Abstract
In pro- and eukaryotic cells, RuvB-like protein 2 (RBL2) resolves Holliday junction recombination intermediates. Here, we identified RBL2 as a suppressor of influenza A virus replication. Human RBL2 appears to interfere with the oligomerization of the viral nucleoprotein, a critical step in the assembly of viral replication complexes.
Influenza A virus is an enveloped virus that belongs to the Orthomyxoviridae family and contains eight negative-sense RNA segments encoding 10 to 11 proteins (16). The RNA polymerase complex consists of three subunits, PB1, PB2, and PA. These polymerase subunits and nucleoprotein (NP), together with the viral RNA (vRNA), form the viral ribonucleoprotein complex (vRNP), which is the minimum component for vRNA replication and transcription.
Although some host factors have now been identified that interact with viral proteins (reviewed in reference 14), relatively little is known about the interplay between virus and host factors. We therefore developed a screening system to identify host proteins in a functional assay. Of the candidates that we identified that regulate virus RNA synthesis, RuvB-like protein 2 (RBL2) was identified as an inhibitor of vRNA synthesis.
RBL proteins (RBL1 and RBL2) are members of the AAA+ (ATPases associated with diverse cellular activities) family of helicases. They share moderate homology with bacterial RuvB, the ATP-dependent motor of the RuvAB complex that drives branch migration of the holiday junction (4). RBL1 and RBL2 are essential for viability in Saccharomyces cerevisiae and Drosophila melanogaster and may have similarly important roles in humans (1, 10).
In mammalian cells, the RBL proteins modulate cellular transformation, signaling, apoptosis, and the response to DNA damage by interacting with proteins such as β-catenin, c-Myc, and ATF2 (1, 2, 21). Moreover, they modulate rRNA processing and small nucleolar RNA maturation (12, 20) and function in complexes such as the chromatin remodeling complex INO80 (18) and the histone acetylase Tip60 complex (9).
Here, we provide evidence that RBL2 inhibits influenza virus replication. The protein interacts with the viral NP protein and interrupts its oligomerization, leading to inhibition of viral polymerase activity.
MATERIALS AND METHODS
Cell culture and viruses.
Human embryonic kidney cells (293T cells and 293 cells) were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% heat-inactivated fetal calf serum and antibiotics. QT6 quail fibrosarcoma cells were maintained in Ham's F12K medium (MP Biomedicals) supplemented with 10% fetal calf serum and 10% tryptose phosphate broth (Sigma). Madin-Darby canine kidney (MDCK) cells were cultured in minimal essential medium containing 5% newborn calf serum and antibiotics. A/WSN/33 virus ([WSN] H1N1) was generated by reverse genetics (15) and propagated in MDCK cells. Viruses were titrated by plaque assay in MDCK cells.
Plasmids.
The PB1, PB2, PA, and NP proteins of the A/Hong Kong/483/97 virus ([HK483] H5N1) were expressed using the pCAGGS vector (7) for the library screen. For all other experiments, these genes were derived from the WSN strain. The three polymerase cDNAs from the WSN strain fused to a c-Myc tag sequence at the 3′ termini and the NP cDNA from the WSN strain fused to a c-Myc tag sequence at the 5′ terminus were also inserted into pCAGGS. pPolI-GFP (where PolI is polymerase I and GFP is green fluorescent protein) drives the synthesis of negative-sense vRNAs comprising the 3′ noncoding region of the neuraminidase (HK483) vRNA, the complementary coding sequence of enhanced GFP (EGFP), and the 5′ noncoding region of the neuraminidase vRNA. Similarly, PolI-Luc drives the transcription of a virus-like RNA expressing luciferase.
Quail RBL2 (qRBL2) was cloned from QT6 cells by using the 5′ RACE System (Invitrogen) according to the manufacturer's instructions. Human RBL2 and RBL1 (hRBL2 and hRBL1, respectively) were cloned from human 293T cells. Briefly, RNA was extracted from these cells by use of an RNeasy Mini Kit (Qiagen). Reverse transcription-PCR (RT-PCR) was performed using an oligo(dT) primer, followed by PCR with gene-specific primers. hRBL2-DN (D299N) (2), which is designed to inactivate helicase activity, was constructed by using site-directed mutagenesis. These PCR products were cloned into the pCAGGS vector and then sequenced.
To assess the interaction between hRBL2 and the viral polymerase in living cells, we used a CoralHue Fluo-Chase kit (Amalgaam). Using the vectors in this kit, we constructed plasmids for the expression of the three polymerases, NP, and hRBL2 fused with the N- or C-terminal portion of Kusabira-Green, resulting in GN and GC fusion proteins (for example, pPB1-GN or pPB1-GC).
Library screening.
Human embryonic kidney 293T cells were transfected with HK483 polymerase and NP protein expression plasmids (note that PB2 possesses glutamic acid at position 627), with a plasmid for the synthesis of a virus-like RNA encoding EGFP (PolI-EGFP) and with the quail QT6 cDNA library. Cells were incubated for 2 days at 33°C and collected, and GFP-expressing cells were sorted by a FACSCalibur instrument (BD). Plasmid DNA was then extracted from the cells and amplified in Escherichia coli. Cells that expressed high levels of GFP underwent selection three times, at which point their plasmid DNA was extracted and sequenced.
Luciferase assay.
Luciferase assays were performed by use of a dual-luciferase reporter assay system (Promega) on a microplate luminometer (Veritas; Turner Biosystems, Sunnyvale, CA), according to the manufacturer's instructions. The internal control for the dual-luciferase assay was pGL4.74(hRluc/TK) (Promega).
Analysis of virus propagation.
To establish virus growth rates, three wells of 293 cells were transfected with the respective plasmid or small interfering RNA (siRNA) and 24 h (plasmid) or 48 h (siRNA) later infected with WSN at a multiplicity of infection (MOI) of 0.05. The cells were maintained at 37°C. At various time points, supernatants were assayed for virus titers by plaque assay in MDCK cells.
Knockdown of hRBL2 by use of siRNA.
siRNA specific to RBL2 (catalog no. HSS116737) was purchased from Invitrogen. At 48 h posttransfection with the siRNA, cells were tested for RBL2 expression levels by Western blot analysis.
Quantification of vRNA products.
To assess viral polymerase activity, cells transfected with protein expression plasmids or siRNAs were infected with WSN at an MOI of 2. At various time points, cells were washed three times with phosphate-buffered saline, after which the RNA was extracted and subjected to RT-PCR with NP gene-specific primers (sequences will be provided upon request) or oligo(dT) primers. The resultant cDNAs were quantified with LightCycler, version 2.0 (Roche), by NP gene-specific primers; beta-actin-specific primers served as an internal control.
Western blot analysis.
To assess hRBL2 expression levels, cells transfected with the respective hRBL2 protein expression plasmid were suspended in Tris-glycine-sodium dodecyl sulfate (SDS) sample buffer (Invitrogen) and subjected to Western blot analysis with anti-hRBL2 (BD Transduction Laboratories) and anti-beta-actin (as an internal control; Sigma) antibodies, according to the manufacturer's instructions. Biotinylated anti-mouse immunoglobulin G (IgG) antibody (Vector) was used as a secondary antibody. Bands were detected using a Vectastain ABC kit (Vector) and ECL Plus Western Blotting Detection Reagents (GE Healthcare); the VersaDoc Imaging System (Bio-Rad) was used to quantify band intensities.
To analyze the expression of the viral M1 protein, cells transfected with protein expression plasmids or siRNA were infected with WSN at an MOI of 3 and incubated at 37°C. At the time points indicated in Fig. 2C or 3C, the cells were washed three times with phosphate-buffered saline and resuspended in Tris-glycine-SDS sample buffer. Western blot analysis was then performed with monoclonal antibodies specific to the M1 protein; beta-actin served as a control. Biotinylated anti-mouse IgG antibody (Vector) was used as a secondary antibody. Bands were detected as described above.
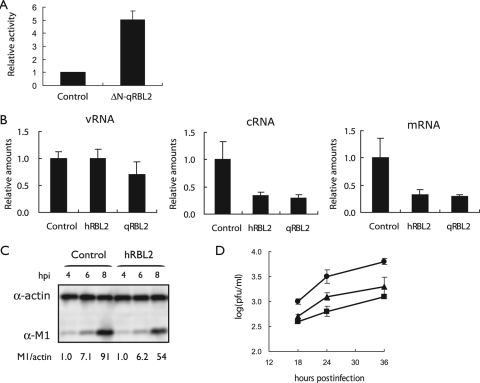
FIG. 2.
Effect of RBL2 expression on influenza virus replication and growth. (A) ΔN-qRBL2 increases influenza virus replication in a minireplicon assay. 293T cells were transfected with plasmids expressing HK483 PB1, PB2-627K, PA, and NP proteins, pPolI-Luc, and ΔN-qRBL2. Twenty-four hours after incubation at 33°C, luciferase expression was detected. (B) Viral polymerase activity in cells expressing hRBL2 or qRBL2. 293 cells were transfected with plasmids expressing qRBL2 or hRBL2 or a control vector. Twenty-four hours later, cells were infected with strain WSN (MOI of 1). Three hours later, RNA was extracted and quantified by real-time RT-PCR with primer sets specific for NP vRNA, cRNA, or mRNA. These values were normalized to beta-actin. The error bars represent standard deviations (n = 3). (C) Viral M1 protein production in cells overexpressing hRBL2. 293 cells expressing hRBL2 or a control vector were infected with strain WSN (MOI of 3). At the indicated hours postinfection (hpi), cell lysates were subjected to Western blot analysis with antibodies against M1 and beta-actin. The values show the ratio of M1 to actin normalized to control cells 4 h after infection. (D) Influenza virus titers in 293 cells overexpressing hRBL2 or qRBL2. 293 cells overexpressing hRBL2 (square), qRBL2 (triangle), or a control vector (circle) were infected with strain WSN (MOI of 0.05). The cells were incubated at 37°C for the indicated time periods. Virus titers in the supernatant were determined by plaque assays in MDCK cells. The error bars represent standard deviations (n = 3). α, anti.
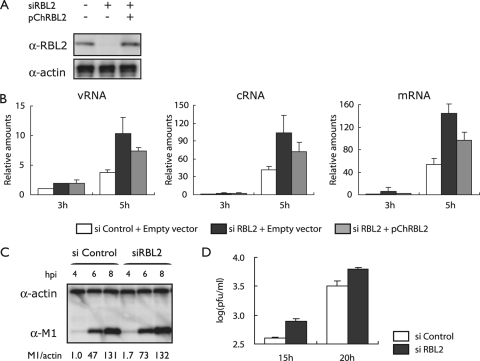
FIG. 3.
Effect of hRBL2 knockdown on influenza virus replication. (A) Knockdown of hRBL2 in 293 cells. 293 cells were transfected with an siRNA specific to hRBL2 (siRBL2) or with a nonspecific control siRNA. Cells were also transfected with a plasmid expressing hRBL2 (pChRBL2) or a control vector. Two days later, hRBL2 expression levels were assessed by Western blot analysis. Beta-actin expression levels served as an internal control. (B) Viral polymerase activity in cells treated with an siRNA to hRBL2. 293 cells were transfected with hRBL2-specific or control siRNAs (si Control) and incubated at 37°C for 48 h. The transfected cells were then infected with strain WSN (MOI of 1). The amounts of vRNA, cRNA, and mRNA were determined at 5 h postinfection as described in the legend of Fig. 2b. (C) Viral M1 protein production in hRBL2 knockdown cells and control cells. Cells were transfected with siRNAs as described above and infected with WSN virus at an MOI of 3. M1 protein levels were assessed as described in the legend of Fig. 2c. (D) Influenza virus titers in hRBL2 knockdown cells and control cells. Cells were treated with siRNAs as described above, infected with WSN virus at an MOI of 0.05, and incubated at 37°C for the indicated time periods. Virus titers were determined in MDCK cells. The error bars represent standard deviations (n = 3). α, anti.
To assess the interaction of hRBL2 with viral polymerase complexes, 293T cells were transfected with plasmids for the expression of hRBL2 (pChRBL2), the viral polymerase proteins (pCAGGS-PB1, -PB2, and -PA), Myc-tagged NP protein, and, in a subset of experiments, a plasmid for the synthesis of virus-like RNA (PolI-GFP). Twenty-four hours later, Myc-tagged proteins were immunoprecipitated with anti-c-Myc agarose (Sigma). Nontagged NP served as a negative control. The beads were resuspended in Tris-glycine-SDS sample buffer, and Western blot analysis was performed with an antibody specific for hRBL2. Biotinylated anti-mouse IgG antibody (Vector) was used as a secondary antibody. Bands were detected as described above.
To assess the interaction of hRBL2 with individual components of the replication machinery, pChRBL2 was cotransfected with plasmids for the synthesis of Myc-tagged polymerase and NP proteins (pCPB1-Myc, pCPB2-Myc, pCPA-Myc, or pCMyc-NP). Immunoprecipitations were performed as described above.
BiFC assays.
To examine the interaction of hRBL2 with viral proteins in living cells, we performed bimolecular fluorescence complementation (BiFC) assays using a CoralHue Fluo-Chase kit (Amalgaam). Briefly, the respective proteins were fused to N- or C-terminal portions of Kusabira-Green, resulting in GN or GC fusion proteins. Pairs of GN and GC fusion proteins were transfected into 293T cells and subjected 12 or 24 h later to luciferase assays or fluorescence-activated cell sorting (FACS) analysis, respectively.
RESULTS AND DISCUSSION
To identify host proteins that regulate viral replication, we developed a screening system in which human embryonic kidney 293T cells are transfected with a plasmid for the synthesis of a virus-like RNA encoding EGFP (PolI-EGFP) and with plasmids for the expression of the influenza A virus polymerase and NP proteins (Fig. 1). The PB2 protein possesses glutamic acid at position 627, which attenuates replication in mammalian systems at 33°C but allows efficient replication in avian systems (19). To identify avian host factors that rescue efficient replication, we cotransfected cells with a cDNA expression library derived from quail QT6 cells. Cells were incubated for 2 days at 33°C, and then GFP-expressing cells were sorted by FACSCalibur (BD). After three rounds of selection, we extracted plasmid DNAs and sequenced them. Among the quail proteins that upregulated viral replication was a truncated qRBL2 protein that lacked 217 N-terminal amino acids (ΔN-qRBL2).
FIG. 1.
Identification of cellular proteins that enhance influenza virus replication. Cells were transfected with plasmids encoding the HK483 (H5N1) PB2, PB1, PA, and NP proteins (the PB2 protein possesses glutamic acid at position 627; PB2-627E). Cells were cotransfected with PolI-EGFP for the synthesis of a virus-like RNA and with an avian (quail cell) cDNA expression library. Cells expressing avian proteins that support efficient replication by PB2-627E in mammalian cells at 33°C produce increased amounts of EGFP. GFP-expressing cells were selected by a FACS cell sorter. After three rounds of selection, plasmid DNA was extracted from the cells and sequenced.
A role for RBL proteins in the regulation of viral infections has not been described to date. However, using a genome-wide RNA interference screen in Drosophila to identify host genes important for influenza virus replication, we found that knockdown of the Drosophila homolog of hRBL2 enhanced reporter gene expression of the influenza virus replicon (6). In addition, Mayer et al. identified hRBL2 as a cellular interaction partner of influenza vRNPs (13). However, neither Hao et al. nor Mayer et al. assessed the biological significance of the RBL2 interaction with influenza vRNPs.
To confirm that ΔN-qRBL2 affects influenza virus RNA replication, we expressed it in human embryonic kidney (293) cells that also expressed the polymerase and NP proteins and a virus-like RNA encoding luciferase. Viral polymerase activity was indeed upregulated upon expression of ΔN-qRBL2 (Fig. 2A). We then overexpressed the full-length hRBL2 and qRBL2 in 293 cells that were subsequently infected with influenza virus strain WSN (H1N1) at an MOI of 1. Assessment of vRNA, cRNA, and mRNA levels by real-time RT-PCR at 3 h postinfection revealed downregulation of polymerase activity upon overexpression of hRBL2 or qRBL2 (Fig. 2B). As a consequence, viral protein synthesis (Fig. 2C) and replication (Fig. 2D) were restricted. These findings indicate that ΔN-qRBL2 (identified in our screen) likely enhances influenza virus replication by acting as a dominant negative protein that competes with the endogenous RBL2 and that RBL2 is a general rather than host-species-specific host factor that suppresses influenza virus replication.
To further demonstrate that RBL2 interferes with influenza virus replication, we knocked it down in 293 cells by use of a specific siRNA (Fig. 3A, left and middle lanes). In these cells, the amounts of viral transcripts were increased relative to control cells (Fig. 3B, compare white and black bars), an effect that was partially reversed upon transfection of RBL2 siRNA-treated cells with a plasmid expressing hRBL2 (pChRBL2) (Fig. 3A, right lane, and B). Further, viral protein production and replication increased in RBL2 siRNA-treated cells (Fig. 3C and D), demonstrating that hRBL2 has an antiviral effect on influenza virus replication.
RBL2 possesses ATPase activity, which is critical for its biological function (2, 4). We therefore assessed a dominant negative form of RBL2 that lacks ATPase activity (hRBL2-DN) (2) for its effects on influenza virus growth. We found that hRBL2-DN interfered with polymerase activity and virus growth at almost the same level as the wild-type protein (data not shown). Hence, the ATPase activity of hRBL2 is not critical for its antiviral effect.
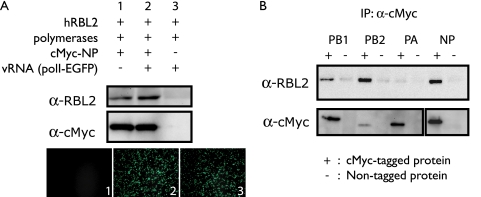
To investigate the interaction of hRBL2 with influenza vRNPs, we expressed hRBL2, the three polymerase subunits, and c-Myc-tagged NP protein in the absence or presence of a virus-like RNA. hRBL2 interacted with the viral replication complex regardless of the presence of vRNA (Fig. 4A). To assess which viral protein interacts with hRBL2, c-Myc-tagged polymerase and NP proteins were separately expressed with hRBL2 and immunoprecipitated with beads coated with an anti-c-Myc antibody. Interestingly, Western blot analysis showed that hRBL2 interacts with three different vRNP components—NP, PB2, and PB1 (Fig. 4B).
FIG. 4.
Interaction of hRBL2 with viral proteins. (A) hRBL2 interacts with viral RNP complexes. The three viral polymerase proteins and c-Myc-tagged NP were expressed in 293T cells in the absence or presence of virus-like RNA (lanes 1 and 2). Nontagged NP was expressed as a negative control (lane 3). One day later, cell lysates were immunoprecipitated with anti-c-Myc beads. Western blot analysis was carried out with anti-hRBL2 and anti-c-Myc antibodies. EGFP expression in transfected cells indicates the functionality of c-Myc-tagged NP protein. (B) hRBL2 interacts with NP, PB2, and PB1. The three polymerase and NP proteins were individually tested for their interaction with hRBL2 as described above. α, anti; IP, immunoprecipitation.
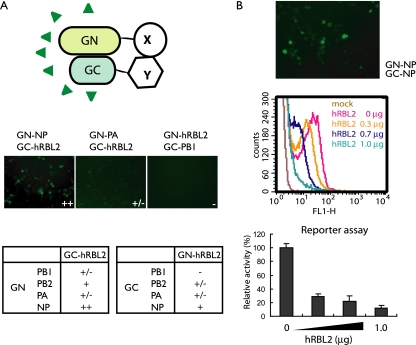
To further study the interaction of hRBL2 with vRNP components, we used a BiFC assay (8, 11) in which the proteins of interest were fused to N-terminal and C-terminal portions of Kusabira-Green (resulting in GN and GC fusion proteins, respectively); interaction of the proteins of interest thus resulted in Kusabira-Green fluorescence (Fig. 5A, upper panel). Strong fluorescence was detected for the hRBL2 interaction with NP (Fig. 5A), whereas the level of fluorescence suggested no interaction or only moderate interaction of hRBL2 and the polymerase proteins in this assay.
FIG. 5.
hRBL2 interferes with NP oligomerization. (A) BiFC assay to assess interactions between hRBL2 and vRNP components. The proteins of interest were fused to the N- or C-terminal portions of Kusabira-Green (GN or GC). Interaction between the proteins of interest thus results in fluorescence. hRBL2 interacts with NP but not with the polymerase subunits. (B) hRBL2 inhibits NP oligomerization in a dose-dependent manner. Cells were transfected with plasmids for the expression of PB2, PB1, PA, pPolI-GFP, and GN-NP and GC-NP. NP-NP interaction was detected by microscopy and FACS analysis (middle panel, red curve). This interaction decreased with increasing amounts of hRBL2 (middle panel). Reporter gene expression in transfected cells indicates the functionality of the GN-NP and GC-NP fusion proteins and the biological significance of hRBL2 interference with NP oligomerization.
NP was found to interact with hRBL2 in both the immunoprecipitation and BiFC assays. To assess if hRBL2 interferes with NP oligomerization, which is important for its biological activities (3, 22), we tested the NP-NP interaction in cells overexpressing hRBL2. As expected, we detected NP-NP interaction in the BiFC assay (Fig. 5B, upper panel); however, hRBL2 overexpression decreased the levels of fluorescence (indicative of NP-NP interaction) in a dose-dependent manner (Fig. 5B, middle panel). Similarly, reporter gene expression from a virus-like RNA was also reduced by hRBL2 expression in a dose-dependent manner (Fig. 5B, lower panel), demonstrating the functionality of NP fusion products and the biological significance of the hRBL2 interference with NP oligomerization. Collectively, these data suggest that hRBL2 disrupts the proper assembly of NP oligomers and, thereby, interrupts its biological activities. In addition to identifying this new host antiviral mechanism, our study also suggests RBL2 as a promising target for antiviral drug development.
Mammalian RBL proteins are implicated in a wide range of cellular activities, including DNA replication and repair, cell cycle progression, and chromatin remodeling (4). Structural analysis revealed that RBL2 and RBL1 can assemble into a double hexameric ring complex in vitro and that the dodecamer is likely one of the functional forms of the proteins (5, 17). Currently, it is not clear whether one of the hexamers contains exclusively RBL2 and the other RBL1 or whether both contain equimolar amounts of RBL2 and RBL1 (4). However, each protein is thought to have specific roles, and some data suggest subunit-specific functions that may not require RBL1/RBL2 complex formation (4). This is in line with findings by Mayer et al., who identified hRBL2, but not hRBL1, as a cellular interaction partner of vRNPs (13). Moreover, we did not detect a role for hRBL1 in influenza virus infection (data not shown). Hence, the inhibitory effect of hRBL2 on influenza virus seems to be a specific function of RBL2 rather than of the RBL1/RBL2 heterooligomer.
Acknowledgments
We thank Susan Watson for editing the manuscript. We also thank H. Mimuro and N. Ishijima for technical support.
This research was supported by a Grant-in-Aid for Specially Promoted Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; by a Grant-in-Aid for JSPS Fellows from the Japanese Society for the Promotion of Science (JSPS); by ERATO (Japan Science and Technology Agency); and by the National Institute of Allergy and Infectious Diseases Public Health Service research grants. S.K. was supported by JSPS Research Fellowships for Young Scientists.
Footnotes
Published ahead of print on 15 April 2009.
REFERENCES
- 1.Bauer, A., S. Chauvet, O. Huber, F. Usseglio, U. Rothbächer, D. Aragnol, R. Kemler, and J. Pradel. 2000. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 196121-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho, S. G., A. Bhoumik, L. Broday, V. Ivanov, B. Rosenstein, and Z. Ronai. 2001. TIP49b, a regulator of activating transcription factor 2 response to stress and DNA damage. Mol. Cell. Biol. 218398-8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elton, D., E. Medcalf, K. Bishop, and P. Digard. 1999. Oligomerization of the influenza virus nucleoprotein: identification of positive and negative sequence elements. Virology 260190-200. [DOI] [PubMed] [Google Scholar]
- 4.Gallant, P. 2007. Control of transcription by Pontin and Reptin. Trends Cell Biol. 17187-192. [DOI] [PubMed] [Google Scholar]
- 5.Gribun, A., K. L. Cheung, J. Huen, J. Ortega, and W. A. Houry. 2008. Yeast Rvb1 and Rvb2 are ATP-dependent DNA helicases that form a heterohexameric complex. J. Mol. Biol. 3761320-1333. [DOI] [PubMed] [Google Scholar]
- 6.Hao, L., A. Sakurai, T. Watanabe, E. Sorensen, C. A. Nidom, M. A. Newton, P. Ahlquist, and Y. Kawaoka. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454890-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 2931840-1842. [DOI] [PubMed] [Google Scholar]
- 8.Hu, C. D., and T. K. Kerppola. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102463-473. [DOI] [PubMed] [Google Scholar]
- 10.Kanemaki, M., Y. Kurokawa, T. Matsu-ura, Y. Makino, A. Masani, K. Okazaki, T. Morishita, and T. A. Tamura. 1999. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 27422437-22444. [DOI] [PubMed] [Google Scholar]
- 11.Kerppola, T. K. 2006. Complementary methods for studies of protein interactions in living cells. Nat. Methods 3969-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King, T. H., W. A. Decatur, E. Bertrand, E. S. Maxwell, and M. J. Fournier. 2001. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 217731-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayer, D., K. Molawi, L. Martínez-Sobrido, A. Ghanem, S. Thomas, S. Baginsky, J. Grossmann, A. García-Sastre, and M. Schwemmle. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagata, K., A. Kawaguchi, and T. Naito. 2008. Host factors for replication and transcription of the influenza virus genome. Rev. Med. Virol. 18247-260. [DOI] [PubMed] [Google Scholar]
- 15.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 969345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palese, P. 2007. Orthomyxoviridae, p. 1647-1689. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 17.Puri, T., P. Wendler, B. Sigala, H. Saibil, and I. R. Tsaneva. 2007. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J. Mol. Biol. 366179-192. [DOI] [PubMed] [Google Scholar]
- 18.Shen, X., G. Mizuguchi, A. Hamiche, and C. Wu. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406541-544. [DOI] [PubMed] [Google Scholar]
- 19.Shinya, K., S. Hamm, M. Hatta, H. Ito, T. Ito, and Y. Kawaoka. 2004. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology 320258-266. [DOI] [PubMed] [Google Scholar]
- 20.Watkins, N. J., I. Lemm, D. Ingelfinger, C. Schneider, M. Hossbach, H. Urlaub, and R. Lührmann. 2004. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell 16789-798. [DOI] [PubMed] [Google Scholar]
- 21.Wood, M. A., S. B. McMahon, and M. D. Cole. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5321-330. [DOI] [PubMed] [Google Scholar]
- 22.Ye, Q., R. M. Krug, and Y. J. Tao. 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 4441078-1082. [DOI] [PubMed] [Google Scholar]