Abstract
The packaging of retroviral genomic RNA (gRNA) requires cis-acting elements within the RNA and trans-acting elements within the Gag polyprotein. The packaging signal ψ, at the 5′ end of the viral gRNA, binds to Gag through interactions with basic residues and Cys-His box RNA-binding motifs in the nucleocapsid. Although specific interactions between Gag and gRNA have been demonstrated previously, where and when they occur is not well understood. We discovered that the Rous sarcoma virus (RSV) Gag protein transiently localizes to the nucleus, although the roles of Gag nuclear trafficking in virus replication have not been fully elucidated. A mutant of RSV (Myr1E) with enhanced plasma membrane targeting of Gag fails to undergo nuclear trafficking and also incorporates reduced levels of gRNA into virus particles compared to those in wild-type particles. Based on these results, we hypothesized that Gag nuclear entry might facilitate gRNA packaging. To test this idea by using a gain-of-function genetic approach, a bipartite nuclear localization signal (NLS) derived from the nucleoplasmin protein was inserted into the Myr1E Gag sequence (generating mutant Myr1E.NLS) in an attempt to restore nuclear trafficking. Here, we report that the inserted NLS enhanced the nuclear localization of Myr1E.NLS Gag compared to that of Myr1E Gag. Also, the NLS sequence restored gRNA packaging to nearly wild-type levels in viruses containing Myr1E.NLS Gag, providing genetic evidence linking nuclear trafficking of the retroviral Gag protein with gRNA incorporation.
The encapsidation of the RNA genome is essential for retrovirus replication. Because the viral genomic RNA (gRNA) constitutes only a small fraction of the total cellular mRNA, a specific Gag-RNA interaction is thought to be required for viral genome packaging (2). The determinants of virus-specific gRNA incorporation include the cis-acting element at the 5′end of the viral gRNA, known as the packaging signal (ψ), and the nucleocapsid (NC) domain of the Gag polyprotein (3, 14, 62). In Rous sarcoma virus (RSV), the NC domain contains basic residues that are required for the recognition of and binding to ψ, as well as two Cys-His motifs that maintain the overall conformation of NC and are essential for RNA packaging (30, 31).
Packaging of gRNA into progeny virions requires that the unspliced viral mRNA be exported from the nucleus. However, cellular proofreading mechanisms ensure that unspliced or intron-containing mRNAs are retained in the nucleus until splicing occurs. Complex retroviruses like human immunodeficiency virus type 1 (HIV-1) overcome this export block of unspliced genomes by encoding the Rev protein, which interacts with a cis-acting sequence in the viral RNA (the Rev-responsive element [RRE]) to facilitate cytoplasmic accumulation of intron-containing viral mRNA (16, 35). The export of the Rev-viral RNA complex is mediated through the interaction of a leucine-rich nuclear export signal (NES) in Rev with the CRM1 nuclear export factor (17, 18, 37, 41). Simple retroviruses do not encode Rev-like regulatory proteins, so other strategies for the export of unspliced viral RNAs are needed. For Mason-Pfizer monkey virus, a cis-acting constitutive transport element induces nuclear export of the unspliced viral RNA in a process mediated by the cellular mRNA nuclear export factor TAP (5, 25, 46, 63). In RSV, an RNA element composed of either of the two direct repeats flanking the src gene mediates the cytoplasmic accumulation of unspliced viral RNA by using host export proteins TAP and Dpb5 (29, 42, 44).
The findings of recent studies suggest that specific RNA export pathways direct viral gRNA to sites of virion assembly (56); for example, HIV-1 gRNA export out of the nucleus by the Rev-RRE-CRM1 complex is required for the proper subcellular localization of Gag and efficient virus particle production (26, 57). In the case of RSV, little is known about the trafficking of the viral RNA destined for virion encapsidation or the effects of the gRNA nuclear export pathway on Gag trafficking and virus particle production. However, we do know that RSV Gag enters the nucleus during infection, owing to nuclear localization signals (NLSs) in the matrix (MA) and NC domains. The nuclear localization of Gag is transient, and export is mediated by a CRM1-dependent NES in the p10 region (6, 52, 53). Thus, it is feasible that Gag may facilitate the nuclear export of the gRNA, either directly or indirectly, to promote particle assembly (53).
In support of this idea, Gag mutants engineered to be more efficiently directed to the plasma membrane than wild-type Gag by the addition of the Src membrane-binding domain (in Myr1E virus) or by the insertion of extra basic residues (in SuperM virus) are not concentrated in nuclei when cells are treated with the CRM1 inhibitor leptomycin B (LMB) (8, 20, 53). Moreover, Myr1E and SuperM virus particles incorporate reduced levels of viral gRNA compared to the levels incorporated by wild-type particles. Thus, there is a correlation between the nuclear transit of Gag and gRNA packaging, although the Myr1E and SuperM viruses may be deficient in gRNA encapsidation because they are transported to the plasma membrane too rapidly (8). To test the hypothesis that the loss of Gag nuclear trafficking is responsible for the gRNA packaging defect, we used a gain-of-function genetic approach whereby a heterologous NLS was inserted into Myr1E Gag, yielding mutant virus Myr1E.NLS. Our results revealed that restoring the nuclear trafficking of Myr1E Gag also restored the incorporation of gRNA into mutant virus particles.
MATERIALS AND METHODS
Plasmids and cells.
All of the RSV proviral constructs contain the RSV Prague C gag gene from pATV8. The plasmids pRC.V8 (10), pRC.Myr1E (45), pGag-GFP(7), pMA-GFP and pMyr1E.MA-GFP (20), pMyr1E.Gag-GFP (53), and pMA.NLS-GFP and pGag.NLS-GFP (21) were described previously. The inserted NLS sequence was based on the bipartite NLS of Xenopus nucleoplasmin (50), although the sequence between the essential basic motifs was mutated to allow screening for positive clones by using digestion with the restriction enzyme BstXI, resulting in the amino acid sequence KRPAATLLAGQAKKK (see Fig. 1). This sequence was inserted between residues 86 and 100 in Gag, a region defined previously to be dispensable for particle assembly and infectivity, by using an SpeI site engineered into the pRC.ΔMA6 plasmid (21, 39). The plasmid pMyr1E.NLS.Gag-GFP, expressing Myr1E.NLS Gag fused to green fluorescent protein (GFP), was created by SstII-SstII restriction fragment exchange between pGag.NLS-GFP and pMyr1E.Gag-GFP (53). SstI-SdaI restriction fragment transfer from pMyr1E.NLS.Gag-GFP into pRC.V8 resulted in the pRC.Myr1E.NLS proviral expression vector. The QuikChange site-directed mutagenesis kit (Stratagene) was utilized to make pMyr1E.KR/AA.Gag-GFP and pMyr1E.KR/AA.KKK/AAA-GFP, and the SstI-SdaI restriction fragment was then ligated into linearized pRC.V8, leading to the construction of the proviral constructs pRC.Myr1E.KR/AA and pRC.Myr1E.KR/AA.KKK/AAA. All plasmids were screened by restriction enzyme digestion and analyzed by dideoxy sequencing to ensure that they contained the appropriate mutations. The quail fibroblast cell line QT6 was maintained as described previously (10).
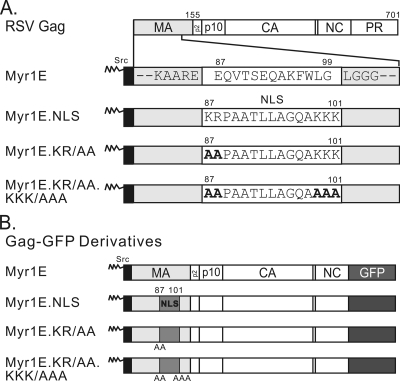
FIG. 1.
Schematic diagram of RSV Gag and mutant proteins. (A) The wild-type Gag protein is depicted at the top, with cleavage products MA (light gray), p2, p10, CA, NC, and PR indicated. Amino acid numbers 155 (the end of MA) and 701 (the end of Gag) are shown above. Gag mutants from the indicated virus constructs are illustrated below, with the focus on the MA domain, which is enlarged for detail. Each mutant contains the 10-amino-acid membrane-binding domain of the v-Src protein (black box), which is myristylated (as indicated by zigzag lines). For Myr1E Gag, the downstream wild-type amino acid sequence between positions 87 and 99 (white box) is shown. Flanking residues (gray boxes) are invariable among all the mutants. The Myr1E.NLS mutant has an insertion based on the bipartite nucleoplasmin NLS sequence located between residues 87 and 101 (white box). Myr1E.KR/AA has replacements of the upstream basic residues (KR) with alanines (bold font), and Myr1E.KR/AA.KKK/AAA has both sets of critical basic residues (KR upstream and KKK downstream) in the NLS changed to alanines. (B) Gag-GFP derivatives have GFP (dark gray box) substituted for the last 7 amino acids of NC and all of PR. Amino acid changes involving the MA domain are identical to those shown above in panel A. The inserted NLS and basic residue mutations are indicated by medium-gray boxes, with the positions of alanine substitutions shown below each box.
Confocal microscopy.
QT6 cells were transiently transfected with 1 μg of plasmid DNA per 0.4 × 106 cells by the calcium phosphate method (19). The medium was changed 4 to 5 h posttransfection, and 11 to 16 h later, cells were washed twice with Tris-buffered saline, fixed with 3.7% paraformaldehyde, stained with 4′,6-diamidino-2-phenylindole (DAPI), and examined using a Leica TCS SP2-AOBS confocal microscope. For drug treatment, cells were incubated for 2 h with 20 nM LMB (Sigma) before imaging. Laser excitations of 488 nm (for enhanced GFP) and 405 nm (for DAPI) were used, and sequential images were obtained at emission ranges of 509 to 525 nm for enhanced GFP and 460 to 490 nm for DAPI. The quantification of nuclear fluorescence was performed as described previously (27). The nuclear fluorescence level was calculated as the total fluorescence intensity of the GFP signal in the nucleus, defined by DAPI staining, divided by the fluorescence intensity of the whole cell, and the pixel intensity in each region was measured using software accompanying the Leica SP2 microscope. Confocal images were exported directly into ImageJ (49) and CorelDrawX3 (Corel Corp., Ottawa, Canada). Any adjustments in intensity were applied equally to all images in a series.
RNase protection assays.
QT6 cells were transfected with wild-type or mutant proviral constructs, and culture medium was collected after 48 h, clarified by low-speed centrifugation, and spun at 126,000 × g through a 25% sucrose cushion to pellet viral particles. The viral pellet was resuspended, aliquots were removed to assay for reverse transcriptase (RT) activity in triplicate, and mean RT values were used to normalize the amount of virus particles in each sample, as described previously (10). Viral RNA was extracted and RNase protection assays were performed as described previously (20). An antisense riboprobe, spanning the 3′ splice acceptor site of the env gene, was transcribed with T7 RNA polymerase in the presence of [32P]CTP and utilized to distinguish between unspliced viral RNA (a 263-nucleotide [nt] protected fragment) and spliced viral RNA (a 183-nt protected fragment) as described previously (21). After separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), radioactive bands were quantitated using a PhosphorImager (Molecular Dynamics).
Radioimmunoprecipitation assays for virus particle release.
Budding was measured using methods described previously (8, 21). Duplicate plates of QT6 cells were transfected by the calcium phosphate method. At 16 h posttransfection, cells were subjected to a 30-min methionine-cysteine starvation period and metabolically labeled with a [35S]methionine-cysteine protein labeling mix (NEN). One set of plates was labeled for 5 min to evaluate intracellular Gag expression, and the other set was labeled for 2.5 h to measure virus particle release (8). Medium was removed, cells and medium were mixed with radioimmunoprecipitation assay buffer, and Gag proteins were immunoprecipitated as described previously (60). Budding efficiency was expressed as the ratio of the CA level present in the medium during the 2.5-h labeling period to the intracellular Gag level detected during the 5-min pulse (8). Quantification was performed using PhosphorImager analysis, and the efficiency of budding for the wild type (RC.V8) was set at 100%.
Infectivity.
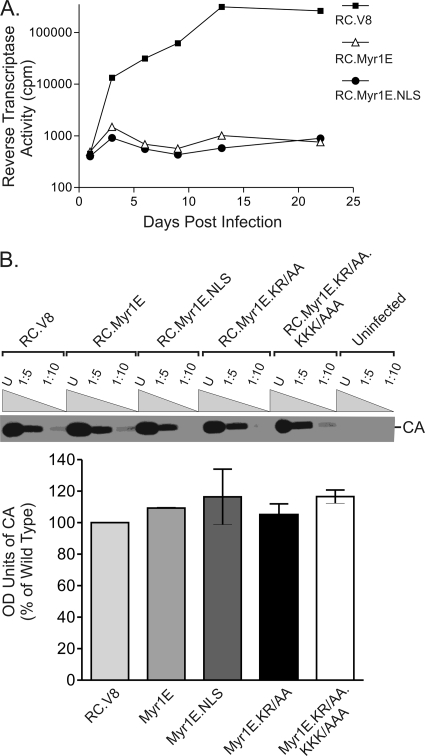
Virus particles were produced following the transfection of QT6 cells with proviral plasmids and pelleted through a sucrose cushion, and samples were normalized according to RT activities as described above. Equivalent amounts of virus particle samples were added to fresh cells, and the cells were passaged every 3 days for 21 days. Before each passage, medium was collected and pelleted through a 25% sucrose cushion at 126,000 × g for 40 min at 4°C and stored at −70°C. At the end of the collection period, RT activity in each sample was measured in triplicate, and mean values are shown (see Fig. 5A) (10).
FIG. 5.
Viral infectivity. (A) Virus particles were collected from the cell culture medium 48 h after transient transfection with wild-type or mutant proviral constructs. Infection was accomplished by adding equivalent numbers of virus particles to fresh QT6 cells for 3 hours. Cells were passaged every 3 days, and samples of culture medium collected at each time point were assayed for RT activity at the end of a 21-day period. (B) The CA-to-RT ratio for each virus was determined by collecting virus particles and normalizing for equivalent particle numbers according to RT activity. The indicated dilutions of normalized virus particle samples (U for undiluted, 1:5 for 5-fold dilutions, and 1:10 for 10-fold dilutions) were loaded onto an SDS-PAGE gel, and Western blot analysis was performed using anti-RSV polyclonal serum. The intensities of CA bands on Western blots were quantitated using ImageJ software, and the results are presented in the graph. The averages of results from three separate experiments along with the standard errors of the means are shown. OD, optical density.
Immunoblot analysis of CA-RT ratio.
QT6 cells were transfected with proviral vectors, virus particles were collected by ultracentrifugation through a sucrose cushion, and samples were normalized by using mean RT activity as described above. Equivalent amounts of particle samples were lysed in sample buffer (125 mM Tris-HCl, pH 6.8, 20% glycerol, 0.5% bromophenol blue, 4% SDS, and 10% β-mercaptoethanol), and viral proteins were separated by SDS-PAGE and transferred onto nitrocellulose. Immunoblotting was performed using anti-RSV rabbit polyclonal antibody (60), followed by secondary antibody anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Sigma), to detect CA by chemiluminescence. Quantification was performed using ImageJ software (49).
RESULTS
To test the hypothesis that nuclear trafficking of RSV Gag facilitates gRNA packaging, a gain-of-function genetic approach was utilized. We took advantage of previously described Gag mutants in which a nuclear transport signal derived from the nucleoplasmin protein was inserted into the MA region of Gag (MA.NLS, consisting of NLS-containing MA alone, and Gag.NLS, comprising the NLS incorporated into the complete RC.V8 Gag polyprotein) (21). The MA.NLS-GFP protein is more highly concentrated in the nucleus than wild-type MA, indicating that the inserted foreign NLS enhances the nuclear trafficking of MA (21). Thus, it was logical to test whether the nucleoplasmin NLS might be strong enough to direct Myr1E Gag to pass through the nucleus transiently before being trafficked to the plasma membrane. We reasoned that if insertion of the additional NLS into Myr1E Gag restored nuclear trafficking, then the gRNA packaging defect of viruses containing Myr1E Gag might be corrected.
Effect of the inserted NLS on nuclear trafficking of Myr1E Gag.
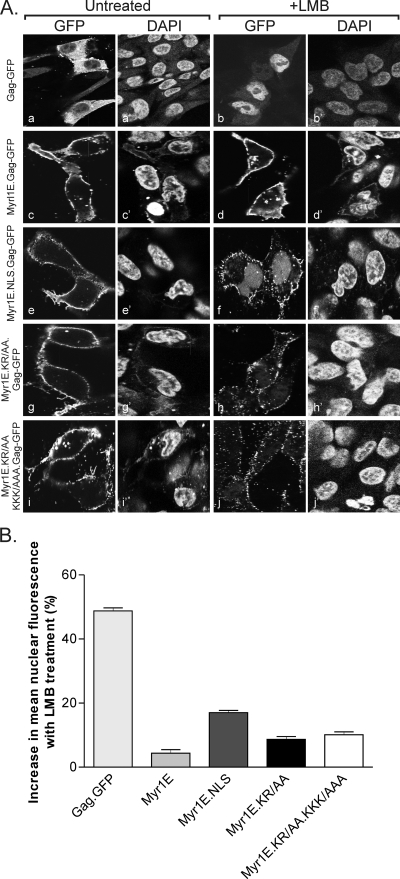
Although MA.NLS-GFP is strongly targeted to the nucleus due to the additional NLS, it remained possible that the Src membrane-targeting signal would be dominant in Myr1E.NLS Gag and override both the endogenous and inserted NLSs in Gag. To determine whether the nucleoplasmin-derived NLS could promote the nuclear trafficking of Myr1E Gag, the subcellular localization patterns of mutant Gag-GFP proteins transiently expressed in avian cells were examined using confocal microscopy (Fig. 1 and 2). Western blot analysis of whole-cell lysates revealed that each of the Gag-GFP fusion proteins was stably expressed, and no free GFP protein was detected (data not shown). Cells were fixed and stained with DAPI to identify nuclei (Fig. 2). As previously reported, wild-type Gag-GFP was located primarily in the cytoplasm and at the plasma membranes of untreated cells (Fig. 2A, panels a and a′) but accumulated within nuclei with the addition of LMB (Fig. 2A, panels b and b′) (53). In contrast, Myr1E Gag-GFP accumulated strongly at the plasma membranes without and with LMB treatment, with no discernible change in the subcellular distribution of the protein (Fig. 2A, panels c and c′ and d and d′, respectively). Although the insertion of the NLS did not alter the localization of Myr1E.NLS Gag-GFP (Fig. 2A, panels e and e′) under steady-state conditions, there was marked accumulation of the protein in the nucleus following incubation with LMB, in addition to the plasma membrane localization (Fig. 2A, panels f and f′).
FIG. 2.
Subcellular distribution of wild-type Gag-GFP and Myr1E Gag-GFP derivatives. (A) Confocal microscopy images of QT6 cells expressing each Gag-GFP fusion protein, as indicated to the left, with matched images of DAPI-stained cells to the right to show the locations of nuclei. Cells treated with 20 nM LMB (+LMB) are pictured to the right. Exposure for the entire figure was optimized using the autoexposure setting in Microsoft Office Picture Manager. (B) Graph showing results from quantitation of the epifluorescence levels of GFP-expressing cells. For each indicated construct, a minimum of 90 cells were scored for nuclear localization as described in Materials and Methods. The mean nuclear fluorescence level for untreated cells was subtracted from that for LMB-treated cells to yield the increase in nuclear fluorescence with LMB treatment. Statistical analysis was per- formed using an unpaired two-tailed t test. The differences in nuclear localization between each mutant and the wild type and between each mutant and the other mutants were significant, with P values of ≤0.0051, except for that between Myr1E.KR/AA and Myr1E.KR/AA.KKK/AAA, which was not statistically significant (P = 0.2791).
As controls for the NLS insertion, alanines were substituted for each of the basic residues in the bipartite NLS that are required for nuclear trafficking activity in the authentic nucleoplasmin sequence (11). In the Myr1E.KR/AA Gag-GFP expression construct, the upstream lysine and arginine codons were mutated to encode alanines (Fig. 1), resulting in a decrease in the degree of nuclear localization in response to LMB treatment (Fig. 2A, compare panels f and h). The replacement of the downstream lysines, generating mutant Myr1E.KR/AAA.KKK/AAA, also reduced the nuclear sequestration of the mutant Gag-GFP compared to that of Myr1E.NLS Gag-GFP, although slight nuclear fluorescence remained detectable (Fig. 2A, panel j).
To measure the degree of nuclear localization for each Gag-GFP fusion protein, the average levels of fluorescence in the nucleus (defined as the nuclear fluorescence divided by the total cell fluorescence) in the absence and presence of LMB treatment was measured. The percent mean nuclear fluorescence in untreated cells was subtracted from the percent mean nuclear fluorescence in treated cells to yield the percent increase in nuclear fluorescence with LMB exposure (Fig. 2B). The percentage of wild-type Gag-GFP localized in the nucleus was 49% higher with LMB treatment, indicating a high degree of sensitivity to the drug. Only 4% more Myr1E Gag-GFP was in the nucleus after exposure to LMB, indicating that very little protein shuttles through the nucleus. The introduction of the heterologous bipartite NLS increased the nuclear fraction of Myr1E.NLS Gag-GFP to 17% in response to LMB. Both NLS basic substitution mutants, Myr1E.KR/AA Gag-GFP and Myr1E.KR/AA.KKK/AAA Gag-GFP, were intermediate in nuclear localization, with 9 and 10% increases in nuclear fluorescence levels, respectively. Therefore, although the basic residue mutants showed diminished nuclear trafficking, there was residual NLS activity as measured by confocal microscopy.
Restoration of gRNA packaging.
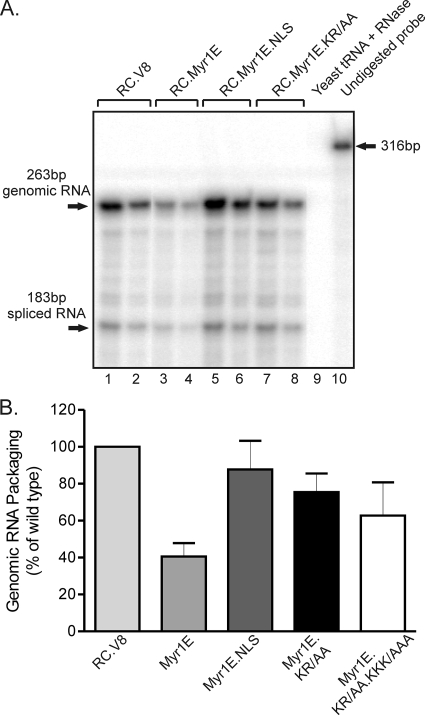
To determine whether increasing the nuclear trafficking of Myr1E Gag would also enhance gRNA packaging, virus-associated RNA levels were measured by performing RNase protection assays of gRNA extracted from equivalent amounts of virus particles normalized using RT activity (10, 45). The packaging efficiency of each mutant was expressed as a ratio to that of the wild-type virus, which was assigned a value of 100% (Fig. 3). As reported previously, Myr1E virus particles contain approximately 40% of the wild-type level of gRNA (P < 0.0001) (Fig. 3) (20, 45). Remarkably, Myr1E.NLS virus particles contained levels of viral RNA nearly equal to wild-type levels (87.7%), indicating the restoration of gRNA packaging in the Myr1E.NLS mutant. The level of viral RNA incorporation for Myr1E.NLS was significantly higher than that for Myr1E (P = 0.0241). The level of gRNA incorporation for basic residue substitution mutant Myr1E.KR/AA was slightly lower than those for Myr1E.NLS and the wild type (75.4% of the wild-type level, indicating a nonsignificant difference), but it was increased compared to Myr1E (P = 0.0114). The level for the double basic knockout mutant Myr1E.KR/AA.KKK/AAA (62.7% of the wild-type level) was intermediate between the wild type and Myr1E, although the differences were not statistically significant. The results for the basic substitution mutant viruses are likely related to the reduction, but not the elimination, of Myr1E.KR/AA and Myr1E.KR/AA.KKK/AAA Gag nuclear localization with LMB treatment (Fig. 2). Therefore, the introduction of a foreign NLS, which enhanced Gag nuclear targeting, also restored gRNA packaging to Myr1E.NLS mutant virus particles.
FIG. 3.
Relative gRNA packaging efficiencies. (A) RNA was extracted from virus particles that had been pelleted through a sucrose cushion, and amounts were normalized according to RT activity to ensure that samples with equivalent particle numbers were analyzed. RNase protection assays were performed using an antisense riboprobe that distinguishes between unspliced viral gRNA (263-nt protected fragment) and spliced viral RNA (183-nt protected fragment). Yeast RNA was digested to control for RNase activity (lane 9). The full-length probe (316 nt) is visualized in the sample to which no RNase was added (lane 10). Undiluted samples (lanes 1, 3, 5, and 7) and a 1:2 dilution of the viral RNA for each construct are shown (lanes 2, 4, 6, and 8), with the name of each virus given above the brackets. The ratios of spliced to total viral RNA species were the same for the wild-type and mutant viruses. (B) The packaging efficiency of wild-type virus was assigned a value of 100%, and the values for the mutant viruses are shown as percentages of that for the wild type. At least four replicates for each virus were performed, and standard errors of the means are shown. Statistical analysis was performed using an unpaired two-tailed t test.
Budding efficiencies of Myr1E-related mutant viruses.
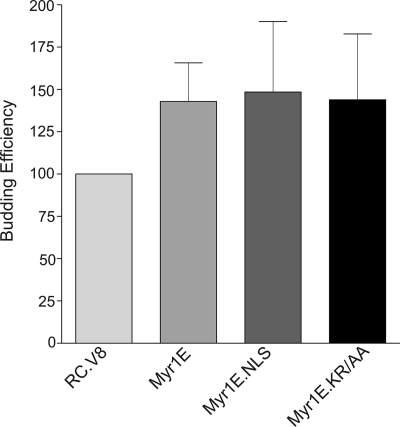
The addition of the Src membrane-targeting domain in mutant Myr1E results in a ∼1.5-fold increase in particle release compared to the wild-type level, presumably due to an increased rate of transport of Myr1E Gag to the plasma membrane (8, 45). If gRNA packaging efficiency is controlled solely by the rapidity of Gag transport to the plasma membrane, then it is plausible to hypothesize that the packaging defect of Myr1E arises from insufficient time to interact with viral gRNA (8). Quantitative budding assays were performed by metabolic radiolabeling using a 5-min pulse and the immunoprecipitation of Gag from the cell lysates, which was compared to the release of Gag (CA) into the medium following a 2.5-h labeling period (8). As shown in Fig. 4, the efficiencies of particle release for Myr1E, Myr1E.NLS, and Myr1E.KR/AA were comparable to that for Myr1E under these conditions.
FIG. 4.
Budding efficiencies of wild-type and mutant viruses. At 16 h following transient transfection with each indicated proviral construct, samples were metabolically labeled with 35[S]methionine for either 5 min (for cell lysates) or 2.5 h (for viral supernatants) and proteins were immunoprecipitated using polyclonal RSV antiserum. The amount of Gag detected by SDS-PAGE was determined using PhosphorImager analysis. Budding efficiency was calculated by comparing the amount of CA released into the medium during the 2.5-h labeling period to the amount of Gag precursor produced within the cells during the 5-min labeling period. The wild-type budding efficiency was assigned a value of 100%, and the values for the mutants were compared to that for the wild type. Each bar represents the mean of results from six to seven independent experiments, and the standard error of the mean is shown for each.
Infectivity defect of mutant viruses in avian cells.
The insertion of a heterologous NLS into Myr1E Gag restored gRNA encapsidation, so we tested whether the increase in gRNA was sufficient to reestablish the infectivity of the replication-incompetent Myr1E virus. Virus particles were collected from QT6 cells transiently transfected with either wild-type or mutant proviral constructs, and the samples were normalized according to exogenous RT activity and incubated with uninfected cells. The abilities of the Myr1E and Myr1E.NLS viruses to spread throughout the cell culture were evaluated by measuring the RT activities present in the medium every 3 days for 3 weeks (Fig. 5A). Both Myr1E and Myr1E.NLS were unable to propagate, whereas the wild-type virus (RC.V8) efficiently infected and replicated in cells. Although the insertion of the nucleoplasmin NLS into Myr1E Gag restored wild-type levels of gRNA packaging, it was not sufficient to restore infectivity. The failure of the increase in gRNA incorporation to restore infectivity to Myr1E.NLS suggests that there are additional blocks to replication. This conclusion is supported by previous findings that Myr1E has a defect in gRNA dimerization (as well as encapsidation) and that RC.V8 with the NLS insertion is blocked at a nuclear step prior to integration (20, 45).
To normalize virus particle samples with respect to infectivity and gRNA packaging, exogenous RT activities were used to compare mutant and wild-type viruses. To ensure that the analysis of RT activity was a valid method of equalizing particle numbers and would not give results skewed by aberrant Gag-Pol incorporation, the ratio of the level of CA protein (a proteolytic product of Gag) to the level of RT was assessed (Fig. 5B). Virus particles were collected from the media of transfected cells, equivalent volumes determined based on RT counts were lysed, and CA was detected by Western blotting. There was no detectable difference in the CA-RT ratios among the wild-type and mutant viruses.
DISCUSSION
The packaging of the retroviral genome is a fundamental yet poorly understood process required for the genesis of infectious viral particles. Critical questions that remain include (i) when during the assembly process does gRNA recognition and selection occur, (ii) where in the cell does Gag initially interact with its RNA genome, and (iii) is the viral gRNA somehow separated from the unspliced viral mRNA meant for translation. In this study, we used mutants of the RSV Gag protein to shed light on these crucial issues and provide support for the proposal of a novel mechanism underlying retroviral encapsidation. We found that restoring nuclear trafficking to a Gag mutant that would otherwise be transported directly to the plasma membrane restored gRNA incorporation to nearly wild-type levels. These results from the genetic gain-of-function assay support the hypothesis that the nuclear localization of RSV Gag is necessary for efficient incorporation of gRNA into virus particles.
RSV Gag is not the only Gag protein with reported nuclear trafficking properties, although Gag nuclear localization is typically transient. For example, a small subset of Mason-Pfizer monkey virus Gag protein is found within the nucleus and in association with the nuclear envelope under steady-state conditions, and a mutant Gag defective in RNA packaging also loses nuclear pore localization (4). In murine leukemia virus (MLV) infection, 18% of the Gag polyprotein within the cell is distributed to the nucleus, and the removal of myristate increases nuclear accumulation. A role for the nuclear fraction of MLV Gag in viral gRNA processing or transport was proposed, but this potential function has not yet been identified (38). HIV Gag was reported previously to undergo transient nuclear localization through a CRM1-dependent NES in MA, although the significance of this finding remains unclear (13). However, deletion of the gRNA-binding domain of HIV-1 Gag (ΔNC) results in partial nuclear localization of Gag, presumably due to a disruption in the Gag-gRNA interactions required for intracellular trafficking (24). Similarly, yeast retrotransposon Ty3 mutants bearing deletions in the gRNA-binding motifs within NC cause Gag3 to accumulate in the nucleus (28). The simian and human foamy virus Gag proteins each contain an NLS in the NC domain, and the nuclear localization of Gag has been implicated in chromatin interaction and proviral integration (54, 58). Thus, although nuclear trafficking of Gag is not unique to RSV, the mechanisms and roles of nuclear transport may vary for different retroviruses.
One of the difficulties associated with studying the nuclear trafficking of retroviral Gag proteins is that only a small subset of the protein appears to be localized to the nucleus under steady-state conditions. Only ∼10 to 20% of RSV Gag is seen in the nucleus under normal conditions; when cells are treated with LMB, Gag becomes concentrated in the nuclei due to the inhibition of CRM1-mediated export (53). In MLV- and Mason-Pfizer monkey virus-infected cells, there are also subsets of Gag proteins found in the nuclei. This small amount of nuclear Gag is consistent with the idea that only a few molecules of Gag would need to enter the nucleus to bind to gRNA because only two genomes are incorporated into each virus particle.
Retroviruses are not unique in hijacking nuclear machinery to transport viral RNAs out of the nucleus, and intriguing parallels can be drawn through comparisons with other viruses. Influenza A viruses encode structural and nonstructural proteins that mediate the nuclear export of the viral ribonucleoprotein complexes (vRNPs) that make up their segmented RNA genomes. Cooperation among vRNP components and M1 (36, 61), nonstructural protein 2/nuclear export protein (40, 43), and nucleoprotein (15) stimulates efficient export of the vRNP from the nucleus and its retention within the cytoplasm. In addition, the E1B 55-kDa and E4 34-kDa proteins of adenovirus facilitate the export of viral mRNAs from the nucleus. Heterodimers of these proteins shuttle in and out of the nuclei in infected cells, and the E4 34-kDa protein interacts directly with CRM1 (12). In the case of herpes simplex virus type 1, the viral nucleocytoplasmic shuttling protein ICP27 selectively binds to intronless herpes simplex virus type 1 mRNAs and promotes their export from the nucleus (47, 51, 51). Thus, it is a common theme for viruses to encode specialized proteins for the export of viral RNAs that may otherwise be trapped within the nucleus, a function similar to that of the Rev protein of HIV-1 (35).
RSV does not have a Rev-like protein, but host factors TAP and Dbp5 facilitate unspliced viral mRNA export for the translation of structural proteins (29). The involvement of Gag in this step is unlikely because the translation of retroviral structural proteins depends on the prior delivery of unspliced viral mRNA to the cytoplasm. However, we can envision a scenario in which the accumulation of Gag in the nucleus serves as a negative regulator of viral mRNA export, causing a switch from translation to assembly that favors the packaging of viral mRNAs as genomes. Alternatively, it remains possible that RSV utilizes two separate export pathways: one mediated by TAP and Dbp5 for the export of unspliced viral RNA destined for translation and another mediated by Gag for the export of gRNA to be packaged into progeny virions. These ideas will be tested in future experiments.
For RSV, we found that providing a strong NLS in cis to a Gag mutant that normally bypasses the nucleus increases gRNA incorporation. These results suggest that interactions between Gag and gRNA that are involved in genome recognition and selection may occur in the nucleus. However, other factors in the nucleus or the inserted NLS itself may indirectly influence packaging through an effect on Gag. For example, it is plausible that in the nucleus Gag undergoes a critical modification (e.g., ubiquitination or sumoylation [23]) needed for gRNA encapsidation or budding. Alternatively, perhaps Gag ferries a nuclear protein involved in genome packaging into the cytoplasm in a manner similar to the mechanism proposed for a mediator of endocytosis, which has an essential nuclear trafficking step prior to plasma membrane localization (22). Additionally, maybe nuclear trafficking results in the delivery of Gag to a perinuclear location which is the actual site of Gag-gRNA interaction. Several perinuclear microdomains, including the pericentrosomal microtubule-organizing center, late endosomal membranes, and nuclear pores, have been suggested as condensation sites where other retroviral Gag proteins initially select the genome for packaging (1, 4, 32, 34, 48). Finally, competition between the nuclear targeting signal and the Src membrane-binding domain may cause Myr1E.NLS to dwell in the cytoplasm longer, increasing the chances of finding the gRNA for packaging.
While the alteration of the nuclear trafficking properties of Myr1E.NLS Gag likely explains the restoration of gRNA packaging, alternative explanations must be considered. It is possible that the insertion of the NLS sequence, which increased the net positive charge by six, enhanced the RNA-binding properties of MA (33, 55). Consistent with this idea, basic residues in the bovine leukemia virus MA domain are involved in specific encapsidation of gRNA (59). However, the restoration of gRNA packaging in Myr1E.NLS is unlikely to be attributable simply to an increase in the net positive charge, because the substitution of neutral for basic residues (in Myr1E.KR/AA and Myr1E.KR/AA.KKK/AAA) did not prevent an increase in gRNA packaging. We found it surprising that the elimination of the basic residues shown previously to be required for nucleoplasmin NLS activity (50) did not completely disrupt the nuclear trafficking of the Myr1E.KR/AAA.KKK/AAA Gag protein. Although the reason for this result is not clear, it is feasible that the inserted sequence changed the conformation of the MA NLS, enhancing its intrinsic nuclear trafficking activity. Alternatively, amino acids flanking the NLS insertion may stabilize the interactions of the Gag mutants with import factors, even in the absence of the basic residues (9, 11).
Based on the collective data, it is intriguing to hypothesize that initial Gag-gRNA interactions occur in (or near) the nucleus, where the genome is selected for packaging. This possibility is attractive because both spliced and unspliced RSV RNAs contain the ψ packaging signal; therefore, the selection of the gRNA from the nuclear pool may favor the encapsidation of genome-length RNAs rather than spliced RNAs. Moreover, the capture of nucleus-localized unspliced viral RNA by RSV Gag would provide a mechanism for separating the gRNA from viral mRNA that is bound to host export factors (TAP and Dbp5) and destined for the ribosome. Whether the genome is marked in some way in the nucleus by association with cellular factors or by localization within a specific subnuclear domain remains an unresolved question. While there are suggestions in the literature that the nuclear localization of Gag proteins of other retroviruses and retrotransposons may contribute to gRNA incorporation, further experimentation using comparative analyses will be needed to identify unifying and divergent features of retroviral genome packaging.
Acknowledgments
This work was supported by Public Health Service grant R01 CA76534 from the National Cancer Institute to L.J.P.
We thank Andrea Beyer and Nicole Gudleski for critical reviews of the manuscript and Eileen Ryan for technical assistance. This research utilized the resources of the Confocal Imaging, DNA Sequencing, Macromolecular Synthesis, and Functional Genomics Core Facilities at the Penn State College of Medicine.
Footnotes
Published ahead of print on 15 April 2009.
REFERENCES
- 1.Basyuk, E., T. Galli, M. Mougel, J. M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5161-174. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214177-218. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz, R. D., A. Ohagen, S. Hoglund, and S. P. Goff. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 696445-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohl, C. R., S. M. Brown, and R. A. Weldon, Jr. 2005. The pp24 phosphoprotein of Mason-Pfizer monkey virus contributes to viral genome packaging. Retrovirology 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray, M., S. Prasad, J. W. Dubay, E. Hunter, K. T. Jeang, D. Rekosh, and M. L. Hammarskjold. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. USA 911256-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butterfield-Gerson, K. L., L. Z. Scheifele, E. P. Ryan, A. K. Hopper, and L. J. Parent. 2006. Importin-β family members mediate alpharetrovirus Gag nuclear entry via interactions with matrix and nucleocapsid. J. Virol. 801798-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callahan, E. M., and J. W. Wills. 2000. Repositioning basic residues in the M domain of the Rous sarcoma virus Gag protein. J. Virol. 7411222-11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callahan, E. M., and J. W. Wills. 2003. Link between genome packaging and rate of budding for Rous sarcoma virus. J. Virol. 779388-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conti, E., and J. Kuriyan. 2000. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure 8329-338. [DOI] [PubMed] [Google Scholar]
- 10.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 694213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dingwall, C., J. Robbins, S. M. Dilworth, B. Roberts, and W. D. Richardson. 1988. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 107841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 164276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupont, S., N. Sharova, C. DeHoratius, C. M. Virbasius, X. Zhu, A. G. Bukrinskaya, M. Stevenson, and M. R. Green. 1999. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature 402681-685. [DOI] [PubMed] [Google Scholar]
- 14.Dupraz, P., and P. F. Spahr. 1992. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J. Virol. 664662-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elton, D., M. Simpson-Holley, K. Archer, L. Medcalf, R. Hallam, J. McCauley, and P. Digard. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75408-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emerman, M., R. Vazeux, and K. Peden. 1989. The rev gene product of the human immunodeficiency virus affects envelope-specific RNA localization. Cell 571155-1165. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82475-483. [DOI] [PubMed] [Google Scholar]
- 18.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 901051-1060. [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara, T., K. Oda, S. Yokota, A. Takatsuki, and Y. Ikehara. 1988. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol. Chem. 26318545-18552. [PubMed] [Google Scholar]
- 20.Garbitt, R. A., J. A. Albert, M. D. Kessler, and L. J. Parent. 2001. trans-acting inhibition of genomic RNA dimerization by Rous sarcoma virus matrix mutants. J. Virol. 75260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garbitt, R. A., K. R. Bone, and L. J. Parent. 2004. Insertion of a classical nuclear import signal into the matrix domain of the Rous sarcoma virus Gag protein interferes with virus replication. J. Virol. 7813534-13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardiner, F. C., R. Costa, and K. R. Ayscough. 2007. Nucleocytoplasmic trafficking is required for functioning of the adaptor protein Sla1p in endocytosis. Traffic 8347-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 182046-2059. [DOI] [PubMed] [Google Scholar]
- 24.Grigorov, B., D. Decimo, F. Smagulova, C. Pechoux, M. Mougel, D. Muriaux, and J. L. Darlix. 2007. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruter, P., C. Tabernero, C. von Kobbe, C. Schmitt, C. Saavedra, A. Bachi, M. Wilm, B. K. Felber, and E. Izaurralde. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1649-659. [DOI] [PubMed] [Google Scholar]
- 26.Jin, J., T. Sturgeon, C. Chen, S. C. Watkins, O. A. Weisz, and R. C. Montelaro. 2007. Distinct intracellular trafficking of equine infectious anemia virus and human immunodeficiency virus type 1 Gag during viral assembly and budding revealed by bimolecular fluorescence complementation assays. J. Virol. 8111226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenney, S. P., T. L. Lochmann, C. L. Schmid, and L. J. Parent. 2008. Intermolecular interactions between retroviral Gag proteins in the nucleus. J. Virol. 82683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen, L. S. Z., N. Beliakova-Bethell, V. Bilanchone, M. Zhang, A. Lamsa, R. DaSilva, G. W. Hatfield, K. Nagashima, and S. Sandmeyer. 2008. Ty3 nucleocapsid controls localization of particle assembly. J. Virol. 822501-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LeBlanc, J. J., S. Uddowla, B. Abraham, S. Clatterbuck, and K. L. Beemon. 2007. Tap and Dbp5, but not Gag, are involved in DR-mediated nuclear export of unspliced Rous sarcoma virus RNA. Virology 363376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, E. G., A. Alidina, C. May, and M. L. Linial. 2003. Importance of basic residues in binding of Rous sarcoma virus nucleocapsid to the RNA packaging signal. J. Virol. 772010-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, E. G., and M. L. Linial. 2000. Yeast three-hybrid screening of Rous sarcoma virus mutants with randomly mutagenized minimal packaging signals reveals regions important for Gag interactions. J. Virol. 749167-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann, M., M. Milev, L. Abrahamyan, X. J. Yao, N. Pante, and A. J. Mouland. 13 March 2009, posting date. Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. J. Biol. Chem. doi: 10.1074/jbc.M808531200. [DOI] [PMC free article] [PubMed]
- 33.Leis, J. P., J. McGinnis, and R. W. Green. 1978. Rous sarcoma virus p19 binds to specific double-stranded regions of viral RNA: effect of p19 on cleavage of viral RNA by RNase III. Virology 8487-98. [DOI] [PubMed] [Google Scholar]
- 34.Levesque, K., M. Halvorsen, L. Abrahamyan, L. Chatel-Chaix, V. Poupon, H. Gordon, L. DesGroseillers, A. Gatignol, and A. J. Mouland. 2006. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic 71177-1193. [DOI] [PubMed] [Google Scholar]
- 35.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338254-257. [DOI] [PubMed] [Google Scholar]
- 36.Martin, K., and A. Helenius. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67117-130. [DOI] [PubMed] [Google Scholar]
- 37.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99677-690. [DOI] [PubMed] [Google Scholar]
- 38.Nash, M. A., M. K. Meyer, G. L. Decker, and R. B. Arlinghaus. 1993. A subset of Pr65gag is nucleus associated in murine leukemia virus-infected cells. J. Virol. 671350-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelle, T. D., and J. W. Wills. 1996. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J. Virol. 702269-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neumann, G., M. T. Hughes, and Y. Kawaoka. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 196751-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neville, M., F. Stutz, L. Lee, L. I. Davis, and M. Rosbash. 1997. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 7767-775. [DOI] [PubMed] [Google Scholar]
- 42.Ogert, R. A., L. H. Lee, and K. L. Beemon. 1996. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J. Virol. 703834-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Neill, R. E., J. Talon, and P. Palese. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17288-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paca, R. E., R. A. Ogert, C. S. Hibbert, E. Izaurralde, and K. L. Beemon. 2000. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J. Virol. 749507-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parent, L. J., T. M. Cairns, J. A. Albert, C. B. Wilson, J. W. Wills, and R. C. Craven. 2000. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J. Virol. 74164-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasquinelli, A. E., R. K. Ernst, E. Lund, C. Grimm, M. L. Zapp, D. Rekosh, M. L. Hammarskjold, and J. E. Dahlberg. 1997. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 167500-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phelan, A., J. Dunlop, and J. B. Clements. 1996. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J. Virol. 705255-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poole, E., P. Strappe, H. P. Mok, R. Hicks, and A. M. Lever. 2005. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site: analysis by confocal microscopy and FRET. Traffic (Copenhagen, Denmark) 6741-755. [DOI] [PubMed] [Google Scholar]
- 49.Rasband, W. S. 2008. ImageJ. National Institutes of Health, Bethesda, MD.
- 50.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64615-623. [DOI] [PubMed] [Google Scholar]
- 51.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheifele, L. Z., E. P. Ryan, and L. J. Parent. 2005. Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J. Virol. 798732-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scheifele, L. Z., R. A. Garbitt, J. D. Rhoads, and L. J. Parent. 2002. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 993944-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schliephake, A. W., and A. Rethwilm. 1994. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 684946-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steeg, C. M., and V. M. Vogt. 1990. RNA-binding properties of the matrix protein (p19gag) of avian sarcoma and leukemia viruses. J. Virol. 64847-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swanson, C. M., and M. H. Malim. 2006. Retrovirus RNA trafficking: from chromatin to invasive genomes. Traffic 71440-1450. [DOI] [PubMed] [Google Scholar]
- 57.Swanson, C. M., B. A. Puffer, K. M. Ahmad, R. W. Doms, and M. H. Malim. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 232632-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tobaly-Tapiero, J., P. Bittoun, J. Lehmann-Che, O. Delelis, M. L. Giron, H. de Thé, and A. Saib. 2008. Chromatin tethering of incoming foamy virus by the structural Gag protein. Traffic 91717-1727. [DOI] [PubMed] [Google Scholar]
- 59.Wang, H., K. M. Norris, and L. M. Mansky. 2003. Involvement of the matrix and nucleocapsid domains of the bovine leukemia virus Gag polyprotein precursor in viral RNA packaging. J. Virol. 779431-9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weldon, R. A., Jr., C. R. Erdie, M. G. Oliver, and J. W. Wills. 1990. Incorporation of chimeric Gag protein into retroviral particles. J. Virol. 644169-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whittaker, G. R., and A. Helenius. 1998. Nuclear import and export of viruses and virus genomes. Virology 2461-23. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., and E. Barklis. 1995. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 695716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zolotukhin, A. S., A. Valentin, G. N. Pavlakis, and B. K. Felber. 1994. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J. Virol. 687944-7952. [DOI] [PMC free article] [PubMed] [Google Scholar]