Advances in sequencing technology have made abundant RNA sequence information available, but the challenge of how to interpret these data remains. The RNA sequence contains many layers of information. RNA sequences code for proteins and small RNAs, such as microRNAs or transacting small interfering RNAs (siRNAs). RNA encodes information about both structure and function. Viral RNA structures, such as riboswitches, internal ribosome entry sites (75), and panhandles (71), regulate the stages of the viral life cycle, including replication (100), transcription (99), splicing (40, 48), aminoacylation (31, 55), translation (12, 75, 98), and encapsidation (27, 60, 71, 84). Because viral RNAs are structurally dynamic, current prediction methods focusing on a single minimum free-energy structure may not always identify functionally relevant structures without additional experimental restraints. Because RNA structure determination is often experimentally difficult despite tremendous advances in RNA crystallography, nuclear magnetic resonance spectroscopy, and chemical modification, RNA structure prediction is an important tool for generating hypotheses about structure-function relationships in RNA. RNA structure prediction can be useful for interpreting or designing mutagenesis experiments, identifying conserved structural features, and designing siRNA strategies. This review will briefly outline the basic ideas and assumptions underlying RNA structure prediction, compare different approaches to RNA structure prediction from a user's perspective, and discuss some applications of RNA structure prediction to viral RNA interference (RNAi) research.
The RNA sequence, or primary structure, determines the secondary structure, or pattern of canonical Watson-Crick pairs forming duplexes and irregular regions, such as loops and single-stranded regions. The secondary structure then determines the tertiary structure or overall three-dimensional shape of the molecule. The tertiary structure contains the interaction sites for proteins, other RNA molecules, carbohydrates, or other small molecules and thus determines the quaternary or higher-order structure. The greater thermodynamic stability of RNA helices than that of tertiary interactions makes the RNA folding process hierarchical (35). For example, the favorable free energy involved in forming two stacked base pairs (0.4 to 0.9 kcal/mol per nucleotide) (105) is larger than the favorable free energy to form tertiary interactions, such as ribose zippers or tetraloop-receptor interactions (0.1 to 0.5 kcal/mol per nucleotide) (25, 97). Note that 1.4 kcal/mol is equivalent to 1 order of magnitude in a binding constant at 37°C. The hierarchical nature of RNA folding makes RNA structure prediction a tractable challenge (92).
WHAT IS RNA STRUCTURE PREDICTION?
Phylogenetic analysis of RNA sequences remains the gold standard of RNA structure prediction. Phylogenetic analysis involves generating an alignment of many RNA sequences and looking for patterns of covariation between two nucleotides. Two nucleotides would covary if, for example, nucleotide 1 was usually a G but sometimes a U, if nucleotide 2 was a C whenever nucleotide 1 was a G, and if nucleotide 2 was an A whenever nucleotide 1 was a U. This pattern of variation maintains the formation of an isosteric Watson-Crick base pair. Phylogenetic analysis requires a good alignment of many RNA sequences over a diverse range of species with enough variation to observe covariation patterns but also enough conservation to establish a good alignment. When regions of 100% conserved nucleotides are shown as large loops in secondary structure diagrams generated by phylogenetic analysis, no information is known about the secondary structure. This does not mean that the nucleotides must necessarily be single stranded. Because RNA-protein interactions and the RNA tertiary structure are also often conserved in evolution, phylogenetic analysis indirectly accounts for such tertiary and quaternary structures. However, sometimes only one or a few sequences are available or the sequences show too little or too much variation for a good alignment. In these cases, computational approaches can generate possible RNA secondary structures.
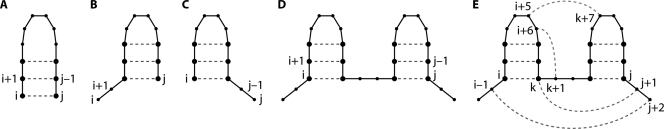
Computational approaches typically use a recursive algorithm to evaluate possible base pairs for a given RNA sequence. There are 1.8N possible structures for any given sequence where N is the number of nucleotides; thus, there are 3.37 × 1025 possible structures for a 100-mer RNA. This is far more structures than even the fastest computer today could ever calculate in anyone's lifetime. All computational approaches make some approximations or simplifications in order to reduce the complexity of the RNA folding problem. In a stochastic context-free grammar (SCFG), the most common approximation in RNA structure prediction programs, nucleotide pairings are evaluated as one of four possibilities: paired with each other, two cases where one nucleotide is paired but the other nucleotide is unpaired, or both nucleotides paired but not to each other (Fig. 1A to D) (29, 72). A recursive algorithm systematically evaluates each possible pair of nucleotides. This approach makes computation of RNA secondary structures practical and reasonable but excludes the possibility of nonnested pairing interactions, such as pseudoknots, kissing hairpins, and base triples (Fig. 1E). All prediction program listed in Tables 1 and 2 are capable of predicting the two hairpins (Fig. 1D) which are compatible with the formation of nonnested pairing interactions and tertiary structures. In addition, several programs specifically address the computational challenges of nonnested pairing and the limitations of SCFG grammars.
FIG. 1.
Four possible relationships between nucleotides i and j. Dots represent nucleotides i < k < j. Lines represent sequential covalent connections. Dashed lines indicate hydrogen-bonded base pairing. (A) i and j are paired. (B) i is unpaired and j is paired. (C) i is paired and j is unpaired. (D) Both i and j are paired but not to each other (29). (E) A nested base pair, such as a base pair between nucleotides i and k in panel E, forms a helical stem. Nonnested interactions, such as a base pair between j+1 and k (a base triple), a base pair between k+1 and i+6 (a pseudoknot), or a base pair between i+5 and k+7 (a kissing hairpin interaction), are not allowed in an SCFG and require a higher-level grammar, more memory, and more run time. A base pair between nucleotides j+2 and i−1 would create a multibranch loop. Consideration of possible multibranch loops requires an additional array. Vienna Websuite and RNAstructure calculate the stabilities of multibranch loops differently. Three-dimensional motifs, such as ribose zippers and tetraloop-receptor interactions, are built from many pairing interactions.
TABLE 1.
Characteristics of programsa
| Program | Access
|
Algorithm
|
Experimental data
|
siRNA design | Pseudoknots | 3D prediction | Unique feature(s) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Free | Code available | Online submission | Windows | Linux | Zuker | Wuchty | McCaskill | Ding and Lawrence | Nussinov | Turner rules | Phylogeny | Restrain as single stranded | Restrain pair | |||||
| mfold | X | X | X | X | X | X | X | X | X | X | First popular folding program | |||||||
| RNAstructure | X | X | X | X | X | X | X | X | X | X | X | X | Advanced chemical modification and SHAPE | |||||
| Vienna | X | X | X | X | X | X | X | X | X | X | X | X | X | RNAalifold, RNAz | ||||
| Sfold | X | X | X | X | X | Advanced siRNA design | ||||||||||||
| RNA STAR | X | X | X | Genetic, Monte Carlo, and greedy algorithms | ||||||||||||||
| PKNOTS | X | X | X | X | X | Pseudoknot grammar | ||||||||||||
| pknotsRG | X | X | X | X | X | X | Focus on three types of pseudoknots | |||||||||||
| NUPACK | X | X | X | X | X | X | Pseudoknot pair probabilities | |||||||||||
| ILM | X | X | X | X | X | X | X | X | Iterative loop matching algorithm | |||||||||
| HotKnots | X | X | X | X | X | X | X | Algorithm to find hot spots | ||||||||||
| ConStruct | X | X | X | X | X | X | X | Alignment editor, user interface | ||||||||||
| Pfold | X | X | X | SCFG training | ||||||||||||||
| CONTRAfold | X | X | X | X | X | X | Conditional training | |||||||||||
| Rosetta | X | X | X | X | X | X | Fragment assembly | |||||||||||
| MC-Sym | X | X | X | X | X | X | X | X | X | X | Nucleotide cyclic motifs | |||||||
X, feature is available; Free, free for academic use; 3D, three dimensional.
TABLE 2.
Websites and key references for programs
| Program | Website | Key references |
|---|---|---|
| mfold | http://mfold.bioinfo.rpi.edu/ | 64, 107-109 |
| RNAstructure | http://rna.urmc.rochester.edu/rnastructure.html | 61, 66, 68 |
| Vienna | http://rna.tbi.univie.ac.at/ | 6, 38, 101, 104 |
| Sfold | http://sfold.wadsworth.org/ | 18, 19, 59 |
| RNA STAR | http://wwwbio.leidenuniv.nl/∼batenburg/STRAbout.html | 1, 39, 41, 91, 96 |
| PKNOTS | http://selab.janelia.org/software.html | 80, 81 |
| pknotsRG | http://bibiserv.techfak.uni-bielefeld.de/download/tools/pknotsrg.html | 77 |
| NUPACK | http://nupack.org/ | 22 |
| ILM | http://cic.cs.wustl.edu/RNA/ | 82, 83 |
| HotKnots | http://www.cs.ubc.ca/labs/beta/Software/HotKnots/ | 47, 78 |
| ConStruct | http://www.biophys.uni-duesseldorf.de/construct3/ | 62, 103 |
| Pfold | http://www.daimi.au.dk/∼compbio/pfold/ | 53, 54 |
| CONTRAfold | http://contra.stanford.edu/contrafold/ | 23, 24 |
| Rosetta | http://www.rosettacommons.org/tiki/tiki-index.php | 14 |
| MC-Sym | http://www.major.iric.ca/MC-Pipeline/ | 63, 74 |
The next step is to rank possible pairs and structures and store this information in what is often called a “fill step.” Free energy is a common ranking system. The free energies of RNA base pair stacks can be measured experimentally. The free energy of a stack of two base pairs accounts for both the hydrogen bonds in Watson-Crick pairs and the interactions between stacked bases, which include electrostatic interactions, dipole-dipole interactions, van der Wals forces, and hydrophobic effects. This smallest unit of an RNA double helix is referred to as a nearest neighbor (7, 32, 93, 105). Expanded nearest neighbors describe irregular or single-stranded regions; for example, the nucleotides in a hairpin loop and the nucleotides in the closing base pair and all the effects of changes in the phosphodiester backbone as the RNA strand makes this hairpin turn would be grouped together as the smallest unit of which to measure the loop free energy. The nearest-neighbor approximation assumes that the free energy of each smallest unit or motif, for example, a stack of base pairs or a hairpin loop, is the same in any secondary structure context. For example, a GNRA tetraloop hairpin would have the same energetic stability in a group I intron, 23S rRNA, or the human immunodeficiency virus (HIV) genome. This approximation is most often reasonable, although non-nearest-neighbor effects have been observed for some noncanonical pairs, especially GU pairs, and single nucleotide bulges (43, 52, 85, 86). This approximation allows the free energy of a secondary structure to be calculated by adding together the energetic stabilities of each smallest unit or motif.
The Turner laboratory members and associates have made extensive free-energy measurements of RNA motifs (32, 49, 66, 67), and these thermodynamic parameters form the core of many RNA structure prediction programs. In order to experimentally measure the free energies of RNA motifs, the UV absorbance of small synthetic oligonucleotide duplexes (typically 8 to 12 nucleotides) is recorded as a function of temperature in an optical melting experiment. One assumption of these experiments is that the RNA exists in only two states, duplex or single stranded; thus, intermediates occur very quickly and do not contribute significantly to the UV absorbance. This assumption often does not hold true for complex motifs, such as large loops, multibranch loops, or pseudoknots, and complicates the thermodynamic analysis of these motifs (17, 33, 65). Differential scanning calorimetry (8) or single-molecule force-extension experiments (57) can measure RNA free energies without using a two-state assumption. The free-energy measurements of hairpin, internal, and bulge loops enable predictions for unmeasured loop sequences, but not every possible sequence of every motif has been measured yet. Loop sequences with surprisingly stable energies are still being discovered. Thus, improvements in the collection of RNA free-energy parameters continue as an active area of research.
Free-energy minimization approaches to RNA structure prediction assume that the lowest free-energy structure will form. Often, this assumption works well; for example, the base pairing in tRNA and group II introns are correctly predicted 83% and 88% of the time, respectively, by using free-energy minimization and the Zuker algorithm (67). However, the lowest free-energy structure may not always be the functional structure. The conformation of the RNA may be determined by folding kinetics rather than thermodynamic free energy (11, 106). Also, no RNA structure prediction program yet incorporates RNA-protein interactions, which can dramatically alter the folding landscape. Many prediction programs also do not consider pseudoknots or other stabilizing RNA tertiary interactions. In addition, a very large number of structures may exist within a small range defined by the error of the free-energy calculation, and sometimes those structures can be very different from one another. This becomes more problematic as the lengths of the RNAs increase. For example, when tRNA (76 nucleotides) and satellite tobacco mosaic virus RNA (1,058 nucleotides) are folded with the Wuchty algorithm in the Vienna suite, which calculates all possible structures within a small energy range, tRNA can form 13 different structures within a 1-kcal/mol range, and the structure most different from the minimum energy structure has 23 nucleotides paired differently. In contrast, satellite tobacco mosaic virus can form 42,768 structures within 1 kcal/mol; the structure most different from the minimum free-energy structure has 560 nucleotides paired differently but is only 0.2 kcal/mol greater in free energy, a free-energy difference well within error of the calculation (see the supplemental material). Additional experimental data that further defines possible RNA conformations can address this problem in RNA structure prediction and becomes even more important for longer RNA sequences.
Free-energy minimization is only one approach to analyzing the array of ranked possible pairs and structures for a given RNA sequence. Many different prediction programs use different algorithms to analyze or “trace back” through this array to generate not only a minimum free-energy structure but also a set of low-energy structures. In addition, experimental data such as chemical modification or phylogenetic covariation can be used in addition to free-energy minimization. Alternatively, possible pairs can be ranked using criteria other than free energy, such as the maximum number of base pairs, or the base pairing can be described using concepts other than SCFGs, such as nucleotide cyclic motifs.
Different prediction programs have developed different strategies for addressing the limitations of RNA structure prediction by free-energy minimization alone. Because even slight differences in calculation methods or the set of thermodynamic parameters can alter the minimum free-energy prediction, comparison of the results from more than one program is a good approach to generating an informed hypothesis about RNA structure and function. Table 1 lists some of these programs and their characteristics, such as access, options for algorithms, and options for including experimental data. Refer to each prediction program's website or primary citation for a complete description of user options and an evaluation of performance. The evaluation criteria and databases of known secondary structures used to evaluate prediction accuracy vary substantially between different research groups and make direct comparisons complex. Results of comparative tests of different RNA structure prediction programs have been reviewed elsewhere (26, 42, 78). Prediction programs aim to predict structures based on a set of rules and assumptions rather than optimize for particular types of structures, although several programs are designed to address the computational difficulties associated with predicting pseudoknotted, nonnested pairing (Fig. 1E). This list is not exhaustive, and the following discussion only highlights some practical considerations for selecting an RNA structure prediction program. The “best” program is the user's decision that depends on the RNA studied, the questions asked, the available experimental data and resources, and the intended applications of the structure prediction.
APPROACHES TO RNA STRUCTURE PREDICTION
mfold and UNAFold.
The popular mfold program has been expanded, updated, and renamed UNAFold (64). The Quikfold option in DINAmelt, the online version of the program, contains the traditional format where a user submits a sequence and chooses RNA or DNA folding, temperature, salt conditions, and a free-energy range for suboptimal structures. UNAFold and mfold use the Zuker algorithms (108, 109) to compute the minimum free-energy structure for a given sequence and systematically sample structures within a percentage of free-energy range to create a set of diverse suboptimal structures. Users can specify nucleotides to be single stranded or in a particular base pair if information from chemical modification, enzymatic probing, compensatory mutations, or phylogenetic covariation is available. The options for hybridization analysis can be applied to the design of siRNA or antisense RNA.
RNAstructure.
RNAstructure 4.6 is a Windows implementation of the Zuker algorithm and includes additional options for other folding algorithms and incorporation of experimental data. The authors of RNAstructure collaborate very closely with the Turner laboratory and keep the most up-to-date thermodynamic parameters (66). The OligoWalk program can be used for siRNA design (61). Two unique ways of incorporating experimental data in the RNA folding is done with Dynalign (42, 68) and chemical modification (15, 66). The Dynalign program computes the lowest free-energy sequence alignment and secondary structure common to two RNA sequences. The two sequences need not be aligned, but the maximum number of sequences to be folded at one time is two. RNAstructure also uses a more-advanced definition of chemical modification restraints for traditional chemical modification reagents and new selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) data (70). Chemical modification reagents, such as dimethyl sulfate, methylate accessible adenine nucleotides and cause stops in reverse transcription primer extension reactions. This type of chemical modification occurs not only at single-stranded adenines but also at adenines in AU pairs that are at the end of a helix, next to a GU pair, or next to bulged nucleotides; and RNAstructure allows for all these possibilities when a nucleotide is marked as chemically modified (66). SHAPE detects flexibility in the RNA backbone that correlates with helical or irregular motifs in secondary structure formation, and these data can be included as a restraint or as a pseudofree energy in ranking possible base pairs (15).
Vienna RNA Websuite.
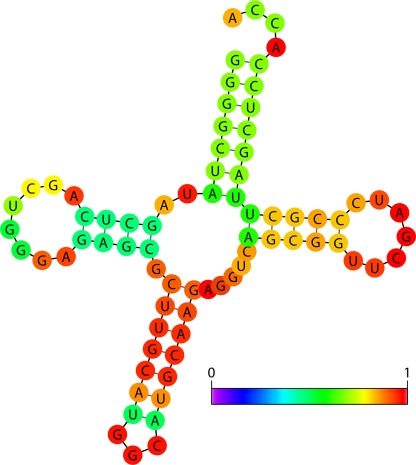
The Vienna RNA Websuite introduced the Wuchty algorithm, developed applications of the McCaskill algorithm, and also offers a wide variety of algorithms and functions. The Wuchty algorithm computes all possible secondary structures within a narrow free-energy range (104). The Wuchty algorithm generates a small but complete set of suboptimal structures that may include some very different secondary structures but also very many highly similar structures. However, structures containing more than one suboptimal region may occur in the Wuchty set of structures but would be absent if the Zuker method for sampling suboptimal structures were used. The McCaskill algorithm evaluates the statistical probabilities of base pairs and secondary structures by using a partition function calculation (69). “Statistical” refers to the thermodynamic and statistical mechanics definition based on a Boltzmann distribution. This information can be overlaid in different colors on the lowest-energy structure and thus provides an estimate of confidence in the predicted structure (Fig. 2). For example, one region of a secondary structure (red) may be exceptionally thermodynamically stable, occur frequently in all suboptimal structures, have a high statistical probability, and thus be more likely to be a correct prediction. In contrast, another region of the secondary structure (purple or dark blue at the left end of the scale near zero probability) may have multiple thermodynamically equivalent possible conformations in the suboptimal structures and a low statistical probability and thus be less clearly a correct prediction.
FIG. 2.
The minimum free-energy secondary structure of tRNAala from Shigella sonnei with the base pair probabilities shown in color. The red pairs have a high probability of forming, green pairs have a medium probability of forming, and blue pairs (none shown) have a low probability of forming. The normalized scale showing zero (dark blue) to one (red) probability is shown on the lower right. Figure produced using Vienna Websuite with default parameters.
The Vienna Websuite includes RNAalifold and RNAz, programs that combine phylogenetic covariation information with thermodynamic stability and base pair statistical probabilities when ranking possible base pairs. RNAalifold accepts any number of aligned sequences in the standard ClustalW and FASTA formats and generates a lowest-energy consensus structure by weighting the ranking of base pairs with covariation data (6). RNAz searches for noncoding RNAs (101) or conserved secondary structure elements within long RNA sequences, such as viral genomes (45, 46), by weighting base pair statistical probabilities and thermodynamic stabilities with covariation data in a combined Z score for possible base pairs. The RNAup and RNAxs functions are used to design siRNA strategies.
Sfold.
The Sfold algorithm uses a unique algorithm to aid in the design of siRNA. The algorithm combines thermodynamic stabilities (67), calculations of target accessibility (59, 88), and empirical rules for efficient siRNA developed by the Zamore, Amgen, and Dharmacon groups (3, 51, 79, 87, 94). The website offers specialized programs for the design of siRNA, antisense RNA, trans-cleaving RNA, and mRNA-microRNA interactions as well as a general program for statistically sampling suboptimal RNA structures. The algorithm uses a partition function calculation and then groups suboptimal structures by similarity (9, 19, 20). The centroid structure is the most-representative structure that is closest in similarity to all the other structures. If the centroid structure is different from the minimum free-energy structure, the centroid structure is often closer to the phylogenetic prediction and contains fewer base pairs, or fewer false-positive base pair predictions, than the minimum free-energy prediction. The point is to show a structure that represents a group of structures rather than a single predicted structure. Many long RNA sequences, such as viral genomes or mRNA, may not have a single structure but instead have a dynamic structure that has some conserved features but also varies and changes, and these many conformations may all exist simultaneously in the cell. Thus, the centroid structure may better describe the overall average prediction for all these conformations and may better show whether a target site is accessible. Sfold also outputs a probability profile of nucleotide accessibility, loop profiles, and an estimate of siRNA potency.
PROGRAMS FOR PREDICTING SECONDARY STRUCTURES WITH PSEUDOKNOTS
RNA STAR.
The problem of predicting pseudoknots (Fig. 1) has inspired a wide variety of solutions and adaptations of existing algorithms. RNA STAR offers three algorithms that simulate the folding of RNA and include pseudoknot structures. The greedy algorithm, based on the idea that maximizing each step of the calculation will produce the optimal result, adds the most stable stem helix onto a growing structure (1). A Monte Carlo algorithm generates a set of random structures and then selects a probability-weighted stem helix to add onto a growing structure (39). The genetic algorithm simulates an evolution of RNA folding by starting with unfolded structures and increasing the fitness criterion, low free energy, in each generation (41, 96). The genetic algorithm allows stem helices to be added or removed and allows “crossovers,” structures containing the best pieces of previous generations. Because these programs are simulations rather than calculations, the output should also be evaluated on how well the program converges to a single structure and the reproducibility of the final structure. The user chooses the number of rounds in the simulation.
PKNOTS and pknotsRG.
PKNOTS uses free-energy minimization and a higher-level grammar that removes the “context-free” restrictions shown in Fig. 1 (80, 81). The computational cost of allowing pseudoknots [O(N6) in time and O(N4) in space], however, makes the program computationally unreasonable for sequences greater than approximately 100 nucleotides. The program pknotsRG restricts the types of possible pseudoknots to three common simple types, with at most two helices, and does not allow complex interactions, such as kissing hairpins or triple helices. This approximation improves the run time [O(N4) in time and O(N2) in space] so that sequences of up to 800 nucleotides are computationally reasonable (77). The O(Nx) function describes the computational complexity and program run time, depending on N, the number of nucleotides in the sequence. The actual time necessary for the computation will also depend on the type of computer used and the processor speed.
NUPACK.
The NUPACK program provides probabilities of pseudoknotted base pair formation (22). These base pair probabilities can predict free energies of pseudoknot stabilities that match experimental values (21). Base pair probabilities are useful for analyses of how pseudoknots may be in equilibrium between two conformations and how mutations can shift that equilibrium. Optical melting experiments and single-molecule pulling experiments can provide measurements of pseudoknot interaction energies (for examples, see references 10, 34, and 76).
ILM.
The ILM program uses an iterative loop matching algorithm to maximize base pairs and allows pseudoknots to form by allowing base pairs to be added or removed in successive rounds (83). The Nussinov algorithm, or maximum loop matching algorithm, is the basic framework for generating a structure with the most possible base pairs (73). The base pairs are ranked using both thermodynamic parameters and covariation data for aligned sequences. ILM requires the RnaViz program (16) to visualize the RNA secondary structure with pseudoknots.
HotKnots.
The HotKnots program takes advantage of the hierarchical nature of RNA secondary and tertiary structure folding (47, 78). A hot spot is a particularly stable branch point in a tree of possible structures. The program generates a tree of structures using the Zuker algorithm and restrains pseudoknot “hot spots” to be single stranded and then allows structures with pseudoknots to form. The HotKnots program is a heuristic that does not guarantee the lowest energy structure but quickly predicts many reasonably good structures.
ConStruct.
The program ConStruct simultaneously aligns multiple RNA sequences and finds a consensus RNA structure, thus combining information on thermodynamic stability and phylogeny (103). Because sequence alignment often requires manual editing, ConStruct offers an interactive user interface that allows manual editing of the alignment. Thus, the alignment can be improved with preliminary information about secondary structure. A new additional step then maximizes base pairing and thus allows for pseudoknot, base triple, and other tertiary interactions.
NONTHERMODYNAMIC APPROACHES
Pfold and CONTRAfold.
Not all folding programs use lowest free energy as the criteria for forming base pairs in an RNA secondary structure. Pfold uses a sequence alignment and a set of base pair probabilities determined from a set of known tRNA and 23S rRNA secondary structures to predict RNA secondary structure (53). CONTRAfold (24) uses a conditional training method and a database of 151 consensus structures from known secondary structures in the Rfam database (36, 37) to generate parameters for RNA folding. The parameters for base stacking interactions are surprisingly proportional to the experimentally measured values. RNA alignment and folding (RAF), an option in CONTRAfold, accepts multiple unaligned sequences and simultaneously aligns the sequences and computes a consensus secondary structure (23).
PREDICTING THREE-DIMENSIONAL STRUCTURE
Rosetta.
FARNA, fragment assembly of RNA using Rosetta software, samples different possible conformations of RNA to predict three-dimensional structure (14). Computational analysis of the crystal structure of the 50S ribosomal subunit of Haloarcula marismortui (4) produced a library of RNA fragments, which is a collection of torsion angles for all possible conformations of three nucleotide pieces, and an energy function, which includes terms for hydrogen bonding, base stacking, compactness, steric clashes, and planar base pairs. The only additional input is the sequence of the RNA to be predicted. The Monte Carlo sampling method limits the length of the RNA to approximately 40 nucleotides. This method accurately predicts the noncanonical regions for 13 out of 20 test cases, including conformations not in the ribosome, such as the base triple in the SL2 stem-loop of the HIV psi packaging signal (2) and the pseudoknot in beet western yellow mosaic virus (30).
MC-Sym.
MC-Sym, macromolecular conformation by symbolic generation, uses a hierarchical approach to three-dimensional structure prediction (74). MC-Fold predicts a secondary structure; MC-Sym predicts a three dimensional structure from a given secondary structure. The program is based on nucleotide cyclic motifs, the smallest nondivisible unit in a graph grammar description of RNA structure which includes backbone torsion angles, base pairing, and base stacking interactions (56, 89). The program also incorporates experimental data from chemical modification, SHAPE, hydroxyl radical footprinting, phylogenetic alignments, and distance restraints from nuclear magnetic resonance spectroscopy. The program is capable of predicting structures of 150 nucleotides and correctly predicts 11 of 13 test sequences, which contain a maximum of 47 nucleotides and include the pseudoknot in yellow leaf virus (13).
APPLICATIONS OF RNA STRUCTURE PREDICTION TO RNAi
One of the most exciting applications of RNA structure prediction is toward understanding and manipulating the ability of small RNAs to regulate gene expression. RNA structure prediction of the secondary structure of the mRNA target, the pre-microRNA, or short hairpin RNA can improve microRNA identification and siRNA design strategies (44, 50, 59, 88, 90). The accessibility of the mRNA target site for base pairing and the energetic cost of rearranging the RNA secondary structure can be important for estimating the effectiveness of the small RNA for regulating gene expression. RNA structure prediction can improve identification of viral sequences that are Dicer substrates to be processed as viral siRNA and also identification of viral siRNA target sites (28, 44, 50, 88, 90, 95). Sequence complementarity alone does not provide sufficient specificity for effective siRNA strategies. Higher-order RNA structure can provide the specificity in molecular recognition to reduce off-target effects (44, 50, 59). RNA structure prediction can also facilitate the interpretation of mutations. For example, in studies of RNAi targeting HIV type 1 (HIV-1) in human cells (102), some resistance mutations in the HIV-1 sequence emerged that were not at the siRNA target site. When the secondary structure prediction of the target site region was recalculated with the resistance mutation sequence, an alternate fold was revealed. Mutations that cause changes in secondary structure in HIV-1 RNA and MS2 bacteriophage RNA occur in forced-evolution experiments in which a deleterious mutation is introduced and the progeny are sequenced to identify compensating mutations (5, 58). Accurate structure prediction methods can thus facilitate future RNAi technologies and viral RNA research.
Supplementary Material
Acknowledgments
The author is supported by grants from the Oklahoma Center for the Advancement of Science and Technology and the Pharmaceutical Research and Manufacturers of America Foundation and by an institutional research grant from the American Cancer Society to the University of Oklahoma Health Sciences Center.
The author thanks Douglas H. Turner and Kay Sheez for critical reading of the manuscript.
Biography
Susan J. Schroeder began exploring the RNA world as an undergraduate in Douglas H. Turner's laboratory at the University of Rochester in New York. She completed her doctoral thesis in the Turner laboratory and studied thermodynamic stabilities of RNA asymmetric internal loops, RNA structure prediction, and nuclear magnetic resonance spectroscopy of a loop in a group I intron active site. As an NIH Ruth L. Kirchstein postdoctoral fellow in Peter B. Moore's laboratory at Yale University, she studied ribosome crystallography, antibiotic resistance, and rRNA mutagenesis. Now an assistant professor in the Chemistry and Biochemistry Department at the University of Oklahoma, Dr. Schroeder focuses on viral RNA structure, function, and energetics. Dr. Schroeder and members of her laboratory study satellite tobacco mosaic virus RNA and prohead RNA from bacteriophage packaging motors in order to improve RNA structure prediction from sequence and discover the fundamental knowledge necessary to solve the RNA folding problem.
Footnotes
Published ahead of print on 15 April 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abrahams, J., M. van den Berg, E. van Batenburg, and C. Pleij. 1990. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 183035-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amarasinghe, G., R. De Guzman, R. Turner, and M. Summers. 2000. NMR structure of the stem-loop SL2 of the HIV-1 psi RNA packaging signal reveals a novel A-U-A base-triple platform. J. Mol. Biol. 299145-156. [DOI] [PubMed] [Google Scholar]
- 3.Amarzguioui, M., and H. Prydz. 2004. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 3161050-1058. [DOI] [PubMed] [Google Scholar]
- 4.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289905-920. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout, B., and A. T. Das. 2009. Virus evolution as a tool to study HIV-1 biology. Methods Mol. Biol. 485436-451. [DOI] [PubMed] [Google Scholar]
- 6.Bernhart, S., I. Hofacker, S. Will, A. Gruber, and P. Stadler. 2008. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics 9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borer, P. N., B. Dengler, I. Tinoco, Jr., and O. C. Uhlenbeck. 1974. Stability of ribonucleic acid-double-stranded helices. J. Mol. Biol. 86843-853. [DOI] [PubMed] [Google Scholar]
- 8.Breslauer, K. J. 1995. Extracting thermodynamic data from equilibrium melting curves for oligonucleotide order-disorder transitions. Methods Enzymol. 259221-241. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho, L. E., and C. E. Lawrence. 2008. Centroid estimation in discrete high-dimensional spaces with applications in biology. Proc. Natl. Acad. Sci. USA 1053209-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, G., J. Wen, and I. Tinoco, Jr. 2007. Single molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA. RNA 132175-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, S., and K. Dill. 2000. RNA folding energy landscapes. Proc. Natl. Acad. Sci. USA 97646-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costantino, D. A., J. S. Pfingsten, R. P. Rambo, and J. S. Kieft. 2008. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 1557-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornish, P., S. Stammler, and D. Giedroc. 2006. The global structures of a wild-type and poorly functional plant luteoviral mRNA pseudoknot are essentially identical. RNA 121959-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, R., and D. Baker. 2007. Automated de novo prediction of native-like RNA tertiary structures. Proc. Natl. Acad. Sci. USA 10414664-14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deigan, K., T. Li, D. Mathews, and K. Weeks. 2009. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA 10697-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Rijk, P., J. Wuyts, and R. De Wachter. 2003. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics 19299-300. [DOI] [PubMed] [Google Scholar]
- 17.Diamond, J., D. Turner, and D. Mathews. 2001. Thermodynamics of three-way multibranch loops in RNA. Biochemistry 406971-6981. [DOI] [PubMed] [Google Scholar]
- 18.Ding, Y., C. Y. Chan, and C. E. Lawrence. 2004. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 32W135-W141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding, Y., and C. E. Lawrence. 2004. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 317280-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding, Y., and C. E. Lawrence. 2001. Statistical prediction of single-stranded regions in RNA secondary structure and application to predicting effective antisense target sites and beyond. Nucleic Acids Res. 291034-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dirks, R. M., and N. A. Pierce. 2004. An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J. Comput. Chem. 251295-1304. [DOI] [PubMed] [Google Scholar]
- 22.Dirks, R. M., and N. A. Pierce. 2003. A partition function algorithm for nucleic acid secondary structure including pseudoknots. J. Comput. Chem. 241664-1677. [DOI] [PubMed] [Google Scholar]
- 23.Do, C. B., C.-S. Foo, and S. Batzglou. 2008. A max-margin model for efficient simultaneous alignment and folding of RNA sequences. Bioinformatics 24i68-i76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Do, C. B., D. A. Woods, and S. Batzglou. 2006. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics 22e90-e98. [DOI] [PubMed] [Google Scholar]
- 25.Doherty, E. A., R. T. Batey, B. Masquida, and J. A. Doudna. 2001. A universal mode of helix packing in RNA. Nat. Struct. Biol. 8339-343. [DOI] [PubMed] [Google Scholar]
- 26.Dowell, R., and S. Eddy. 2004. Evaluation of several lightweight stochastic context-free grammars for RNA secondary structure prediction. BMC Bioinformatics 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Souza, V., and M. F. Summers. 2004. Structural basis for packaging the dimeric genome of Moloney murine leukemia virus. Nature 431586-590. [DOI] [PubMed] [Google Scholar]
- 28.Du, Q.-S., C.-G. Duan, Z.-H. Zhang, Y.-Y. Fang, R.-X. Fang, Q. Xie, and H.-S. Guo. 2007. DCL4 targets Cucumber mosaic virus satellite RNA at novel secondary structures. J. Virol. 819142-9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eddy, S. R. 2004. How do RNA folding algorithms work? Nat. Biotechnol. 221457-1458. [DOI] [PubMed] [Google Scholar]
- 30.Egli, M., G. Minasov, L. Su, and A. Rich. 2002. Metal ions and flexibility in a viral RNA pseudoknot at atomic resolution. Proc. Natl. Acad. Sci. USA 994302-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Felden, B., C. Florentz, A. McPherson, and R. Giege. 1994. A histidine accepting tRNA-like fold at the 3′ end of satellite tobacco mosaic virus RNA. Nucleic Acids Res. 222882-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freier, S. M., R. Kierzek, J. A. Jaeger, N. Sugimoto, M. H. Caruthers, T. Neilson, and D. H. Turner. 1986. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. USA 839373-9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giedroc, D., C. Theimer, and P. Nixon. 2000. Structure, stability, and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J. Mol. Biol. 298167-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green, L., C. Kim, C. Bustamante, and I. Tinoco, Jr. 2008. Characterization of the mechanical unfolding of RNA pseudoknots. J. Mol. Biol. 375511-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenleaf, W., K. Frieda, D. Foster, M. Woodside, and S. Block. 2008. Direct observation of hierarchical folding in single riboswitch aptamers. Science 319630-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths-Jones, S., A. Bateman, M. Marshall, A. Khanna, and S. R. Eddy. 2003. Rfam: an RNA family database. Nucleic Acids Res. 31439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffiths-Jones, S., S. Moxon, M. Marshall, A. Khanna, S. R. Eddy, and A. Bateman. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33D121-D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruber, A. R., R. Lorenz, S. H. Bernhart, R. Neubock, and I. L. Hofacker. 2008. The Vienna RNA Websuite. Nucleic Acids Res. 36W70-W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gultyaev, A. P. 1991. The computer simulation of RNA folding involving pseudoknot formation. Nucleic Acids Res. 192489-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gultyaev, A. P., H. A. Heus, and R. C. Olsthoorn. 2007. An RNA conformational shift in recent H5N1 influenza A viruses. Bioinformatics 23272-276. [DOI] [PubMed] [Google Scholar]
- 41.Gultyaev, A. P., F. H. van Batenburg, and C. W. Pleij. 1995. The computer simulation of RNA folding pathways using a genetic algorithm. J. Mol. Biol. 25037-51. [DOI] [PubMed] [Google Scholar]
- 42.Harmanci, A., G. Sharma, and D. Mathews. 2007. Efficient pairwise RNA structure prediction using probabilistic alignment constraints in Dynalign. BMC Bioinformatics 8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He, L., R. Kierzek, J. SantaLucia, Jr., A. E. Walter, and D. H. Turner. 1991. Nearest-neighbor parameters for GU mismatches. Biochemistry 3011124-11132. [DOI] [PubMed] [Google Scholar]
- 44.Hofacker, I. 2007. How microRNAs choose their targets. Nat. Genet. 391191-1192. [DOI] [PubMed] [Google Scholar]
- 45.Hofacker, I., M. Fekete, C. Flamm, M. Huynen, S. Rauscher, P. Stolorz, and P. Stadler. 1998. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 263825-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hofacker, I., and P. Stadler. 1999. Automatic detection of conserved base pairing patterns in RNA virus genomes. Comput. Chem. 23401-414. [DOI] [PubMed] [Google Scholar]
- 47.Jabbari, H., A. Condon, and S. Zhao. 2008. Novel and efficient RNA secondary structure prediction using hierarchical folding. J. Comput. Biol. 15139-163. [DOI] [PubMed] [Google Scholar]
- 48.Jablonski, J. A., E. Buratti, C. Stuani, and M. Caputi. 2008. The secondary structure of the human immunodeficiency virus type 1 transcript modulates viral splicing and infectivity. J. Virol. 828038-8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaeger, J., D. Turner, and M. Zuker. 1989. Improved predictions of secondary structures for RNA. Proc. Natl. Acad. Sci. USA 867706-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kertesz, M., N. Iovino, U. Unnerstall, U. Gaul, and E. Segal. 2007. The role of site accessibility in microRNA target recognition. Nature Genetics 391278-1284. [DOI] [PubMed] [Google Scholar]
- 51.Khvorova, A., A. Reynolds, and S. D. Jayasena. 2003. Functional siRNas and miRNas exhibit strand bias. Cell 115209-216. [DOI] [PubMed] [Google Scholar]
- 52.Kierzek, R., M. E. Burkard, and D. H. Turner. 1999. Thermodynamics of single mismatches in RNA duplexes. Biochemistry 3814214-14223. [DOI] [PubMed] [Google Scholar]
- 53.Knudsen, B., and J. Hein. 2003. Pfold: RNA secondary structure grammar prediction using stochastic context-free grammars. Nucleic Acids Res. 313423-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knudsen, B., and J. Hein. 1999. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics 15446-454. [DOI] [PubMed] [Google Scholar]
- 55.Koenig, R., S. Barends, A. Gultyaev, D. Lesemann, H. Vetten, S. Loss, and C. Pleij. 2005. Nemesia ring necrosis virus: a new tyomovirus with genomic RNA having a histylatable tobamovirus-like 3′ end. J. Gen. Virol. 861827-1833. [DOI] [PubMed] [Google Scholar]
- 56.Lemieux, S., and F. Major. 2006. Automated extraction and classification of RNA tertiary structure cyclic motifs. Nucleic Acids Res. 342340-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li, P. T., J. Vieregg, and I. Tinoco, Jr. 2008. How RNA unfolds and refolds. Annu. Rev. Biochem. 7777-100. [DOI] [PubMed] [Google Scholar]
- 58.Licis, N., and J. van Duin. 2006. Structural constraints and mutational bias in the evolutionary restoration of a severe deletion in RNA phage MS2. J. Mol. Evol. 63314-329. [DOI] [PubMed] [Google Scholar]
- 59.Long, D., R. Lee, P. Williams, C. Chan, V. Ambros, and Y. Ding. 2007. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 14287-294. [DOI] [PubMed] [Google Scholar]
- 60.Loo, L., R. Guenther, S. Lommel, and S. Franzen. 2007. Encapsidation of nanoparticles by red clover necrotic mosaic virus. J. Am. Chem. Soc. 12911111-11117. [DOI] [PubMed] [Google Scholar]
- 61.Lu, Z., and D. Mathews. 2008. OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 36W104-W108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luck, R., S. Graf, and G. Steger. 1999. ConStruct: a tool for thermodynamic controlled prediction of conserved secondary structure. Nucleic Acids Res. 274208-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Major, F., M. Turcotte, D. Gautheret, G. Lapalme, E. Fillion, and R. Cedergren. 1991. The combination of symbolic and numerical computation for three-dimensional modeling of RNA. Science 2531255-1260. [DOI] [PubMed] [Google Scholar]
- 64.Markham, N. R., and M. Zuker. 2008. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 4533-31. [DOI] [PubMed] [Google Scholar]
- 65.Mathews, D. H., and D. H. Turner. 2002. Experimentally derived nearest-neighbor parameters for the stability of RNA three-and four-way multibranch loops. Biochemistry 41869-880. [DOI] [PubMed] [Google Scholar]
- 66.Mathews, D. H., M. D. Disney, J. L. Childs, S. J. Schroeder, M. Zuker, and D. H. Turner. 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 1017287-7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288911-940. [DOI] [PubMed] [Google Scholar]
- 68.Mathews, D. H., and D. H. Turner. 2002. Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol. 317191-203. [DOI] [PubMed] [Google Scholar]
- 69.McCaskill, J. 1990. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 291105-1119. [DOI] [PubMed] [Google Scholar]
- 70.Merino, E. J., K. A. Wilkinson, J. L. Coughlan, and K. M. Weeks. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 1274223-4231. [DOI] [PubMed] [Google Scholar]
- 71.Mir, M., B. Brown, B. Hjelle, W. Duran, and A. Panganiban. 2006. Hantavirus N protein exhibits genus-specific recognition of the viral RNA panhandle. J. Virol. 8011283-11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nussinov, R., and A. Jacobson. 1980. Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc. Natl. Acad. Sci. USA 776309-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nussinov, R., G. Pieczenik, J. Griggs, and D. Kleitman. 1978. Algorithms for loop matchings. SIAM J. Appl. Math. 3568-82. [Google Scholar]
- 74.Parisien, M., and F. Major. 2008. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 45251-55. [DOI] [PubMed] [Google Scholar]
- 75.Pfingsten, J., and J. Kieft. 2008. RNA Structure-based ribosome recruitment: lessons from the Dicistroviridae intergenic region IRESes. RNA 141255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Puglisi, J., J. Wyatt, and I. Tinoco, Jr. 1988. A pseudoknotted RNA oligonucleotide. Nature 331283-286. [DOI] [PubMed] [Google Scholar]
- 77.Reeder, J., and R. Giegerich. 2004. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics 5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ren, J., B. Rastegari, A. Condon, and H. Hoos. 2005. HotKnots: heuristic prediction of RNA secondary structures including pseudoknots. RNA 111491-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reynolds, A., D. Leake, Q. Boese, S. Scaringe, W. Marshall, and A. Khvorova. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22326-330. [DOI] [PubMed] [Google Scholar]
- 80.Rivas, E., and S. Eddy. 2000. The language of RNA: a formal grammar that includes pseudoknots. Bioinformatics 16334-340. [DOI] [PubMed] [Google Scholar]
- 81.Rivas, E., and S. R. Eddy. 1999. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol. 2852053-2068. [DOI] [PubMed] [Google Scholar]
- 82.Ruan, J., G. Stormo, and W. Zhang. 2004. ILM: a web server for predicting RNA secondary structures with pseudoknots. Nucleic Acids Res. 32W146-W149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruan, J., G. Stormo, and W. Zhang. 2004. An iterated loop matching approach to the prediction of RNA secondary structures with pseudoknots. Bioinformatics 2058-66. [DOI] [PubMed] [Google Scholar]
- 84.Schneemann, A. 2006. The structural and functional role of RNA in icosahedral virus assembly. Annu. Rev. Microbiol. 6051-67. [DOI] [PubMed] [Google Scholar]
- 85.Schroeder, S. J., and D. H. Turner. 2000. Factors affecting the thermodynamics stability of small asymmetric internal loops in RNA. Biochemistry 399257-9274. [DOI] [PubMed] [Google Scholar]
- 86.Schroeder, S. J., and D. H. Turner. 2001. Thermodynamic stabilities of internal loops with GU closing pairs in RNA. Biochemistry 4011509-11517. [DOI] [PubMed] [Google Scholar]
- 87.Schwarz, D. S., G. Hutvagner, T. Du, Z. Xu, N. Aronin, and P. D. Zamore. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115199-208. [DOI] [PubMed] [Google Scholar]
- 88.Shao, Y., C. Chan, A. Maliyekkel, C. Lawrence, I. Roninson, and Y. Ding. 2007. Effect of target secondary structure on RNAi efficiency. RNA 131631-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.St-Onge, K., P. Thibault, S. Hamel, and F. Major. 2007. Modeling RNA tertiary structure motifs by graph-grammars. Nucleic Acids Res. 351726-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tafer, H., S. L. Ameres, G. Obernosterer, C. A. Gebeshuber, R. Schroeder, J. Martinez, and I. L. Hofacker. 2008. The impact of target accessibility on the design of effective siRNAs. Nat. Biotechnol. 26578-583. [DOI] [PubMed] [Google Scholar]
- 91.Taufer, M., A. Licon, R. Araiza, D. Mireles, F. van Batenburg, A. Gultyaev, and M. Leung. 2009. PseudoBase++: an extension of PseudoBase for easy searching, formatting, and visualization of pseudoknots. Nucleic Acids Res. 37D127-D135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tinoco, I., Jr., and C. Bustamante. 1999. How RNA folds. J. Mol. Biol. 293271-281. [DOI] [PubMed] [Google Scholar]
- 93.Tinoco, I., Jr., O. C. Uhlenbeck, and M. D. Levine. 1971. Estimation of secondary structure in ribonucleic acids. Nature 230362-367. [DOI] [PubMed] [Google Scholar]
- 94.Ui-Tei, K., Y. Naito, F. Takahashi, T. Haraguchi, H. Ohki-Hamazaki, A. Juni, R. Ueda, and K. Saigo. 2004. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 32936-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Uzilov, A. V., J. M. Keegan, and D. H. Mathews. 2006. Detection of non-coding RNAs on the basis of predicted secondary structure formation free energy change. BMC Bioinformatics 7173-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Batenburg, F., A. Gultyaev, and C. Pleij. 1995. An APL-programmed genetic algorithm for the prediction of RNA secondary structure. J. Theor. Biol. 174269-280. [DOI] [PubMed] [Google Scholar]
- 97.Vander Meulen, K., J. Davis, T. Foster, M. Record, Jr., and S. Butcher. 2008. Thermodynamics and folding pathway of tetraloop receptor-mediated RNA helical packing. J. Mol. Biol. 384702-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van Lipzig, R., A. Gultyaev, C. Pleij, M. van Montagu, M. Cornelissen, and F. Meulewaeter. 2002. The 5′ and 3′ extremities of the satellite tobacco necrosis virus translational enhancer domain contribute differentially to stimulation of translation. RNA 8229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang, S., L. Mortazavi, and K. A. White. 2008. Higher-order RNA structural requirements and small-molecule induction of tombusvirus subgenomic mRNA transcription. J. Virol. 823864-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang, S., and K. A. White. 2007. Riboswitching on RNA virus replication. Proc. Natl. Acad. Sci. USA 10410406-10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Washietl, S., I. Hofacker, and P. Stadler. 2005. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. USA 1022454-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Westerhout, E. M., M. Ooms, M. Vink, A. T. Das, and B. Berkhout. 2005. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 33796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wilm, A., K. Linnenbrink, and G. Steger. 2008. ConStruct: improved construction of RNA consensus structures. BMC Bioinformatics 9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wuchty, S., W. Fontana, I. L. Hofacker, and P. Schuster. 1999. Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers 49145-165. [DOI] [PubMed] [Google Scholar]
- 105.Xia, T., J. SantaLucia, Jr., M. E. Burkard, R. Kierzek, S. J. Schroeder, X. Jiao, C. Cox, and D. H. Turner. 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 3714719-14735. [DOI] [PubMed] [Google Scholar]
- 106.Zarrinkar, P., and J. Williamson. 1994. Kinetic intermediates in RNA folding. Science 265918-924. [DOI] [PubMed] [Google Scholar]
- 107.Zuker, M. 2003. Mfold server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 24448-52. [DOI] [PubMed] [Google Scholar]
- 109.Zuker, M., and P. Stiegler. 1981. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 9133-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.