Abstract
Macro domains (also called “X domains”) constitute a protein module family present in all kingdoms of life, including viruses of the Coronaviridae and Togaviridae families. Crystal structures of the macro domain from the Chikungunya virus (an “Old World” alphavirus) and the Venezuelan equine encephalitis virus (a “New World” alphavirus) were determined at resolutions of 1.65 and 2.30 Å, respectively. These domains are active as adenosine di-phosphoribose 1″-phosphate phosphatases. Both the Chikungunya and the Venezuelan equine encephalitis virus macro domains are ADP-ribose binding modules, as revealed by structural and functional analysis. A single aspartic acid conserved through all macro domains is responsible for the specific binding of the adenine base. Sequence-unspecific binding to long, negatively charged polymers such as poly(ADP-ribose), DNA, and RNA is observed and attributed to positively charged patches outside of the active site pocket, as judged by mutagenesis and binding studies. The crystal structure of the Chikungunya virus macro domain with an RNA trimer shows a binding mode utilizing the same adenine-binding pocket as ADP-ribose, but avoiding the ADP-ribose 1″-phosphate phosphatase active site. This leaves the AMP binding site as the sole common feature in all macro domains.
The Togaviridae virus family comprises two positive-sense RNA virus genera, viz., Alphavirus and Rubivirus (52). The genus Alphavirus contains at least 28 viruses and has a worldwide distribution, even though each virus has a local distribution. Some alphaviruses are not known to cause illness, but others can cause severe disease in higher eukaryotes, in particular, humans and horses. Alphaviruses present in the “Old World” principally cause arthritis and skin rashes, whereas alphaviruses of the “New World” may cause severe and even fatal encephalitis. Most have vertebrate hosts, principally mammals, birds, or fish, and are transmitted by mosquitoes, although other hematophagous arthropods such as lice or mites can be vectors of some alphaviruses (37).
Although known for decades as a virus that produces sporadic human outbreaks in Africa, India, Southeast Asia, and the Philippines, Chikungunya virus (CHIKV) unexpectedly emerged as a major arbovirus pathogen in many islands of the Indian Ocean in 2005, and, more recently (2007) in Singapore and Australia. The virus was also introduced into northern Italy in 2007, where it established localized outbreaks. In urban areas, the virus is mainly transmitted by the mosquito Aedes aegypti but an adaptive mutation facilitated transmission by another urban mosquito, Aedes albopictus, in La Réunion island in 2005 as well as other places (7). Epidemics are sporadic, but a large part of the population can be infected, as was the case in La Réunion island in 2006, where one-third of the population (∼250,000 people) was infected, leading to 237 deaths. CHIKV symptoms in humans include fever, rash, and severe arthritis, which usually disappear after 1 week, but, in some cases, arthritis can persist for more than 1 year (18). Venezuelan equine encephalitis virus (VEEV) is a New World alphavirus, present in the United States and Central and South America. Different subtypes of VEEV exist, some of which cause epidemics while others are zoonotic. In a few cases, VEEV causes encephalitis in humans, particularly children (40). This virus is also pathogenic for horses, with an observed case fatality rate of around 80 to 90%. No vaccine or drugs are licensed for treatment against alphaviruses.
The 5′ region of the positive-stranded RNA [(+) RNA] genome encodes four nonstructural proteins (nsP1, nsP2, nsP3, nsP4), and the 3′ region encodes the three major structural proteins (the capsid and two envelope proteins). The nsP1 protein is a membrane-associated protein that bears methyltransferase and guanylyltransferase activities. It is involved in the capping of the (+) RNA genome (1). The nsP2 protein is made of three domains, the first containing helicase, RNA triphosphatase, and nucleoside triphosphatase activities (39, 53), whereas the second and third domains are a papaine-like protease and a nonfunctional methyltransferase, respectively (47). Moreover, nsP2 contains a nuclear localization sequence which allows 50% of the translated nsP2 to be translocated into the nucleus (35). The nsP4 protein contains the RNA-dependent RNA polymerase, involved in genome replication and transcription (42). The functions, roles, and activities of the nsP3 protein are less well understood. Although it is involved in the transcription process at an early stage of the infection (51), no precise function or activity has been attributed to this protein. It is made of two domains, the first one being a unique macro domain (described below) located in the conserved N-terminal region. The C-terminal region is less conserved and is phosphorylated in up to 16 positions on serines and threonines (22, 25, 50). The role of phosphorylation is not well documented, but deletion of the phosphorylated residues decreases the RNA synthesis level (49). Moreover, the absence of phosphorylation on nsP3 in variants of the alphavirus Semliki Forest virus (SFV) decreases viral pathogenicity, and the absence of the C terminus of nsP3 alters SFV neurovirulence (48). The C terminus of nsP3 is thus thought to have a nonessential regulatory role.
The first 160 residues of the N-terminal region of nsP3, known as the macro domain, have been initially identified based on sequence similarity analysis between alphavirus, rubivirus, and coronavirus (15). The domain was named the “X domain,” referring to a domain with an unknown function conserved in these viruses. Subsequent sequence analysis revealed that this domain is remarkably conserved in all kingdoms of life (2): 1,081 domains showing similarity to X domains are currently indexed in the SMART database (24). They may represent a protein or a single domain in a larger protein, and the domain can also exist in duplicate or triplicate in the same protein (21). In particular, this domain is present in a variant of histone H2A. This variant is called macroH2A, and its difference from the conventional histone H2A is the presence of an additional domain called “macro,” which shows similarity to X domains. Consequently, all X domains have also been called macro domains.
In viruses, macro domains exist in alphaviruses and in viruses related to the genus Alphavirus, such as rubella virus (genus Rubivirus) and hepatitis E virus (HEV) (genus Hepevirus). Coronavirus and Torovirus, which belong to the Coronaviridae family, are the only other viral genera containing a macro domain, located in their large multifunctional nsp3 protein, which is otherwise unrelated to alphavirus nsP3.
The crystal structures of several macro domains have been determined in archaebacteria (Archaeoglobus fulgidus) (2), eubacteria (Escherichia coli and Thermus thermophilus), and eukaryotes (Saccharomyces cerevisiae, Rattus norvegicus, and Homo sapiens) (4, 19, 20). The crystal structure of the macro domain of the coronavirus responsible for severe acute respiratory syndrome (SARS-CoV) has also been determined (10, 27, 43). The structural conservation between these structures is remarkable and they have been defined as a family in the SCOP database (30). This structural conservation suggests an important role in the biology of their host organism, but this role remains elusive so far.
An enzymatic activity was first discovered for the yeast macro domain, which acts as an ADP-ribose 1″-phosphate phosphatase with a somewhat low turnover constant of 1.7 min−1 (28). This activity is involved in the downstream processing of ADP-ribose 1″-phosphate, a side product of cellular pre-tRNA splicing, thus controlling the metabolism of ADP-ribose 1″-phosphate or other ADP-ribose derivatives with known regulatory functions in the cell. This activity has also been reported for A. fulgidus (17), SARS-CoV (10, 43), and human CoV (HCoV) (38), but was at the limit of detection for the alphavirus SFV (10). The affinity of ADP-ribose for the SARS-CoV enzyme was low (dissociation constant [Kd] of 52.7 μM) (43), and the pathway in which ADP-ribose 1″-phosphate phosphatase is putatively involved remains elusive. A recent study showed that ADP-ribose binding is not a common property to all macro domains. Indeed, the group 3 CoV macro domain does not bind ADP-ribose, whereas a group 1 CoV macro domain binds it with a dissociation constant Kd of 29 μM (36).
In the case of A. fulgidus, poly(ADP-ribose) (PAR) binding has been reported (17). Such a binding property has also been identified in CoVs (10), showing that some macro domains, in particular viral macro domains, are able to bind long, negatively charged polymers. The role of PAR binding could be related to a cellular response to viral infection. Indeed, in response to inflammation or stress, the nuclear enzyme PAR polymerase-1 (PARP-1) promotes PAR synthesis (5). Alphavirus infection can induce PARP-1 activation (31) which leads to the depletion of ATP and NAD+ present in the cell and apoptosis-inducing factor (AIF)-induced apoptosis.
In this article, we present the biochemical, enzymatic, and structural analysis of two alphavirus macro domains, one from CHIKV, a representative of the Old World alphaviruses, the other from VEEV, a New World alphavirus present in the Americas. The crystal structures of the CHIKV and VEEV macro domains in complex with ADP-ribose show essentially the same positioning of the ligand as that seen in the SARS-CoV macro domain structure. The crystal structure of the CHIKV macro domain in complex with adenosine-containing short RNAs shows that the adenosine binding site is generally conserved in macro domains. Moreover, binding of the short RNA is enhanced by the presence of positively charged patches present at the surface of the protein. The latter patches, located in the immediate vicinity of the ADP-ribose binding crevice, are significantly more charged than that of the SARS-CoV macro domain.
MATERIALS AND METHODS
Oligonucleotides.
RNA (AAAAAAAAAGCUACC, AAA, UUU, GGGGGG, UCGGGGGCUGGC) and DNA (AAAGCCAAAAA) oligonucleotides were purchased from Dharmacon.
Expression and purification of alphavirus macro domains.
The cDNAs corresponding to two alphavirus macro domains from the nsP3 protein of CHIKV (strain Ross, amino acids [aa] 1 to 160 of nsP3) and VEEV (strain P676, aa 1 to 160) were cloned into the pDest14 plasmid using the “Gateway” cloning procedure (Invitrogen). A hexa-histidine sequence tag was fused at either the N-terminal end (CHIKV) or the C-terminal-end (VEEV). An incomplete factorial expression screening in E. coli was performed for each construct in order to design the best expression conditions required for scale-up, as previously described (3). The proteins (wild type and mutants) were then produced under the following conditions: (i) with E. coli Rosetta (DE3)(pLysS) (Novagen) cells at 25°C in SB medium (Athena Enzymes) overnight after induction with 500 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) for the CHIKV macro domain and (ii) with E. coli Rosetta (DE3)(pLysS) cells at 37°C in 2YT medium for 4 h after induction with 500 μM IPTG for the VEEV macro domain. Cells were harvested by centrifugation at 2,800 × g. Cell pellets were then resuspended in 50 mM Tris buffer, 300 mM NaCl, 10 mM imidazole, 0.1% Triton, and 5% glycerol (pH 8.0). Lysozyme (0.25 mg/ml), phenylmethylsulfonyl fluoride (1 mM), DNase I (2 μg/ml), and EDTA-free protease cocktail (Roche) were added before performing a sonication step. The lysates were centrifuged at 12,000 × g for 45 min, and the supernatant was recovered. The recombinant proteins were purified using the Akta Xpress fast protein liquid chromatography system (GE Healthcare) as follows. The first purification step (immobilized metal affinity chromatography) was performed on a 5-ml His prep column (GE Healthcare). The proteins were eluted with 50 mM Tris buffer (pH 8.0) containing 300 mM NaCl and 500 mM imidazole. The purification was then refined by a size exclusion chromatography step on a preparative Superdex 200 column (GE Healthcare) preequilibrated in a buffer designed for its ability to keep the protein soluble and stable (14): 10 mM Tris (pH 7.5) plus 300 mM NaCl for the CHIKV macro domain and 10 mM Bicine (pH 8.5) plus 50 mM NaCl for the VEEV macro domain. Proteins were then concentrated up to 14.5 mg/ml and 14 mg/ml using a Vivaspin 10-kDa molecular-mass-cutoff centrifugal concentrator (Vivascience) for the CHIKV and VEEV macro domains, respectively. The Eastern equine encephalitis virus (EEEV) macro domain was obtained from EEEV cDNA by the same procedure described above and purified to homogeneity following the same protocol as that of the CHIKV macro domain. It was used in phosphatase assays (see Fig. 4). Its crystal structure will be reported elsewhere.
FIG. 4.
Detection by TLC of ADP-ribose 1″-P, the product of the ADP-ribose phosphatase activity. Control lanes (ADP-ribose 1″-phosphate and ADP-ribose) and reaction lanes with corresponding enzymes and D10A, N24A, and Y114A mutants are indicated. The EEEV lane corresponds to the purified EEEV macro domain designed and produced in the same manner as the CHIKV macro domain. The yeast macro domain protein Poa1p serves as a positive control.
A seleno-methionine-substituted protein was used to determine the CHIKV macro domain structure. The protein was expressed according to standard conditions of methionine biosynthesis pathway inhibition (9) and purified following the same procedure as that of the native protein.
Crystallization.
Initial crystallization trials were set up for CHIKV and VEEV macro domains with a nano-drop dispenser (Honeybee; Genomic Solutions) in 96-well sitting drop plates (Greiner Bio One) using three commercial crystallization kits: Structure Screen combination, Stura footprints (Molecular Dimensions Limited), and Nextal SM1 (Qiagen).
The initial crystallization conditions of the CHIKV protein were further optimized by the hanging drop vapor diffusion method in Linbro plates by mixing 3 μl of protein solution with 2 μl of reservoir solution. Crystals grew in a mixture of 46% polyethylene glycol 600 (PEG 600) and 100 mM HEPES (pH 7.4). For the VEEV macro domain, initial hits were optimized by mixing 100 nl of protein and 100 nl of reservoir solution using the sitting drop vapor diffusion method. Optimized crystallization conditions of VEEV macro domain were 18% PEG 3350 plus 11 mM sodium acetate. Diffraction intensities were recorded on different beamlines (Table 1) at the European Synchrotron Radiation Facility (Grenoble, France) and at SOLEIL Synchrotron (Gif-sur-Yvette, France). Integration of the different datasets was performed using MOSFLM (23) or XDS (16). Scaling and merging of the intensities were carried out using programs from the CCP4 suite (6) or XSCALE (16), depending on the data sets. Statistics are provided in Table 1.
TABLE 1.
Crystallization analysis data for CHIKV and VEEV
| Parameter | Result ford:
|
|||||
|---|---|---|---|---|---|---|
| CHIKV
|
VEEV
|
|||||
| Native | Selenomethio-nylated protein | Soaked with ADP-ribose | Soaked with RNA | Native | Cocrystallized with ADP-ribose | |
| Beamline | ESRF ID14-EH2 | ESRF ID23-1 | ESRF ID23-1 | ESRF ID14-EH2 | ESRF ID23-1 | SOLEIL PROXIMA1 |
| Space group | P31 | P31 | P31 | P31 | I4 | P212121 |
| Cell dimensions (Å) | a = b = 87.06, c = 84.49 | a = b = 86.81, c = 84.70 | a = b = 87.96, c = 84.17 | a = b = 86.82, c = 81.32 | a = b = 129.63, c = 42.49 | a = 74.00, b = 87.0, c = 105.00 |
| Wavelength (Å) | 0.9330 | 0.9792 | 0.8856 | 0.9330 | 0.974 | 0.980 |
| Resolution range (Å) | 35.00-1.65 (1.74-1.65) | 35.00-1.80 (1.90-1.80) | 35.00-1.90 (2.00-1.90) | 30.00-2.00 (2.11-2.00) | 35.00-2.30 (2.42-2.30) | 30.00-2.60 (2.70-2.60) |
| Total no. of reflections | 493,231 (71,649) | 2,000,862 (237,557) | 310,123 (38,681) | 213,703 (30,275) | 108,812 (16,349) | 109,136 (10,819) |
| No. of unique reflections | 86,209 (12,610) | 64,424 (8,446) | 57,403 (8,387) | 46,206 (6,793) | 15,851 (2,317) | 20,549 (2,014) |
| Completeness (%) | 100 (100) | 97.3 (87.3) | 99.9 (99.9) | 99.6 (100.0) | 99.2 (100.0) | 99.2 (89.2) |
| I/Σ(I) | 18.1 (3.2) | 37.4 (6.1) | 15.7 (2.4) | 17.4 (2.8) | 9.0 (5.4) | 15.74 (4.72) |
| Rsym (%)a | 5.2 (46.8) | 8.8 (50.2) | 7.2 (55.4) | 5.2 (52.5) | 13.9 (26.4) | 10.4 (49.7) |
| Multiplicity | 5.7 (5.7) | 31.1 (28.1) | 5.4 (4.6) | 4.6 (4.5) | 6.9 (7.1) | 5.3 (5.4) |
| Anomalous completeness (%) | 97.1 (86.1) | |||||
| Anomalous multiplicity | 15.4 (14.0) | |||||
| R (%)b | 16.6 | 17.0 | 21.5 | 20.3 | 19.7 | |
| Rfree (%)c | 19.6 | 20.1 | 26.0 | 26.1 | 27.5 | |
| rmsd bond length (Å) | 0.012 | 0.013 | 0.014 | 0.013 | 0.010 | |
| rmsd angle (°) | 1.347 | 1.447 | 1.398 | 1.348 | 1.433 | |
| Protein Data Bank no. | 3GPG | 3GPO | 3GPQ | 3GQE | 3GQO | |
Rsym = Σ |I− <I> |/Σ I, where I is the observed intensity and <I> is the average intensity. Values in parentheses refer to the highest-resolution shell.
R = Σ ‖Fo|− |Fc‖/Σ|Fo|.
Rfree is calculated as R, but on 5% of all reflections that are never used in crystallographic refinement.
Values in parentheses refer to the highest-resolution shell. ESRF, European Synchrotron Radiation Facility.
Structure determination.
The structure of the CHIKV macro domain was determined using the single-wavelength anomalous dispersion method (SAD) on a 1.80-Å data set collected at the peak of the selenium absorption edge from a seleno-methionine derivative crystal. Location of all the 16 selenium atoms (4 selenium atoms in each of the 4 molecules of the asymmetric unit) was performed using SHELXD (44). Phases and figures of merit were calculated using SHELXE (45). The excellent quality of the map allowed the program ARPWARP to build residues 2 to 160 for molecules A and B and to partially build molecules C and D. Refinement was performed against a 1.65-Å resolution data set using REFMAC (6), Buster-TNT (41), and COOT (11). Refinement statistics are listed in Table 1. The VEEV macro domain was determined by molecular replacement with the program PHASER (29) using the CHIKV macro domain as a model. Crystals diffracted to a 2.30-Å resolution for the VEEV protein. Refinement was performed using REFMAC, Buster-TNT, and COOT.
Crystal soaking and cocrystallization experiments.
CHIKV and VEEV macro domain crystals were soaked for 16 h with ADP-ribose (4 mM) (Sigma). The CHIKV macro domain was soaked for 16 h with PAR (1.3 mM in the ADP-ribose monomer) (Biomol International LP.), short RNA oligonucleotides AAA (2 to 10 mM) (Dharmacon) and UUUUUU (2 to 10 mM) (Dharmacon), and the DNA oligonucleotide AAAGCCAAAAA (10 mM) (Dharmacon). Crystals were transferred in drops containing the mother liquor solution and the ligand. Cocrystallization of the VEEV macro domain with ADP-ribose (5 mM) was performed in 0.2 M ammonium sulfate, 0.1 M sodium cacodylate (pH 6.5), and 30% PEG 8000.
Site-directed mutagenesis.
Based on the CHIKV macro domain structure, four point mutations were made using Stratagene's Quickchange site-directed mutagenesis. Single-amino-acid substitutions to alanine were performed in the CHIKV macro domain for D10, N24, H67, and Y114.
Thermal denaturation shift assay.
Several potential ligands were tested at 2 mM each (ADP-ribose, ADP-glucose, ATP, ADP, AMP, S-adenosyl-homocysteine, manganese chloride [MnCl2], magnesium chloride [MgCl2], NAD+, and GDP using the quantitative PCR machine ICycler IQ (Bio-Rad) according to the previously described thermal shift assay (12). Briefly, the CHIKV macro domain (final concentration of 3 mg/ml in the size exclusion chromatography buffer described above) was mixed with the ligand and a SYPRO orange solution in concentrations recommended by the manufacturer (Invitrogen) in a final volume of 25 μl. Incremental steps of temperature from 20 to 90°C were applied to the samples. The denaturation of the protein was monitored by following the increase of the fluorescence emitted by the probe that binds the exposed hydrophobic regions of the denatured protein. A melting temperature (Tm) can be calculated as the mid-log of the transition phase from the native to the denatured protein using a Boltzmann model. ADP-ribose titration from 0 to 2.5 mM was then assessed on the CHIKV (wild type and D10A single mutant), VEEV and SARS-CoV macro domains at 1 mg/ml in 10 mM HEPES plus 150 mM NaCl (pH 7.5).
ITC.
For isothermal titration calorimetry (ITC), ADP-ribose binding to CHIKV and VEEV macro domains was measured using a Microcal MCS isothermal titration calorimeter. Experiments were carried out at 25°C in a buffer containing 10 mM HEPES plus 150 mM NaCl (pH 7.5). The protein concentration in the cell was 20 μM, whereas the ADP-ribose concentration in the syringe was 300 μM. Heats of dilution were measured by injecting the ligand into the protein solution. The recorded heat curve was subtracted from the experimental curves prior to data analysis. Titration curves were fitted using MicroCal Origin software, assuming one set of sites and enthalpy changes (ΔH), equilibrium constants (Kd), and stoichiometry were extracted.
PAR binding assay.
The PAR (1‴-2″ branched polymer of ADP-ribose linked by 1″-2′ glycosidic bonds), was synthesized by auto-poly-ADP-ribosylation of PARP-1 in a reaction volume of 400 μl using 4 U of human PARP-1 (Sigma), 150 μM NAD+, and 40 μCi of [32P]NAD+ (GE Healthcare). After 2 h of incubation at 24°C, the reaction was stopped by dilution (30-fold) with the slot blot buffer (10 mM Tris [pH 7.5], 300 mM NaCl, 0.05% Tween 20). PAR binding on the macro domains of several alphaviruses (CHIKV, VEEV, Sindbis virus, and SFV) as well as SARS-CoV was tested. Various amounts of each protein (2,000, 1,000, 500, 250, 125, 62.5, 31.25, 15.63, 7.813, 3.906, 1.953, and 0.9766 pmol) were blotted on a nitrocellulose membrane (Schleicher & Schuell) using a slot blot apparatus (Bio-Rad). Bovine serum albumin (BSA) was also included as a negative control. The membrane was then incubated for 1 h in the PAR preparation and then washed extensively in slot blot buffer (five times in 100 ml), and the membrane-bound fraction of PAR was analyzed using photostimulated plates and an FLA3000 fluorescent image analyzer (Fuji).
RNA binding assay.
Fifty microliters of RNA (AAAAAAAAAGCUACC; 100 μM) was labeled using 25 U of T4 polynucleotide kinase (New England Biolabs), 100 μM ATP, and 50 μCi [γ-32P]ATP (GE Healthcare). After 30 min at 37°C, the reaction was stopped by incubating the mixture for 10 min at 70°C. RNA binding to the CHIKV macro domain native and mutants was tested as follows: 2,000, 1000, 500, 250, 125, 62.5, 31.25, 15.63, 7.813, 3.906, 1.953, and 0.9766 pmol of protein were blotted onto a nitrocellulose membrane (Schleicher & Schuell) using a slot blot apparatus (Bio-Rad). BSA and hepatitis C virus RNA-dependent RNA polymerase (HCVpol) were also included as negative and positive controls, respectively. The radiolabeled RNA was diluted (300×) in blotting buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 0.05% Tween 20). The membrane supporting the blotted proteins was incubated for 1 h in this preparation at room temperature. The membrane was washed five times in 100 ml of blotting buffer, and the membrane-bound fraction of RNA was analyzed using photostimulated plates and an FLA3000 fluorescent image analyzer (Fuji).
ADP-ribose 1″-phosphate phosphatase assay.
ADP-ribose-1″-phosphate phosphatase activity was detected with thin-layer chromatography (TLC) assays for the CHIKV (wild type and mutants), EEEV, and VEEV macro domains. The yeast protein Poa1p was used as a positive control. First, ADP-ribose 1″-phosphate was produced by incubating 7 mM ADP-ribose 1″-2″-cyclic phosphate with cyclic phosphodiesterase (15 ng/ml) for 3 h at 28°C. Then, 1 μl of the reaction was mixed with 1 μl of 45 μM of the macro domain tested and 1 μM of 60 mM MES (morpholineethanesulfonic acid) (pH 5.0). The reaction mixture was incubated for 1 h at 28°C. Two microliters of the reaction mixture was then spotted onto a polyethyleneimine-F (PEI-F) cellulose TLC plate, and the plate was developed at room temperature in 150 mM NaCl and 150 mM sodium formate (pH 3.0). The spots were detected from the fluorescent background under a UV lamp emitting at a 254-nm wavelength (10).
Protein structure accession number.
The coordinates of CHIKV macro domain structure, apo form, in complex with ADP-ribose and with RNA AAA have been deposited in the Protein Data Bank under accession no. 3GPG, 3GPO, and 3GPQ, respectively. Those of the VEEV macro domain structure apo form and in complex with ADP-ribose have been deposited in the Protein Data Bank under accession no. 3GQE and 3GQO.
RESULTS
Protein production and crystallization.
A cDNA construct encompassing aa 1 to 160 of the alphavirus nsP3 macro domain was designed based on available macro domain crystal structures (2, 4, 19, 20, 43), secondary structures, and disorder predictions. CHIKV and VEEV nsP3 macro domains were selected in order to compare macro domains from New and Old World alphaviruses. These constructs were expressed in E. coli and purified, and crystals were obtained for these two proteins. Molecular replacement techniques using the closest structural homologue (E. coli macro domain, identity of 29.9% in 1 to 160 aa for the CHIKV macro domain) were unsuccessful. The crystal structure of the CHIKV macro domain was determined using the SAD technique in conjunction with seleno-methionylated protein crystals that diffracted to 1.80 Å. It was then refined against a native data set at a 1.65-Å resolution. The CHIKV crystals belong to the space group P31, and four molecules are present in the asymmetric unit. VEEV macro domain structure in an apo form was then determined at a 2.30-Å resolution using molecular replacement and the CHIKV macro domain as a template. The VEEV crystals of the apo form belong to the space group I4, with two molecules in the asymmetric unit.
Overall structure of alphavirus macro domains and comparison with other macro domains.
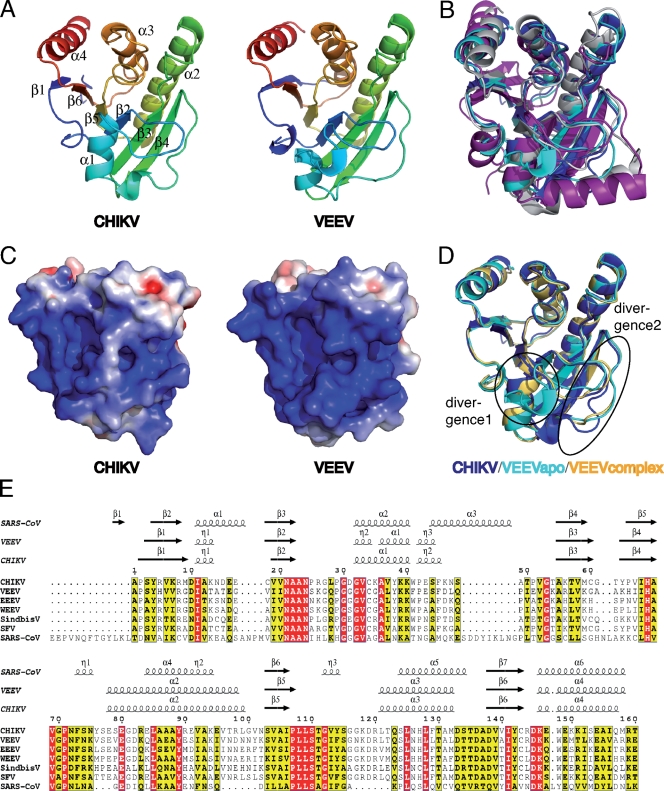
The structures of the alphavirus macro domain consist of a central twisted six-stranded β sheet surrounded by three helices on one side and one on the other (Fig. 1A). The core β sheet is well conserved within the existing structures of macro domains from all origins also. Positions of α helices are also well conserved, even if a deletion is present in alphavirus macro domains near residue 48 (in CHIKV macro domain; the numbering of CHIKV will be used throughout), leading thus to the absence of one helix present in other macro domains (Fig. 1E, α3 in SARS-CoV). Moreover, the α helix 2 (aa 78 to 100) is longer in alphavirus macro domains than in other macro domains (Fig. 1B and E). The loops connecting the secondary structure elements are variable in sequence and structure when compared to non-alphavirus macro domain structures available, in particular the loops between the β3-β4, β4-α2, and β5-α3 elements. The root mean square deviations (rmsds) between the CHIKV macro domain and other reported macro domain structures are comprised between 2.01 Å and 2.97 Å on 160 aa (sequence identity between 21.9 and 30.0%). Remarkably, the closest structure is the E. coli macro domain and not the SARS-CoV macro domain present in the nsp3 protein product of the SARS-CoV orf1a replicase polyprotein (10, 46). The rmsds between the Cα of SARS-CoV and alphavirus macro domains are between 2.80 and 2.97 Å on 160 aa. Structural differences between SARS-CoV and the alphavirus macro domain are consistent with the differences observed in all of the known macro domain crystal structures. Moreover, the SARS-CoV macro domain is longer at the N terminus and the additional 13 residues allow the formation of a seventh β strand in the central β sheet. Therefore, from a structural point of view, there is no obvious cluster of related viral macro domain structures.
FIG. 1.
Structures of the macro domains from the alphaviruses CHIKV and VEEV. (A) Representation of CHIKV and VEEV macro domains in a purple-to-red gradient (from N terminus to C terminus). Secondary structure elements are labeled on the CHIKV macro domain. (B) Superposition of representations of the CHIKV, VEEV, SARS-CoV, and E. coli macro domains, colored, respectively, in dark blue, cyan, purple, and white. (C) Electrostatic surface potential presented between −4 and 4 kT/e for the CHIKV and VEEV macro domains. The potential was generated using the PDB2PQR server (8) and Adaptative Poisson-Boltzmann Solver (APBS) plug-in in Pymol (http://www.pymol.org). (D) Representation of the superposition between the macro domains from CHIKV (apo form in dark blue), VEEV (apo form in cyan and complexed with ADP-ribose in light orange). The two main divergences between the three structures are indicated and circled. (E) Sequence alignment of the macro domains studied. They belong to the genus Alphavirus, except for SARS-CoV. Residues in red boxes are strictly conserved, while those in yellow boxes are conserved by at least four out of seven viruses aligned. Secondary structure elements from the viral macro domain crystal structures obtained are represented above the alignment.
Electrostatic surface analysis of alphavirus macro domains reveals a highly positively charged patch located both in the crevice, previously defined as an ADP-ribose 1″-phosphate phosphatase active site (17) and at its periphery. The other face of the protein, located far from the active site, is negatively charged (not shown). This bimodal charge distribution is much more pronounced in alphavirus macro domains than in other macro domain structures.
Between the two alphavirus macro domain structures, the sequence identity is 57% and the crystal structures are also very similar, the rmsd between CHIKV and VEEV macro domains being 0.91 Å on 160 aa (Fig. 1A and D). Interestingly, the main structural divergence appears between residues 30 to 37 when comparing CHIKV and VEEV macro domains in the apo form (Fig. 1D, divergence 1). Some of these residues, located close to the active site, are involved in RNA and ADP-ribose binding (see below). This structural difference could thus impact substrate binding differentially within the alphaviruses (see Discussion). Interestingly, the corresponding loop in other available macro domain structures is very well conserved and corresponds to the position of the CHIKV macro domain loop. In addition, two loops located far from the active site, from positions 48 to 52 and 62 to 65, are different between the CHIKV and VEEV macro domains (Fig. 1D, divergence 2). These loops are located far from the active site. The divergence of these loops does not originate from crystal packing. Indeed, a crystal form obtained for the VEEV macro domain in complex with ADP-ribose (see below) shows a similar position of these loops despite a different packing.
Biochemical and structural basis of ADP-ribose binding.
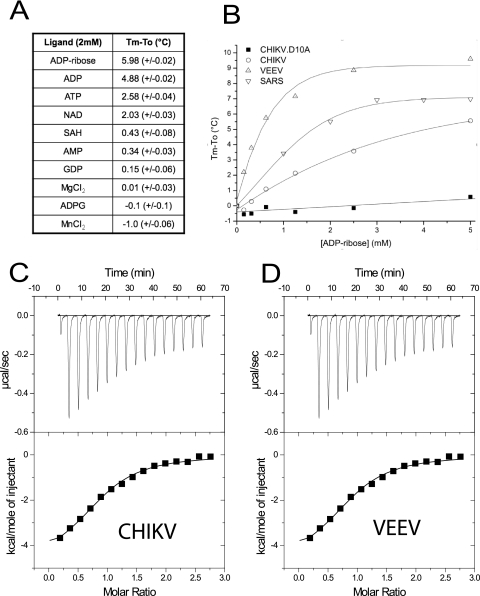
Since several macro domains were reported as ADP-ribose binding proteins (17), the binding of ADP-ribose and other putative substrates was tested on the CHIKV macro domain using a thermal shift assay following the strategy developed previously (33). The Tm of the protein is measured using SYPRO orange fluorescence, which increases when this dye binds to hydrophobic core of the protein upon thermal denaturation. The melting temperature with the ligand (Tm) is compared to the Tm of the protein alone (To) in order to assess ligand binding. The ligands tested on the CHIKV macro domain can be divided into two groups (Fig. 2A). First of all, ADP-ribose, ADP, ATP, and NAD+ shift significantly the Tm of the protein to higher values (between 2 and 6°C). This first group contains an ADP moiety. A ribose at the distal position contributes to stabilizing the macro domain, better than either nothing (for ADP), a phosphate (for ATP), a nicotinamide group (for NAD+), or a glucose (for ADP-glucose). The other ligands have either a poor effect or no effect on the thermal melting resistance, suggesting that they do not bind the protein. Altogether, these results indicate that the CHIKV macro domain has a specificity for (i) an adenine rather than a guanine, (ii) two phosphate groups rather than either one or three, and (iii) a ribose at the distal position.
FIG. 2.
ADP-ribose binding. (A) Thermal denaturation shift at a given ligand concentration (2 mM) for the CHIKV macro domain. Tm is the melting temperature of the protein in the presence of the ligand, and To is the melting temperature of the protein alone. (B) ADP-ribose titration using the thermal denaturation shift assay on CHIKV (wild type and D10A mutant), VEEV, and SARS-CoV macro domains. (C) ITC assay of ADP-ribose binding to the CHIKV macro domain. (Upper panel) ITC raw data of ADP-ribose binding to the CHIKV and VEEV macro domains. (Lower panel) ADP-ribose binding isotherm derived from raw data. (D) Same experiment as in panel C but using the VEEV macro domain.
Both CHIKV and VEEV macro domains bind ADP-ribose, although they exhibit a quite different thermal denaturation shift profile (Fig. 2B). The large Tm-To observed for the VEEV macro domain will find a structural explanation (see Fig. 1D and Fig. 3B for details). When the D10 position is changed to alanine, the CHIKV macro domain loses its ability to bind ADP-ribose (Fig. 2B; see the structural rationale for the selection of this mutant below).
FIG. 3.
Structural basis for ADP-ribose binding. (A) ADP-ribose binding site of the CHIKV macro domain. On the left side is a representation of the CHIKV macro domain with helices, strands, and loops colored, respectively, in red, yellow, and green. ADP-ribose is displayed in sticks with carbons in yellow, oxygens in red, nitrogens in blue, and phosphorus in orange. The Fo − Fc difference map, contoured at 3σ, was calculated at a 1.80-Å resolution from a model in which the ligand was omitted. On the right side is an electrostatic surface representation of the CHIKV macro domain in complex with ADP-ribose. The electrostatic potential is shown between −8 and 8 kT/e and has been generated as in Fig. 1C. The ADP-ribose molecule is shown as on the left side of panel A. (B) The ADP-ribose binding site presented with the same orientation as in panel A. The CHIKV macro domain in complex with ADP-ribose is shown in the upper part, the VEEV macro domain in complex with a Bicine molecule originating from the buffer is presented in the middle part, and the VEEV macro domain in complex with ADP-ribose is shown in the lower part. ADP-ribose and Bicine are shown in cyan. Residues interacting with ADP-ribose in the CHIKV macro domain are shown in blue, and mutated residues are indicated with an asterisk. Corresponding residues in VEEV are colored in blue. Hydrogen bonds between the ligand and the protein are shown in black dotted lines. The loops containing residues 30 to 37 are shown in yellow in CHIKV and VEEV macro domains: their conformations diverged particularly between residues 32 and 35.
The ADP-ribose binding efficacy was then assessed by ITC. The Kds for CHIKV (Fig. 2C) and VEEV (Fig. 2D) macro domains are 5 ± 0.4 μM and 3.9 ± 0.65 μM, respectively. Compared to the existing data, the binding affinity of both CHIKV and VEEV macro domains for ADP-ribose is almost the same as that of the yeast protein Poa1p (32) and 5 and 10 times stronger than that of SARS-CoV and HEV (10). In contrast, the macro domain of SFV, the only other virus analyzed in this way, does not appear to bind ADP-ribose, although protein instability might have impaired precise measurement (32).
The structural basis of ADP-ribose specificity was revealed by soaking and cocrystallization with ADP-ribose for CHIKV and VEEV macro domains, respectively. ADP-ribose binds to a crevice, previously defined as the ADP-ribose 1″-phosphate phosphatase active site (17, 28), in a similar manner to the two alphaviruses' macro domains. The crevice is located at the top of the β strands 2, 4, and 5 and surrounded by the loops connecting β2-α1 and β5-α3 (Fig. 1A and 3A). ADP-ribose lies in a slightly bent conformation comparable to that seen in other known macro domain-ADP-ribose complexes. The specificity for the adenosine base detected by the thermal shift assay (Fig. 2) is at least in part provided by the D10 residue. The adenine moiety is selectively hydrogen bonded via its N6 nitrogen to the D10 side chain (Fig. 3B). Interestingly, this residue is conserved in most macro domain sequences but not Sindbis virus (Fig. 1E). However, in the latter virus, the N10 residue could engage the carbonyl group of its side chain amide into a similar hydrogen bond with a N6 adenine. The D residue at this position was shown to be responsible for the adenosine specificity in A. fulgidus (17). In the CHIKV macro domain, the N6 nitrogen is also hydrogen bonded to R144. In addition, G32 contributes to the binding of adenine. The 3′-OH of the ADP-ribose proximal ribose (the adenosine ribose) provides a hydrogen bond to T111 (Fig. 3B). Several water molecules also interact with the proximal ribose. The phosphate binding site involves the main chain NH groups of the residues G112, V113, and Y114 for the CHIKV macro domain and G112, I113, and F114 for the VEEV macro domain (Fig. 3B). These residues define a positively charged pocket which is likely to bind bulky negatively charged groups. The distal ribose of ADP-ribose is coordinated by the Y/F114 side chain and also makes hydrogen bonds with residues N24 and D/G31 in the main chain in CHIKV and VEEV, respectively. These residues interacting with ADP-ribose are moderately conserved in alphavirus macro domains (Fig. 1E). A noticeable amino acid polymorphism in the ADP-ribose binding pocket is the isoleucine and phenylalanine in some alphaviruses, such as VEEV, in lieu of V113 and Y114 as in CHIKV (Fig. 1E). Even if these changes do not modify the conformation of the ADP-ribose bound in the CHIKV and VEEV macro domain (Fig. 3B), they are likely to promote differences in ADP-ribose binding affinity between them (see Discussion). This binary complex structure allowed the selection of mutants in order to probe ligand-protein interactions. The CHIKV macro domain mutant D10A expectedly showed a reduced thermal shift compared to the same assay with the native protein (Fig. 2B).
No noticeable conformational change occurs in the CHIKV macro domain between the apo and ADP-ribose bound forms. On the contrary, in VEEV macro domain, the loop containing residues 30 to 37 changes its conformation when it binds to ADP-ribose, and adopts a conformation similar to the corresponding loop the CHIKV macro domain (Fig. 1D and 3B). Additionally, residue R114 adopts a different rotamer. These rearrangements are necessary in order to allow coordination of the adenine and the proximal ribose of the ADP-ribose. Furthermore, the residue N24 adopts two different rotamers, depending on the asymmetric unit of the molecule. These modifications are probably related to the differences in thermal denaturation shifts yet lead to similar measured equilibrium constants around 4 to 5 μM. Moreover, these observed rearrangements could explain the absence of ADP-ribose binding in VEEV crystals soaked with ADP-ribose despite the affinity of this domain for ADP-ribose as seen using ITC. Furthermore, in the VEEV macro domain crystal, one molecule of the asymmetric unit contains a Bicine molecule (Fig. 3B), likely originating from the gel filtration buffer. This Bicine molecule could have prevented ADP-ribose binding in the soaking experiment but corroborates the proposition that this pocket is able to accommodate negatively charged groups.
ADP-ribose 1″-phosphate phosphatase activity.
ADP-ribose 1″-phosphate phosphatase was the first activity detected for the yeast Poa1p macro domain based on a genome-wide search of this specific activity (28). In viruses, this activity has already been detected for SARS-CoV, SFV, and HEV macro domains (10) but was associated with a poor turnover constant. In the case of alphaviruses, the SFV ADP-ribose 1″-phosphate phosphatase activity is at the limit of detection (10). This observation, together with our ADP-ribose complexes presented above, prompted us to test this activity for the alphavirus macro domains studied here. CHIKV and VEEV macro domains showed an activity comparable to that of yeast Poa1p macro domain (Fig. 4). To get insight into the characterization of the residues involved in catalytic activities, we tested whether the residues involved in ADP-ribose binding in the crystal structure were indeed able to alter the ADP-ribose 1″-phosphate phosphatase activity. The CHIKV macro domain N24A and Y114A mutants were inactive, while the D10A mutant showed only a decreased activity under the conditions used (Fig. 4). As pinpointed by our structural model, N24 is vicinal to the electrophilic center, i.e., the 1″ phophosphorus of the ADP-ribose 1″ phosphate. The decrease but not annihilation, of activity promoted by D10A is indeed consistent with a role in binding, and not catalysis, as D10 is hydrogen bonded to the ADP-ribose adenine (Fig. 3B), far away from the 1″ phophosphorus of the ADP-ribose 1″ phosphate. In contrast, N24 could well be involved in the phosphatase reaction, as in the case of the SARS-CoV macro domain (10). N24 is correctly positioned to activate a water molecule, which in turn could promote a nucleophilic attack on the phosphorus of the ADP-ribose 1″ phosphate. The distance of 6 Å between N24 and the 1″ position of the ribose could be consistent with such a reaction, leaving enough space for both a water molecule and the phosphate.
PAR binding.
Conserved positive patches on the electrostatic surface of the protein (Fig. 1C) suggest possible binding of longer negatively charged chains such as PAR or RNA. Macro domain binding to PAR has already been shown for A. fulgidus and SARS-CoV macro domains (10, 17). The difficulty of obtaining homogeneous PAR in sufficient quantities inhibited our attempts to obtain a crystal structure of an alphavirus macro domain in complex with PAR. However, we tested PAR binding for several alphavirus macro domains in comparison with SARS-CoV PAR binding using a slot-blot assay. All alphavirus macro domains tested (CHIKV, VEEV, Sindbis virus, and SFV) showed stronger PAR binding than SARS-CoV macro domain (Fig. 5) (10). The alanine single-amino-acid mutants described above were assayed for PAR binding. The mutants did not induce a significant decrease in PAR binding either (not shown). This indicates that any such single mutation is not sufficient to destabilize the interaction with a PAR chain significantly.
FIG. 5.
Slot blot PAR binding assay of macro domains from CHIKV, VEEV, Sindbis virus, SARS-CoV, and SFV. BSA was used as a negative control. PAR was synthezised using PARP-1 and [32P]NAD+, diluted, and used as a probe to label immobilized proteins (2,000 to 0.98 pmol) blotted onto a nitrocellulose membrane as described in Materials and Methods.
RNA binding.
Since alphavirus are single-stranded RNA viruses and macro domains are able to bind long negatively charged chains, it had been hypothesized that a viral macro domain could also bind RNA (21, 34). Neuvonen and Ahola (32) have recently shown that several viral macro domains are able to bind PAR as well as RNA. The interaction between RNA and the CHIKV macro domain was studied using a slot blot assay in conjunction with the HCV RNA-dependent RNA polymerase serving as a positive control. Single-stranded RNA binding was indeed detected using a wide variety of RNA oligonucleotides of unrelated sequences. It was detected not only for the wild-type CHIKV macro domain but also for the D10A, N24A, and Y114A mutants (Fig. 6A). No clear difference in binding affinity appeared when compared to wild type. This means that as in the case of PAR, either the RNA binding site does not correspond to the ADP-ribose binding site or a single-amino-acid change has no drastic impact on RNA binding, as previously hypothesized in the case of PAR binding.
FIG. 6.
RNA binding assays. (A) Slot blot RNA binding assay with wild-type and mutant CHIKV macro domains using the 5′-32P-labeled RNA AAAAAAAAAGCUACC as a probe to label immobilized proteins. The amount of proteins blotted onto the membrane ranges from 2,000 to 0.98 pmol, as indicated. (B) Surface representation of the CHIKV macro domain in complex with the RNA AAA. Proposed nt 1, 2, and 3 (I, II, and III, respectively) are circled. The electrostatic potential is shown between −6 and 6 kT/e and has been generated as in Fig. 1C. (C) RNA binding site. RNA is shown in green and interacting residues in blue, with oxygens in red. H-bonds are indicated with dotted lines.
Structural basis of RNA binding.
The structural basis of RNA binding was then assessed by soaking crystals of the CHIKV macro domain with several small RNAs (see Materials and Methods) and one DNA (AAAGCCAAAAA). Only small adenine-containing RNA or DNA gave a significant extra density upon analysis of the soaked crystals. In these cases, a density was observed within the hydrophobic crevice that binds ADP-ribose. The AMP was virtually superimposable to the adenosine moiety observed in the binary complexes with ADP-ribose. However, the visible density differs between the molecules of the asymmetric unit (data not shown). In contrast, in the case of the AAA RNA, two molecules of the asymmetric unit show a clear density corresponding to the soaked ligand. This density corresponds to a bent RNA that is partially bound in the crevice corresponding to the ADP-ribose binding site (Fig. 6B and C).
Interestingly, an adenine base of the RNA is bound in a similar fashion to that of the ADP-ribose adenine. The RNA is partly disordered at its extremities. Due to steric constraints, a 3′ phosphate cannot be positioned near the loop linking β5 to α3 (containing residues 111 to 113; Fig. 6B). Thus, the first nucleotide (nt 1 [at the 5′ end]) of the RNA is located near this latter loop (Fig. 6B). Only the 3′ phosphate of nt 1 is visible, together with the entire nt 2 and nt 3.
The binding of nt 1 is promoted by coordination of its phosphate with the main chain nitrogen group of residues G32, G112, and V113 (Fig. 6C). The 2′-O position of the nt 2 ribose is coordinated by W148 and C143. As in the case of ADP-ribose, specificity for adenosine of nt 2 is mediated by the same hydrogen bond between D10 and the adenine N6. The adenine of nt 2 is also coordinated by residues G32 and R144. The phosphate of nt 3 makes hydrogen bonds with nitrogen groups of residues R144 and D145, whereas the ribose of nt 3 interacts with D145 and W148. The adenine of nt 3 does not make any interaction with the protein. Therefore, we conclude that the CHIKV macro domain exhibits an RNA binding pocket encompassing an AMP binding pocket identical to that used for ADP-ribose binding.
DISCUSSION
We have determined two novel alphavirus macro domain crystal structures and shown that these domains can bind ADP-ribose, RNA, and PAR. The crystal structures of the alphavirus macro domains show a high degree of conservation with other available macro domain structures (2, 4, 10, 19, 20, 27, 43). Surprisingly, the macro domain that shows the highest sequence and structural similarity to alphavirus macro domain is that of E. coli and not coronavirus. A direct reflection of this observation is that alphaviruses exhibit a much higher affinity for both ADP-ribose and PAR than coronaviruses do. This might suggest that the macro domain module has been acquired twice during evolution for different purposes within the viral world or that the difference of macro domain function between alphavirus and coronavirus has promoted divergence. This point attracts attention to the fact that one should not automatically assume that these macro domains perform the same role, still as elusive, in coronaviruses and alphaviruses. Along these lines, Neuvonen and Ahola (32) have reported a comparative study of viral and cellular macro domains and concluded that the function of a given macro domain might not directly illuminate the function of any other related macro domain.
ADP-ribose binding and ADP-ribose 1″-phosphate phosphatase activity, shown in most of the macro domains studied, are also detected in alphaviruses (10, 17, 28, 43). The level of activity is comparable to that of other tested viral macro domains (Fig. 4) (10, 32), except for that of SFV, which appears to be lower, assuming that protein stability is not interfering with activity measurements (32).
Using mutagenesis experiments, we have confirmed the binding determinants of the ADP-ribose molecule. The D10 residue conserved in all but one macro domain plays a central role in adenine specificity, but not in ADP-ribose 1″-phosphate phosphatase activity that is dependent on N24 and Y114 residues. We show here that the CHIKV macro domain also binds RNA. The crystal structure of the CHIKV macro domain in complex with the AAA trimer RNA allows the identification of residues involved in substrate binding. This D10 residue is also involved in the specific recognition of adenines, in the same manner as in the case of ADP-ribose. The RNA is only partially bound in the ADP-ribose binding site. However, even if the D10A substitution is able to suppress ADP-ribose binding, the CHIKV D10A macro domain is still able to bind longer adenine-containing polymers, such as RNA or PAR. This indicates that either binding of PAR/RNA involves more than a single AMP unit or RNA/PAR binds to another site rather than the ADP-ribose binding cleft.
A notable structural characteristic of macro domains is the presence of positively charged residues close to the ADP-ribose binding site, these basic patches being particularly pronounced in the alphavirus macro domain. Since macro domains from different species bind PAR and RNA (10, 17, 32), these characteristics suggest that residues of these patches could be involved in the binding of such negatively charged substrates longer than ADP-ribose. In agreement with this localized enhanced basicity present in alphavirus macro domains, our results reveal a higher binding affinity to these substrates in alphaviruses than in coronaviruses. Also, since random sequence RNAs bind quite well to the macro domains of different alphaviruses, the basic surface (Fig. 1C) might represent another unspecific, negatively charged polymer binding site distinct from the AMP binding crevice.
Although many RNA oligonucleotides of unrelated sequences bind to the macro domain, it is remarkable that a single AMP remains well defined at all electronic densities, remaining the sole ligand moiety common to all macro domains so far. Together with the possible RNA or PAR binding site outside the ADP-ribose binding cleft on the basic patches, it is tempting to speculate that the viral macro domains work with another yet-to-be defined AMP-containing substrate. Recent results have also proposed this possibility when CoV macro domains were compared and found to bind ADP-ribose very differently: although structurally very close, a group 3 CoV macro domain does not bind ADP-ribose, whereas a group 1 CoV macro domain binds it with a Kd of 29 μM (36).
There are a number of investigational avenues regarding the role of the viral macro domains. First, alphavirus infection can induce synthesis of a large quantity of PAR upon PARP-1 activation (31). This leads to depletion of ATP and NAD+ present in the cell and induces activation of the AIF, which in turn promotes apoptosis. The role of the viral macro domain could thus be related to the binding of PAR and to the modification of the cellular response to viral infection. It remains to be investigated if and how PAR, which is synthesized by the PARP-1 in the nucleus, could interact directly with alphavirus macro domain, the latter having been detected only in the cytoplasm.
Second, the binding of RNA certainly requires further biochemical studies using the replication complex of these viruses. Indeed, the macro domain, being part of a large replicase, could serve either as a non-sequence-specific RNA recruitment factor or adenine-containing RNA recruitment factor in order to provide the RNA template to the neighboring nonstructural proteins.
Third, there are other areas of investigation that have been suggested to depend on the function of the macro domain, such as inflammation (13). It is also possible that functions described in the second and third points above might be combined since the coronavirus macro domain is located in the same nsp3 as the papain-like proteinase domain, which has been shown to possess deubiquitinylating and interferon antagonistic activity (26).
Our work provides a structural basis with which to begin to address these questions in alphaviruses, for which replicons and infectious recombinant clones exist and which might prove less demanding than equivalent studies using much larger CoV genomes.
Acknowledgments
We thank Violaine Lantez, Karen Dalle, and Nadège Brisbarre for technical support, as well as the European and SOLEIL Synchrotron Radiation Facility staffs for help with crystallographic data collection. A.E.G. thanks Dmitriy Samborsky and Alexander Kravchenko for administrating the Viralis software.
This work was supported in part by the VIZIER integrated project (LSHG-CT-2004-511960) of the European Union 6th Framework, the Direction Générale de l'Armement, and the Agence Nationale de la Recherche (CHIKVIRULENCE grant).
Footnotes
Published ahead of print on 22 April 2009.
REFERENCES
- 1.Ahola, T., and L. Kaariainen. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 92507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, M. D., A. M. Buckle, S. C. Cordell, J. Lowe, and M. Bycroft. 2003. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 330503-511. [DOI] [PubMed] [Google Scholar]
- 3.Berrow, N. S., K. Bussow, B. Coutard, J. Diprose, M. Ekberg, G. E. Folkers, N. Levy, V. Lieu, R. J. Owens, Y. Peleg, C. Pinaglia, S. Quevillon-Cheruel, L. Salim, C. Scheich, R. Vincentelli, and D. Busso. 2006. Recombinant protein expression and solubility screening in Escherichia coli: a comparative study. Acta Crystallogr. D Biol. Crystallogr. 621218-1226. [DOI] [PubMed] [Google Scholar]
- 4.Chakravarthy, S., S. K. Y. Gundimella, C. Caron, P.-Y. Perche, J. R. Pehrson, S. Khochbin, and K. Luger. 2005. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 257616-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiarugi, A., and M. A. Moskowitz. 2002. Cell biology. PARP-1—a perpetrator of apoptotic cell death? Science 297200-201. [DOI] [PubMed] [Google Scholar]
- 6.Collaborative Computational Project, No. 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol. Crystallogr. 50760-763. [DOI] [PubMed] [Google Scholar]
- 7.de Lamballerie, X., E. Leroy, R. N. Charrel, K. Ttsetsarkin, S. Higgs, and E. A. Gould. 2008. Chikungunya virus adapts to tiger mosquito via evolutionary convergence: a sign of things to come? Virol. J. 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolinsky, T. J., P. Czodrowski, H. Li, J. E. Nielsen, J. H. Jensen, G. Klebe, and N. A. Baker. 2007. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35W522-W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doublie, S. 1997. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276523-530. [PubMed] [Google Scholar]
- 10.Egloff, M.-P., H. Malet, Á. Putics, M. Heinonen, H. Dutartre, A. Frangeul, A. Gruez, V. Campanacci, C. Cambillau, J. Ziebuhr, T. Ahola, and B. Canard. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 808493-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 602126-2132. [DOI] [PubMed] [Google Scholar]
- 12.Ericsson, U. B., B. M. Hallberg, G. T. Detitta, N. Dekker, and P. Nordlund. 2006. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal. Biochem. 357289-298. [DOI] [PubMed] [Google Scholar]
- 13.Eriksson, K. K., L. Cervantes-Barragán, B. Ludewig, and V. Thiel. 2008. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 8212325-12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geerlof, A., J. Brown, B. Coutard, M. P. Egloff, F. J. Enguita, M. J. Fogg, R. J. Gilbert, M. R. Groves, A. Haouz, J. E. Nettleship, P. Nordlund, R. J. Owens, M. Ruff, S. Sainsbury, D. I. Svergun, and M. Wilmanns. 2006. The impact of protein characterization in structural proteomics. Acta Crystallogr. D Biol. Crystallogr. 621125-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26795-800. [Google Scholar]
- 17.Karras, G. I., G. Kustatscher, H. R. Buhecha, M. D. Allen, C. Pugieux, F. Sait, M. Bycroft, and A. G. Ladurner. 2005. The macro domain is an ADP-ribose binding module. EMBO J. 241911-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy, A. C., J. Fleming, and L. Solomon. 1980. Chikungunya viral arthropathy: a clinical description. J. Rheumatol. 7231-236. [PubMed] [Google Scholar]
- 19.Kumaran, D., S. Eswaramoorthy, F. W. Studier, and S. Swaminathan. 2005. Structure and mechanism of ADP-ribose-1″-monophosphatase (Appr-1″-pase), a ubiquitous cellular processing enzyme. Protein Sci. 14719-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kustatscher, G., M. Hothorn, C. Pugieux, K. Scheffzek, and A. G. Ladurner. 2005. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 12624-625. [DOI] [PubMed] [Google Scholar]
- 21.Ladurner, A. G. 2003. Inactivating chromosomes: a macro domain that minimizes transcription. Mol. Cell 121-3. [DOI] [PubMed] [Google Scholar]
- 22.Lastarza, M. W., A. Grakoui, and C. M. Rice. 1994. Deletion and duplication mutations in the C-terminal nonconserved region of Sindbis virus nsP3: effects on phosphorylation and on virus replication in vertebrate and invertebrate cells. Virology 202224-232. [DOI] [PubMed] [Google Scholar]
- 23.Leslie, A. G. W. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EACMB Newsl. Protein Crystallogr. 26.
- 24.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, G. P., M. W. La Starza, W. R. Hardy, J. H. Strauss, and C. M. Rice. 1990. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 179416-427. [DOI] [PubMed] [Google Scholar]
- 26.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Ménard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 7915199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malet, H., K. Dalle, N. Bremond, F. Tocque, S. Blangy, V. Campanacci, B. Coutard, S. Grisel, J. Lichiere, V. Lantez, C. Cambillau, B. Canard, and M. P. Egloff. 2006. Expression, purification and crystallization of the SARS-CoV macro domain. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62405-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martzen, M. R., S. M. McCraith, S. L. Spinelli, F. M. Torres, S. Fields, E. J. Grayhack, and E. M. Phizicky. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 2861153-1155. [DOI] [PubMed] [Google Scholar]
- 29.McCoy, A. J. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 6332-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247536-540. [DOI] [PubMed] [Google Scholar]
- 31.Nargi-Aizenman, J. L., C. M. Simbulan-Rosenthal, T. A. Kelly, M. E. Smulson, and D. E. Griffin. 2002. Rapid activation of poly(ADP-ribose) polymerase contributes to Sindbis virus and staurosporine-induced apoptotic cell death. Virology 293164-171. [DOI] [PubMed] [Google Scholar]
- 32.Neuvonen, M., and T. Ahola. 2009. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J. Mol. Biol. 385212-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pantoliano, M. W., E. C. Petrella, J. D. Kwasnoski, V. S. Lobanov, J. Myslik, E. Graf, T. Carver, E. Asel, B. A. Springer, P. Lane, and F. R. Salemme. 2001. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 6429-440. [DOI] [PubMed] [Google Scholar]
- 34.Pehrson, J. R., and R. N. Fuji. 1998. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 262837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peränen, J., M. Rikkonen, P. Liljeström, and L. Kääriäinen. 1990. Nuclear localization of Semliki Forest virus-specific nonstructural protein nsP2. J. Virol. 641888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piotrowski, Y., G. Hansen, A. L. Boomars-van der Zanden, E. J. Snijder, A. E. Gorbalenya, and R. Hilgenfeld. 2009. Crystal structures of the X-domains of a group-1 and a group-3 coronavirus reveal that ADP-ribose binding may not be a conserved property. Protein Sci. 186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers, A. M., A. C. Brault, Y. Shirako, E. G. Strauss, W. Kang, J. H. Strauss, and S. C. Weaver. 2001. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 7510118-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Putics, Á., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 7912721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rikkonen, M., J. Peränen, and L. Kääriäinen. 1994. ATPase and GTPase activities associated with Semliki Forest virus nonstructural protein nsP2. J. Virol. 685804-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivas, F., L. A. Diaz, V. M. Cardenas, E. Daza, L. Bruzon, A. Alcala, O. De la Hoz, F. M. Caceres, G. Aristizabal, J. W. Martinez, D. Revelo, F. De la Hoz, J. Boshell, T. Camacho, L. Calderon, V. A. Olano, L. I. Villarreal, D. Roselli, G. Alvarez, G. Ludwig, and T. Tsai. 1997. Epidemic Venezuelan equine encephalitis in La Guajira, Colombia, 1995. J. Infect. Dis. 175828-832. [DOI] [PubMed] [Google Scholar]
- 41.Roversi, P., E. Blanc, C. Vonrhein, G. Evans, and G. Bricogne. 2000. Modelling prior distributions of atoms for macromolecular refinement and completion. Acta Crystallogr. D Biol. Crystallogr. 561316-1323. [DOI] [PubMed] [Google Scholar]
- 42.Rubach, J. K., B. R. Wasik, J. C. Rupp, R. J. Kuhn, R. W. Hardy, and J. L. Smith. 2009. Characterization of purified Sindbis virus nsP4 RNA-dependent RNA polymerase activity in vitro. Virology 384201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saikatendu, K. S., J. S. Joseph, V. Subramanian, T. Clayton, M. Griffith, K. Moy, J. Velasquez, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure 131665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider, T. R., and G. M. Sheldrick. 2002. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 581772-1779. [DOI] [PubMed] [Google Scholar]
- 45.Sheldrick, G. M. 2002. Macromolecular phasing with SHELXE. Z. Kristallogr. 217644-650. [Google Scholar]
- 46.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strauss, E. G., R. J. De Groot, R. Levinson, and J. H. Strauss. 1992. Identification of the active site residues in the nsP2 proteinase of Sindbis virus. Virology 191932-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuittila, M. T., M. G. Santagati, M. Röyttä, J. A. Määttä, and A. E. Hinkkanen. 2000. Replicase complex genes of Semliki Forest virus confer lethal neurovirulence. J. Virol. 744579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vihinen, H., T. Ahola, M. Tuittila, A. Merits, and L. Kaariainen. 2001. Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J. Biol. Chem. 2765745-5752. [DOI] [PubMed] [Google Scholar]
- 50.Vihinen, H., and J. Saarinen. 2000. Phosphorylation site analysis of Semliki forest virus nonstructural protein 3. J. Biol. Chem. 27527775-27783. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Y.-F., S. G. Sawicki, and D. L. Sawicki. 1994. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J. Virol. 686466-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weaver, S. C., T. K. Frey, H. V. Huang, R. M. Kinney, C. M. Rice, J. T. Roehrig, R. E. Shope, and E. G. Strauss. 2005. Togaviridae, p. 999-1008. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. VIIIth Report of the ICTV. Elsevier/Academic Press, London, United Kingdom.
- 53.Wengler, G. 1993. The NS 3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology 197265-273. [DOI] [PubMed] [Google Scholar]