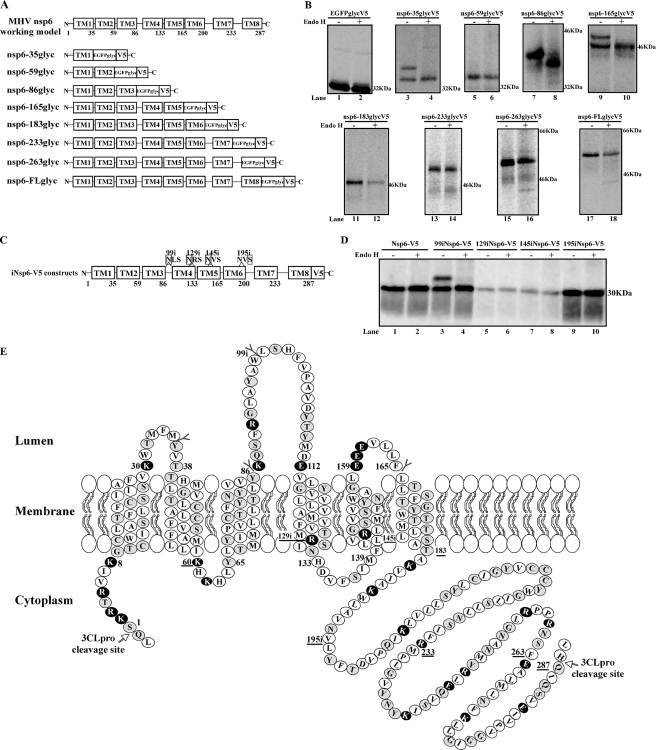
Determining the topology of nsp6 by the use of EGFPglyc and insertion of glycosylation consensus sites. (A) Schematics of a working topology model of MHV nsp6 (obtained from our consensus experiments) and nsp6-EGFPglycV5 fusion constructs generated for endo H assay. (B) Metabolic labeling and endo H treatment of nsp6-EGFPglycV5 fusion proteins. The nsp6-EGFPglycV5 fusion proteins expressed in transfected BsrT7 cells were radiolabeled from 20 to 22 h posttransfection, and then cell lysates were subjected to immunoprecipitation with V5 antibody, treated with endo H or left untreated, separated by 12.5% SDS-PAGE, and analyzed by autoradiography. (C) Map of plasmid DNA construct showing the sites of inserted glycosylation acceptor consensus sequences (NXS). The locations of glycosylation insertion in the nsp6-V5 construct are represented, with the amino acid number at the site of insertion. (D) Metabolic labeling and endo H analysis of glycosylation sequence insertion expression constructs of nsp6-V5. The plasmid DNAs (iNsp6-V5 constructs) were transfected and analyzed as described for panel B. (E) MHV-A59 nsp6 topology model, summarizing the results of EGFPglycV5 and glycosylation sequence insertion experiments. Amino acid positions indicated by the symbol “Y” were glycosylated and were positive by endo H assay, whereas those positions tested but found not glycosylated and negative by endo H assay are depicted by solid black horizontal lines. The inserted glycosylation acceptor sequence positions precede the letter i. Selected charged residues are shown in white characters on a black background. K, lysine residues; R, arginine residues; E, glutamic acid residues.
We found that fusion protein products expressed from the reporter constructs (nsp6-35glycV5, nsp6-86glycV5, and nsp-165glycV5) were glycosylated, as shown by sensitivity to endo H treatment, indicating that the C-terminal end of these chimeric proteins must extend into the ER lumen (Fig. 3B, lanes 4, 8, and 10). In contrast, the remaining reporter constructs were not sensitive to endo H treatment; therefore, the C-terminal end of the chimeric constructs must extend into the cytoplasm (Fig. 3B, lanes 6, 12, 14, 16, and 18). Thus, these results indicate the presence of three luminal loops in nsp6. Identical results were obtained when we used PNGaseF (data not shown), which indicates that the lack of endo H sensitivity was not attributable to the protein transiting through the Golgi body, thereby rendering the protein insensitive to endo H treatment.
To further investigate nsp6 topology in detail, we exploited a glycosylation sequence insertion mutagenesis approach (7) to create acceptor sequences in the region between amino acids 86 and 200 of nsp6 by the use of site-directed mutagenesis as described in reference 32 in order to independently investigate the topology, since bioinformatics predictions of the TM domains within this region differ (Fig. 2). Consensus glycosylation acceptor sites (NXS) were generated at four sites in the nsp6-V5 plasmid backbone by introducing single-codon insertions as depicted in Fig. 3C. All the glycosylation insertion constructs were expressed and analyzed by use of the endo H assay as described above. As expected, the parental nsp6-V5 protein is not glycosylated and did not show a mobility shift after endo H treatment (Fig. 3D, lanes 1 and 2). In contrast, expression of 99iNsp6-V5 revealed evidence of endo H sensitivity (Fig. 3D, lanes 3 and 4), indicating the ER luminal localization of the N99 introduced into MHV nsp6. This result is in agreement with those obtained with the nsp6-86glycV5 construct that is also endo H sensitive (Fig. 3B, lanes 7 and 8). The insertion of glycosylation acceptor sequences at other sites yielded endo H-negative results (lanes 6, 8, and 10), indicating the possibility that the introduced NX(S/T) motifs (i) are localized in the cytosol, (ii) are localized within the membrane, or (iii) are not used, as the glycosylation site is not at least 12 amino acids away from the end of the preceding TM and 14 amino acids away from the beginning of the following TM (12 + 14 rule), thus rendering it inaccessible for glycosylation (1, 7, 30). Our results confirm and extend the results of a recent study (22) in which authors were unable to resolve whether TM6 or TM7 acted as the final TM domain. Our results indicate that TM6 is the final TM domain for MHV nsp6. We propose a topology model of MHV-A59 nsp6 in Fig. 3E which is in accordance with the distribution of positively charged residues (positive inside rule; reviewed in reference 31), depicting the higher number of lysine and arginine residues facing the cytosolic side of the membrane and the majority of charged residues excluded from the TM domain. Taken together, the results presented above are consistent with a six-TM domain model of MHV nsp6. This report provides new information on the membrane topology of nsp6 and provides potential clues with respect to the assembly of the coronavirus replication complex.
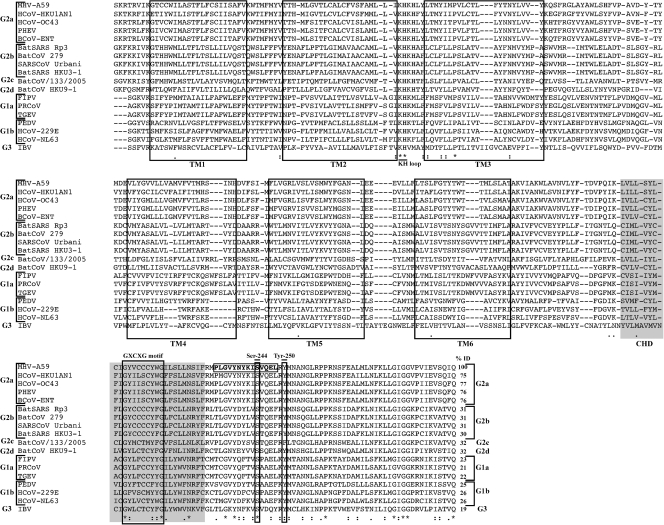
To determine whether the experimentally determined six-TM-spanning domain topology of MHV-A59 is conserved among coronaviruses, we performed MUSCLE (10) and ClustalW (29) multiple sequence alignment of nsp6 amino acid sequences representing group 1, group 2, and group 3 coronaviruses obtained from PATRIC (http://patric.vbi.vt.edu/) (27). The most striking observations were the amino acid sequence conservation in the C terminus of all nsp6 proteins and the conservation in the hydrophobicities within the putative TM domains (Fig. 4). This analysis revealed several conserved sites that may be important for the function of nsp6. We designated the conserved region between TM2 and TM3 the “KH loop” because of the invariant lysine and histidine residues that are present in the cytosolic loop (Fig. 3E), although the function of these amino acids is not yet known. We also designated the hydrophobic region in the C-terminal tail the “conserved hydrophobic domain” (Fig. 4). We speculate that cysteine residue(s) within the region we designated the “conserved G(X)C(X)G motif” may be modified by palmitoylation, indicating that this region of nsp6 may have important functions in establishing protein-protein or protein-membrane interactions during the assembly of the viral replication complex. Additionally, for the MHV-A59 nsp6 protein, the NetPhosK 1.0 server (http://www.cbs.dtu.dk/services/NetPhosK/) predicted serine and tyrosine residues (serine 244 and tyrosine 250; see Fig. 4) at the C-terminal region as sites of possible phosphorylation by epidermal growth factor receptor kinase and protein kinase C, respectively. Both predicted sites are highly conserved in all coronavirus nsp6 proteins (Fig. 4). Overall, our analysis revealed conserved features in the nsp6 C-terminal region whose importance in viral replication can be investigated using a coronavirus reverse genetics system.
FIG. 4.
Multiple sequence alignment (MSA) and percent sequence identity of coronavirus nsp6. The nsp6 amino acid sequences of 18 different coronaviruses were obtained from PATRIC (http://patric.vbi.vt.edu/) and aligned using MUSCLE and ClustalW software. The experimentally determined TM domains of MHV-A59 nsp6 were used as a reference for alignment. Unshaded boxes indicate the conserved TM domains that aligned with other coronavirus nsp6 sequences; the conserved hydrophobic domain (CHD) predicted by all the topology programs is indicated by gray shading. The residues of the peptide against which the nsp6 antibody was raised are boxed, with residue designations shown in boldface. Putative sites for palmitoylation (cysteine residue[s]) within the GXCXG motif) and phosphorylation (serine 244 and tyrosine 250 in MHV-A59 nsp6) are indicated. Percent identity (% ID) values are indicated. In MSA, the following notations were used: asterisk indicate invariant amino acids, colons indicate highly similar amino acids, and dots indicate similar amino acids. HCoV, human coronavirus; PHEV, porcine hemagglutinating encephalomyelitis virus; BCoV, bovine coronavirus; BatSARS, bat severe acute respiratory syndrome coronavirus; BatCoV, bat coronavirus; SARSCoV, severe acute respiratory syndrome coronavirus; FIPV, feline infectious peritonitis virus; PRCoV, porcine respiratory coronavirus; TGEV, transmissible gastroenteritis virus; PEDV, porcine epidemic diarrhea virus; IBV, infectious bronchitis virus.
Acknowledgments
This work was supported by Public Health Service research grants AI 060915 (to S.C.B.) and HHSN2662040035C (to B.S.).
Footnotes
Published ahead of print on 22 April 2009.
REFERENCES
- 1.Angeles Juanes, M., J. C. Igual, and M. C. Bano. 2008. Membrane topology and post-translational modification of the Saccharomyces cerevisiae essential protein Rot1. Yeast 2593-106. [DOI] [PubMed] [Google Scholar]
- 2.Baker, S. C., and M. R. Denison. 2008. Cell biology of nidovirus replication complexes, p. 103-113. In S. Perlman, T. Gallagher, and E. J. Snijder (ed.), Nidoviruses. ASM Press, Washington, DC.
- 3.Baker, S. C., C. K. Shieh, L. H. Soe, M. F. Chang, D. M. Vannier, and M. M. Lai. 1989. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 633693-3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonilla, P. J., S. A. Hughes, and S. R. Weiss. 1997. Characterization of a second cleavage site and demonstration of activity in trans by the papain-like proteinase of the murine coronavirus mouse hepatitis virus strain A59. J. Virol. 71900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 743379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchholz, U. J., S. Finke, and K.-K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, J. C., and R. A. F. Reithmeier. 2007. Scanning N-glycosylation mutagenesis of membrane proteins. Methods 41451-459. [DOI] [PubMed] [Google Scholar]
- 8.Clementz, M. C., A. Kanjanahaluethai, T. E. O'Brien, and S. C. Baker. 2008. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 375118-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denison, M. R., S. A. Hughes, and S. R. Weiss. 1995. Identification and characterization of a 65-kDa protein processed from the gene 1 polyprotein of the murine coronavirus MHV-A59. Virology 207316-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edgar, R. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 763697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham, R. L., and M. R. Denison. 2006. Replication of murine hepatitis virus is regulated by papain-like proteinase 1 processing of nonstructural proteins 1, 2, and 3. J. Virol. 8011610-11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanjanahaluethai, A., and S. C. Baker. 2001. Processing of the replicase of murine coronavirus: papain-like proteinase 2 (PLP2) acts to generate p150 and p44. Adv. Exp. Med. Biol. 494267-273. [DOI] [PubMed] [Google Scholar]
- 14.Kanjanahaluethai, A., and S. C. Baker. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J. Virol. 747911-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanjanahaluethai, A., Z. Chen, D. Jukneliene, and S. C. Baker. 2007. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 361391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanjanahaluethai, A., D. Jukneliene, and S. C. Baker. 2003. Identification of the murine coronavirus MP1 cleavage site recognized by papain-like proteinase 2. J. Virol. 777376-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, X. T., Y. Q. Lu, and M. R. Denison. 1996. Intracellular and in vitro-translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology 222375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, X. T., A. C. Sims, and M. R. Denison. 1998. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J. Virol. 722265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, S., S. Kastner, J. Krijnse-Locker, S. Buhler, and R. Bartenschlager. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2k-regulated manner. J. Biol. Chem. 2828873-8882. [DOI] [PubMed] [Google Scholar]
- 21.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oostra, M., M. C. Hagemeijer, M. van Gent, C. P. Bekker, E. G. te Lintelo, P. J. Rottier, and C. A. de Haan. 2008. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 8212392-12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oostra, M., E. G. te Lintelo, M. Deijs, M. H. Verheije, P. J. Rottier, and C. A. de Haan. 2007. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 8112323-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piñón, J. D., R. R. Mayreddy, J. D. Turner, F. S. Khan, P. J. Bonilla, and S. R. Weiss. 1997. Efficient autoproteolytic processing of the MHV-A59 3C-like proteinase from the flanking hydrophobic domains requires membranes. Virology 230309-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao, P. V., and T. M. Gallagher. 1998. Mouse hepatitis virus receptor levels influence virus-induced cytopathology. Adv. Exp. Med. Biol. 440549-555. [DOI] [PubMed] [Google Scholar]
- 26.Schiller, J. J., A. Kanjanahaluethai, and S. C. Baker. 1998. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder, E. E., N. Kampanya, J. Lu, E. K. Nordberg, H. R. Karur, M. Shukla, J. Soneja, Y. Tian, T. Xue, H. Yoo, F. Zhang, C. Dharmanolla, N. V. Dongre, J. J. Gillespie, J. Hamelius, M. Hance, K. I. Huntington, D. Jukneliene, J. Koziski, L. Mackasmiel, S. P. Mane, V. Nguyen, A. Purkayastha, J. Shallom, G. Yu, Y. Guo, J. Gabbard, D. Hix, A. F. Azad, S. C. Baker, S. M. Boyle, Y. Khudyakov, X. J. Meng, C. Rupprecht, J. Vinje, O. R. Crasta, M. J. Czar, A. Dickerman, J. D. Eckart, R. Kenyon, R. Will, J. C. Setubal, and B. W. S. Sobral. 2007. PATRIC: The VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 35D401-D406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sparks, J. S., X. Lu, and M. R. Denison. 2007. Genetic analysis of murine hepatitis virus nsp4 in virus replication. J. Virol. 8112554-12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Geest, M., and J. S. Lolkema. 2000. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol. Mol. Biol. Rev. 6413-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Heijne, G. 2007. The membrane protein universe: what's out there and why bother? J. Intern. Med. 261543-557. [DOI] [PubMed] [Google Scholar]
- 32.Zheng, L., U. Baumann, and J. L. Reymond. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32e115. [DOI] [PMC free article] [PubMed] [Google Scholar]