Abstract
The ability to traverse an intact nuclear envelope and productively infect nondividing cells is a salient feature of human immunodeficiency virus type 1 (HIV-1) and other lentiviruses, but the viral factors and mechanism of nuclear entry have not been defined. HIV-1 integrase (IN) is implicated to play a role in the nuclear import of the virus, but the cellular pathway for IN trafficking and the role of IN in mediating the nuclear import of viral particles are unknown. Using a semipermeabilized cell assay, we observed that the nuclear import of IN was not the result of passive diffusion but occurred independently of cytosolic factors, metabolic energy, and the classical receptor-mediated, Ran-dependent import pathways. To determine if IN enters the nucleus by interacting with the nucleopore complex (NPC), we found that IN bound directly with the FxFG-rich C-terminal domain of nucleoporin 153 (NUP153C). When added in excess to the import assay, NUP153C inhibited the nuclear import of IN. Known binding partners of NUP153C competed with IN for binding with NUP153 and also inhibited the nuclear import of IN. In cultured cells, overexpression of NUP153C reduced the infectivity of an HIV-derived vector by interfering with the nuclear translocation of the viral cDNA. These results support a functional role for the IN-NUP153 interaction in HIV-1 replication and suggest that HIV-1 subviral particles gain access to the nucleus by interacting directly with the NPC via the binding of particle-associated IN to NUP153C.
Partitioning of the eukaryotic genome into a separate cellular compartment is important for the regulation of cellular events, ranging from gene expression to cell cycle progression and cell activation. Controlling access to the genome may also serve to provide genetic stability and function as a potential deterrent to invading pathogens. Indeed, for many viruses that replicate in the nucleus, the nuclear envelope represents a significant barrier to establishing a productive infection. Circumvention and appropriation of nuclear import factors and pathways are strategies that viruses use to gain access to the nucleus (44).
For retroviruses, nuclear import of the viral genome for the purpose of integration is a critical step in the viral life cycle (15). After cell entry and uncoating, the newly reverse transcribed viral genome exists with viral and host cell proteins in a large nucleoprotein complex termed the preintegration complex (PIC), which represents the subviral particle imported to the nucleus (reviewed in references 54 and 83). For most retroviral PICs, access to the nucleus takes place during cell division and the concomitant breakdown of the nuclear envelope. In contrast, the members of the lentivirus and alpharetrovirus subfamilies are able to productively infect nondividing cells, or cells arrested in the G1 phase of the cell cycle, indicating that lentiviral and alpharetroviral PICs are able to traverse an intact nuclear envelope (37, 48, 80).
The pathway and mechanism by which the lentiviral PIC crosses the nuclear envelope of an interphase cell are not clearly defined. Some components of the PIC have been hypothesized to confer a karyophilic property to the complex by bridging an interaction between the complex and cellular nuclear import factors (reviewed in reference 83). The identity or nature of such a factor is unknown, but viral integrase (IN) is a likely candidate due to its own karyophilic property and close association with the PIC (17, 47). In addition to IN, viral proteins matrix (MA) and Vpr, as well as the reverse transcription intermediate cDNA flap, are viral factors that have been studied for their role in PIC nuclear import. However, viruses mutated for both MA and Vpr are still capable of infecting nondividing cells (41, 51), and a definitive role for the cDNA flap in the nuclear import of the complex remains to be demonstrated (31, 56). More recently, the viral capsid protein (CA) has been reported to be the primary determinant for the nuclear import of human immunodeficiency virus type 1 (HIV-1) (30, 100, 102, 103). However, a role for CA in nuclear import may reflect the importance of the proper uncoating of the core particle prior to nuclear translocation (101) as opposed to that of a mechanism for movement of the complex across the nuclear envelope. Taken together, these findings suggest a central role for IN in the nuclear import of the PIC.
Many studies have shown that lentiviral IN is karyophilic (6, 13, 27, 28, 41, 46, 73, 88, 89, 91, 97), but other reports indicate that the nuclear accumulation of IN depends on the cellular factor LEDGF/p75 (58, 62, 91). Subsequent studies found that LEDGF/p75 is critical for integration and target site selection and not critical for nuclear entry of HIV-1 (23, 33, 57, 82). Conflicting data also exist regarding the pathway that IN takes to the nucleus. The nuclear import of HIV-1 IN has been reported to result from an interaction between IN and the importin α/importin β (imp α/β) heterodimer (6, 41, 46) and between IN and importin 7 (38). Other studies dispute these findings and demonstrate that HIV-1 IN is imported to the nucleus via a nonclassical pathway that requires ATP but not soluble nuclear import receptors (27). Genetic studies that used infection assays to characterize the role of IN in PIC nuclear import have been confounded by the fact that, in addition to the nuclear import of the PIC (13, 88), IN mutations affect other essential replication steps during infection, including the packaging and processing of the viral polyprotein (16, 74) and reverse transcription (59, 88, 98, 105).
We sought to define the nuclear import pathway of IN by studying its nuclear import characteristics in the context of a well-described in vitro nuclear import assay (1, 65). The results presented here describe a mechanism of nuclear entry for HIV-1 IN that takes place through the nucleopore complex (NPC) independent of soluble nuclear import factors and the Ran-GTP cycle. Our findings indicate that an interaction between HIV-1 IN and the FxFG-rich C-terminal domain of NUP153 (NUP153C), a constituent of the NPC, is central to IN's nuclear accumulation. Supporting a functional role for NUP153 in the replication of HIV-1, we observed that the overexpression of cytoplasmic NUP153C in 293T cells reduced the infectivity of an HIV-based vector.
MATERIALS AND METHODS
Purification of C-terminal FLAG-tagged HIV-1 IN.
An expression construct encoding pNL4-3-derived HIV-1 IN expressed in frame with a C-terminal FLAG epitope (Stratagene) was generated by amplifying the HIV-1 IN gene in its entirety, using the primers 5′EcorIHIN and 3′XhoIHIN (all primer sequences are available upon request) as the forward and reverse primers, respectively. The PCR fragment was cloned into the TOPO TA cloning vector (Invitrogen Corporation, Carlsbad, CA) and sequenced by the UCLA Sequencing Core Facility using the ABI 3700 DNA analyzer (PE Applied Biosystems, Foster City, CA). The IN gene was then released from the TOPO vector by digestion with EcoRI and XhoI and ligated with the mammalian expression vector pCMV-Tag 4A (Stratagene). Next, primers 5′NdeI HIN and 3′BamHIstpFlag were used to PCR amplify the entire IN-FLAG gene segment for subsequent ligation with the pT7-7 bacterial expression vector, prepared by digestion with NdeI and BamHI. This ligation resulted in the generation of an expression construct encoding HIV-1 IN in frame with an N-terminal His tag and C-terminal FLAG epitope (IN-FLAG). IN-FLAG was expressed in BL21-CodonPlus Escherichia coli as a His-tagged protein. Purification of the tagged protein and subsequent removal of the His tag were performed as described previously (4, 81).
In vitro nuclear import assay using digitonin semipermeabilized cells.
The nuclear import assay was carried out as previously described (1, 65). HeLa cells were grown as a monolayer on glass coverslips (22 mm2; Corning) in Dulbecco's modified Eagle medium (Mediatech) containing 10% fetal bovine serum, 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Mediatech). When the cells were approximately 70% confluent, they were put on ice, the medium was aspirated, and the cells were washed once with 1 ml of transport buffer (20 mM HEPES [pH 7.5]; 110 mM potassium acetate; 2 mM magnesium acetate; 0.5 mM EGTA; 1 mM dithiothreitol; 1 μg/ml of the protease inhibitors aprotinin, leupeptin, and pepstatin; 0.5 mM phenylmethanesulfonyl fluoride). Cells were then incubated in transport buffer containing 35 μg/ml of digitonin detergent (20 mg/ml in dimethyl sulfoxide) for 5 min at 4°C and subsequently washed with fresh transport buffer. To deplete cells of any residual nuclear import factors that might have remained closely associated with the NPC or been retained in the nucleus after permeabilization, cells were pretreated with an ATP-regenerating system (ATP-RS; 0.4 mM ATP, 8 units/ml creatine phosphokinase, and 2 mM creatine phosphate). This was accomplished by placing the slides containing the monolayer cell side down onto 50 μl of transport buffer containing the components of the ATP-RS and incubating the reaction at 30°C for 15 min. The ATP-RS pretreatment was stopped by placing the reactions on ice and adding 500 μl of cold transport buffer. The coverslips were removed from the wells, and their edges were blotted on tissue paper (Kimwipe) before the slides were inverted cell side down onto 50 μl of a nuclear import reaction. Import reactions were allowed to proceed for 30 min at 30°C in a humidified chamber.
A typical nuclear import reaction contained either 50 μl of transport buffer or 50 μl of HeLa cytosolic extract (∼5 mg/ml total protein) along with a complete ATP-RS. Purified bovine serum albumin (BSA) molecules previously labeled with tetramethyl rhodamine isothiocyanate (TRITC) and cross-linked with a classical nuclear localization signal (NLS) sequence (NLS-BSA-TRITC) and IN-FLAG were added to the import reaction at final concentrations of 1.0 and 0.7 μM, respectively. Where indicated, adenylyl-imidodiphosphate (AMP-PNP; Sigma) or guanylyl-imidodiphosphate (GMP-PNP; Sigma) was used at a final concentration of 2.5 mM. To deplete the cells of ATP before the nuclear import reaction, cells were pretreated with 1 μl of apyrase (2 U/μl; Sigma) in 50 μl of transport buffer for 15 min at 30°C. To inhibit active nuclear import mediated by the NPC, cells were pretreated with wheat germ agglutinin (WGA; 50 μg/ml) in transport buffer for 15 min at 30°C. For competition experiments, nuclear transport factor 2 (NTF2) (0.8 mg/ml; Sigma), imp β (0.7 mg/ml; Sigma), or synthetic peptides (6 mg/ml; Sigma) representing either a wild-type or mutant simian virus 40 (SV40) NLS (65) were added to the nuclear import reactions at the indicated concentrations.
Import assays were terminated by placing the cells on ice and adding 300 μl of cold transport buffer. The nuclear import of NLS-BSA-TRITC and enhanced green fluorescent protein (EGFP) was directly visualized in the permeabilized cells using fluorescence microscopy. Indirect immunofluorescence using the monoclonal M2 anti-FLAG antibody (Sigma) was used to determine the subcellular localization of IN-FLAG.
Indirect immunofluorescence and fluorescence microscopy.
For import reactions containing IN-FLAG, cells were further permeabilized after fixation with 0.5% Triton X-100 in phosphate-buffered saline (PBS). Cells were washed with PBS, and coverslips were removed from the last wash step, inverted onto 75 μl of a blocking solution containing 0.2% cold water fish gelatin (Sigma) and 0.2% Tween 20 (Sigma) in PBS, and incubated for 30 min at room temperature. Coverslips were then removed from the blocking solution, and the edge was blotted on a Kimwipe and subsequently inverted onto 60 μl of the primary antibody solution composed of a 1:500 dilution of the anti-FLAG M2 monoclonal antibody (Sigma) in blocking solution. Incubation with the primary antibody was allowed to proceed for a minimum of 2 h. Coverslips were then washed, cell side up, three times with 2 ml PBS for 8 min with gentle rocking. After the last wash step, coverslips were inverted onto 60 μl of the secondary antibody solution composed of a 1:500 dilution of goat anti-mouse immunoglobulin G conjugated with fluorescein isothiocyanate (FITC; Zymed) in blocking solution. Incubation with the secondary antibody took place in the dark for a minimum of 1 h at room temperature. Cells were then washed and mounted onto slides, as described above. Import reactions were visualized by fluorescence microscopy using an Olympus BX60 fluorescence microscope. For each field of view, images from the DAPI (4′,6-diamidino-2-phenylindole) (nuclear subcellular compartment) and TRITC (NLS-BSA-TRITC) or FITC (IN-FLAG) channels were collected.
Quantitative analysis of nuclear import reactions.
The nuclear import of IN-FLAG and NLS-BSA-TRITC was quantified using NIH ImageJ software (http://rsb.info.nih.gov/ij/). In the semipermeabilized system, we quantified only the total fluorescence originating from nuclear accumulation of the import protein and did not express the level of nuclear import as a ratio of nuclear to cytoplasmic fluorescence. This is because the level of fluorescence from the import protein contained in the cytoplasm of the semipermeabilized cells varied due to events not directly related to nuclear import, such as movement of the import protein from the cytoplasm into extracellular space during the wash step and the presence or absence of cytosolic factors interacting with the import substrate. Briefly, DAPI channel images were used to identify and locate nuclei. The nucleus of each cell to be analyzed was outlined using the DAPI channel image. The coordinates of the nucleus were then transferred to an image containing the same field of view but had been visualized using either the TRITC (NLS-BSA-TRITC) or FITC (IN-FLAG) channel. The average pixel intensity from the defined nuclear region was then calculated. Under each reaction condition, the number of nuclei analyzed was ≥50. Values represent average pixel intensities for all nuclei analyzed under each set of reaction conditions and are expressed as a percentage of the control reaction, which was set at 100%.
GST-NUP153C pulldown assay.
The glutathione S-transferase (GST)-NUP153C expression vector, which encodes NUP153C (amino acid residues 896 to 1475) as a C-terminal fusion protein with GST, was generously provided by Larry Gerace at The Scripps Research Institute and was purified as described previously (67). Briefly, expression of GST-NUP153C in CodonPlus cells was induced with 0.4 mM isopropyl-d-1-thiogalactopyranoside for 4 h at 37°C. Cells were collected by centrifugation and resuspended in lysis buffer containing 1 μg/ml each of leupeptin, pepstatin, and aprotinin and 0.5 mM EDTA and lysed by sonication. The GST-NUP153C fusion protein was purified using a glutathione-agarose resin, according to the manufacturer's instructions (Pharmacia). The purified protein was dialyzed against PBS containing 10% glycerol and protease inhibitors, as described above, and stored at −80°C.
The pulldown assay was performed as previously described (67, 79). For each pulldown experiment, 50 μl of glutathione-agarose beads (Pierce) was washed twice in sterile, double-distilled H2O and then once in binding buffer (transport buffer containing 0.5 mg/ml digitonin [diluted from a stock solution of 200 mg/ml in dimethyl sulfoxide]). Beads were resuspended in 500 μl fresh binding buffer, and 6 μg of GST-NUP153C was added to the bead suspension. Binding of GST-NUP153C to the beads was performed at 4°C with gentle rocking for a minimum of 2 h. After binding, the beads were pelleted by centrifugation, and the supernatant was aspirated. Beads were washed three times with 500 μl binding buffer. Loaded beads were resuspended in fresh binding buffer and incubated for at least 2 h with 33 pmol IN-FLAG under different reaction conditions. The beads were then washed five times as described above with 500 μl binding buffer. After the last wash, beads were pelleted and resuspended in 100 μl sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis sample buffer (62.5 mM Tris-HCl [pH 6.8], 25% [vol/vol] glycerol, 10% [wt/vol] SDS, 0.5% [wt/vol] bromophenol blue, 5% [vol/vol] β-mercaptoethanol), boiled for 5 min, and subsequently analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting with the anti-FLAG M2 monoclonal antibody, according to the manufacturer's protocol.
Cloning and transient expression of NUP153C.
The DNA fragment encoding NUP153C was obtained by PCR amplification, using the NUP153C-GST expression construct as the template and 5′XhoINUP153C and 3′MluINUP153C as the forward and reverse primers, respectively. After sequence confirmation, the PCR product was digested with XhoI and MluI and cloned into the eukaryotic expression vector pIRES (Clontech) that was previously digested with XhoI and MluI. To monitor the transfection efficiency of the NUP153C-expressing construct, the red fluorescence protein HcRedI (Clontech) was cloned into the multiple cloning site downstream of the internal ribosomal entry site present in the pIRES vector. HcRedI was PCR amplified from the pHcRedI plasmid using forward primer 5′XbaIHcRedI and reverse primer 3′NotIHcRedI. The HcRedI-encoding fragment was sequenced, digested with XbaI and NotI, and ligated with the NUP153C-containing pIRES vector to form pNIHR.
Virus preparation.
Viral stocks were prepared by transfection of monolayers of 293T cells in 75-cm2 flasks with 4 μg each of plasmid DNA encoding the vesicular stomatitis virus glycoprotein (VSV-G) envelope and an HIV-derived reporter virus (NLEGFPBglVprX) expressing EGFP (3). Culture supernatants were collected 36 and 48 h after transfection and filtered through a 45-μm tube top filter apparatus (Corning). Filtered supernatants were then treated with 2 U of RNase-free DNase I (Amersham Pharmacia) per ml of viral stock at 37°C for 45 min and then pelleted by ultracentrifugation at 100,000 × g for 2 h at 4°C. Viral pellets were resuspended in PBS and stored at −80°C until use. The virus titer was determined by an enzyme-linked immunosorbent assay (Coulter Inc.) against the HIV-1 CA (p24) antigen.
Infection assay for evaluating the role of NUP153 in viral replication.
Typically, 5 × 105 293T cells were transferred into a single well of a six-well plate in Dulbecco's modified Eagle medium containing 10% fetal bovine serum, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. After 24 h, or when the culture reached approximately 70% confluence, cells were transfected with pNIHR or with an empty vector using PolyFect (Qiagen), according to the manufacturer's protocol. At 8 h posttransfection, cells were transduced at a multiplicity of infection (MOI) of 0.5 with NLEGFPΔBglVprX virus pseudotyped with VSV-G (3). Twenty-four hours posttransduction, cells were washed once with PBS and fixed with 0.2% paraformaldehyde in PBS for 1 h at 4°C. After 1 h, the fixative was removed, and cells were washed once with PBS and then resuspended in PBS at a final concentration of 1 × 106 cells/ml. Cells were analyzed by flow cytometry using a BD LSR I flow cytometer (Becton-Dickinson), with a 488-nm argon laser for sample excitation. Live cells were gated using forward versus side scatter plots, and standard fluorescence filter sets for Texas Red and green fluorescent protein were used to detect HcRedI-positive (pNIHR-transfected) and EGFP-positive (vector-transduced) cells, respectively.
DNA extraction and PCR analysis of viral DNA.
Genomic DNA was extracted from approximately 5 × 105 cells that were previously transfected and infected with the NLEGFPΔBglVprX virus using the DNeasy blood and tissue kit (Qiagen). The DNA concentration was measured by absorbance at 260 nm. Viral DNA integrated into the genomic DNA of the infected cells was quantified using a fluorescence-monitored two-step PCR assay, as described previously (85). In the first round of the PCR, proviral DNA sequences were amplified with an outward-facing Alu primer, AluI, that anneals within conserved regions of Alu repeat elements and an HIV-1 long terminal repeat (LTR)-specific primer, LM667 (anneals to nucleotide positions 494 to 516). The 5′ end of the LTR-specific primer contains a phage lambda-specific heel sequence. Alu-LTR sequences were amplified from 20 ng genomic DNA in a 20-μl reaction mixture containing 20 mM Tris-HCl (pH 8.4), 1.5 mM MgCl2, 0.8 mM deoxynucleoside triphosphates, 50 mM KCl, 100 nM AluI primer, 300 nM LM667 primer, and 0.5 U of Platinum Taq DNA polymerase (Invitrogen). The PCR cycle conditions used were as follows: a denaturation step of 5 min at 95°C, then 20 cycles of amplification (94°C for 30 s, 58°C for 30 s, and 72°C for 3 min), and extension at 72°C for 10 min. In the second round of the PCR, 1 μl of the first PCR product was added to a 50-μl reaction containing 1× QuantiTect SYBR green PCR master mix, 300 nM of an internal LTR primer LR (anneals to nucleotide positions 622 to 599), and 300 nM of the lambda-specific primer λT. The PCR protocol included a denaturation step of 15 min at 95°C followed by 40 cycles of 30 s at 95°C, 30 s at 58°C, and 30 s at 72°C and was run on a Bio-Rad iCycler iQ real-time PCR machine.
To detect total viral cDNA (integrated and nonintegrated), the 5′ primer NLR9005 annealing to the nef gene and the 3′ primer 620bk annealing to the R region of the LTR were used to amplify an approximately 600-bp fragment. Nonintegrated nuclear forms of the viral cDNA present in the DNA extracts were detected by first using 5′NL8950 and 3′NL910R to generate an outer PCR product that was subsequently used as the template in a nested PCR using the 5′ primer NLR9005 described above and the 3′ primer Efg that anneals to sequences downstream of the left LTR. This primer combination results in an approximately 700-bp product, representing the 1-LTR circular form of HIV-1 cDNA found primarily in the nucleus (71). The relative abundance of 1- versus 2-LTR circles (ratio of 9:1) (18) and our experimental conditions of a low MOI and DNA extraction of paraformaldehyde-fixed cells made the 1-LTR circular form of viral cDNA a more convenient marker for nuclear import. Due to the rapid loss of the linear viral cDNA at the time of the analysis (24 h postinfection), it is unlikely that nonintegrated, linear viral cDNA would contribute significantly to the quantitation of the 1-LTR circle (19). This was further confirmed by diluting linearized vector DNA into genomic DNA extracted from nontransfected, uninfected 293T cells. Linearized vector DNA concentrations that resulted in a detectable 1-LTR signal were greater than that of 20 ng vector DNA/μl. Because this concentration of linearized vector DNA is far in excess of the amount of viral cDNA expected in the infected cell samples, we conclude that linearized viral cDNA did not contribute to the 1-LTR signal detected in our samples.
RESULTS
Nuclear import of FLAG-tagged HIV-1 IN in the semipermeabilized cell assay.
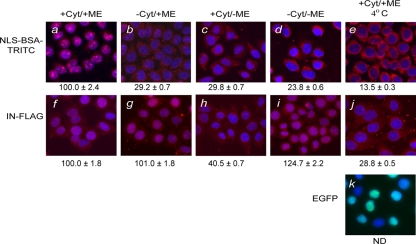
We first used the well-established semipermeabilized cell assay to examine the factors and conditions required for the nuclear import of HIV-1 IN. Consistent with previously published results, the NLS-BSA-TRITC control substrate was imported into the nuclei of semipermeabilized cells in the presence of cytosolic extract and the ATP-RS (Fig. 1a). The import substrate bearing the mutant form of the NLS peptide displayed no karyophilic property under identical import conditions, illustrating the validity and specificity of the nuclear import reaction (data not shown). In the absence of the cytosolic extract, which contains the soluble import factors, the import substrate remained in the cytoplasm of the permeabilized cells, and no discrete signal could be detected in the nucleus (Fig. 1b). To test the energy requirement during nuclear import, ATP was depleted by omitting the ATP-RS and adding the nonhydrolyzable ATP analog AMP-PNP and apyrase, which depletes the permeabilized cells of triphosphate nucleotides by hydrolyzing both GTP and ATP to their diphosphate derivatives. In the absence of ATP, which in turn caused a loss of Ran-GTP (78), the control substrate NLS-BSA-TRITC failed to accumulate in the nucleus (Fig. 1c). As expected, a loss of nuclear accumulation of the import protein was also observed when the assay was carried out in the absence of cytosolic extract and ATP (Fig. 1d). To further demonstrate that the control substrate NLS-BSA-TRITC entered the nucleus by an active process and was not the result of passive diffusion, we carried out the import assay at 4°C (Fig. 1e). At low temperature, the active import process is appreciably suppressed, while passive diffusion continues to occur, albeit at a lower rate (84). As expected, no accumulation of the import protein was observed when the reaction was carried out at 4°C, and the substrate signal was distributed throughout the cytoplasm. The control reaction with purified recombinant EGFP illustrates the specificity of the low-temperature inhibition of active import (compare Fig. 1e and k); at 27 kDa, EGFP passively diffused into the nuclei of semipermeabilized cells even at a low temperature (94).
FIG. 1.
The nuclear import of HIV-1 IN is independent of cytosolic factors and metabolic energy. Nuclear import requirements of NLS-BSA-TRITC (a to e) and FLAG-tagged IN (f to j) were characterized using the semipermeabilized cell assay in the presence (+) or absence (−) of cytosolic extract (Cyt) and metabolic energy (ME) at 30°C (a to d and f to i) or at 4°C (e, j, and k). Quantitation for the level of nuclear import observed under each reaction condition was based on ≥50 nuclei. Complete reactions for NLS-BSA-TRITC (a) and HIV-1 IN (f) were set to 100%, and the levels of nuclear import observed (b to e and g to j) were calculated as percentages of the corresponding complete reactions in NLS-BSA-TRITC (a) and IN (f), respectively. Values indicated below the images are the means ± SEM for three experiments and represent the total fluorescence emanating from the import protein localized to the nucleus. NLS-BSA-TRITC was visualized directly by fluorescence microscopy. FLAG-tagged HIV-1 IN was visualized by indirect immunofluorescence using a monoclonal antibody directed at the FLAG epitope, followed by a fluorescently labeled secondary antibody. For uniformity, the fluorescence signal of both HIV-1 IN and NLS-BSA-TRITC was pseudocolored as red, while the blue fluorescence represents the DAPI-counterstained nuclei. Purified recombinant EGFP (27 kDa) was used as a control protein for passive diffusion under import conditions that restricted active transport, and its fluorescence signal was not pseudocolored. ND, not determined.
We applied the same analyses to purified FLAG-tagged IN. Fusing IN with the FLAG epitope was intended to be used for the sensitive and reliable detection of IN without significantly altering the nature of the protein. In vitro integration assays demonstrated that FLAG-tagged IN retained 3′-end processing and strand transfer activities comparable to those of untagged IN (data not shown), supporting that the FLAG tag had no discernible effect on the functional properties of IN. For simplicity, the FLAG-tagged IN is referred to hereafter as IN. The nuclear import of IN occurred in the presence of both the ATP-RS and cytosolic extracts (Fig. 1f). However, in contrast to NLS-BSA-TRITC, the nuclear import of IN was observed in the absence of cytosolic factors (compare Fig. 1g and b). In the presence of cytosolic factors but in the absence of ATP, the import of IN was substantially reduced (Fig. 1h), suggesting the requirement of renewable energy. However, high levels of IN accumulated in the nucleus in the absence of both nuclear import factors and the ATP-RS (Fig. 1i), indicating that the energy cofactor was not essential to the nuclear import of IN. To ensure that no residual ATP might be contributing to the nuclear import of IN, we pretreated the permeabilized cells with apyrase and included AMP-PNP in the import reaction. As expected, NLS-BSA-TRITC failed to accumulate in the nucleus under these conditions, but IN continued to be efficiently imported into the nucleus under the same conditions (data not shown). These results support our conclusion that HIV-1 IN does not require the ATP-RS for nuclear import. The apparent inhibitory effect of adding cytosolic extract without metabolic energy on IN nuclear import is discussed in more detail below (see Fig. 4A) and was likely due to the presence of bound import factors and import complexes at the NPC. These bound factors and complexes were unable to undergo the final step of release into the nucleus in the absence of the energy cofactor (61).
FIG. 4.
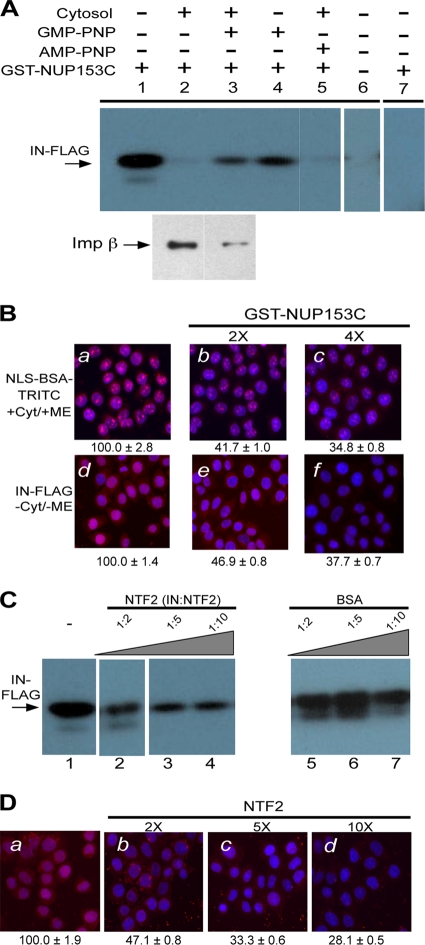
HIV-1 IN interacts with NUP153C. (A) HIV-1 IN binds to NUP153C. GST-NUP153C (60 pmol) was loaded onto glutathione-agarose beads and incubated with FLAG-tagged HIV-1 IN (33 pmol; lanes 1 to 5) or with FLAG-tagged FIV IN (33 pmol; lane 7) in binding buffer. Lane 6 shows a control reaction in which NUP153C was omitted. Binding experiments were carried out in the absence (lanes 1, 4, 6, and 7) or presence (lanes 2, 3, and 5) of the HeLa cytosolic extract and with (lanes 3 and 4) or without (lanes 1, 2, 5, 6, and 7) GMP-PNP. AMP-PNP is included as a specificity control for the stimulatory effect of GMP-PNP on the binding of HIV-1 IN to GST-NUP153C in the presence of cytosol (lane 5). Imp β contained in the HeLa cytosolic extract that was bound to NUP153C in the absence (bottom, lane 2) or presence (bottom, lane 3) of GMP-PNP was probed with a monoclonal anti-imp β antibody (Sigma). Arrows indicate the position corresponding to FLAG-tagged HIV-1 IN (top, lanes 1 to 6), FLAG-tagged FIV IN (top, lane 7), and imp β (bottom, lanes 2 and 3). (B) NUP153C inhibits the nuclear import of NLS-BSA-TRITC and HIV-1 IN. Nuclear import reactions for NLS-BSA-TRITC and HIV-1 were carried out under the indicated reaction conditions and as described in the legend to Fig. 1, except that GST-NUP153C was added at a molar ratio of 2:1 (panels b and e) or 4:1 (panels c and f) to the import protein. Quantitation of nuclear import for ≥50 nuclei from each reaction is indicated below the images and reported as the percentage of the corresponding reaction carried out in the absence of excess GST-NUP153C, as shown in panels a (NLS-BSA-TRITC) and d (IN-FLAG). (C) HIV-1 IN binding to NUP153C is inhibited by NTF2. GST pulldown reactions were carried out in the absence of the cytosolic extract (lane 1) and as described above for panel A, except that purified NTF2 was included as a competitive inhibitor for IN binding to NUP153C (lanes 2 to 4). Purified BSA was used as a control (lanes 5 to 7). Ratios indicate the molar excess of competitor protein to IN. (D) NTF2 inhibits the nuclear import of HIV-1 IN. The nuclear import reaction for HIV-1 IN was carried out in the absence of cytosol and metabolic energy, as described in the legend to Fig. 1, except that NTF2 was added as a competitor (panels b to d) to the import reaction at the indicated excesses. Quantitation of nuclear import for ≥50 nuclei from each reaction is indicated below the images and reported as the percentage of the corresponding reaction carried out in the absence of excess NTF2, as shown in panel a. Values are the means ± SEM of three experiments, and symbols have the same connotations as described in the legend to Fig. 1.
To confirm that passive diffusion was not responsible for the nuclear accumulation of HIV-1 IN, the nuclear import assay was also carried out at 4°C (Fig. 1j). IN (32 kDa) exists predominately in multimeric form as a dimer, a tetramer, or even a higher-order oligomer (29, 34, 42, 53). Since the cutoff size for passive diffusion across the NPC is estimated to be 60 kDa (reviewed in references 49, 60, and 84), the molecular weight of such multimeric forms of IN would preclude them from efficient passive diffusion across the NPC. However, IN monomers would still be small enough to undergo passive diffusion and accumulate in the nucleus as a result of binding to chromosomal DNA or cellular nuclear proteins such as LEDGF/p75 (21). The lack of nuclear accumulation for IN at 4°C strongly suggested that IN did not passively diffuse into the nucleus. These findings led us to conclude that nuclear entry for HIV-1 IN is via a novel and nonclassical nuclear import pathway.
Effects of a nonhydrolyzable GTP analog, GMP-PNP, on the nuclear import of HIV-1 IN.
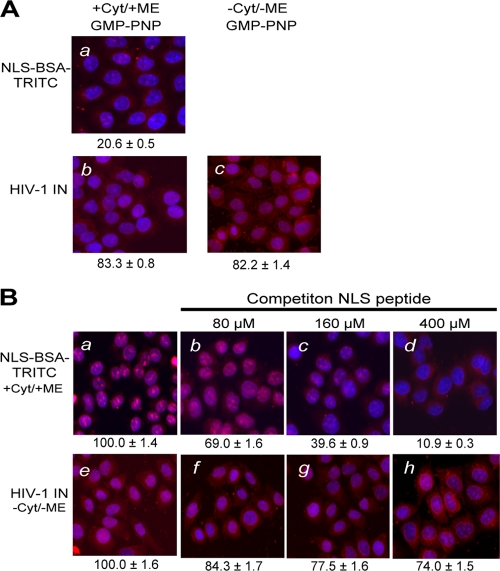
To directly test the role of GTP in the nuclear import of HIV-1 IN, we added GMP-PNP, a nonhydrolyzable analog of GTP, to the import reaction. Binding of GMP-PNP to Ran blocks Ran's intrinsic GTPase activity and sequesters Ran in a form that is unable to support receptor-mediated nuclear import (64). As expected, when GMP-PNP was included in reactions containing cytosol and the ATP-RS, NLS-BSA-TRITC failed to import to the nucleus (Fig. 2A, panel a) (66). In contrast, GMP-PNP had no inhibitory effect on the nuclear import of IN, regardless of whether the reaction was carried out in the presence of cytosolic factors and the ATP-RS (Fig. 2A, panel b) or in their absence (Fig. 2A, panel c). The result provided further evidence for a nonconventional nuclear import pathway for HIV-1 IN.
FIG. 2.
Inhibitors of classical nuclear import pathways do not affect HIV-1 IN nuclear import. (A) Differential effects of the nonhydrolyzable GTP analog GMP-PNP on the nuclear import of NLS-BSA-TRITC and HIV-1 IN. GMP-PNP was included in the import reaction at a final concentration of 2.5 mM. Quantitation of nuclear import observed under each reaction condition was done as described in the legend to Fig. 1 and expressed as percentages of the corresponding reactions without GMP-PNP, as shown in Fig. 1a (NLS-BSA-TRITC) and Fig 1f and i (HIV-1 IN). (B) Differential effects of excess SV40 NLS peptide. Excess purified peptide corresponding to the NLS of the SV40 large T antigen was included in the nuclear import reactions at the indicated concentrations. Quantitation of nuclear import observed under each reaction condition was done as described in the legend to Fig. 1 and expressed as percentages of the corresponding reactions without the peptide competitor, as shown in panels a (NLS-BSA-TRITC) and e (HIV-1 IN). Symbols have the same connotations as described in the legend to Fig. 1. Values are the means ± SEM for three experiments.
HIV-1 IN nuclear import is independent of the imp α/β pathway.
Nuclear import of the NLS-BSA-TRITC conjugate is directed by the NLS sequence derived from the SV40 large T antigen and depends on the imp α/β nuclear import pathway. Although our earlier results (Fig. 1 and 2A), as well as those reported by others (27), indicated that HIV-1 IN nuclear import was independent of the imp α/β pathways, other groups have reported that HIV-1 IN nuclear import is mediated by an imp α/β heterodimer (41) or by imp α alone (6). To probe the potential role of imp α/β in the nuclear import of HIV-1 IN, we carried out competition experiments to determine the effect of excess purified NLS peptide on the nuclear import of HIV-1 IN. If IN is imported by the imp α/β pathway, we would expect to see a reduction in the level of nuclear import in the presence of excess peptide.
The nuclear import of NLS-BSA-TRITC, which served as a positive control, was disrupted by excess NLS peptide (Fig. 2B). The inhibition was concentration dependent (Fig. 2B, panels b to d), and no inhibition was observed when the import reaction was done in the presence of excess mutant NLS peptide (data not shown). Consistent with our results shown in Fig. 1 suggesting that HIV-1 IN is imported into the nucleus independent of imp α and imp β, the presence of excess NLS peptide did not appreciably affect the nuclear accumulation of IN (Fig. 2B, panels f to h).
WGA inhibits the nuclear import of HIV-1 IN.
Proteins imported into the nucleus by the imp α/β pathway pass through the NPC as a result of transient but specific interactions between imp β and FxFG motifs present on nucleoporins comprising the NPC (39, 96). Proteins imported to the nucleus by alternative receptor-mediated pathways (i.e., M9 and transportin) also depend upon interactions between the import receptor and the NPC (12, 79). For proteins that demonstrate receptor-independent nuclear import, the NPC still provides the passageway to the nucleus presumably as a result of direct interactions between the import protein and the NPC (63, 95, 104). WGA is a lectin that binds to posttranslational modifications of N-acetylglucosamine present on nucleoporins and acts to inhibit active nuclear import (39). To assess the role of the NPC in the nuclear import of IN, permeabilized cells were treated with WGA (50 μg/ml) for 15 min at 30°C prior to carrying out the nuclear import reaction. Cells pretreated with WGA did not support the nuclear import of NLS-BSA-TRITC and IN, and these proteins demonstrated a predominantly perinuclear fluorescence pattern (Fig. 3a and b). Since WGA specifically inhibits active transport through the NPC and has no effect on passive diffusion, purified recombinant EGFP was used as a positive control, which was still able to accumulate in the nucleus (Fig. 3c). These results indicate that despite the fact that HIV-1 IN is not imported to the nucleus via the classical pathway, the NPC is an essential element of the import mechanism.
FIG. 3.
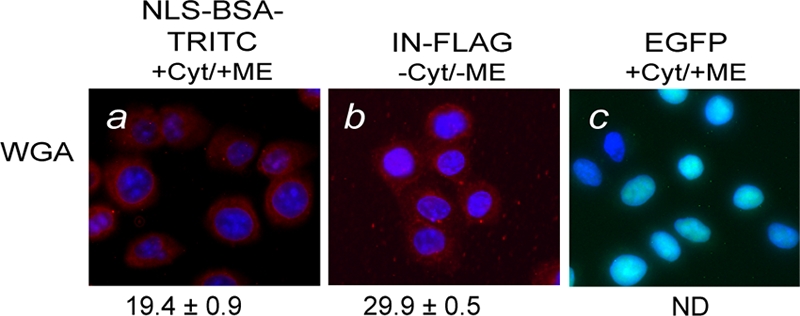
WGA inhibits the nuclear import of NLS-BSA-TRITC and HIV-1 IN. Nuclear import reactions for NLS-BSA-TRITC and HIV-1 IN were carried out as described in the legend to Fig. 1, except that the cells were pretreated with WGA for 15 min at 30°C. Assay conditions are described along the top of the images, and the quantitation method and symbols are used as described in the legend to Fig. 1. Values are expressed as mean percentages ± SEM of the corresponding reaction carried out in the absence of WGA, as shown in Fig. 1a (NLS-BSA-TRITC) and Fig. 1i (HIV-1 IN). The fluorescence signal for EGFP (c) was not pseudocolored as red.
HIV-1 IN binds directly to nucleoporin NUP153.
The observation that IN enters the nucleus through the NPC, but in a receptor- and energy-independent manner, suggested that IN may interact directly with nucleoporins. Other proteins that demonstrate receptor-independent nuclear import have been shown to bind directly to the nucleoporin NUP153 (63, 95, 104), suggesting that NUP153 is a target for diverse nuclear import pathways. To test the possibility that HIV-1 IN interacts with NUP153, we performed pulldown assays using a fusion protein containing GST and the C-terminal region of NUP153. We chose this region of NUP153 for the pulldown assay because it is directly involved in the trafficking of import complexes into the nucleus as well as the exporting of mRNA. In contrast, the N-terminal domain of NUP153 contains motifs that are important for targeting the protein to the NPC, while the central zinc-binding motif is involved in nuclear membrane disassembly during mitosis and Ran binding (reviewed in reference 9). The C-terminal region of NUP153 is rich in FxFG repeats and interacts with imp α/β nuclear import complexes just before their disassembly by Ran-GTP binding (66). Additionally, NUP153C has a flexible morphology and displays variable topology in the NPC (26, 69, 70), and at times, it is even exposed to the cytoplasmic side of the nuclear envelope (36).
Purified IN was incubated with GST-NUP153C bound to glutathione-agarose beads and then analyzed by immunoblotting to determine the level of IN binding. IN was pulled down with GST-NUP153C (Fig. 4A, lane 1) but failed to pull down in the presence of GST alone (Fig. 4A, lane 6) or when HIV-1 IN was replaced by purified FLAG-tagged feline immunodeficiency virus (FIV) IN (FIN) (Fig. 4A, lane 7), indicating that the direct binding of HIV-1 IN to NUP153C is a specific interaction. The lack of direct binding between FIN and NUP153C also suggested that the mechanism of nuclear import for FIN is distinct from that of HIV-1 IN (97) and is consistent with the observation that FIN nuclear import in the semipermeabilized cell assay was cytosol and energy dependent (data not shown).
When IN was incubated with GST-NUP153C in the presence of a 1:10 dilution of the same cytosol used in the semipermeabilized cell assay, binding of IN to GST-NUP153C was reduced (Fig. 4A, lane 2). The inhibitory effect of cytosol on the binding of IN to GST-NUP153C was likely due to the presence of import complexes in the cytosol that competed with IN for binding sites on GST-NUP153C, similar to the reduction of IN nuclear import by adding cytosolic extract in the semipermeabilized cell nuclear import assay (Fig. 1h). Consistent with this explanation was the observation that addition of GMP-PNP to the pulldown assay restored binding of IN to GST-NUP153C in the presence of cytosolic extract (Fig. 4A, lane 3). Binding of GMP-PNP by cytosolic Ran would act constitutively to dissociate import complexes that would otherwise compete with IN for binding sites on GST-NUP153C (66, 79). This was supported by the observation that less cytosolic imp β was bound to GST-NUP153C in the presence of GMP-PNP (Fig. 4A, bottom, compare lanes 2 and 3). The specificity of the action of GMP-PNP was illustrated by the inability of a nonhydrolyzable ATP analog, AMP-PNP, to restore a similar level of binding to GST-NUP153C (Fig. 4A, lane 5) as well as the failure of GMP-PNP alone to stimulate binding of IN to GST-NUP153C (Fig. 4A, lane 4). In summary, these results showed the ability of HIV-1 IN to directly interact with GST-NUP153C and support our conclusion that the mechanism of import for HIV-1 IN involves a direct and specific interaction with the nucleoporin.
Nuclear import of HIV-1 IN in the semipermeabilized cell assay is inhibited by excess GST-NUP153C.
To determine whether the binding of IN to GST-NUP153C has any functional consequences for the nuclear import of IN, we included GST-NUP153C in the semipermeabilized cell assay. As a positive control, we chose NLS-BSA-TRITC because the NUP153C region is also able to bind imp α/β import complexes. In the presence of both cytosolic extract and the ATP-RS, a twofold-higher excess of GST-NUP153C reduced nuclear accumulation of NLS-BSA-TRITC by approximately 60% (Fig. 4B, compare panels a and b). When the import reaction was carried out in the presence of a fourfold-higher excess of GST-NUP153C, the nuclear import of NLS-BSA-TRITC was further reduced to 34.8% of the original level (Fig. 4B, compare panels a and c). These results are consistent with previous studies demonstrating that the GST-NUP153C fragment can inhibit classical nuclear import pathways (79).
For HIV-1 IN, the GST-NUP153C competition reaction was carried out in the absence of both cytosolic extract and the ATP-RS, conditions that supported maximal IN nuclear import (Fig. 1). When a twofold-higher excess of GST-NUP153C was included in the reaction, a 53.1% reduction in the nuclear import of IN was observed (Fig. 4B, compare panels d with e). When a fourfold-higher excess of GST-NUP153C was included, the nuclear import of IN decreased further to 37.7% (Fig. 4B, compare panels d with f). The result suggested that GST-NUP153C can compete with endogenous NUP153 or other components of the NPC for binding to IN and inhibit the nuclear accumulation of IN.
Known NUP153C-binding proteins compete with HIV-1 IN for binding to GST-NUP153C and disrupt the nuclear import of IN.
To further characterize the binding of IN with GST-NUP153C, we performed competition pulldown experiments using proteins known to interact with NUP153C. NTF2 is a 15-kDa protein that transports cytoplasmic Ran-GDP into the nucleus, where it undergoes nucleotide exchange to regenerate nuclear Ran-GTP (77). NTF2 is enriched at the nuclear envelope and interacts with nucleoporins that contain FxFG repeats, including NUP153 (25, 68). When included in the pulldown assay with IN, excess NTF2 disrupted binding of IN to GST-NUP153C in a concentration-dependent manner (Fig. 4C, lanes 2 to 4). As a negative control, reactions containing similar amounts of BSA instead of NTF2 showed little or no inhibition of IN binding to GST-NUP153C (Fig. 4C, lanes 5 to 7). The result suggested that NUP153C has overlapping binding sites for IN and NTF2.
Since NTF2 blocks the binding of IN with NUP153, we would predict that in the presence of excess NTF2, the nuclear import of IN would be reduced due to competition with NTF2 for access to NUP153-binding sites. To test this hypothesis, excess NTF2 was added to the semipermeabilized cell assay under the condition that supported the maximal nuclear import of IN (Fig. 4D). In the absence of cytosolic extract and the ATP-RS, a twofold-higher excess of NTF2 inhibited the nuclear import of IN to 47.1% of the level otherwise observed under the same import condition (Fig. 4D, panel b). The levels of import were further reduced to 33.3% (Fig. 4D, panel c) and 28.1% (Fig. 4D, panel d) of the original level when NTF2 was present at a 5- and 10-fold-higher excess over IN, respectively.
Imp β, another NUP153-interacting protein, also inhibited the nuclear import of IN in the semipermeabilized cell assay (data not shown). The inhibitory effect of imp β further indicates that this import receptor is not required for IN nuclear import and supports our conclusion that import receptors actually compete with HIV-1 IN for nuclear import, presumably by competing for binding sites at the NPC.
Inhibition of early viral replication steps by NUP153C.
To evaluate the biological relevance of the NUP153C interaction with HIV-1 IN, we transiently expressed the NUP153C fragment in 293T cells and then challenged the transfected cells with VSV-G-pseudotyped HIV-1 that encodes EGFP in the viral genome. Cells transduced with this viral vector are EGFP positive after the viral genome successfully undergoes nuclear import, integration, and expression. If NUP153C interacts with the incoming viral particle, we predict that overexpression of NUP153C would exert an inhibitory effect on the nuclear import of the incoming virus by competing with its binding to endogenous NUP153. As a result of such an interaction, the subsequent steps of integration and expression from the viral genome would also be inhibited, resulting in a reduction in the number of EGFP-positive cells.
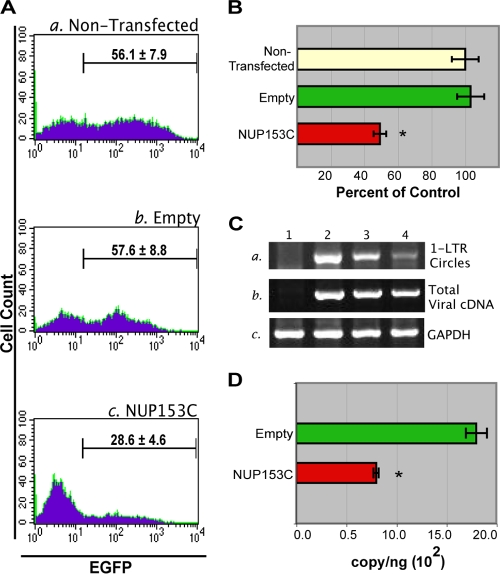
293T cells transfected either with an expression construct encoding NUP153C or with an empty vector, as well as nontransfected 293T cells, were transduced with the pseudotyped EGFP reporter virus 8 h posttransfection at an MOI of 0.5. Twenty-four hours posttransduction (32 h posttransfection), the cells were collected and analyzed by flow cytometry to quantitate the number of transduced, EGFP-positive cells. 293T cells that were nontransfected and transfected with an empty vector demonstrated transduction rates of 56.1% and 57.6%, respectively (Fig. 5A, panels a and b). In comparison, only 28.6% of the cells transfected with the NUP153C expression construct were EGFP positive (Fig. 5A, compare panel c with panels a and b). Therefore, in the presence of NUP153C, the infectivity of the HIV-based vector, as measured by EGFP-positive cells, was significantly reduced to 51.0% of the empty vector control (P = 0.0002) (Fig. 5B). The level of inhibition in transgene expression is expected, given that only 38% of the cells were transfected with the NUP153C expression construct, as demonstrated by a cotransfection control (data not shown).
FIG. 5.
Infectivity of a lentiviral-based vector is inhibited by overexpressing NUP153C. (A) The level of EGFP transgene expression from an HIV-1-based vector is reduced in the presence of NUP153C. Nontransfected 293T cells (panel a) and cells transfected with an empty plasmid (panel b) or with a plasmid encoding NUP153C (panel c) were challenged with a VSV-G-pseudotyped HIV-1-based vector encoding EGFP at 8 h posttransfection. Twenty-four hours postinfection, the cells were collected and analyzed by flow cytometry for EGFP expression. Histograms are representative of results under the indicated experimental conditions. The number of EGFP-positive cells under each condition is expressed as the mean ± SEM for at least three experiments. (B) Graphical representation of EGFP expression normalized to the nontransfected control. An asterisk indicates a significant difference from the empty vector control (P = 0.0002). (C) NUP153C expression inhibits the nuclear import of viral cDNA. DNA was extracted from the same cell populations that were analyzed by flow cytometry in panel A and was subjected to PCR using primers to amplify the 1-LTR circle form of viral DNA (top), total viral DNA (middle), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (bottom) from genomic DNA. Lane 1 shows nontransduced 293T cells. Lane 2 shows 293T cells transduced with the VSV-G-pseudotyped HIV-1-based vector encoding EGFP. Lane 3 shows 293T cells transfected with an empty vector 8 h before transduction with the HIV-1 vector. Lane 4 shows 293T cells transfected with NUP153C 8 h prior to transduction with the HIV-1 vector. (D) NUP153C expression inhibits viral cDNA integration. Alu-PCR was used to quantify the copy number of provirus integrated in cells transfected with either an empty vector (green bar; n = 3) or the NUP153C-expressing vector (red bar; n = 9). The values are expressed as the copy number of provirus/ng genomic DNA. An asterisk indicates a significant difference from the empty vector control (P = 0.0002).
To confirm that the overexpression of NUP153C reduces the infectivity of the HIV-1-derived vector by interfering with the nuclear import of the viral cDNA, we used a divergent PCR strategy to detect the nuclear forms of viral cDNA contained in the infected samples. We found that the nontransfected, empty vector-transfected, and NUP153C-transfected samples produced similar levels of late reverse transcription products, which reflects the level of total viral DNA (Fig. 5C, middle, lanes 2 to 4). This result indicated that NUP153C did not inhibit infection with the vector by altering reverse transcription of the viral genome. As a marker for the level of PIC nuclear import, we measured the 1-LTR circle form of viral cDNA that accumulates in the nucleus following infection as a result of homologous recombination between the left and right viral LTRs. The 1-LTR circles were amplified using divergent forward and reverse primers that annealed to sites in nef and gag, respectively (71). Similar levels of 1-LTR circles were found for the nontransfected and empty vector-transfected samples (Fig. 5C, top, lanes 2 and 3). In contrast, the NUP153C-transfected sample demonstrated a reduced level of the 1-LTR form of viral cDNA (Fig. 5C, top, lane 4), suggesting that the nuclear translocation of the viral cDNA was inhibited in the presence of NUP153C. These results showed that the overexpression of NUP153C inhibits viral replication and implicates NUP153 as a cellular protein that plays an important role in the nuclear trafficking of the viral genome.
As further evidence that the overexpression of NUP153C abrogates nuclear translocation of the viral genome, we used real-time Alu-PCR to quantitate the level of provirus formation in the infected cells (86). Our results demonstrate a significant reduction (P = 0.0002) in the number of integration events in the cells expressing NUP153C compared to that of the cells transfected with the empty vector (Fig. 5D).
DISCUSSION
We have used an in vitro nuclear import assay to better characterize the nuclear import pathway of HIV-1 IN, which may help define the import mechanism of the PIC. We observed that HIV-1 IN is actively transported into the nuclei of semipermeabilized cells independent of the classical receptor-mediated nuclear import pathway that requires both cytosolic extract and ATP. Although the nuclear transport of IN is distinct from the classical import pathway, the mechanism is still temperature sensitive, indicating that the protein does not passively diffuse across the nuclear envelope. In addition to being temperature sensitive, WGA effectively inhibited the nuclear accumulation of IN, indicating a role for the NPC. In further support of a role for constituents of the NPC in IN nuclear import, we demonstrated an interaction between IN and NUP153C.
The direct interaction between IN and NUP153 offers a plausible mechanism by which the NPC promotes the nuclear transport of IN in the absence of import factors. The FxFG-rich C-terminal domain of NUP153 interacts not only with imp α/β import complexes (reviewed in reference 9) but also with a subset of proteins that undergo import receptor-independent nuclear entry, including the transcription factors PU.1 (104), Stat1 (63), Smad2 (99), and the kinase ERK2 (95). The active nuclear import of IN via NUP153 may also shed light on the mechanism for PIC nuclear import by providing a contact between the subviral particle and the NPC, the gateway to the nucleus in the interphase cell (reviewed in reference 24).
It is important to acknowledge that the relevance of in vitro assays using recombinant, purified proteins can be compromised by potential differences between the purified IN and the IN contained in the PIC. However, the importance of the IN-NUP153 interaction in the nuclear import of the PIC is supported by the observation that overexpression of NUP153C inhibits infection by an HIV-1 vector. When overexpressed, the subcellular distribution of NUP153C is cytoplasmic (10), which would allow the nucleoporin fragment to exert its negative effect on the viral life cycle by interacting with the incoming viral particle and interfering with its trafficking into the nucleus. In support of this interpretation, we find that both nuclear accumulation of the viral cDNA and provirus integration are reduced in cells expressing NUP153C (Fig. 5). Consistent with our data, two independent screening studies using small interfering RNA (siRNA) knockdown identified NUP153 as a cellular factor that functions during the early steps of viral replication (14, 50). Furthermore, siRNA depletion of NUP153 results in decreased nuclear transport of the viral cDNA (50).
We postulate that the direct interaction between PIC-associated IN and NUP153C is a key event in the nuclear import of the virus. Such a role for NUP153 in PIC nuclear import is consistent with its role in the transport of various cargos between the cytoplasm and nucleus. NUP153C contains numerous FxFG repeats that function to bind to diverse nuclear import complexes via conserved motifs present on import receptor proteins (11). During nucleocytoplasmic transport, the repeat sequences are thought to attribute a dual function to the NPC, termed “selective gating” (55). Selective gating refers to the ability of the FxFG repeats to provide a transient but specific binding platform for import complexes during passage through the NPC while, at the same time, functioning as a physical barrier to restrict passage of macromolecules in the absence of import receptor binding (reviewed in reference 24). Although direct evidence describing the movement of import complexes through the NPC is lacking, there are descriptive comparisons of import complexes “stepping” (75) and “sliding” (61, 72) over FxFG repeats. Alternatively, the “selective phase” model (40, 76) suggests that FxFG domains form a gel-like barrier through which only receptor-associated cargos can dissolve. In both models, the FxFG repeats of NUP153 and other nucleoporins present a formidable entropic barrier for passage through the NPC and entry into the nucleus. Thus, the ability of the PIC-associated IN to interact with the FxFG repeats of NUP153 may serve to overcome this barrier and promote the transport of the viral cDNA into the nucleus.
While our model highlights the potential role of NUP153 in the nuclear import of HIV-1, it does not formally exclude the possible role of other nucleoporins in this step of the viral life cycle. Transport of a large complex like the PIC may very well require multiple contacts between PIC-associated factors and NPC constituents. In addition to NUP153, the siRNA screening study identified several nucleoporins that are essential host factors for HIV-1 replication (14). Whether the finding indicates that the overall integral structure of the NPC is essential for nuclear entry of the virus or that the subviral complex targets these nucleoporins directly is not known. It is also worth noting that the nucleoporins NUP107, NUP133, and NUP160, identified as essential host factors for HIV-1 replication (14, 50), interact or form complexes with NUP153 (93). Another nucleoporin that may also be involved in the active nuclear import of HIV-1 PICs is NUP98, a GLFG repeat-containing nucleoporin (32). Additionally, NUP62 and NUP358 are FxFG nucleoporins that are known to make significant contact with import complexes (12) and may also be targeted by the PIC during nuclear translocation. Conversely, it is possible that interaction between NUP153 and the PIC may comprise more than one viral component. The viral protein Vpr has also been shown to interact with NUP153 amino acids 447 to 634 in the protein's central domain (92), but the role of this interaction in the nuclear transport of Vpr or the functional significance in viral replication has not been described. Also, given the finding that deletion of Vpr from the virus fails to abrogate infection in nondividing cells (51), the Vpr-NUP153 interaction may not represent a key aspect of the nuclear import process for the PIC.
Recent reports have suggested that CA and uncoating of the viral core particle are essential to the nuclear import of the PIC (30, 100, 102, 103). Since HIV-1 CA is not reported to be karyophilic, these findings suggest that proper uncoating of the core particle may be essential to the appropriate presentation of the karyophilic determinant within the PIC. In our model, we propose that IN is the PIC constituent that facilitates an interaction with NUP153 to achieve the nuclear import of the subviral particle. In the event of aberrant uncoating, the ability of IN to interact with NUP153 may be altered and the import of the complex inhibited. The important role of uncoating in viral replication has been demonstrated for diverse viruses (reviewed in reference 43). As exemplified in the nuclear import step of adenovirus type 2 (Ad2) DNA (87), docking of the viral particle at the NPC triggers particle dissociation and release of the viral DNA for subsequent nuclear import. While some Ad2 capsid proteins translocate into the nucleus during the process of dissociation, nuclear import signals found on proteins associated with the Ad2 DNA facilitate the nuclear import of the nucleoprotein complex. A similar scenario may be relevant to the nuclear import of HIV-1, as localization of uncoating has been placed at the NPC (5). Whether proteins associated with the NPC can function as positive factors for uncoating HIV-1 is not known.
Although several studies have shown that lentiviral INs are karyophilic (6, 13, 27, 28, 41, 46, 73, 88, 89, 97), some reports observe the nuclear exclusion of HIV-1 IN in the absence of LEDGF/p75 (58, 62, 91), suggesting that LEDGF/p75 functions as a nuclear factor that promotes the nuclear accumulation of IN. In light of our data, we propose that LEDGF/p75 provides a mechanism by which IN can be released from NUP153 into the nucleoplasm. Protein substrates of the receptor-mediated nuclear import pathways are unloaded in the nucleus as a result of Ran-GTP binding to the imp β component of the transport complex. Because nuclear import of IN is independent of the Ran-GTP/Ran-GDP cycle, we would not predict a Ran-GTP-dependent dissociation of IN from NUP153 in the nucleus. An alternative model for dissociating IN from NUP153 and releasing IN into the nucleus would be competitive binding to IN by LEDGF/p75. In the context of the PIC, such a role for LEDGF/p75 would be compatible with the finding that LEDGF/p75 tethers the PIC to chromatin via its interaction with PIC-associated IN (33, 82).
The important role of NUP153 in our import model for HIV-1 IN may also explain the inconsistencies reported previously regarding the requirement of import receptors for the nuclear import of IN (6, 27, 41, 46). The C-terminal domain of NUP153 that interacts with IN is unique among nucleoporins in that it displays variable topology within the NPC (36). Depending on the transport status of the pore, the localization of NUP153C can range from the nuclear basket to the cytoplasmic face of the NPC (69, 70). Increased transport activity through the NPC results in increasing exposure of NUP153C to the cytoplasm (reviewed in reference 9). A previous report (41) observed that imp α/β stimulating the nuclear import of IN may not be the result of a direct interaction between IN and the import receptors. One alternative explanation is that adding these import receptors indirectly causes an overall stimulation of transport activity (69), thereby leading to increased exposure of NUP153C at the cytoplasmic face of the NPC and facilitating IN′s interaction with the nucleoporin for nuclear import.
Another important aspect of a functional role for NUP153 in the nuclear import of the PIC is the intimate relationship between this nucleoporin and mRNA export (reviewed in reference 9). The cytoplasmic exposure of the C-terminal domain of NUP153 is increased as a result of active transport of mRNA from the nucleus to the cytoplasm (7, 8, 45, 69, 70). Conversely, the inhibition of transcription can constrain NUP153C to the nuclear side of the NPC. These observations together with the growing evidence that pore proteins may couple the NPC to chromosomal regions active in transcription (reviewed in references 2, 20) argue that NUP153 plays a role in nuclear transport that extends beyond shuttling cargos between the nucleus and cytoplasm. In the context of our model of HIV-1 PIC nuclear import, the conformational characteristics of NUP153's C-terminal domain (i.e., cytoplasmic exposure) as related to its function in mRNA export may contribute to the preferential import of the PIC through an NPC that is exporting mRNA from a transcriptionally active region of the genome. One possible consequence is that the trafficking of the PIC may be biased to transcriptionally active sites in the genome for integration.
Also noteworthy in the context of mRNA biogenesis and the nuclear import of the PIC is the identification of transportin 3 (TNPO3), a nuclear transport receptor, as a positive factor for PIC nuclear import (14, 22). It is proposed that TNPO3 promotes the nuclear import of the PIC by binding to PIC-associated IN (22). Based on its role as a transport receptor for regulators of mRNA splicing factors (35, 52), TNPO3 may represent a cellular factor that acts in concert with the NPC to comprise the pathway of nuclear entry for the PIC. Alternatively, TNPO3 may promote the nuclear import of regulators of mRNA splicing that are required for the splicing and expression of another cellular protein that promotes the nuclear import of the PIC. Another possible explanation is that TNPO3 knockdown may result in altered splicing and processing of mRNA for export (90), which in turn interferes with the functional and conformation status of the NPC, leading to reduced transport of the virus through the NPC. Further experiments are required to better understand the role of TNPO3 and the relationship among IN, TNPO3, and NUP153 during the nuclear import of HIV-1.
Our study reveals a novel nuclear import pathway for IN and suggests that the direct interaction between IN and the nucleoporin NUP153 plays a pivotal role in the nuclear entry of the PIC during viral replication. Our report brings to light an overlooked factor potentially regulating the nuclear import of the PIC, namely, the functional role of the NPC. Of particular interest are the structural and conformational characteristics of the nucleoporins that are related to the transport function of NPC and how they may affect the nuclear import of the HIV-1 PIC in nondividing cells.
Acknowledgments
We thank Larry Gerace (The Scripps Research Institute) for helpful discussions and the GST-NUP153C plasmid, Mary S. Moore (Ross University) for NLS-BSA-TRITC, Irvin Chen for the HIV-1-based vector, Xin Liu and Hong Wu for the use of the fluorescence microscope, and Rick Bushman (University of Pennsylvania School of Medicine) for comments on the manuscript.
Flow cytometry was performed at the UCLA Jonsson Comprehensive Cancer Center (JCCC) and the Center for AIDS Research Flow Cytometry Core Facility, supported by National Institutes of Health (NIH) awards CA16042 and AI28697 and by the JCCC, the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA. This work was supported by NIH grant CA68859 to S.A.C. C.L.W. was a recipient of a Cota-Robles Fellowship and a Dissertation Year Fellowship award from the UCLA Graduate Division.
Footnotes
Published ahead of print on 15 April 2009.
REFERENCES
- 1.Adam, S. A., R. Sterne-Marr, and L. Gerace. 1992. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 21997-110. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar, A., and S. M. Gasser. 2007. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8507-517. [DOI] [PubMed] [Google Scholar]
- 3.An, D. S., Y. Xie, and I. S. Chen. 2001. Envelope gene of the human endogenous retrovirus HERV-W encodes a functional retrovirus envelope. J. Virol. 753488-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appa, R. S., C. G. Shin, P. Lee, and S. A. Chow. 2001. Role of the nonspecific DNA-binding region and alpha helices within the core domain of retroviral integrase in selecting target DNA sites for integration. J. Biol. Chem. 27645848-45855. [DOI] [PubMed] [Google Scholar]
- 5.Arhel, N. J., S. Souquere-Besse, S. Munier, P. Souque, S. Guadagnini, S. Rutherford, M. C. Prevost, T. D. Allen, and P. Charneau. 2007. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 263025-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armon-Omer, A., A. Graessmann, and A. Loyter. 2004. A synthetic peptide bearing the HIV-1 integrase 161-173 amino acid residues mediates active nuclear import and binding to importin alpha: characterization of a functional nuclear localization signal. J. Mol. Biol. 3361117-1128. [DOI] [PubMed] [Google Scholar]
- 7.Ball, J. R., C. Dimaano, A. Bilak, E. Kurchan, M. T. Zundel, and K. S. Ullman. 2007. Sequence preference in RNA recognition by the nucleoporin Nup153. J. Biol. Chem. 2828734-8740. [DOI] [PubMed] [Google Scholar]
- 8.Ball, J. R., C. Dimaano, and K. S. Ullman. 2004. The RNA binding domain within the nucleoporin Nup153 associates preferentially with single-stranded RNA. RNA 1019-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ball, J. R., and K. S. Ullman. 2005. Versatility at the nuclear pore complex: lessons learned from the nucleoporin Nup153. Chromosoma 114319-330. [DOI] [PubMed] [Google Scholar]
- 10.Bastos, R., A. Lin, M. Enarson, and B. Burke. 1996. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 1341141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayliss, R., T. Littlewood, and M. Stewart. 2000. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 10299-108. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Efraim, I., and L. Gerace. 2001. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J. Cell Biol. 152411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 71025-1035. [DOI] [PubMed] [Google Scholar]
- 14.Brass, A. L., D. M. Dykxhoorn, Y. Benita, N. Yan, A. Engelman, R. J. Xavier, J. Lieberman, and S. J. Elledge. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319921-926. [DOI] [PubMed] [Google Scholar]
- 15.Brown, P. O. 1997. Integration, p. 161-203. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Bukovsky, A., and H. Gottlinger. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 706820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 906125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7631-634. [DOI] [PubMed] [Google Scholar]
- 19.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 763739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117427-439. [DOI] [PubMed] [Google Scholar]
- 21.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278372-381. [DOI] [PubMed] [Google Scholar]
- 22.Christ, F., W. Thys, J. De Rijck, R. Gijsbers, A. Albanese, D. Arosio, S. Emiliani, J. C. Rain, R. Benarous, A. Cereseto, and Z. Debyser. 2008. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 181192-1202. [DOI] [PubMed] [Google Scholar]
- 23.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 111287-1289. [DOI] [PubMed] [Google Scholar]
- 24.Cook, A., F. Bono, M. Jinek, and E. Conti. 2007. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76647-671. [DOI] [PubMed] [Google Scholar]
- 25.Cushman, I., B. R. Bowman, M. E. Sowa, O. Lichtarge, F. A. Quiocho, and M. S. Moore. 2004. Computational and biochemical identification of a nuclear pore complex binding site on the nuclear transport carrier NTF2. J. Mol. Biol. 344303-310. [DOI] [PubMed] [Google Scholar]
- 26.Daigle, N., J. Beaudouin, L. Hartnell, G. Imreh, E. Hallberg, J. Lippincott-Schwartz, and J. Ellenberg. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 15471-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Depienne, C., A. Mousnier, H. Leh, E. Le Rouzic, D. Dormont, S. Benichou, and C. Dargemont. 2001. Characterization of the nuclear import pathway for HIV-1 integrase. J. Biol. Chem. 27618102-18107. [DOI] [PubMed] [Google Scholar]
- 28.Depienne, C., P. Roques, C. Creminon, L. Fritsch, R. Casseron, D. Dormont, C. Dargemont, and S. Benichou. 2000. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260387-395. [DOI] [PubMed] [Google Scholar]
- 29.Deprez, E., P. Tauc, H. Leh, J. F. Mouscadet, C. Auclair, and J. C. Brochon. 2000. Oligomeric states of the HIV-1 integrase as measured by time-resolved fluorescence anisotropy. Biochemistry 399275-9284. [DOI] [PubMed] [Google Scholar]
- 30.Dismuke, D. J., and C. Aiken. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 803712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 7612087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ebina, H., J. Aoki, S. Hatta, T. Yoshida, and Y. Koyanagi. 2004. Role of Nup98 in nuclear entry of human immunodeficiency virus type 1 cDNA. Microbes Infect. 6715-724. [DOI] [PubMed] [Google Scholar]
- 33.Emiliani, S., A. Mousnier, K. Busschots, M. Maroun, B. Van Maele, D. Tempe, L. Vandekerckhove, F. Moisant, L. Ben-Slama, M. Witvrouw, F. Christ, J. C. Rain, C. Dargemont, Z. Debyser, and R. Benarous. 2005. Integrase mutants defective for interaction with LEDGF/P75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 28025517-25523. [DOI] [PubMed] [Google Scholar]
- 34.Engelman, A., F. D. Bushman, and R. Craigie. 1993. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 123269-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erkmann, J. A., E. J. Wagner, J. Dong, Y. Zhang, U. Kutay, and W. F. Marzluff. 2005. Nuclear import of the stem-loop binding protein and localization during the cell cycle. Mol. Biol. Cell 162960-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fahrenkrog, B., B. Maco, A. M. Fager, J. Koser, U. Sauder, K. S. Ullman, and U. Aebi. 2002. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 140254-267. [DOI] [PubMed] [Google Scholar]
- 37.Fassati, A. 2006. HIV infection of non-dividing cells: a divisive problem. Retrovirology 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fassati, A., D. Gorlich, I. Harrison, L. Zaytseva, and J. M. Mingot. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 223675-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finlay, D. R., D. D. Newmeyer, T. M. Price, and D. J. Forbes. 1987. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 104189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frey, S., and D. Gorlich. 2007. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130512-523. [DOI] [PubMed] [Google Scholar]
- 41.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of non-dividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 949825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao, K., S. L. Butler, and F. Bushman. 2001. Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. EMBO J. 203565-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greber, U. F., and A. Fassati. 2003. Nuclear import of viral DNA genomes. Traffic 4136-143. [DOI] [PubMed] [Google Scholar]
- 44.Greber, U. F., and M. Fornerod. 2005. Nuclear import in viral infections. Curr. Top. Microbiol. Immunol. 285109-138. [DOI] [PubMed] [Google Scholar]
- 45.Griffis, E. R., B. Craige, C. Dimaano, K. S. Ullman, and M. A. Powers. 2004. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol. Biol. Cell 151991-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hearps, A. C., and D. A. Jans. 2006. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta-dependent mechanism. Biochem. J. 398475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikeda, T., H. Nishitsuji, X. Zhou, N. Nara, T. Ohashi, M. Kannagi, and T. Masuda. 2004. Evaluation of the functional involvement of human immunodeficiency virus type 1 integrase in nuclear import of viral cDNA during acute infection. J. Virol. 7811563-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katz, R. A., J. G. Greger, and A. M. Skalka. 2005. Effects of cell cycle status on early events in retroviral replication. J. Cell. Biochem. 94880-889. [DOI] [PubMed] [Google Scholar]
- 49.Kiseleva, E., M. W. Goldberg, J. Cronshaw, and T. D. Allen. 2000. The nuclear pore complex: structure, function, and dynamics. Crit. Rev. Eukaryot. Gene Expr. 10101-112. [PubMed] [Google Scholar]
- 50.Konig, R., Y. Zhou, D. Elleder, T. L. Diamond, G. M. Bonamy, J. T. Irelan, C. Y. Chiang, B. P. Tu, P. D. De Jesus, C. E. Lilley, S. Seidel, A. M. Opaluch, J. S. Caldwell, M. D. Weitzman, K. L. Kuhen, S. Bandyopadhyay, T. Ideker, A. P. Orth, L. J. Miraglia, F. D. Bushman, J. A. Young, and S. K. Chanda. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 13549-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kootstra, N. A., and H. Schuitemaker. 1999. Phenotype of HIV-1 lacking a functional nuclear localization signal in matrix protein of gag and Vpr is comparable to wild-type HIV-1 in primary macrophages. Virology 253170-180. [DOI] [PubMed] [Google Scholar]
- 52.Lai, M. C., R. I. Lin, S. Y. Huang, C. W. Tsai, and W. Y. Tarn. 2000. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 2757950-7957. [DOI] [PubMed] [Google Scholar]
- 53.Lee, S. P., J. Xiao, J. R. Knutson, M. S. Lewis, and M. K. Han. 1997. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry 36173-180. [DOI] [PubMed] [Google Scholar]
- 54.Li, M., and R. Craigie. 2006. Virology: HIV goes nuclear. Nature 441581-582. [DOI] [PubMed] [Google Scholar]
- 55.Lim, R. Y., B. Fahrenkrog, J. Koser, K. Schwarz-Herion, J. Deng, and U. Aebi. 2007. Nanomechanical basis of selective gating by the nuclear pore complex. Science 318640-643. [DOI] [PubMed] [Google Scholar]
- 56.Limón, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 7612078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Llano, M., D. T. Saenz, A. Meehan, P. Wongthida, M. Peretz, W. H. Walker, W. Teo, and E. M. Poeschla. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314461-464. [DOI] [PubMed] [Google Scholar]
- 58.Llano, M., M. Vanegas, O. Fregoso, D. Saenz, S. Chung, M. Peretz, and E. M. Poeschla. 2004. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 789524-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu, R., A. Limon, E. Devroe, P. A. Silver, P. Cherepanov, and A. Engelman. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 7812735-12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lyman, S. K., and L. Gerace. 2001. Nuclear pore complexes: dynamics in unexpected places. J. Cell Biol. 15417-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 27833528-33539. [DOI] [PubMed] [Google Scholar]
- 63.Marg, A., Y. Shan, T. Meyer, T. Meissner, M. Brandenburg, and U. Vinkemeier. 2004. Nucleocytoplasmic shuttling by nucleoporins Nup153 and Nup214 and CRM1-dependent nuclear export control the subcellular distribution of latent Stat1. J. Cell Biol. 165823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Melchior, F., T. Guan, N. Yokoyama, T. Nishimoto, and L. Gerace. 1995. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. J. Cell Biol. 131571-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore, M. S., and G. Blobel. 1992. The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell 69939-950. [DOI] [PubMed] [Google Scholar]
- 66.Moroianu, J., G. Blobel, and A. Radu. 1996. Nuclear protein import: Ran-GTP dissociates the karyopherin alphabeta heterodimer by displacing alpha from an overlapping binding site on beta. Proc. Natl. Acad. Sci. USA 937059-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakielny, S., S. Shaikh, B. Burke, and G. Dreyfuss. 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 181982-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paschal, B. M., and L. Gerace. 1995. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J. Cell Biol. 129925-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paulillo, S. M., E. M. Phillips, J. Koser, U. Sauder, K. S. Ullman, M. A. Powers, and B. Fahrenkrog. 2005. Nucleoporin domain topology is linked to the transport status of the nuclear pore complex. J. Mol. Biol. 351784-798. [DOI] [PubMed] [Google Scholar]
- 70.Paulillo, S. M., M. A. Powers, K. S. Ullman, and B. Fahrenkrog. 2006. Changes in nucleoporin domain topology in response to chemical effectors. J. Mol. Biol. 36339-50. [DOI] [PubMed] [Google Scholar]
- 71.Pauza, C. D., and J. Galindo. 1989. Persistent human immunodeficiency virus type 1 infection of monoblastoid cells leads to accumulation of self-integrated viral DNA and to production of defective virions. J. Virol. 633700-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peters, R. 2005. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic 6421-427. [DOI] [PubMed] [Google Scholar]
- 73.Pluymers, W., P. Cherepanov, D. Schols, E. De Clercq, and Z. Debyser. 1999. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258327-332. [DOI] [PubMed] [Google Scholar]
- 74.Quillent, C., A. M. Borman, S. Paulous, C. Dauguet, and F. Clavel. 1996. Extensive regions of pol are required for efficient human immunodeficiency virus polyprotein processing and particle maturation. Virology 21929-36. [DOI] [PubMed] [Google Scholar]
- 75.Rexach, M., and G. Blobel. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83683-692. [DOI] [PubMed] [Google Scholar]
- 76.Ribbeck, K., and D. Gorlich. 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 212664-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ribbeck, K., G. Lipowsky, H. M. Kent, M. Stewart, and D. Gorlich. 1998. NTF2 mediates nuclear import of Ran. EMBO J. 176587-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwoebel, E. D., T. H. Ho, and M. S. Moore. 2002. The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J. Cell Biol. 157963-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shah, S., and D. J. Forbes. 1998. Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr. Biol. 81376-1386. [DOI] [PubMed] [Google Scholar]
- 80.Sherman, M. P., and W. C. Greene. 2002. Slipping through the door: HIV entry into the nucleus. Microbes Infect. 467-73. [DOI] [PubMed] [Google Scholar]
- 81.Shibagaki, Y., and S. A. Chow. 1997. Central core domain of retroviral integrase is responsible for target site selection. J. Biol. Chem. 2728361-8369. [DOI] [PubMed] [Google Scholar]
- 82.Shun, M.-C., N. K. Raghavendra, N. Vandegraaff, J. E. Daigle, S. Hughes, P. Kellam, P. Cherepanov, and A. Engelman. 2007. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 211767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suzuki, Y., and R. Craigie. 2007. The road to chromatin—nuclear entry of retroviruses. Nat. Rev. Microbiol. 5187-196. [DOI] [PubMed] [Google Scholar]
- 84.Talcott, B., and M. S. Moore. 1999. Getting across the nuclear pore complex. Trends Cell Biol. 9312-318. [DOI] [PubMed] [Google Scholar]
- 85.Tan, W., Z. Dong, T. A. Wilkinson, C. F. Barbas III, and S. A. Chow. 2006. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J. Virol. 801939-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tan, W., K. Zhu, D. J. Segal, C. F. Barbas III, and S. A. Chow. 2004. Fusion proteins consisting of human immunodeficiency virus type 1 integrase and the designed polydactyl zinc finger protein E2C direct integration of viral DNA into specific sites. J. Virol. 781301-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trotman, L. C., N. Mosberger, M. Fornerod, R. P. Stidwill, and U. F. Greber. 2001. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 31092-1100. [DOI] [PubMed] [Google Scholar]
- 88.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 744795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsurutani, N., J. Yasuda, N. Yamamoto, B. I. Choi, M. Kadoki, and Y. Iwakura. 2007. Nuclear import of the preintegration complex is blocked upon infection by human immunodeficiency virus type 1 in mouse cells. J. Virol. 81677-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Valencia, P., A. P. Dias, and R. Reed. 2008. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. USA 1053386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vanegas, M., M. Llano, S. Delgado, D. Thompson, M. Peretz, and E. Poeschla. 2005. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 1181733-1743. [DOI] [PubMed] [Google Scholar]
- 92.Varadarajan, P., S. Mahalingam, P. Liu, S. B. Ng, S. Gandotra, D. S. Dorairajoo, and D. Balasundaram. 2005. The functionally conserved nucleoporins Nup124p from fission yeast and the human Nup153 mediate nuclear import and activity of the Tf1 retrotransposon and HIV-1 Vpr. Mol. Biol. Cell 161823-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vasu, S., S. Shah, A. Orjalo, M. Park, W. H. Fischer, and D. J. Forbes. 2001. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155339-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei, X., V. G. Henke, C. Strubing, E. B. Brown, and D. E. Clapham. 2003. Real-time imaging of nuclear permeation by EGFP in single intact cells. Biophys. J. 841317-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Whitehurst, A. W., J. L. Wilsbacher, Y. You, K. Luby-Phelps, M. S. Moore, and M. H. Cobb. 2002. ERK2 enters the nucleus by a carrier-independent mechanism. Proc. Natl. Acad. Sci. USA 997496-7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wolff, B., M. C. Willingham, and J. A. Hanover. 1988. Nuclear protein import: specificity for transport across the nuclear pore. Exp. Cell Res. 178318-334. [DOI] [PubMed] [Google Scholar]
- 97.Woodward, C. L., Y. Wang, W. J. Dixon, H. Htun, and S. A. Chow. 2003. Subcellular localization of feline immunodeficiency virus integrase and mapping of its karyophilic determinant. J. Virol. 774516-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu, X., H. Liu, H. Xiao, J. A. Conway, E. Hehl, G. V. Kalpana, V. Prasad, and J. C. Kappes. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 732126-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu, L., Y. Kang, S. Col, and J. Massague. 2002. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFbeta signaling complexes in the cytoplasm and nucleus. Mol. Cell 10271-282. [DOI] [PubMed] [Google Scholar]
- 100.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 785670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamashita, M., and M. Emerman. 2006. Retroviral infection of non-dividing cells: old and new perspectives. Virology 34488-93. [DOI] [PubMed] [Google Scholar]
- 102.Yamashita, M., and M. Emerman. 2005. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 1e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamashita, M., O. Perez, T. J. Hope, and M. Emerman. 2007. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 31502-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhong, H., A. Takeda, R. Nazari, H. Shio, G. Blobel, and N. R. Yaseen. 2005. Carrier-independent nuclear import of the transcription factor PU. 1 via RanGTP-stimulated binding to Nup153. J. Biol. Chem. 28010675-10682. [DOI] [PubMed] [Google Scholar]
- 105.Zhu, K., C. Dobard, and S. A. Chow. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 785045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]