Abstract
The ability of interferon (IFN) to induce the expression of antiviral genes, and therefore suppress viral infection, is dependent on the activity of cellular suppressors. The Ras/MEK pathway is one of these cellular suppressors, since the activation of Ras/MEK permits viral replication in the presence of alpha IFN (IFN-α). Here, we have investigated the mechanism by which activated Ras/MEK inhibits the IFN-α response. We found that the induction of antiviral proteins in response to IFN-α was impaired in Ras-transformed NIH 3T3 (RasV12) cells. The inhibition of the Ras/MEK pathway restored the IFN-mediated induction of antiviral genes, indicating that activated Ras interrupts the IFN pathway upstream of antiviral gene transcription. Indeed, the IFN-induced phosphorylation of signal transducer and activator of transcription 1 (STAT1) and STAT2 was inhibited in RasV12 cells compared to that of vector control cells. In addition, we found that the total amount of STAT2 was reduced in RasV12 cells. To determine if the impaired IFN-α response can be rescued by restoring the overall level of STAT2, we overexpressed STAT2 in RasV12 cells. The IFN-α-induced phosphorylation of STAT1 and STAT2, as well as the expression of antiviral protein, were restored, and IFN-induced antiviral protection was partially restored. Moreover, we demonstrated that the downregulation of STAT2 levels by Ras/MEK was mediated at the transcriptional level. Thus, the activation of the Ras/MEK pathway reduces the amount of STAT2 available for propagating the IFN signal, resulting in the impairment of the IFN-α-induced antiviral response.
The cellular antiviral state mediated by type I interferon (IFN) is the most important host defense mechanism occurring at the early stage of virus infection (15, 42, 45). IFN binds to the IFN-α receptor (IFNAR), which consists of two subunits, IFNAR1 and IFNAR2 (34). The binding of IFN leads to the heterodimerization of the two subunits and the subsequent phosphorylation of two tyrosine kinases, Janus kinase 1 (Jak1) and tyrosine kinase 2 (Tyk2), which are associated with the intracellular domains of the IFNAR (42, 43). Phosphorylated Jak1 and Tyk2, in turn, phosphorylate signal transducer and activator of transcription 1 (STAT1) and STAT2, which are downstream transcriptional factors located in the cytoplasm (9). Once phosphorylated, STAT1 and STAT2 form a trimeric complex with the DNA binding protein, IFN regulatory factor 9, termed IFN-stimulated gene factor 3 (ISGF3) (20, 27). ISGF3 then translocates to the nucleus, where it binds to specific promoter elements of IFN-inducible genes (the IFN-stimulated response element) and induces the expression of hundreds of IFN-inducible genes that have antiviral and immunoregulatory functions (10, 14). However, IFN does not always induce the antiviral response effectively. The efficacy of IFN can be limited by anti-IFN proteins encoded in viral genomes or by host cellular suppressors regulating IFN signaling (24, 28, 50). Even IFN-sensitive viruses (not armed with anti-IFN genes) cause significant diseases in humans and animals (31), indicating that cellular suppressors of the IFN pathway are important in defining susceptibility to infection and viral tropism.
Ras is a membrane-bound GTP binding protein that is essential for the regulation of several biological processes, including proliferation, transformation, and differentiation (6, 11). It is believed that Ras plays multiple roles in the promotion of viral replication (1, 2, 12, 23, 29, 32, 37, 38). The deregulation of Ras is a common target of several oncolytic viruses (1, 4, 7, 12). Activated Ras has been implicated in the negative regulation of the IFN response. The activation of K-Ras suppresses the IFN-γ-activated sequence-mediated transcription of IFN-γ in human cancer cells (22). When BALB/c-3T3 cells are transfected with viral oncogene (v-Ras), the induction of major histocompatibility complex class I by IFN-α is inhibited (35). It also has been reported that the antiviral protein PKR is not fully functional in cells with activated Ras (4, 30, 47). Finally, we and other researchers have demonstrated that the activation of Ras and its downstream elements, Raf and mitogen-activated protein kinase kinase (MEK), suppress the IFN-α-induced antiviral responses (3, 33). IFN normally acts to protect cells from virus infection. However, in NIH 3T3 cells expressing constitutively activated Ras/MEK, viruses are able to replicate efficiently even in the presence of IFN (3). Noser et al. (33) also reported the interaction between the Ras/MEK and IFN pathways in human cancer cell lines. These two studies demonstrate that the Ras/MEK pathway is a cellular suppressor of the IFN pathway and that the suppression of the IFN response by activated Ras can be a common mechanism that is exploited by some oncolytic viruses.
Until now, it was unknown which component of the IFN pathway is inhibited by the activated Ras/MEK pathway. Here, we demonstrate that the activation of Ras/MEK reduces the total amount of STAT2 by suppressing STAT2 transcription, leading to the impairment of STAT2 activation and, therefore, impairment in establishing the antiviral state.
MATERIALS AND METHODS
Cells, viruses, and reagents.
Murine fibroblast cell lines, NIH 3T3 and L929 cells, were obtained from the American Type Culture Collection. All cell lines used in this study were maintained in high-glucose Dulbecco's modified essential medium (DMEM) (Invitrogen, Burlington, Ontario, Canada) with 10% fetal bovine serum (FBS) (Cansera, Etobicoke, Ontario, Canada). The activated Ras mutant (H-Ras) construct in the pBABE retroviral vector and mouse STAT2 construct in the MSCV retroviral vector were generously provided by Patrick Lee (Dalhousie University, Halifax, Canada) (32) and David Farrar (University of Texas Southwestern Medical Center, Dallas) (13), respectively. The retroviral vectors containing the mutant genes were transfected into Bosc23 packaging cells by using Superfect (Qiagen, Valencia, CA) according to the manufacturer's instructions. Supernatants containing retroviruses were harvested at 48 h after transfection, filtered, and stored at −70°C. NIH 3T3 cells infected with pBABE retroviruses were selected with 2 μg of puromycin/ml (Sigma-Aldrich, St. Louis, MO) for 2 weeks. The cells infected with an MSCV retrovirus were purified by flow-cytometric sorting based on green fluorescent protein (GFP). After purification, we confirmed that more than 95% of the cells expressed GFP. Vesicular stomatitis virus (VSV) (Indiana strain; provided by J. C. Bell, University of Ottawa, Ottawa, Canada) (46) was amplified, and its titer was determined, using L929 cells. Recombinant mouse IFN-α and actinomycin D (ActD) were obtained form Sigma-Aldrich, and recombinant mouse IFN-γ was from Peprotech (Rocky Hill, NJ). U0126, PD98059, SL327 (MEK1/2 inhibitor), MG-132, and cycloheximide (CHX) were purchased from Calbiochem (La Jolla, CA). Two RNA interference (RNAi) oligonucleotides corresponding to nucleotide sequences of H-Ras (5′-CCA CUA UAG AGG AUU CCU ACC GGA A-3′ [positions 289 to 314] and 5′-CCU GUG UGU GUU UGC CAU AAC AAC-3′ [positions 422 to 446]) were synthesized by Invitrogen. RNAi oligonucleotides to ERK1 (sc-29308) and ERK2 (sc-35336) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and mixed 1:1 for the efficient targeting of both ERK1 and ERK2. Two nontargeting control RNA oligonucleotides comprised of random nucleotide sequences were obtained from Invitrogen and Dharmacon (Lafayette, CO) and were used as controls for nonspecific effects due to the transfection of duplex RNA. Antibodies to phosphorylated STAT1, total STAT1, phosphorylated STAT2, total STAT2, and phosphorylated ERK1/2 were obtained from Upstate (Lake Placid, NY), VSV-G protein was from Alpha Diagnostic (San Antonio, TX), total ERK was from Santa Cruz Biotechnology, β-catenin was from BD Bioscience (Mississauga, ON), actin was from Sigma-Aldrich, and Ras was from Cell Signaling Technology (Beverely, MA). The antibody to 2′5′-oligoadenylate synthetase (OAS) was provided by Yoshihiro Sokawa (Kyoto Institute of Technology, Kyoto, Japan) (52).
Cell culture and virus infection.
Cells were plated in 6-well plates (for Western blot analysis and reverse transcription-PCR [RT-PCR]), 24-well plates (for the virus progeny assay), or 10-cm dishes (for Northern blot analysis). For RNAi treatment, the cells at 40 to 50% confluence were washed twice with phosphate-buffered saline (PBS) and then incubated with a transfection mixture containing DMEM without antibiotics, 10% FBS, 20 μg/ml Lipofectamine RNAiMAX (Invitrogen), and the oligonucleotides (final concentration range, 25 pmol). The transfection was repeated after 24 h for the greater suppression of target genes. For virus infection, the cells were untreated or were pretreated with IFN-α (500 U/ml) for 16 h and then challenged with VSV at a multiplicity of infection of 1 PFU/cell. The supernatant was harvested for a progeny virus assay. The viral concentrations of supernatants from the triplicate wells were determined by plaque assay as described before (17).
Northern blot analysis.
cDNA probes used for Northern blot analysis were generated using the following primers: OAS1a (5′-ATT ACC TCC TTC CCG ACA CC-3′ and 5′-GCA TCA GGA GGT GGA GTT TG-3′), Mx1 (5′-AAA CCT GAT CCG ACT TCA CTT CC-3′ and 5′-TCT TCT TCT CTC TGG TGT CAC TC-3′), and glyceraldehyde-3-phosphatase dehydrogenase (GAPDH) (5′-GGG TGG AGC CAA ACG GGT CA-3′ and 5′-TGC GAC TTC AAC AGC AAC TCC-3′). RNA was extracted from cells with TRIzol (Invitrogen). Ten micrograms of each RNA sample was denatured in Northern Max Formaldehyde Load Dye (Ambion Inc., Austin, TX) at 60°C for 10 min and then fractionated in 1% agarose gels containing 2.2 M formaldehyde gel and 1× 3-(N-morpholino) propansulfonic acid (MOPS) buffer in 1× MOPS-0.22 M formaldehyde running buffer. RNA was transferred overnight to a nylon membrane (Hybond-XL; Amersham Biosciences, Little Chalfont, Buckinghamshire, England) by upward capillary action using 2× saline sodium citrate (SSC; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer and then cross-linked to the membrane. cDNA probes were labeled with 32CTP using a Megaprime DNA labeling kit (Amersham Biosciences) and purified with a Nick column containing G50 Sephadex (Amersham Biosciences). Membranes were prehybridized in hybridization buffer (Ambion) for 1 h at 42°C. Hybridizations were performed overnight at 42°C. The membranes were washed at 42°C twice with 2× SSC-0.1% sodium dodecyl sulfate (SDS) and twice with 0.2× SSC-0.1% SDS, and then they were exposed to Kodak BioMax XAR scientific imaging film (Amersham Biosciences). The quantification of band intensities was carried out using Kodak molecular imaging software (Eastman Kodak Company).
RT-PCR.
RNA (0.5 μg) from each sample was reverse transcribed to cDNA with a random primer using the first-strand cDNA synthesis kit from Amersham Biosciences (Piscataway, NJ). PCRs using the cDNA were performed with Taq polymerase (Invitrogen) using optimal annealing temperatures as determined empirically for each primer set. PCR products were visualized by ethidium bromide staining after electrophoresis on a 1% agarose gel. Primer sequences are the following: STAT2 forward, 5′-GTC TTC AGA CCC CCA TCA GA-3′; STAT2 reverse, 5′-CTG CCT TCC TGG AGT CTC AC-3′; OAS forward, 5′-GGT TGG AGT GCC AAT GAA GT-3′; OAS reverse, 5′-ACT GTT GGG GGA CAA GAC AG-3′; STAT1 forward, 5′-TGG TGA AAT TGC AAG AGC TG-3′; STAT1 reverse, 5′-GGT CTG CGT TCA GAC CTC TC-3′; GAPDH forward, 5′-GGG TGG AGC CAA ACG GGT CA-3′; GAPDH reverse, 5′-GGA GGT GCT GTT GAA GTC GCA-3′; 18S forward, 5′-GTT GGT GGA GCG ATT TGT CT-3′; and 18S reverse, 5′-GGC CTC ACT AAA CCA TCC AA-3′.
Quantitative RT-PCR.
Quantitative PCR was performed in triplicate using the TaqMAN gene expression assay for STAT2 or 18S and analyzed on an ABI PRISM 7000 (Applied Biosystems, Foster City, CA). STAT2 mRNA levels were normalized to 18S levels, and then the relative amounts of STAT2 compared to those of the control were determined.
Western blot analysis.
Cells were washed in PBS and lysed in PBS containing 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 μg/ml aprotinin, 100 μg/ml phenylmethylsulfonyl fluoride, and 1% phosphatase inhibitor cocktail (Sigma). Protein samples were cleared of debris by centrifugation. The protein concentration was determined by the Bradford method (Bio-Rad, Mississauga, Ontario, Canada). The samples were subjected to 8 to 12% SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Bio-Rad). The membrane was blocked with 5% skim milk in TBS (20 mM Tris and 137 mM NaCl [pH 7.3]) containing 0.1% Tween 20 and then incubated with the primary antibodies listed above. After being washed, the membrane was incubated with peroxidase-conjugated goat anti-mouse immunoglobulin G or anti-rabbit immunoglobulin G (Santa Cruz), and specific bands were detected by ECL (Amersham, Baie d'Urfe, Quebec, Canada) as described previously (18). The quantification of band intensities was carried out using Kodak molecular imaging software.
Statistical analysis.
Statistical analyses were conducted with one-way analysis of variance (ANOVA).
RESULTS
Activation of the Ras/MEK pathway interferes with IFN-α-induced transcriptional responses.
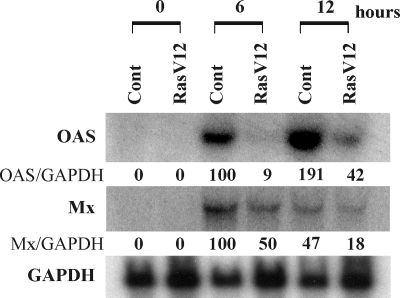
To determine whether activated Ras suppresses the induction of antiviral genes by IFN-α, vector control cells and cells that stably express constitutively activated Ras (RasV12) (3) were stimulated with IFN-α (500 U/ml) for 0, 6, and 12 h. At each time point, RNA samples were extracted for the Northern blot analysis of the IFN-α-induced antiviral genes OAS and Mx (myxovirus resistance gene), as well as for the housekeeping gene GAPDH (Fig. 1). In the absence of IFN-α stimulation, there was no expression of any antiviral gene. At 6 and 12 h after IFN-α treatment, the induction of OAS and Mx mRNA was observed in both control and RasV12 cells. Importantly, the expression levels of OAS and Mx mRNA upon IFN-α stimulation were lower in RasV12 cells than in vector control cells.
FIG. 1.
Activation of Ras interrupts induction of antiviral genes by IFN-α. Vector control cells (Cont) and RasV12 cells were stimulated with IFN-α (500 U/ml) for 0, 6, and 12 h. Northern blot analysis for OAS, Mx, and GAPDH mRNA was performed. The density ratios of OAS (OAS/GAPDH) and Mx (Mx/GAPDH) to GAPDH are shown as percentages normalized to values for vector control cells stimulated with IFN-α for 6 h.
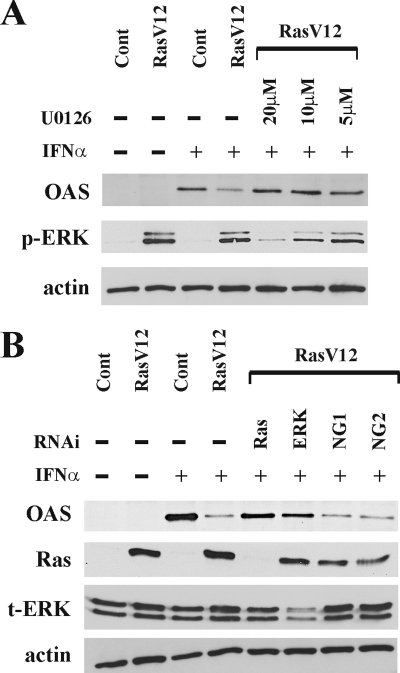
In our previous study, we determined that the Raf/MEK pathway downstream of Ras was responsible for the interruption of the IFN response (3). Therefore, we further investigated whether the inhibition of the Ras/MEK pathway could rescue the IFN-α transcriptional responses in Ras-activated cells. The protein expression level of the antiviral protein OAS was determined by Western blot analysis in RasV12 cells with the MEK inhibitor U0126 (Fig. 2A). Without IFN-α stimulation, there was no expression of OAS in vector control cells or RasV12 cells. Twenty-four hours after IFN-α stimulation, we observed a higher induction of OAS protein in vector control cells than in RasV12 cells. OAS induction was restored by pretreating RasV12 cells with U0126 for 16 h prior to IFN-α stimulation in a dose-dependent manner.
FIG. 2.
Induction of OAS by IFN-α is dependent on activation of the Ras/MEK pathway. (A) Vector control cells (Cont), RasV12 cells, and RasV12 cells pretreated with U0126 (20, 10, or 5 μM) for 16 h were stimulated with IFN-α for 24 h. Western blot analysis for OAS, phosphorylated ERK1/2 (p-ERK), and actin was performed. (B) Vector control cells (Cont), RasV12 cells, and RasV12 cells treated with RNAi to Ras, ERK1/2, or random nucleotide sequences from Invitrogen (NG1) or from Dharmacon (NG2) were stimulated with IFN-α for 24 h. The Western blot analysis for OAS, Ras, total ERK1/2 (t-ERK), and actin is shown.
To further confirm the involvement of the Ras/MEK/ERK pathway in inhibiting the induction of the antiviral protein OAS by IFN-α, Ras or ERK1/2 expression was knocked down by RNAi in RasV12 cells (Fig. 2B). The knockdown of the transfected active mutant H-Ras gene or endogenous ERK1/2 in RasV12 cells restored the induction of OAS after IFN-α stimulation. The efficient knockdown of the active mutant Ras or ERK1/2 was confirmed by Western blotting to be Ras or ERK1/2, respectively. To ensure that the RNAi results are not due to off-target effects, RasV12 cells were transfected with two independent, control RNA oligonucleotides (NG1 and NG2) that had been obtained from different sources. The control RNAi cells had reduced OAS induction in response to IFN-α. Taken together, these results clearly demonstrate that the induction of antiviral genes by IFN-α is negatively regulated by the activation of the Ras/MEK pathway.
The Ras/MEK pathway inhibits activation of STAT proteins induced by IFN-α.
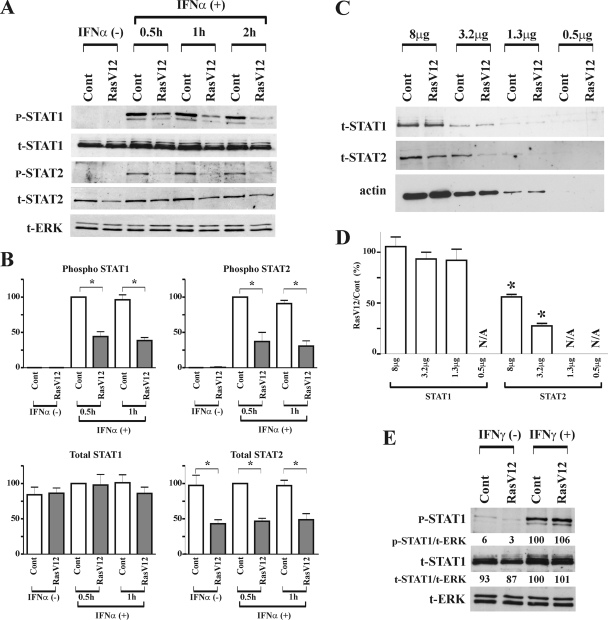
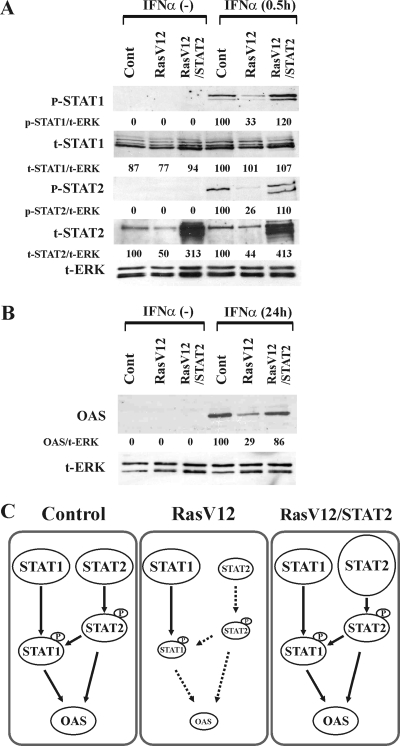
To identify the upstream element(s) of the IFN pathway targeted by the Ras/MEK pathway, we examined the activation status of STAT1 and STAT2, which directly mediate the IFN-α transcriptional response. At 0.5, 1, and 2 h after IFN-α stimulation, protein samples from vector control cells and RasV12 cells were analyzed by Western blotting (Fig. 3A). In the absence of IFN-α, the phosphorylation of STAT1 and STAT2 was not observed in vector control cells or RasV12 cells. While the phosphorylation of STAT1 and STAT2 occurred in both cells following IFN-α stimulation, the levels of phosphorylation were lower in RasV12 cells than in vector control cells at each time point. Interestingly, we also found that the total amount of STAT2, but not STAT1, was decreased in RasV12 cells. These results were found to be statistically significant based on a densitometry analysis of three independent experiments (Fig. 3B). To further confirm the reduction of STAT protein levels in RasV12 cells, we performed Western blot analysis of STAT1 and STAT2 levels on serially diluted samples (Fig. 3C and D). While STAT2 levels are markedly reduced in RasV12 cells, STAT1 levels are similar in vector control and RasV12 cells regardless of the amount of protein loaded (Fig. 3C). Supporting the conclusion that STAT1 levels are not altered in RasV12 cells, we found that IFN-γ induced a similar amount of STAT1 phosphorylation in both vector control cells and RasV12 cells (Fig. 3E).
FIG. 3.
Activation of Ras inhibits phosphorylation of STAT1 and STAT2 induced by IFN-α. (A) Vector control cells (Cont) and RasV12 cells were stimulated with IFN-α (500 U/ml) for 0, 0.5, 1, and 2 h. Western blot analysis for phosphorylated STAT1 (p-STAT1), total STAT1 (t-STAT1), phosphorylated STAT2 (p-STAT2), total STAT2 (t-STAT2), and total ERK (t-ERK) was performed. Representative images of three independent experiments are shown. (B) The total and phosphorylated amounts of STAT1 and STAT2 were quantified by densitometry analysis. The density ratios of t-STAT1, p-STAT1, t-STAT2, and p-STAT2 to t-ERK are shown as percentages normalized to values for vector control cells stimulated with IFN-α for 0.5 h. The results are means ± standard errors of the means (SEM) from three independent experiments. *, P < 0.05 by one-way ANOVA. (C) Decreasing amounts of total protein were analyzed by Western blot analysis for levels of t-STAT1 and t-STAT2 and for actin (as a loading control). (D) The total amounts of STAT1 and STAT2 were quantified by densitometry analysis. The levels of t-STAT1 and t-STAT2 were normalized to that for actin, and the relative amount of STAT protein in RasV12 compared to that of control cells is depicted graphically. The results are means ± SEM from three independent experiments. N/A, not applicable; *, P < 0.05 by one-way ANOVA. (E) Activation of Ras does not inhibit phosphorylation of STAT1 induced by IFN-γ. Vector control cells (Cont) and RasV12 cells were stimulated with IFN-γ (50 ng/ml) for 0.5 h. The density ratios of phosphorylated STAT1 (p-STAT1/t-ERK) and total STAT1 (t-STAT1/t-ERK) to total ERK are shown as percentages normalized to values for vector control cells stimulated with IFN-γ.
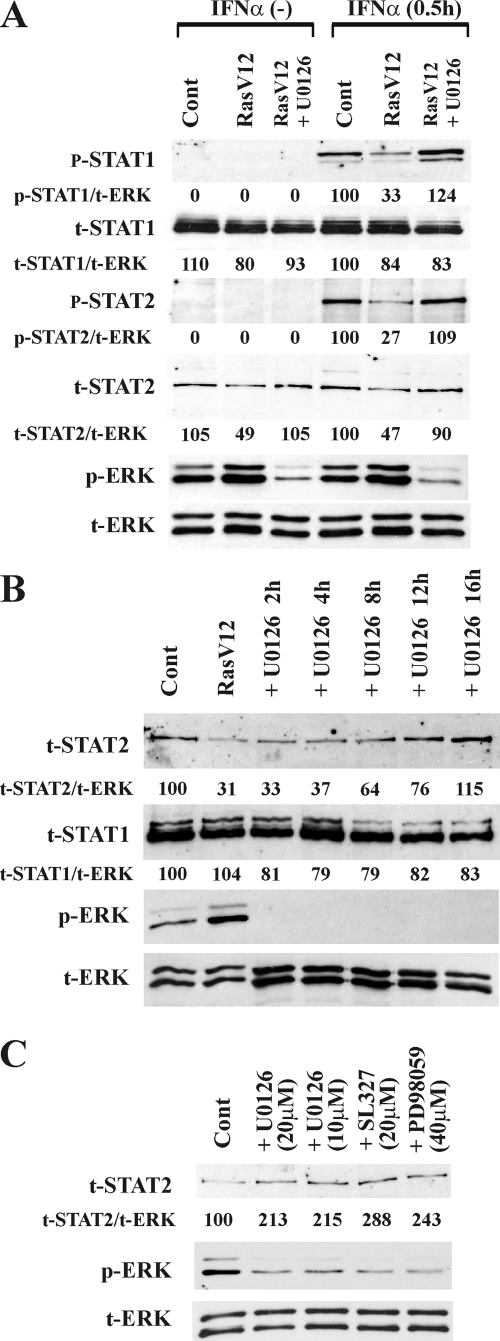
We next determined whether the inhibition of the Ras/MEK pathway could rescue the activation of the STAT proteins induced by IFN-α in RasV12 cells. RasV12 cells were pretreated with U0126 for 16 h and then stimulated with IFN-α (Fig. 4A). In response to IFN-α stimulation, the levels of phosphorylated STAT1 and STAT2 were restored to the same level as that in vector control cells. In addition, the total amount of STAT2 also was restored to the same level as that of the control after treatment with U0126 in the absence of IFN-α. The knockdown of the active mutant Ras by RNAi in RasV12 cells also restored both the total level of STAT2 and the degree of STAT2 phosphorylation in response to IFN-α stimulation (see Fig. S1 in the supplemental material). To examine the time course of the restoration of total STAT2 by U0126, RasV12 cells were incubated with U0126 for increasing amounts of time (Fig. 4B). The level of STAT2 was increased starting at 8 h after U0126 treatment and reached a level equivalent to that observed in vector control cells by 16 h. In contrast to STAT2, the level of STAT1 was maintained consistently regardless of the activation levels of the Ras/MEK pathway. Of note, treatment with U0126 in these experiments did not affect cell viability (97.4% ± 0.4% viability in untreated cells versus 98.1% ± 0.3% viability in cells treated with U0126 for 40 h; n = 3; data not shown). To determine whether Ras/MEK regulates the expression level of STAT2 in cells in the absence of constitutively active Ras/MEK, vector control NIH 3T3 cells were treated with the MEK inhibitor U0126, PD98059, or SL327 for 16 h (Fig. 4C). All of the inhibitors increased the expression level of STAT2 protein, indicating that physiological levels of active MEK also influence the basal level of STAT2 in normal cell lines.
FIG. 4.
Inhibition of the Ras/MEK pathway restores STAT signaling in RasV12 cells. (A) Activation of STAT1 and STAT2 by IFN-α is restored in RasV12 cells treated with the MEK inhibitor U0126. Vector control cells (Cont) and RasV12 cells left untreated or pretreated with U0126 (20 μM) for 16 h were stimulated with or without IFN-α (500 U/ml) for 0.5 h. The density ratios of phosphorylated STAT1 (p-STAT1), total STAT1 (t-STAT1), phosphorylated STAT2 (p-STAT2), and total STAT2 (t-STAT2) to total ERK (t-ERK) are shown as percentages normalized to values for IFN-α-stimulated vector control cells. (B) Inhibition of MEK restores the total level of STAT2 in Ras-activated cells. Vector control cells (Cont) and RasV12 cells were left untreated or were treated with U0126 (20 μM) for 2, 4, 8, 12, and 16 h. The density ratios of t-STAT2 and t-STAT1 to t-ERK are shown as percentages normalized to values for vector control cells. (C) Inhibition of MEK increases the total level of STAT2 in vector control cells. Vector control cells were treated with U0126, SL327, or PD98059 for 16 h. Western blot analysis for p-STAT1, t-STAT1, p-STAT2, t-STAT2, phosphorylated ERK-1/2 (p-ERK), and t-ERK was performed.
These results clearly demonstrate that the activation of the Ras/MEK pathway suppresses the phosphorylation of STAT1 and STAT2, which is mediated by IFN-α. There are two possible mechanisms underlying the reduced phosphorylation of STAT proteins: (i) decreased overall levels of STAT2 limit the phosphorylation of STAT2 and, subsequently, of STAT1, as shown in other experimental systems (25, 39), or (ii) the disruption of signal propagation from upstream elements of the IFN pathway, such as JAK1, Tyk2, or IFN receptors.
Introduction of STAT2 restores IFN-α-induced antiviral responses in RasV12 cells.
If the resistance of RasV12 cells to IFN-α is caused by the insufficient availability of STAT2, the introduction of STAT2 into the cells should restore the responsiveness to IFN-α. To test this hypothesis, RasV12 cells overexpressing STAT2 (RasV12/STAT2) were generated by the infection of an MSCV retrovirus containing the mouse STAT2 gene, followed by cell sorting based on GFP fluorescence. As shown in Fig. 5A, RasV12/STAT2 cells clearly express a high level of total STAT2. IFN-α stimulation resulted in the phosphorylation of STAT1 and STAT2 proteins in RasV12/STAT2 cells at levels comparable to those of vector control cells, while STAT phosphorylation remained suppressed in RasV12 cells (Fig. 5A). This experiment also demonstrates, as reported previously (21), that STAT1 is not phosphorylated in the absence of phosphorylated STAT2. Moreover, the induction of the antiviral protein OAS in RasV12/STAT2 cells was equivalent to that in vector control cells 24 h after IFN-α stimulation (Fig. 5B). These results demonstrate that the insufficient availability of STAT2 is one of the underlying mechanisms for the unresponsiveness to IFN in Ras-activated cells, resulting in the impaired phosphorylation of the STAT proteins and the decreased induction of antiviral proteins (Fig. 5C). The degrees of STAT1 and STAT2 phosphorylation were similar in vector control cells and RasV12/STAT2 cells, even though the total levels of STAT2 obviously were different. This indicates that the upstream signals, which regulate the phosphorylation of STAT1 and STAT2, are equivalent in control and RasV12 cells stimulated with IFN-α.
FIG. 5.
Introduction of STAT2 into RasV12 cells restores the IFN response. (A) Restoration of STAT1 and STAT2 phosphorylation. Vector control cells (Cont), RasV12 cells, and RasV12/STAT2 cells were left unstimulated or were stimulated with IFN-α (500 U/ml) for 0.5 h. The density ratios of phosphorylated STAT1 (p-STAT1/t-ERK), total STAT1 (t-STAT1/t-ERK), phosphorylated STAT2 (p-STAT2/t-ERK), and total STAT2 (t-STAT2/t-ERK) to total ERK (t-ERK) are shown as percentages normalized to values for IFN-α-stimulated vector control cells. (B) Restoration of OAS induction at 24 h after IFN stimulation. The density ratios of OAS (OAS/t-ERK) to total ERK are shown as percentages normalized to values for IFN-α-stimulated vector control cells. Western blot analysis for p-STAT1, t-STAT1, p-STAT2, t-STAT2, OAS, and t-ERK was performed. (C) Diagram depicting the activation of STAT signaling in vector control cells, RasV12 cells, and RasV12/STAT2 cells stimulated with IFN.
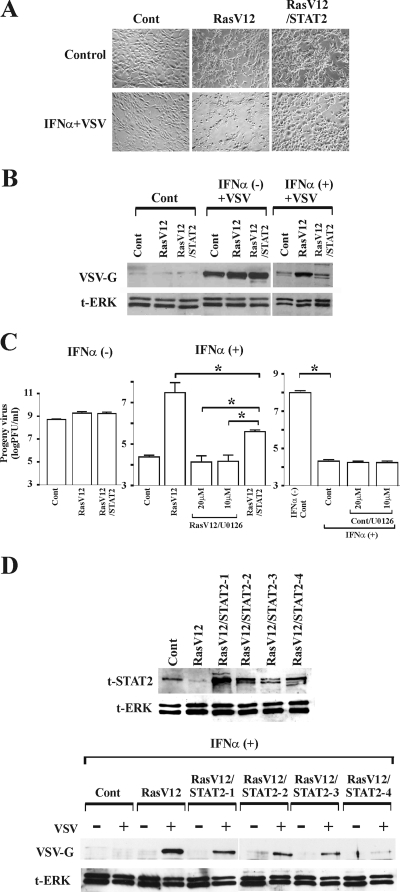
We next investigated whether the introduction of STAT2 could restore the ability of IFN-α to protect RasV12 cells from virus infection. Vector control cells, RasV12 cells, and RasV12/STAT2 cells were left untreated or were pretreated with IFN-α for 16 h and then challenged with VSV. As expected, despite the presence of IFN-α, RasV12 cells clearly exhibited cytopathic effects (Fig. 6A). In contrast, VSV infection was blocked in vector control cells and RasV12/STAT2 in the presence of IFN-α. In the absence of IFN-α, similar amounts of viral proteins were observed in the three cells; however, treatment with IFN-α inhibited viral protein synthesis more efficiently in vector control cells and RasV12/STAT2 cells than in RasV12 cells (Fig. 6B). Similarly, while there was no difference in viral progeny in the absence of IFN-α, significantly less virus progeny was produced in RasV12/STAT2 cells and vector control cells than in RasV12 cells in the presence of IFN-α (Fig. 6C). However, the introduction of STAT2 did not completely restore the ability of IFN-α to prevent progeny virus production in RasV12 cells. In contrast, when the Ras/MEK pathway was inhibited by U0126 treatment in RasV12 cells, IFN-α was as effective in inducing the antiviral response as it was in vector control cells. The addition of U0126 did not further increase the ability of IFN-α to inhibit virus production in vector control cells. This is likely because IFN-α alone is able to efficiently induce the maximum antiviral response. To determine if the amount of STAT2 overexpression correlated with the ability of STAT2 to restore the IFN response, we examined the responsiveness to IFN-α of four independently generated RasV12/STAT2 cell lines (Fig. 6D). We found that higher levels of STAT2 expression did not cause greater reduction in viral infection, suggesting that a threshold level of STAT2 is required to reestablish the IFN-induced antiviral response in RasV12 cells.
FIG. 6.
IFN-α protects RasV12/STAT2 cells from virus infection independently of STAT2 overexpression levels. Cytopathic effects (A), viral protein synthesis (B), and progeny virus production (C) in vector control cells (Cont), RasV12 cells, and RasV12/STAT2 cells at 24 h after VSV infection are shown. The cells were pretreated with U0126 for 16 h when indicated, then left untreated or treated with IFN-α (500 U/ml) for 16 h, followed by challenge with VSV (multiplicity of infection, 1). Viral protein synthesis was determined by Western blot analysis for VSV-G protein and total ERK (t-ERK). *, P < 0.01 by one-way ANOVA (n = 3). (D) Viral protein synthesis was determined in four additional, independently generated pools of RasV12/STAT2 cells (STAT2-1, STAT2-2, STAT2-3, and STAT2-4) as described above.
These results conclusively demonstrate that the reduction of STAT2 levels by Ras/MEK is one of the mechanisms underlying the decreased responsiveness of RasV12 cells to IFN-α.
STAT2 is regulated at the transcriptional level.
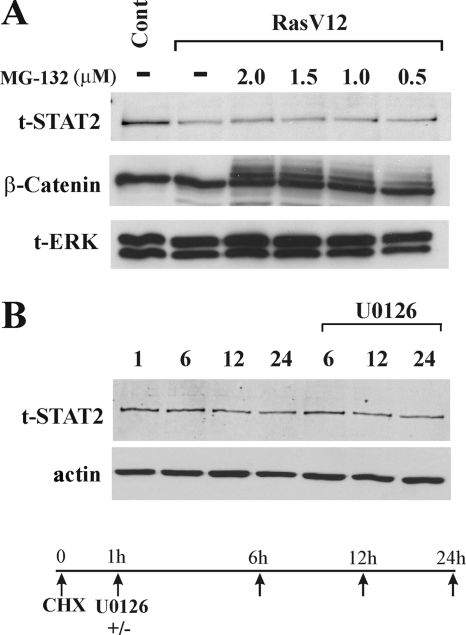
The reduction in STAT2 levels in RasV12 cells could occur at the protein level or the mRNA level. To determine if activated Ras causes the degradation of STAT2 protein, RasV12 cells were treated with the proteosome inhibitor MG132. While the levels of ubiquinated β-catenin were stabilized in RasV12 cells by MG132, STAT2 protein expression levels were not affected (Fig. 7A). To determine if STAT2 levels are regulated by proteasome-independent protein degradation pathways, we treated RasV12 cells with CHX to inhibit overall protein synthesis and analyzed the degradation of STAT2 by Western blotting in the presence or absence of U0126 (Fig. 7B). We found that the inhibition of Ras/MEK did not affect the rate of STAT2 degradation in RasV12 cells. Therefore, the degradation of STAT2 protein is not the cause of its downregulation in cells with an activated Ras/MEK pathway.
FIG. 7.
Reduction of STAT2 levels in RasV12 cells is not due to protein degradation. (A) Treatment of RasV12 cells with the proteosome inhibitor MG132 does not restore total levels of STAT2. RasV12 cells were treated with MG132 (0 to 2 μM) for 6 h, followed by Western blot analysis for total STAT2 (t-STAT2), β-catenin, and total ERK (t-ERK). (B) Degradation of STAT2 protein in RasV12 cells is not altered by U0126 treatment. RasV12 cells were pretreated with 20 μg/ml CHX for 1 h, followed by treatment with U0126 (20 μM) for the indicated times. Western blot analysis for total-STAT2 and actin was performed.
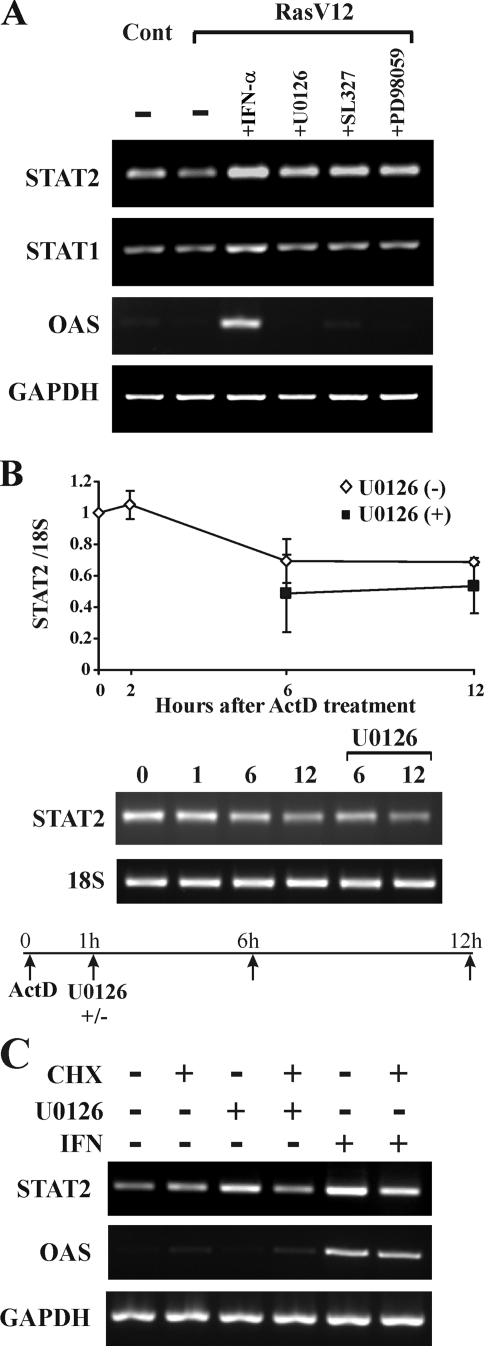
We then investigated the effect of the Ras activation on STAT2 mRNA levels to determine if changes to the level of mRNA are responsible for changes in protein expression. We found that the inhibition of MEK in RasV12 cells by U0126 treatment increased the expression of STAT2 mRNA (Fig. 8A). This effect was not due to nonspecific effects of U0126, since two other MEK inhibitors, SL327 and PD98059, also increased STAT2 mRNA levels. In addition, while IFN-α activated the transcription of STAT2, STAT1, and OAS mRNA, which are known IFN-inducible genes, MEK inhibitors did not induce STAT1 or OAS mRNA. These data suggest that the activation of MEK suppresses the level of STAT2 mRNA, but that this is not a general mechanism that affects other genes that can be induced by IFN-α.
FIG. 8.
Ras/MEK pathway controls the transcription of STAT2 mRNA. (A) Inhibition of MEK in RasV12 cells increases the expression of STAT2 mRNA. Vector control and RasV12 cells were treated with IFN-α (500 U/ml), U0126 (20 μM), SL327 (20 μM), or PD98059 (40 μM) for 6 h. Levels of STAT2, STAT1, OAS, and GAPDH mRNA were determined by RT-PCR. (B) Degradation of STAT2 mRNA in RasV12 cells is not altered by U0126 treatment. RasV12 cells were pretreated with 4 μM ActD for 1 h, followed by treatment with U0126 (20 μM). STAT2 and 18S mRNA levels were determined by RT-PCR (right) and quantitative RT-PCR (left). Data are means ± standard errors of the means from three independent experiments. (C) Protein synthesis is required for the derepression of STAT2 mRNA transcription. RasV12 cells were pretreated with 20 μg/ml CHX for 1 h and then treated with U0126 (20 μM) or IFN (500U/ml) for 6 h. STAT2, OAS, and GAPDH mRNA levels were determined by RT-PCR.
The increased levels of STAT2 mRNA in response to MEK inhibition could be due to a decreased rate of mRNA degradation or increased rate of transcription. To resolve this question, we treated cells with ActD, a general inhibitor of transcription, and monitored STAT2 mRNA levels with RT-PCR and quantitative RT-PCR (Fig. 8B). We found that the inhibition of MEK did not change the rate or degree of STAT2 mRNA degradation. Therefore, we can conclude that active transcription, and not the regulation of mRNA degradation, is responsible for the increase in STAT2 mRNA levels in response to MEK inhibition.
We then asked whether the derepression of STAT2 transcription in response to the inhibition of MEK is due to a direct effect on a transcriptional regulator of STAT2 or indirectly via the synthesis of another protein. RasV12-expressing cells were pretreated with CHX, followed by incubation with U0126 or IFN (Fig. 8C). We found that CHX impaired the increase in STAT2 levels in response to U0126. Moreover, the IFN-induced increase in STAT2 mRNA levels was partially blocked by CHX treatment. In contrast, the induction of OAS expression by IFN-α does not require additional protein synthesis. These data demonstrate that the inhibition of MEK induces the rapid expression of a transcriptional regulator that upregulates STAT2 expression.
DISCUSSION
Intact STAT signaling is vital for maintaining the robust induction of the antiviral responses induced by type I IFN. Here, we demonstrate that the Ras/MEK pathway controls STAT2 transcription, which results in the suppression of the IFN-α response. Many viruses express anti-IFN proteins as a strategy to antagonize the host antiviral system directly (26, 16). Such viral anti-IFN proteins may target different levels of the IFN pathway, including the inhibition of IFN synthesis, the disruption of Jak-STAT signaling, and interference with antiviral protein function (41). There are several viral proteins known to specifically suppress STAT2 function, either via proteasome-mediated degradation, the inhibition of nuclear translocation, or decreased expression levels (19, 40). However, this is the first study to show that the activation of a cellular pathway suppresses type I IFN antiviral activity by specifically targeting STAT2 expression at the transcriptional level.
Even viruses that do not encode anti-IFN proteins and remain sensitive to IFN in vitro can cause significant diseases in humans and animals (31), indicating the existence of other strategies for viruses to evade the IFN system in vivo. One such strategy is to selectively replicate in cells that are less sensitive to IFN due to elevated activities of the cellular suppressors. The cellular activity of the Ras/MEK pathway may influence cellular susceptibility to IFN and underlie host susceptibility to virus and/or organ tropism of viruses. Viruses also may convert host cells into IFN-resistant cells by activating the Ras/MEK pathway during infection.
While the phosphorylation of both STAT1 and STAT2 were equally suppressed in Ras-activated cells upon IFN-α stimulation (Fig. 3), the introduction of unphosphorylated STAT2 into the cells rescued the phosphorylation of STAT1 as well as that of STAT2 (Fig. 5). It is known that the phosphorylation of STAT1 is dependent on the presence of phosphorylated STAT2 in response to type I IFN signaling (21, 25, 39). The stimulation of IFN-α induces the phosphorylation of IFN-α receptor 1 on Tyr466, which recruits unphosphorylated STAT2 to the receptor. STAT2 phosphorylated by the Jak kinases in turn provides a docking site for unphosphorylated STAT1 to then induce STAT1 phosphorylation. IFN-γ, but not IFN-α, can induce the phosphorylation of STAT1 in STAT2-deficient U6A cells (25). Similarly to our findings, the phosphorylation of STAT1 induced by IFN-α in U6A cells was restored fully upon transfection with STAT2 cDNA. In addition, activated Ras/MEK did not suppress the phosphorylation of STAT1 induced by IFN-γ, which does not require STAT2 to induce STAT1 phosphorylation (Fig. 3E). Therefore, these results suggest that the decreased phosphorylation of STAT1 in Ras-activated cells stimulated with IFN is likely a secondary event caused by insufficient phosphorylated STAT2.
A previous report found that the overexpression of K-Ras in human cancer cells resulted in the decreased expression of both STAT1 and STAT2 via the phosphatidylinositol-3-kinase pathway (22). In contrast, we found that STAT2, but not STAT1, expression is reduced in RasV12 compared to the level for vector control cells (Fig. 3), which was restored upon the inhibition of the Ras/MEK pathway by U0126 or RNAi (Fig. 4; also see Fig. S1 in the supplemental material). Consistently with the changes in STAT2 protein expression, STAT2 mRNA levels also were reduced in RasV12 cells and restored by the treatment of MEK inhibitors (Fig. 8). STAT1 mRNA expression, however, was not changed in control cells, RasV12 cells, or RasV12 cells treated with U0126 (Fig. 8). In addition, the phosphorylation of STAT1 by IFN-γ, which is independent of STAT2 levels, was unaffected by the expression of RasV12 (Fig. 3E). These results suggest that the Ras/MEK pathway regulates STAT2, but not STAT1, in the mice fibroblast system, while the Ras activation of phosphatidylinositol-3-kinase reduces both STAT1 and STAT2 expression in human cancer cells (22).
IFNs signal to STAT2 via IFN receptors and Jak1/Tyk2. Therefore, it is possible that the suppression of the IFN pathway upstream of STAT2 by activated Ras/MEK contributes to the impaired phosphorylation of STAT2. However, our evidence suggests that this is not the case. First, we examined phosphatase activity directed against STAT2 in control and RasV12 cells (see Fig. S2A in the supplemental material). While we found that the inhibition of overall tyrosine phosphatase activity by treating RasV12 cells with orthovanadate resulted in a modest increase in the level of phosphorylated STAT2, it was not restored to the level of IFN-α-stimulated control cells. In addition, the direct determination of phosphatase activity toward immunopurified phosphorylated STAT2 revealed that RasV12 cells contain less phosphatase activity against phosphorylated STAT2 than vector control cells (see Fig. S2B in the supplemental material). Second, we examined the expression level of negative regulators of IFN-α signaling, including the suppressors of cytokine signaling 1 through 4, the SH2 domain-containing protein tyrosine phosphatase 1 (Shp-1) and Shp-2, and protein inhibitors of activated STAT1 through STAT4 (see Fig. S3 in the supplemental material) (8, 36, 51). We found that only the Shp-1 phosphatase had higher expression in RasV12 cells; however, there was no reduction in Shp-1 mRNA levels upon U0126 treatment, suggesting that Shp-1 expression is regulated by other elements downstream of Ras but not by the Ras/MEK pathway. Also, the phosphorylation of STAT2 was restored by the introduction of STAT2 into RasV12 cells to the same level as that of the control cells (Fig. 5A), indicating that the activation of the upstream kinases Jak1 and Tyk2 are equivalent and regulated similarly in both cell types. Lastly, there was no difference observed in the expression level of IFN-α/β receptor between vector control and RasV12 cells (data not shown). Therefore, the evidence to date suggests that the regulation of components upstream of STAT2 by Ras/MEK does not play a major role in suppressing the IFN-α-induced phosphorylation of STATs in RasV12 cells. However, since some contribution by upstream elements cannot be excluded, this possibility will be examined in future studies.
Here, we demonstrate that the downregulation of STAT2 in response to the activated Ras/MEK pathway is one mechanism used to interrupt the antiviral response induced by IFN-α. However, the Ras/MEK pathway also suppresses the antiviral response induced by IFN through other mechanisms. We observed an almost complete recovery from the Ras-induced inhibition of IFN-α-dependent STAT1 and STAT2 phosphorylation and antiviral protein OAS induction in RasV12/STAT2 cells (Fig. 5). In contrast, the ability of IFN-α to suppress progeny virus production was not fully restored in Ras/STAT2 cells, while IFN-α inhibited viral replication in RasV12 cells treated with U0126 to the same extent as that in vector control cells (Fig. 6C). These observations suggest that activated Ras/MEK suppresses the transcription of other genes that mediate IFN antiviral effects or have additional suppressive effects on cell death or the release of viral particles that are not rescued by overexpression of STAT2 and likely independent of ISGF3 activation. Interestingly, Noser et al. (33) reported that MxA expression was increased in response to the inhibition of the Ras/MEK pathway in some human cancer cell lines, suggesting that the Ras/MEK pathway suppresses the expression of multiple IFN-inducible genes.
We found that the inhibition of MEK in RasV12 cells increased STAT2 transcription (Fig. 8), while the stability of STAT2 protein (Fig. 7) or mRNA (Fig. 8) was unaffected. Furthermore, we found that the derepression of STAT2 transcription by the inhibition of MEK requires de novo protein synthesis. This indicates that Ras/MEK represses the expression of a transcription factor that normally promotes the constitutive expression of STAT2. This may be accomplished by either (i) the MEK-mediated repression of a transcriptional activator or (ii) the MEK-mediated activation of a transcriptional repressor. CHX treatment suppresses the IFN-induced transcription of certain IFN-inducible genes, such as guanylate binding protein 1 (GBP1) and GBP2, indicating that de novo protein synthesis is required for their maximal induction by IFN (5, 49). Similarly, we found in this study that STAT2, but not OAS, induction by IFN was partially inhibited by CHX treatment, suggesting that the induced expression of another transcriptional factor cooperates with ISGF3 to achieve maximal STAT2 induction. It would be of interest to determine if the same transcriptional factor also is upregulated by the inhibition of MEK, which then could synergize with the IFN-mediated activation of ISGF3.
In conclusion, we have demonstrated that the downregulation of STAT2 function is one of the ways by which the Ras/MEK pathway can reduce the expression of antiviral proteins and, subsequently, the ability of IFN-α to promote the resistance of host cells to virus infection. It is notable that treatment with any of the MEK inhibitors alone is sufficient to restore STAT2 levels in cells with constitutively active Ras/MEK as well as with physiological levels of active Ras/MEK. Small-molecule inhibitors of MEK recently have progressed to clinical trials of cancer treatment and are proven to have low toxicity in vivo (44, 48). Determining if the combined treatment of a MEK inhibitor with IFNs enhances the potent antiviral, antitumor, and immunoregulatory activities of type I IFNs warrants further study.
Supplementary Material
Acknowledgments
This work was supported by grants (to K.H.) from the Canadian Institutes for Health Research (CIHR). K.H. is a recipient of a New Investigator Award from the CIHR. S.L.C. was supported by a postdoctoral award from the CIHR and the Newfoundland and Labrador Department of Innovation, Trade, and Rural Development.
We thank John Bell, David Farrar, Patrick Lee, and Yoshihiro Sokawa for their generous gifts of constructs, cell lines, antibodies, and viruses. We thank Dianne Codner, Julie Whitten, Yumiko Komatsu, and Theerawat Pongnopparat for technical support.
Footnotes
Published ahead of print on 22 April 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Balachandran, S., M. Porosnicu, and G. N. Barber. 2001. Oncolytic activity of vesicular stomatitis virus is effective against tumors exhibiting aberrant p53, Ras, or myc function and involves the induction of apoptosis. J. Virol. 753474-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber, S. A., L. Bruett, B. R. Douglass, D. S. Herbst, M. C. Zink, and J. E. Clements. 2002. Visna virus-induced activation of MAPK is required for virus replication and correlates with virus-induced neuropathology. J. Virol. 76817-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battcock, S. M., T. W. Collier, D. Zu, and K. Hirasawa. 2006. Negative regulation of the alpha interferon-induced antiviral response by the Ras/Raf/MEK pathway. J. Virol. 804422-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann, M., I. Romirer, M. Sachet, R. Fleischhacker, A. Garcia-Sastre, P. Palese, K. Wolff, H. Pehamberger, R. Jakesz, and T. Muster. 2001. A genetically engineered influenza A virus with ras-dependent oncolytic properties. Cancer Res. 618188-8193. [PubMed] [Google Scholar]
- 5.Briken, V., H. Ruffner, U. Schultz, A. Schwarz, L. F. Reis, I. Strehlow, T. Decker, and P. Staeheli. 1995. Interferon regulatory factor 1 is required for mouse Gbp gene activation by gamma interferon. Mol. Cell. Biol. 15975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S. L., R. Khosravi-Far, K. L. Rossman, G. J. Clark, and C. J. Der. 1998. Increasing complexity of Ras signaling. Oncogene 171395-1413. [DOI] [PubMed] [Google Scholar]
- 7.Cascalló, M., G. Capella, A. Mazo, and R. Alemany. 2003. Ras-dependent oncolysis with an adenovirus VAI mutant. Cancer Res. 635544-5550. [PubMed] [Google Scholar]
- 8.Croker, B. A., H. Kiu, and S. E. Nicholson. 2008. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cell Dev. Biol. 19414-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davie, J. R., and V. A. Spencer. 2001. Signal transduction pathways and the modification of chromatin structure. Prog. Nucleic Acid Res. Mol. Biol. 65299-340. [DOI] [PubMed] [Google Scholar]
- 10.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 9515623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhillon, A. S., S. Hagan, O. Rath, and W. Kolch. 2007. MAP kinase signalling pathways in cancer. Oncogene 263279-3290. [DOI] [PubMed] [Google Scholar]
- 12.Farassati, F., A. D. Yang, and P. W. Lee. 2001. Oncogenes in Ras signalling pathway dictate host-cell permissiveness to herpes simplex virus 1. Nat. Cell Biol. 3745-750. [DOI] [PubMed] [Google Scholar]
- 13.Farrar, J. D., J. D. Smith, T. L. Murphy, S. Leung, G. R. Stark, and K. M. Murphy. 2000. Selective loss of type I interferon-induced STAT4 activation caused by a minisatellite insertion in mouse Stat2. Nat. Immunol. 165-69. [DOI] [PubMed] [Google Scholar]
- 14.Fu, X. Y., D. S. Kessler, S. A. Veals, D. E. Levy, and J. E. Darnell, Jr. 1990. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc. Natl. Acad. Sci. USA 878555-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes. Infect. 4647-655. [DOI] [PubMed] [Google Scholar]
- 16.Grandvaux, N., B. R. Tenover, M. J. Servant, and J. Hiscott. 2002. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 15259-267.12015460 [Google Scholar]
- 17.Hirasawa, K., A. Kim, H. S. Han, J. Han, H. S. Jun, and J. W. Yoon. 2003. Effect of p38 mitogen-activated protein kinase on the replication of encephalomyocarditis virus. J. Virol. 775649-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirasawa, K., S. G. Nishikawa, K. L. Norman, T. Alain, A. Kossakowska, and P. W. K. Lee. 2002. Oncolytic reovirus against ovarian and colon cancer. Cancer Res. 621696-1701. [PubMed] [Google Scholar]
- 19.Horvath, C. M. 2004. Silencing STATs: lessons from paramyxovirus interferon evasion. Cytokine Growth Factor Rev. 15117-127. [DOI] [PubMed] [Google Scholar]
- 20.Horvath, C. M., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1996. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 166957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Improta, T., C. Schindler, C. M. Horvath, I. M. Kerr, G. R. Stark, and J. E. Darnell, Jr. 1994. Transcription factor ISGF-3 formation requires phosphorylated Stat91 protein, but Stat113 protein is phosphorylated independently of Stat91 protein. Proc. Natl. Acad. Sci. USA 914776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klampfer, L., J. Huang, G. Corner, J. Mariadason, D. Arango, T. Sasazuki, S. Shirasawa, and L. Augenlicht. 2003. Oncogenic Ki-ras inhibits the expression of interferon-responsive genes through inhibition of STAT1 and STAT2 expression. J. Biol. Chem. 27846278-46287. [DOI] [PubMed] [Google Scholar]
- 23.Kong, X., H. San Juan, A. Behera, M. E. Peeples, J. Wu, R. F. Lockey, and S. S. Mohapatra. 2004. ERK-1/2 activity is required for efficient RSV infection. FEBS Lett. 55933-38. [DOI] [PubMed] [Google Scholar]
- 24.Krebs, D. L., and D. J. Hilton. 2000. SOCS: physiological suppressors of cytokine signaling. J. Cell Sci. 1132813-2819. [DOI] [PubMed] [Google Scholar]
- 25.Leung, S., S. A. Qureshi, I. M. Kerr, J. E. Darnell, Jr., and G. R. Stark. 1995. Role of STAT2 in the alpha interferon signaling pathway. Mol. Cell. Biol. 151312-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12143-156. [DOI] [PubMed] [Google Scholar]
- 27.Levy, D. E., D. S. Kessler, R. Pine, and J. E. Darnell, Jr. 1989. Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 31362-1371. [DOI] [PubMed] [Google Scholar]
- 28.Liu, B., J. Liao, X. Rao, S. A. Kushner, C. D. Chung, D. D. Chang, and K. Shuai. 1998. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 9510626-10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo, H., B. Yanagawa, J. Zhang, Z. Luo, M. Zhang, M. Esfandiarei, C. Carthy, J. E. Wilson, D. Yang, and B. M. McManus. 2002. Coxsackievirus B3 replication is reduced by inhibition of the extracellular signal-regulated kinase (ERK) signaling pathway. J. Virol. 763365-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mundschau, L. J., and D. V. Faller. 1992. Oncogenic ras induces an inhibitor of double-stranded RNA-dependent eukaryotic initiation factor 2 alpha-kinase activation. J. Biol. Chem. 26723092-23098. [PubMed] [Google Scholar]
- 31.Muñoz, A., and L. Carrasco. 1984. Action of human lymphoblastoid interferon on HeLa cells infected with RNA-containing animal viruses. J. Gen. Virol. 65377-390. [DOI] [PubMed] [Google Scholar]
- 32.Norman, K. L., K. Hirasawa, A. D. Yang, M. A. Shields, and P. W. Lee. 2004. Reovirus oncolysis: the Ras/RalGEF/p38 pathway dictates host cell permissiveness to reovirus infection. Proc. Natl. Acad. Sci. USA 10111099-11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noser, J. A., A. A. Mael, R. Sakuma, S. Ohmine, P. Marcato, L. P. Wk, and Y. Ikeda. 2007. The RAS/Raf1/MEK/ERK signaling pathway facilitates VSV-mediated oncolysis: implication for the defective interferon response in cancer cells. Mol. Ther. 151531-1536. [DOI] [PubMed] [Google Scholar]
- 34.Novick, D., B. Cohen, and M. Rubinstein. 1994. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell 77391-400. [DOI] [PubMed] [Google Scholar]
- 35.Offermann, M. K., and D. V. Faller. 1990. Effect of cellular density and viral oncogenes on the major histocompatibility complex class I antigen response to gamma-interferon in BALB-c/3T3 cells. Cancer Res. 50601-605. [PubMed] [Google Scholar]
- 36.Palvimo, J. J. 2007. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem. Soc. Trans. 351405-1408. [DOI] [PubMed] [Google Scholar]
- 37.Planz, O., S. Pleschka, and S. Ludwig. 2001. MEK-specific inhibitor U0126 blocks spread of Borna disease virus in cultured cells. J. Virol. 754871-4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pleschka, S., T. Wolff, C. Ehrhardt, G. Hobom, O. Planz, U. R. Rapp, and S. Ludwig. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 3301-305. [DOI] [PubMed] [Google Scholar]
- 39.Qureshi, S. A., S. Leung, I. M. Kerr, G. R. Stark, and J. E. Darnell, Jr. 1996. Function of Stat2 protein in transcriptional activation by alpha interferon. Mol. Cell. Biol. 16288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramaswamy, M., L. Shi, S. M. Varga, S. Barik, M. A. Behlke, and D. C. Look. 2006. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology 344328-339. [DOI] [PubMed] [Google Scholar]
- 41.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 891-47. [DOI] [PubMed] [Google Scholar]
- 42.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindler, C. 1999. Cytokines and JAK-STAT signaling. Exp. Cell Res. 2537-14. [DOI] [PubMed] [Google Scholar]
- 44.Sebolt-Leopold, J. S. 2000. Development of anticancer drugs targeting the MAP kinase pathway. Oncogene 196594-6599. [DOI] [PubMed] [Google Scholar]
- 45.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55255-281. [DOI] [PubMed] [Google Scholar]
- 46.Stojdl, D. F., B. Lichty, S. Knowles, R. Marius, H. Atkins, N. Sonenberg, and J. C. Bell. 2000. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 6821-825. [DOI] [PubMed] [Google Scholar]
- 47.Strong, J. E., M. C. Coffey, D. Tang, P. Sabinin, and P. W. Lee. 1998. The molecular basis of viral oncolysis: usurpation of the Ras signaling pathway by reovirus. EMBO J. 173351-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, N., and J. Lyons. 2005. Recent progress in targeting the Raf/MEK/ERK pathway with inhibitors in cancer drug discovery. Curr. Opin. Pharmacol. 5350-356. [DOI] [PubMed] [Google Scholar]
- 49.Tnani, M., and B. A. Bayard. 1999. Evidence for IRF-1-dependent gene expression deficiency in interferon unresponsive HepG2 cells. Biochim. Biophys. Acta 145159-72. [DOI] [PubMed] [Google Scholar]
- 50.Wormald, S., and D. J. Hilton. 2004. Inhibitors of cytokine signal transduction. J. Biol. Chem. 279821-824. [DOI] [PubMed] [Google Scholar]
- 51.Xu, D., and C. K. Qu. 2008. Protein tyrosine phosphatases in the JAK/STAT pathway. Front. Biosci. 134925-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto, A., A. Iwata, Y. Koh, S. Kawai, S. Murayama, K. Hamada, S. Maekawa, S. Ueda, and Y. Sokawa. 1998. Two types of chicken 2′,5′-oligoadenylate synthetase mRNA derived from alleles at a single locus. Biochim. Biophys. Acta 1395181-191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.