Abstract
The outcome of a viral infection is regulated in part by the complex coordination of viral and host interactions that compete for the control and optimization of virus replication. Severe acute respiratory syndrome coronavirus (SARS-CoV) intimately engages and regulates the host innate immune responses during infection. Using a novel interferon (IFN) antagonism screen, we show that the SARS-CoV proteome contains several replicase, structural, and accessory proteins that antagonize the IFN pathway. In this study, we focus on the SARS-CoV papain-like protease (PLP), which engages and antagonizes the IFN induction and NF-κB signaling pathways. PLP blocks these pathways by affecting activation of the important signaling proteins in each pathway, IRF3 and NF-κB. We also show that the ubiquitin-like domain of PLP is necessary for pathway antagonism but not sufficient by itself to block these pathways regardless of the enzymatic activity of the protease. The potential mechanism of PLP antagonism and its role in pathogenesis are discussed.
The outcome of a viral infection is mediated, in part, through the complex interplay of viral and cellular components that coordinate the innate immune response. During virus entry and replication, the innate immune response machinery senses the invading virus and activates a cascade of signaling pathways that ultimately concludes with the translation of several hundred antiviral proteins to convert the intracellular environment into a suboptimal context for replication (32). In response to this powerful selective environment, viruses have evolved strategies to disable the host's innate immune arsenal and optimize the intracellular environment for efficient virus replication and release. Screens for interferon (IFN) antagonist activity of various viruses have shown that most viruses, including all highly pathogenic human viruses tested, attempt to modulate the innate immune response early in infection (25).
The innate immune response is a series of signaling cascades that are activated once a foreign pathogen is detected in the host (34, 45). The response is initiated by cytoplasmic protein sensors, such as RIG-I (retinoic acid-inducible gene I) and MDA5, which bind to by-products of viral entry and replication in the cell (2, 28, 51, 59). During viral infection, RIG-I and MDA5 bind to viral single-stranded RNA and/or double-stranded RNA (dsRNA) and signal through a protein called MAVS (mitochondrial antiviral signaling protein) (51, 59). MAVS activation leads to the activation of IKKi and TBK1, the kinases which phosphorylate IRF3 to induce its dimerization and import into the nucleus (34). Once phosphorylated by the kinases, IRF3 dimerizes and is imported into the nucleus. Nuclear IRF3, in complex with several other proteins, induces type I IFN to alert neighboring cells of the infection. Once IRF3 induces transcription of IFN-β, this powerful cytokine is secreted from the cells. The NF-κB pathway is also activated via these mediators as well. Homologous to the IRF3 signaling pathway, kinases play a key role in activating the NF-κB pathway as well. The IKKα, IKKβ, and IKKγ kinases phosphorylate the NF-κB inhibitor IκΒα. After IκΒα is phosphorylated, it is targeted for degradation, which allows NF-κB to be imported into the nucleus. There it induces IFN-β and complexes with IRF3 to induce antiviral genes. The STAT1 signaling pathways are activated via the binding of type I (IFN-β), II (IFN-γ), and III (IFN-λ) IFN binding to their receptors on the surface of cells. This binding initiates either the formation of the STAT1/STAT2/IRF9 complex (called ISGF3) after type I or type III binding or STAT1 homodimerization after type II IFN binding to receptors. STAT1 complex formation leads to its import into the nucleus and activation of antiviral genes. The regulation of these signaling cascades is crucially important for the survival of the host. Disregulation of these events can lead to increased infection and disease.
Many viruses encode proteins that block, modulate, or slow several important signaling pathways that are involved in induction or amplification of the innate immune response (3, 7, 10, 19, 35, 37, 38, 49, 55, 56). The Ebola virus encodes the VP35 and VP24 proteins that block the dsRNA sensing and STAT1 signaling pathways, respectively (7, 43). Coxsackievirus B3 encodes the 3a protein, which blocks almost total protein secretion during infection (9). The severe acute respiratory syndrome coronavirus (SARS-CoV) encodes several innate immune antagonists, including the open reading frame 6 (ORF6) protein, nucleocapsid, ORF3b, and nonstructural protein 1 (NSP1), which collectively antagonize the IRF3, JAK/STAT, and/or NF-κB signaling pathway (18, 29, 53). Although each pathogen targets similar signaling pathways, they antagonize host communication networks by different mechanisms using distinctly different viral proteins.
SARS is an often fatal disease which emerged from China during the fall of 2002 (15, 30). The disease quickly spread across Asia, Europe, and North America. At the resolution of the outbreak, more than 8,000 people had been infected, resulting in ∼800 deaths and economic losses in the tens of billions worldwide (23). The disease was caused by a new human CoV (HCoV), named the SARS-CoV (21). The virus likely originated from bats, as bat CoVs are pervasive, and several are close relatives of the SARS-CoV in humans and civets (31, 39). Thus, resurgence of SARS from zoonotic sources remains a distinct possibility, making further understanding of the mechanisms regulating pathogenesis and virulence of high significance (14, 54).
A previous report demonstrated that the papain-like protease (PLP) of the SARS-CoV is an IFN antagonist (13). PLP was shown to block phosphorylation and activation of IRF3, thereby antagonizing IFN-β induction. Mechanistically, a direct interaction between PLP and IRF3 was hypothesized to sterically hinder the activation of IRF3. Finally, it was also reported that PLP did not antagonize the NF-κB signaling pathway and that mutations in the catalytic site of PLP did not affect its antagonism.
In this work, we develop a novel IFN antagonist screen to identify several new potential IFN antagonists encoded in the SARS-CoV genome and then focus on the mechanism of action of one of them, PLP. In contrast to the studies detailed above, we will show through in vivo expression studies and biochemical studies involving purified proteins that PLP does not directly bind to IRF3 or perturb the phosphorylation of IRF3 by kinases and that PLP blocks the NF-κB signaling pathway via stabilization of the NF-κB inhibitor, IκBα. Finally, our work identifies a role for the active site of PLP and establishes that the ubiquitin-like (UBL) domain of PLP is necessary, but not sufficient, for antagonism of IRF3 function. Importantly, deletion of the PLP UBL domain did not alter intrinsic proteolytic or deubiquitinating (DUB) activities. Finally, the HCoV-NL63 virus but not mouse hepatitis virus (MHV) PLP was able to block IRF3 and NF-κB signaling, demonstrating strain-specific variation among members of the family Coronaviridae.
MATERIALS AND METHODS
Cloning of SARS-CoV ORFs into VRPs.
SARS-CoV ORFs were PCR amplified from the SARS-CoV molecular clone (57) and inserted into the Venezuelan equine encephalitis (VEE) plasmid VR21 downstream of the 26S promoter by overlap PCR (4). For most replicase proteins, an ATG start codon was added at the N terminus of each coding region and a stop codon was added at the C-terminus. Briefly, PCRs were performed with Expand Long Taq (Roche Molecular Biochemicals) in 30 cycles of 94°C for 30 s, 55°C for 30 s, and extensions at 68°C for 1 min. The initial amplicon, which was used in the construction of all of the constructs, was generated with primers 5′nsp4Sw (5′-GATTGAGGCGGCTTTCGGCG) and 3′26S (5′-TTAATTAAGTCAATCGGCGCGCCCTTGGCGGACTAGACTATGTC) using pVR21 as a template. The second amplicon was made using the primers shown in Table 1 that flanked the SARS-CoV cistron of interest and which included overlap sequences with the 3′26S primer. These amplicons were then fused together by overlap PCR with the forward 5′NSL4sw primer and the reverse primer for each ORF to produce a single amplicon and then cloned into VR21 using the digestion scheme described above.
TABLE 1.
Primers and plasmids used in this study
| Primer or plasmid | Sequence
|
|
|---|---|---|
| Forwarda | Reverse | |
| VEE primers | ||
| ORF3a | GTCTAGTCCGCCAAGATGGATTTGTTTATGAGAT | GATCGGCGCGCCTTACAAAGGCACGCTAGTAGTCG |
| ORF6 | GTCTAGTCCGCCAAGATGTTTCATCTTGTTGACT | GATCGGCGCGCCTTATGGATAATCTAACTCCATAG |
| ORF8b | GTCTAGTCCGCCAAGATGTGCTTGAAGATCCTTG | GATCGGCGCGCCTTAATTTGTTCGTTTATTTAAAA |
| Spike | GTCTAGTCCGCCAAGATGTTTATTTTCTTATTAT | GATCGGCGCGCCTTATGTGTAATGTAATTTGACAC |
| N | GTCTAGTCCGCCAAGATGTCTGATAATGGACCCC | GATCGGCGCGCCTTATGCCTGAGTTGAATCAGCAG |
| Bat ORF6 | GTCTAGTCCGCCAAGATGTTTCATCTAGTTGACT | GATCGGCGCGCCTTATGGATAATCTAACTCCATA |
| NS1 | GTCTAGTCCGCCAAGATGATGGATCCAAACACTGT | GATCGGCGCGCCTCAAACTTCTGACCTAATTGTTCC |
| NSP1 | GTCTAGTCCGCCAAGATGGAGAGCCTTGTTCTTGG | GATCGGCGCGCCTTAACCTCCATTGAGCTCACGAG |
| NSP5 | GTCTAGTCCGCCAAGATGAGTGGTTTTAGGAAAAT | GATCGGCGCGCCTTATTGGAAGGTAACACCAGAGC |
| NSP7 | GTCTAGTCCGCCAAGATGTCTAAAATGTCTGACGT | GATCGGCGCGCCTTACTGAAGAGTAGCACGGTTAT |
| NSP8 | GTCTAGTCCGCCAAGATGGCTATTGCTTCAGAATT | GATCGGCGCGCCTTACTGTAGTTTAACAGCTGAGT |
| NSP10 | GTCTAGTCCGCCAAGATGGCTGGAAATGCTACAGA | GATCGGCGCGCCTTACTGCATCAAGGGTTCGCGGA |
| NSP12 | GTCTAGTCCGCCAAGATGAGGCTGTAGGTGCTTGT | GATCGGCGCGCCTTATTGTAATGTAGCCACATTGC |
| NSP15 | GTCTAGTCCGCCAAGATGAGTTTAGAAAATGTGGC | GATCGGCGCGCCTTATTGTAGTTTTGGGTAGAAGG |
| NSP16 | GTCTAGTCCGCCAAGATGGCAAGTCAAGCGTGGCA | GATCGGCGCGCCTTATTAGTTGTTAACAAGAATAT |
| PLP | GTCTAGTCCGCCAAGATGGGTGACAAAATTGTGTAC | GATCGGCGCGCCTTACGACACAGGCTTGATGGTTG |
| Expression plasmids | ||
| PLP WT | GAATTCACCATGGAGGTTAAGACTATAAAAGTG | CCCGGGCTTGATGGTTGTAGTGTAAGATGTTTCCT |
| UBL only | GAATTCACCATGGAGGTTAAGACTATAAAAGTG | CCCCGGGTAGTACAAAGAAAGTCTTACCCTCA |
| PLPΔUBL | GAATTCACCATGCCTAGTGATGACACACTACGTAGTGAAGCTTT | CCCCGGGTAGTACAAAGAAAGTCTTACCCTCA |
| MHV PLP1 | GAATTCACCATGTCTATCTTGGATGAGCTTCAAAC | ACTGCCCGGGCTTTTCAGCTATAGCACCTGCAACACCT |
| MHV PLP2 | GAATTCACCATGTTGGATGATGATGCTCGTGTCTTT | ACGTCCCGGGCGATAAATCTGGCTTATACTCCACACAC |
| NL63 PLP | GAATTCACCATGGTTGTAGAGAGTAATGTTATGGAT | CCCGGGTGCACCAGTATCAAGTTTATCCATAACAGA |
| NSP2/3 cleavage | GAATTCACCATGCAGTGTATACGTGGCAAGGAGCAG | CCCGGGGTTGGTAAGGAGATCAGAAACTGGT |
| PLP W94A | ACCACACAAAGAAAgcGAAATTTCCTCAAG | |
| PLP C112A | ATGGGCTGATAACAATgcaTATTTGTCTAG | |
| PLP D287A | CCCTCTATCGTATTGcCGGAGCTCACCTTA | |
Added ATG start codons are underlined. Lowercase letters represent mutated bases.
Expression plasmids.
ORFs were cloned into the CAGGS/GFP (green fluorescent protein) or CAGGS/HA (hemagglutinin) vector for expression in HEK293T cells as previously described by our group (29). Amplicons were produced using the primers shown in Table 1. For each construct, an ATG start codon was added as the first codon but no stop codon was included at the 3′ terminus of each ORF. Rather, an HA or GFP tag was fused in frame to each ORF. The amplicons and vector were digested with EcoRI/XmaI fragments for cloning, and all constructs were verified by sequence analysis. Expression of each plasmid was checked by Western blotting with either anti-HA (Sigma H3663) or anti-GFP (Clontech 632459) antibodies after expression in 293T cells for 24 h.
Antibodies.
For all Western blot experiments, anti-HA (Sigma H3663), anti-GFP (Clontech 632459), anti-V5 (Invitrogen R960), antiactin (Santa Cruz I-19), and anti-Flag (Sigma F7425) were used. Phospho-specific anti-IRF3 antibody was provided by John Hiscott. To test VEE replicon particle (VRP) expression of SARS-CoV proteins, antibodies were generated from mice in our laboratory. VRPs expressing each protein were inoculated in the footpad. At 2 weeks after inoculation, mice were boosted with the same VRP. At 4 weeks after the boost, mice were bled and serum was collected for use in these assays. For Western blots, serum was precleared by incubation of 25 μl of serum on a monolayer of MA104 cells in a single well of a 24-well plate. At 2 h after incubation, serum was removed from the well and used for Western blotting at a concentration of 1:500. This procedure was used for all antibodies shown in Fig. S1 in the supplemental material, with the exception of anti-NSP5 and anti-E. Anti-NSP5 antibodies were used directly without preclearance, and anti-E antibody was kindly provided by Luis Enjuanes.
IFN bioassay and IFN-β RT-PCR.
MA104 cells were infected with infectious clone SARS (icSARS) or Sendai virus (SeV) at a multiplicity of infection (MOI) of 5. Medium was removed from the cells for type I IFN bioassay, and intracellular RNA was extracted from the cultures with Trizol at different times postinfection. The bioassay has been described previously for use with human type I IFN and in A549 cells, and the methods were described more fully by Shabman et al. (48). Briefly, the pH of the medium was lowered to 2 with 2 N HCl and incubated at 4°C overnight to kill the activity of any other IFNs and chemokines which may alter affects of the assay. IFNs are very stable at low pH, while other proteins will be denatured. The pH of the medium was then adjusted to pH 7 with NaOH. One hundred microliters of the medium was then added to each well of a 96-well plate containing A549 cells and incubated for 24 h. Encephalomyocarditis virus was then added at an MOI of 5, and the cultures were incubated for 24 h before scoring the plates for cytopathic effect (CPE). CPE was scored by comparing medium-treated cells to a standard curve of IFN-β-treated cells.
Intracellular RNA was converted to cDNA with Superscript II (Invitrogen, Carlsbad, CA) using random hexamers for cDNA synthesis, and the products were then used for a reverse transcription-PCR (RT-PCR) using primer pairs specific for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and IFN-β. The GAPDH-specific primers were 5′ GTCTTCACCACCATGGAGAAGGCTGGGGCT 3′ and 5′ ACAGCCTTGGCAGCGCCAGTAGAGGCAGGG 3′, while the IFN-β primers were 5′ GACGCCGCATTGACCATCTA 3′ and 5′ CCTTAGGATTTCCACTCTGACT 3′, respectively.
Immunoprecipitations.
HA- and Flag-tagged plasmids were transfected into 293T cells as described below. After 24 h of expression, cells were treated with lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40), the extract was centrifuged at 13,200 rpm for 10 min at 4°C, and the supernatant was removed. EZ View Red anti-Flag M2 affinity gel beads (F2426; Sigma, St. Louis, MO) were washed three times in 500 μl of lysis buffer per 100 μl of beads, and 25 μl of washed beads was added to 100 μl of protein extract and rotated overnight at 4°C. For HA immunoprecipitations, protein G Dynabeads (Invitrogen) were mixed with mouse monoclonal anti-HA antibody for 2 h on a rotating wheel at 4°C. After incubation, the bead-antibody mixture was washed three times with lysis buffer and used for further immunoprecipitations. Twenty-five microliters of protein G/HA antibody-treated beads was used for each immunoprecipitation. One hundred microliters of protein extract was mixed with 100 μl of protein extract and rotated overnight at 4°C. The mixture was then washed three times with lysis buffer at 4°C. The protein extract-bead mixture was then resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer before boiling and electrophoresis. For V5 immunoprecipitations, an identical procedure to HA immunoprecipitations was used with the exception that anti-V5 antibody (Invitrogen R960) was used in place of anti-HA.
Luciferase assays.
To analyze the induction of IFN-β- or NF-κB-induced genes, a luciferase reporter assay was used in 293T cells. Briefly, an expression construct containing the luciferase ORF and either the IFN-β promoter (IFN-β/luciferase) or a promoter containing three copies of the NF-κB binding site (NF-κB luciferase) was cotransfected with either a GFP control plasmid or the designated PLP plasmid. Additionally, all transfections contained a plasmid encoding Renilla luciferase under the control of the constitutive SV40 promoter (SV40/Renilla luciferase). Transfections of reporter plasmids into 293T cells were performed with the Fugene6 transfection reagent as directed by the manufacturer (Roche). Twenty-four hours posttransfection, the cells were lysed and assayed for luciferase expression using the Dual-Glo luciferase reagent (Promega) per the manufacturer's instructions using a luminometer. The resulting readout was analyzed by dividing the firefly luciferase induction under each condition by the Renilla luciferase expression in each well. This controls for transfection variation from well to well. The ratio of experimental to mock treatment is graphed in each figure. Lysates were also assayed by Western blotting for expression of the transfected proteins (see the supplemental material). All three wells of each triplicate experiment were combined, and 10 μl of the resulting lysates was analyzed by Western blotting with the designated antibodies.
IRF3173-426, IRF31-426, and PLP purifications.
Four liters of Escherichia coli BL21(DE3) cells (Novagen) containing plasmid pGEX6P-1-IRF3173-426 was grown in LB supplemented with 50 μg/ml of carbenicillin at 25°C to an optical density at 600 nm of 0.6. To induce expression, the cultures were treated with IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 0.6 mM and then grown for an additional 4 h under the above conditions. Cells were pelleted by centrifugation (3,800 × g, 30 min, 4°C) and resuspended in 100 ml phosphate-buffered saline (PBS) supplemented with 2 mg DNase I, 5 mg lysozyme, and three Roche complete mini EDTA-free protease inhibitor cocktail tablets. Cells were sonicated on ice using a 600-W model VCX ultrasonicator. The cell debris was pelleted by centrifugation (40,900 × g, 30 min, 4°C), and the clarified cell lysate was loaded onto a 5-ml GSTrap column (GE Healthcare) equilibrated with PBS and maintained at 4°C. Unbound protein was washed from the column with a 10× column volume (CV) isocratic PBS wash. Protein was eluted with 50 ml of elution buffer (50 mM Tris [pH 8.0], 15 mM glutathione). Fractions containing glutathione-S-transferase (GST)-tagged IRF3173-426, as judged by SDS-PAGE analysis, were pooled, concentrated, and exchanged into cleavage buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol) using 10-kDa molecular mass cutoff Millipore Centricon filter devices. The concentration of the 5-ml pool was determined by the Bradford assay, and 15 U of PreScission protease (GE Healthcare) per mg of GST-IRF3173-426 was then added to the pool and incubated for 16 h at 4°C. The cleaved protein sample was diluted to 7 ml with PBS and loaded onto the GSTrap column, which was equilibrated with PBS and maintained at 4°C. The column flow-through, expected to contain detagged IRF3, was collected and saved. The column was washed with 12 ml of PBS, and the flow-through was again collected and saved. All samples were analyzed by SDS-PAGE, and those containing detagged IRF3 were concentrated and exchanged into Mono-Q load buffer (20 mM Tris [pH 7.5], 10 mM β-mercaptoethanol) using 10-kDa molecular mass cutoff Millipore Centricon filter devices. The concentrated protein was loaded onto an 8-ml Mono-Q 10/100 GL column (GE Healthcare) equilibrated with load buffer and maintained at 4°C. Following loading of the column, unbound protein was washed from the column with a 2× CV of load buffer. A 20× CV gradient from 0% to 100% Mono-Q elution buffer (20 mM Tris [pH 7.5], 250 mM NaCl) was used to elute purified IRF3. Fractions containing IRF3173-426, as judged by SDS-PAGE analysis, were pooled and concentrated to 1 mg/ml. Glycerol was added to the protein to 10% (vol/vol). Aliquots of protein were frozen on dry ice and then stored at −80°C.
IRF31-426 was purified in the same manner as IRF3173-426, with the exception that Rosetta2(DE3)(pLysS) cells (Novagen) were used and LB was supplemented with 50 μg/ml of carbenicillin and 37 μg/ml of chloramphenicol. Lysozyme was also eliminated from the lysis step.
For SARS-CoV PLP purification, the catalytic core of SARS-CoV PLP was purified as previously described (6).
Native gel analysis.
Native gel analysis consisted of incubation of various amounts and combinations of proteins under native conditions and then analyzing their migration by native PAGE. IKKi, IKKβ, and TBK1 kinases were purchased from Invitrogen, while IκBα and NF-κB (p50) were purchased from Biomol International. Incubations generally consisted of 10- to 20-μl mixtures containing 5 to 20 μg of each protein, alone or combined with another protein, in 20 mM Tris (pH 7.5). Incubation mixtures were kept on ice for 10 to 15 min before an equal volume of 2× native gel sample buffer (125 mM Tris [pH 6.8], 20% glycerol, 0.01 mg/ml bromophenol blue) was added. Typically, entire samples were loaded onto Tris-HCl 4 to 20% gradient or 10% acrylamide gels equilibrated in running buffer (25 mM Tris, 190 mM glycine) at 4°C.
Cross-linking experiments.
Proteins were cross-linked according to the protocol supplied with the bis(sulfosuccinimidyl)suberate (BS3) cross-linking reagent (Pierce). Prior to cross-linking, all proteins were dialyzed into buffer containing 20 mM HEPES (pH 7.5) using Slide-A-Lyzer minidialysis tubes (Pierce). In each dialysis tube, 100 μl of each purified protein was dialyzed into 1 liter of 20 mM HEPES (pH 7.5) for 2 h at 4°C with stirring in order to remove Tris buffer, which would react with the cross-linking reagent. Each 30-μl cross-linking reaction mixture contained 20 μM of each protein, either alone or with another protein, and 1.5 mM freshly prepared BS3. Reaction mixtures were incubated at room temperature for 35 min and then quenched with 1 μl of 1 M Tris (pH 7.5) followed by an additional 15-min incubation. Ten microliters of 4× nonreducing SDS-PAGE sample buffer was added to each sample. Twenty microliters of each sample was then loaded onto a 12.5% SDS-PAGE gel.
IKKi and TBK1 phosphorylation of IRF3.
Reactions to test the effect of purified PLP on the phosphorylation of purified IRF3173-426 by IKKi (Invitrogen) were performed in 30-μl reaction mixtures at room temperature for 1 h. Reaction mixtures contained 10 μg of purified IRF3173-426, 1× Z′-LYTE kinase buffer (Invitrogen), 0.7 μg of IKKi, 330 μM ATP, and 20 μg of PLP. One reaction mixture lacked ATP and PLP, and another lacked PLP. Fifteen to 20 μl of each reaction was analyzed by SDS-PAGE. The same experiment was performed with 0.54 μg of TBK1 (Invitrogen) as the kinase. For native gel analysis, 10 μl of each sample was diluted with 10 μl of 2× native gel sample buffer, and samples were run on a Tris-HCl 4 to 20% gradient gel under native conditions as described above.
IKKβ phosphorylation of IκBα.
Phosphorylation experiments were performed as described above for IRF3, but with 6 μg of IκBα (Biomol) as the substrate and 0.82 μg of IKKβ (Invitrogen) as the kinase.
RESULTS
VRP-based IFN antagonist screen.
The Venezuelan equine encephalitis virus (VEE), an alphavirus, has been engineered to produce replication-deficient particles (called VRPs), which encode a foreign transgene under the control of the endogenous viral 26S subgenomic mRNA promoter (4). As VEE and VRP vectors induce robust type I IFN responses following infection (47, 52), expression of an IFN antagonist from the 26S promoter may downregulate VRP-induced IFN responses in cell culture (1).
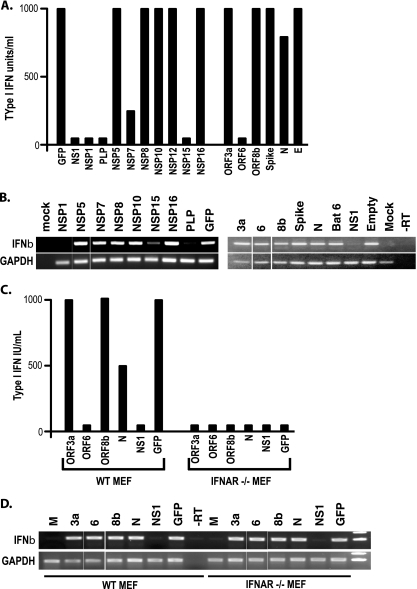
To test this hypothesis, we inserted several SARS-CoV structural and nonstructural ORFs, accessory ORFs, and the influenza virus NS1 genes into VEE vectors and isolated high-titer VRPs. The VRPs expressed robust levels of each of the SARS replicase, accessory, and structural proteins (see Fig. S1 in the supplemental material). Cultures of MA104 cells were infected with each VRP, and the induction and secretion of type I IFN were measured by IFN bioassay (Fig. 1A and B). When analyzing the accessory ORFs of SARS-CoV for their ability to block type I IFN expression, the data demonstrated that SARS-CoV ORF6 protein, and to a lesser extent the N protein, reduced the accumulation of IFN in the media of infected cells, along with NS1 of influenza virus, a known IFN antagonist (20) (Fig. 1A). Additionally, we were able to assay for IFN-β mRNA induction by RT-PCR. Concordant with the reported literature, the influenza virus NS1 gene blocked induction of IFN-β mRNA during VRP infection but ORF6- and N-expressing VRPs did not. While the RT-PCR assay is not quantitative, VRP/NS1-infected cells show a clear lack of IFN-β mRNA by RT-PCR while VRP/ORF6-infected cells show a clear presence of IFN-β mRNA. We have previously shown that ORF6 protein antagonizes STAT1 signaling by blocking STAT1's nuclear import, and several groups have implicated the CoV N protein as an antagonist as well, supporting the robustness of this assay (18, 29, 56).
FIG. 1.
VRP-based screen for IFN antagonists. (A) MA104 cells were infected with VRPs expressing the identified ORFs at an MOI of 5 for 24 h. Type I IFN was quantified from supernatant of the infections using a type I IFN bioassay. Shown is the average of three experiments, each in triplicate. (B) RNA was isolated from the same infections as in panel A and used for RT-PCR of IFN-β and GAPDH expression. (C) MEFs from wild-type (WT) 129 mice and IFNAR−/− mice were infected with the identified VRPs and analyzed by bioassay (C) and RT-PCR for IFN-β and GAPDH expression (D).
To determine whether the IFN bioassay was measuring the accumulation of type I IFN that was amplified via the JAK/STAT pathway or just the initially IRF3-induced IFN-β being secreted from the cell, we isolated mouse embryonic fibroblasts (MEFs) from IFN-α/β receptor-knockout (IFNAR−/−) mice. VRPs expressing GFP efficiently induced the secretion of IFN-β in wild-type MEFs (Fig. 1C and D). In contrast, infection of IFNAR−/− MEFs induced IFN-β mRNA synthesis but did not accumulate significant amounts of IFN-β in the media, as measured by bioassay. These data demonstrate that the VRP-based IFN bioassay screen provides an effective IFN-β amplification pathway for the detection of IFN antagonists.
Identification of SARS-CoV replicase protein IFN antagonists.
The VRP IFN antagonism assay identified ORF6 and N as IFN antagonists, consonant with previous reports indicating their role in IFN antagonism (18, 29). Next, the VRP system was used to screen the NSPs (also called replicase proteins) encoded in SARS-CoV ORF1a and ORF1b for IFN antagonism activity. While several NSPs were unable to be stably cloned into VRPs either due to their large size (NSP3) or inherent instability (NSP4, -13, and -14); NSP1, NSP2, NSP5, NSP6 to -12, NSP15, and NSP16 were efficiently cloned and expressed from VRP-infected MA104 cells (see Fig. S1 in the supplemental material). However, VRPs expressing NSP2, NSP9, ORF3b, ORF7a, and M were also stable, but none demonstrated IFN antagonist ability in these assays. However, transgene expression could not be shown due to inadequate antibodies for Western blot analyses and has been excluded from the analysis. In VRP-infected MA104 cells, the IFN bioassay identified NSP1, NSP7, NSP15, and the PLP domain of NSP3 as probable IFN antagonists. Interestingly, NSP1, NSP15, and PLP also effectively reduced IFN-β mRNA induction, while NSP7 expression induced normal levels of IFN-β mRNA (Fig. 1B). Previous studies using overexpression plasmids and reporter assays had only implicated NSP1 and NSP3 as potential antagonists, demonstrating the ability of the VRP assay to detect novel antagonist activities (53). These data suggest that NSP1, NSP7, NSP15, and PLP may be SARS-CoV-encoded IFN antagonists. NSP7 and NSP15 are currently being analyzed in our lab. Below we analyze the IFN antagonist properties of PLP.
SARS-CoV PLP blocks the IRF3 pathway.
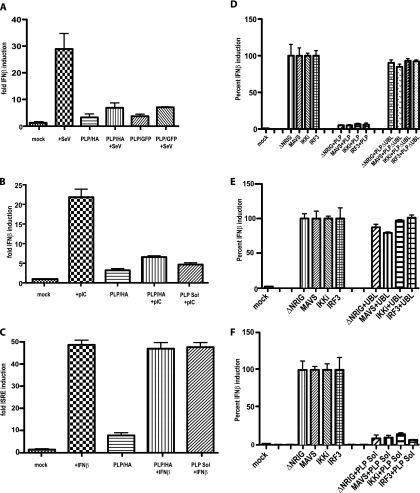
Previous groups have reported conflicting results concerning the mechanism of PLP antagonism of IFN expression (13, 53). Based on the VRP assay, our data (Fig. 1B) supported these earlier findings that had suggested that PLP was blocking the IRF3-based IFN induction pathway. Consequently, we determined whether PLP affected IFN-β promoter expression from an IFN-β/luciferase reporter plasmid, using dsRNA and SeV as inducers (Fig. 2A and B). Briefly, 293T cells were transfected with an IFN-β/luciferase reporter plasmid and either a GFP-encoding control plasmid or a plasmid expressing HA-tagged PLP or GFP-tagged PLP, as well as a control plasmid expressing Renilla luciferase under the SV40 promoter as a transfection control. At 24 h posttransfection, cells were retransfected with 2 μg poly(I-C) per well (a mimic of dsRNA) or infected with SeV. Six hours after treatment, the cultures were analyzed for the amount of luciferase expressed from the IFN-β promoter. Equal amounts of lysates were also used for Western blotting analysis to ensure equal expression for each transfection (see Fig. S2A in the supplemental material). We found that GFP- and HA-tagged PLP were able to inhibit poly(I-C)- or SeV-induced IFN-β induction. We do not believe that PLP expression is affecting SeV infection, since at 48 h postinfection, SeV-infected wells are showing a CPE, while uninfected cultures show no CPE. This demonstrates that SeV replicates to comparable levels in the transfected and untransfected cells. Using a V5-tagged plasmid (PLP Sol) identical to that used by Devaraj et al. (13), we found that these plasmids act the same as the HA- and GFP-tagged PLP constructs. Equal amounts of lysates were also used for Western blotting analysis to ensure equal expression for each transfection (see Fig. S2B in the supplemental material). Consonant with our earlier findings and reports in the literature (13), PLP expression blocked poly(I-C) and SeV induction of gene expression from the IFN-β promoter (Fig. 2A and B).
FIG. 2.
SARS PLP is an IFN antagonist. (A) HA- and GFP-tagged PLP expression plasmids were transfected into 293T cells with an IFN-β/luciferase construct. At 24 h posttransfection, cells were infected with SeV for 6 h and then luciferase was assayed to quantitate the level of induction of IFN-β transcription. (B) Using the same assay as in panel A, poly(I-C) was used as the inducer of IFN-β transcript. (C) To analyze whether PLP affected JAK/STAT signaling, PLP was transfected into cells with an ISRE/luciferase plasmid and incubated for 24 h. At 24 h, cells were treated with IFN-β protein for 6 h and then assayed for the induction of the ISRE promoter. We do not see a block in Jak/STAT signaling. (D) To identify where in the IFN-β induction pathway PLP is acting, cells were transfected with the IFN-β/luciferase reporter and plasmids containing either GFP, N-RIG, MAVS, IKKi or IRF3. The level of induction of each was set at 100%. Each plasmid was also transfected with HA-tagged PLP. Note that PLP blocked induction by each plasmid, signifying that PLP is not inhibiting signaling events upstream of IRF3. On the right, PLPΔUBL was used instead of PLP in the same experiments. Note that PLPΔUBL does not inhibit any of the inducers. (E) The UBL of PLP was used by itself to see if it blocked IFN induction via Ν-RIG, MAVS, IKKi, or IRF3. The UBL of PLP does not block the induction of IFN-β by these proteins. (F) Using the previously published (13) PLP construct, we find that it does block induction of IFN-β when each of the IRF3 signaling pathway proteins is expressed.
PLP lacks activity against the JAK/STAT1 pathway.
To determine whether PLP blocked the JAK/STAT pathway, which is activated following the IFN-β interaction via the IFN-α/β receptor, we used a plasmid containing the IFN-stimulatable response element (ISRE) promoter element fused to the firefly luciferase ORF (ISRE/luciferase). Treatment of cells with IFN-β protein induces strong expression of luciferase in cells transfected with the ISRE/luciferase construct. At 24 h after cotransfection of PLP with the ISRE/luciferase plasmid, cells were treated with IFN-β for 6 h, which induced robust expression of luciferase in the presence or absence of PLP (Fig. 2C). Equal amounts of lysates were also used for Western blotting analysis to ensure equal expression for each transfection (see Fig. S2C in the supplemental material). These data demonstrate that PLP does not affect the JAK/STAT pathway.
PLP blocks IRF3 activation.
To identify the cellular targets associated with PLP antagonism, we analyzed whether PLP was inhibiting the IRF3 activation pathway (Fig. 2D). Individual components of the IRF3 signaling pathway (RIG-I, MAVS, IKKi, and IRF3) can be used to stimulate IFN-β gene induction. The overexpression of each induces IFN-β, and by using an IFN-β/luciferase plasmid as a reporter, we can assay changes in the induction of IFN-β. When plasmids expressing the IRF3 pathway components N-RIG, MAVS, IKKi, and IRF3 are transfected into cells, IFN-β transcription was induced (left third of Fig. 2D). Next, 293T cells were cotransfected with plasmids expressing N-RIG, MAVS, IKKi, or IRF3 with either a GFP-encoding control plasmid or an HA-tagged PLP plasmid. In Fig. 2D, we demonstrate robust induction of IFN-β luciferase expression following cotransfection with GFP control plasmid. In contrast, cotransfection of PLP with each pathway component resulted in a dramatic reduction in IFN-β reporter gene expression. Equal amounts of lysates were also used for Western blotting analysis to ensure equal expression for each transfection (see Fig. S2D in the supplemental material). These data suggest that PLP blocks the virus-sensing pathway at the level of IRF3 activation and/or signaling to the nucleus, in agreement with earlier reports (13).
PLP inhibits the phosphorylation of IRF3.
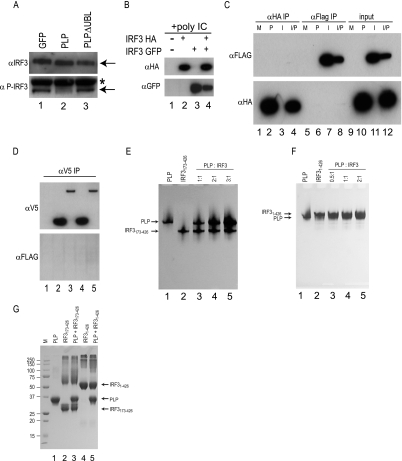
Activation of IRF3 is achieved by phosphorylation of IRF3 via the kinases IKKi and TBK1 (17). To determine if PLP antagonizes IRF3 phosphorylation, we transfected a plasmid expressing the IKKi kinase into 293T cells in both the presence and absence of HA-tagged PLP. IKKi overexpression should phosphorylate IRF3, leading to its activation, nuclear import, and the subsequent induction of IFN-β transcription. As previously reported, IKKi expression robustly phosphorylated IRF3, which was visualized using a phospho-specific, IRF3 antibody as a probe (Fig. 3A). Importantly, PLP expression blocked the phosphorylation of IRF3 while having no effect on total IRF3 protein levels. These data suggest that PLP is targeting an activity upstream of IRF3 that affects IRF3 phosphorylation.
FIG. 3.
Mechanism of PLP inhibition. (A) The effect of PLP on the phosphorylation of IRF3 was assayed by Western blotting. 293T cells were transfected with a plasmid expressing IKKi and either GFP, PLP, or PLPΔUBL for 24 h. Protein was analyzed by Western blotting with either anti-IRF3 antibody (αIRF3) or anti-phospho-IRF3 antibody (αP-IRF3). The arrows signify the specific bands in each lane, and the asterisk denotes a background band from the phospho-IRF3 antibody. (B) HA-tagged IRF3 and GFP-tagged IRF3 were either singly transfected or cotransfected into 293T cells to test for coimmunoprecipitation conditions. At 24 h posttransfection, 500 ng poly(I-C) was added for 6 h to induce IRF3 homodimerization and IFN-β. Lanes 2 and 3 show input extract blotted with anti-HA (αHA) or anti-GFP (αGFP) antibodies. The lysates from cotransfected cultures were used for immunoprecipitation with anti-HA antibody (αHA IP). The resulting immunoprecipitation is shown in lane 4. Note that HA- and GFP-tagged IRF3 was able to be immunoprecipitated with anti-HA antibody. (C) IRF3 and PLP were expressed either individually or together in 293T cells to identify if they bound each other in the cell. HA-tagged PLP and Flag-tagged IRF3 were transfected, and the extracts were used in immunoprecipitation experiments. In lanes 1 to 4, anti-HA antibody was used to immunoprecipitate the proteins. In lanes 5 to 8, anti-Flag antibody (αFLAG) was used for immunoprecipitation, and in the right section of the gel, 5% of the input for the immunoprecipitation was run in lanes 9 to 12. The top panel was visualized with anti-Flag antibody for the Western blot, and the bottom panel used anti-HA for the Western blot. M, mock transfection; P, PLP transfections only; I, IRF3 transfections only; and I/P, IRF3 and PLP transfected together. (D) V5-tagged PLP from Devaraj et al. (13) was used in IRF3 immunoprecipitations. Identical conditions were used as in panel B, but anti-V5 antibodies (αV5) were used for the pull down of V5-tagged protein complexes. Lane 1 is mock-immunoprecipitated extract. Lane 2 is V5 PLP Sol alone, lane 3 is V5-PLP TM alone, lane 4 is V5-PLP Sol cotransfected with Flag-tagged IRF3, and lane 5 is V5-PLP TM cotransfected with Flag-tagged IRF3. The top panel shows a Western blot of the immunoprecipitated extracts with anti-V5 antibody (αV5 IP), and the bottom panel is an identical Western blot with anti-Flag antibody. (E) Interactions between purified PLP and IRF3173-416 (E) or IRF31-426 (F) are shown as analyzed by 10% native-PAGE gels run at 4°C at pH 8.5. Proteins were incubated in different ratios, as indicated above the gels, for 10 min and then diluted with 2× sample buffer before being loaded onto the gels. Locations of the individual proteins are indicated by arrows to the left of the gels. (G) Cross-linking experiments with the cross-linking agent BS3 were performed between purified PLP and the two forms of purified IRF3. Following a 30-min incubation at room temperature in the presence of BS3, the protein mixtures were quenched and then analyzed by SDS-PAGE. Arrows to the right of the gel indicate the locations of the three individual proteins incubated without other proteins. Molecular mass marker (M) sizes are shown to the left of the gel in kDa.
PLP does not inhibit the in vitro phosphorylation of IRF3.
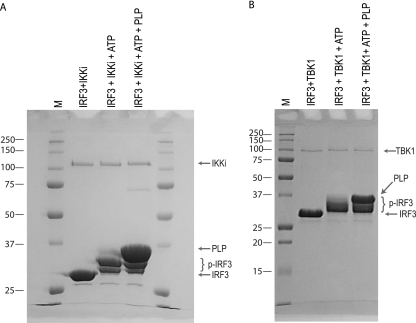
To determine if PLP interferes with the kinase activity of either of the upstream kinases, IKKi or TBK1, their activities were studied in an in vitro kinase assay using purified components. Purified IRF3 and each kinase were incubated in the absence and presence of purified PLP. SDS-PAGE analysis of the reaction products revealed that both IKKi and TBK1 catalyze robust phosphorylation of IRF3, as indicated by the more slowly migrating bands of the IRF3 protein (Fig. 4A and B). The presence of PLP in these reactions had little if any effect on the level of phosphorylation, suggesting that PLP does not directly antagonize the phosphorylation status of IRF3.
FIG. 4.
PLP does not inhibit the in vitro phosphorylation of IRF3. Purified IRF3173-416 was incubated with IKKi (A) and TBK1 (B) and ATP to induce phosphorylation and then analyzed by SDS-PAGE. The same reactions were carried out in the presence of PLP to determine if PLP interferes with IRF3 phosphorylation. Locations of nonphosphorylated IRF3, phosphorylated IRF3 (p-IRF3), PLP, IKKi, and TBK1 are indicated by arrows or brackets to the right of the gels. Molecular mass marker (M) sizes are shown to the left of the gels in kDa.
IRF3 and PLP do not directly interact.
In contrast to our findings noted above, a recent study reported that a direct interaction between IRF3 and PLP was associated with a block in phosphorylation of IRF3 by IKKi or TBK1 (13). To reconfirm whether a direct interaction occurs within cells, a Flag-tagged IRF3 construct and HA-tagged PLP were singly or doubly expressed in 293T cells. After transfection, the protein complexes were extracted in NP40-containing buffer and the membrane fractions were removed. Supernatant was assayed for both Flag-tagged IRF3 and HA-tagged PLP, and both proteins were found to be robustly expressed in the transfected cultures (Fig. 3C, input). Anti-Flag tag and anti-HA antibodies were used in coimmunoprecipitation experiments to directly evaluate an IRF3-PLP interaction. After coimmunoprecipitation with either the anti-FLAG or anti-HA antibodies, Western blotting of the immunoprecipitated proteins demonstrated that IRF3 and PLP were not bound in complexes despite robust levels of Flag-tagged IRF3 or HA-tagged PLP expression (Fig. 3C). Therefore, no IRF3/PLP complexes were detected in these assays. To demonstrate that IRF3 coimmunoprecipitation was possible under these extraction conditions, HA-tagged and Flag-tagged IRF3 were either singly or dually transfected into 293T cells. At 24 h posttransfection, cells were transfected with poly(I-C) as for the previous transfections, and 6 h later, lysates were harvested. Lysates were immunoprecipitated under conditions identical conditions to those described for PLP and IRF3, and the resulting immunoprecipitated protein was analyzed by Western blotting using anti-HA and anti-Flag antibodies (Fig. 3B). We found that Flag-tagged IRF3 was able to successfully immunoprecipitate HA-tagged IRF3 under these conditions, demonstrating that the lack of IRF3/PLP binding is not due to experimental or technical protocols. We also performed coimmunoprecipitation with the exact plasmids used in the studies of Devaraj et al. (13): V5-tagged cytoplasmic PLP and PLP containing the normal downstream transmembrane domain (PLP TM). Using the identical procedure described above, we immunoprecipitated V5-tagged PLPs alone or cotransfected with Flag-tagged IRF3. IRF3 was not coimmunoprecipitated with either PLP (Fig. 3D). Therefore, our data do not support a direct interaction between PLP and IRF3 in the context of transfected cells.
To probe the potential interaction of PLP and IRF3 using purified components in vitro, PLP and two versions of human IRF3, including a full-length protein (IRF31-426) and IRF3 protein lacking its DNA-binding domain (IRF3173-416), were purified to homogeneity and then analyzed for a potential intermolecular interactions by native gel electrophoresis. Neither the full-length nor truncated IRF3 proteins, at different protein/protein molar ratios, formed a complex with PLP, as no larger-molecular-mass bands were visible on the gel (Fig. 3E and F). To investigate the potential that a PLP/IRF3 complex may be too weak to withstand native gel electrophoresis, the proteins were incubated and then cross-linked with a reagent that covalently links lysine side chains that are in close proximity. In the presence of cross-linker, both versions of IRF3 cross-link with themselves, as they are known to form dimers (Fig. 3G); however, no additional higher-order bands representing a PLP/IRF3 complex were observed when PLP was added to the incubation. Both the native gel and cross-linking experiments corroborate our earlier finding that PLP is not directly binding to IRF3.
PLP blocks NF-κB induction.
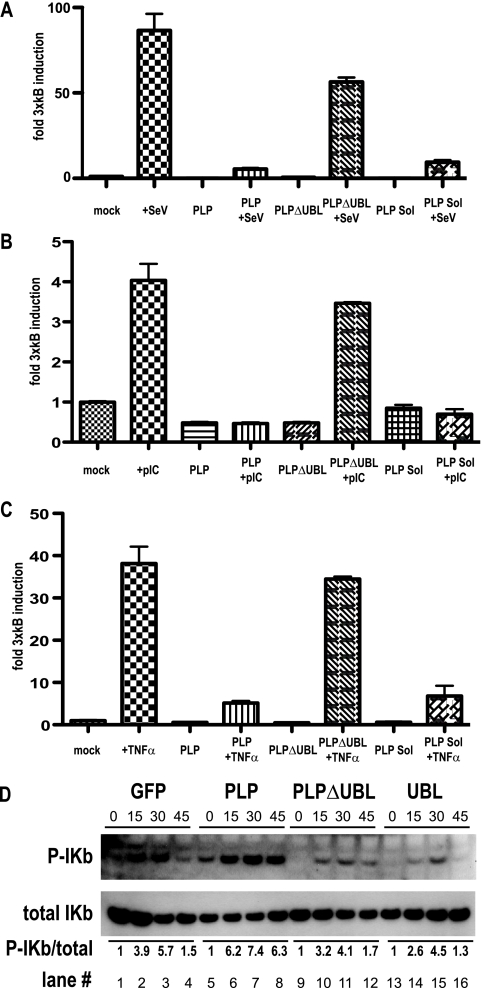
The NF-κB pathway is another important signaling cascade that regulates innate immune responses. Similar to components in the IRF3 pathway, phosphorylation plays a central role in the induction of NF-κB-induced genes. Because PLP was affecting the IRF3 pathway by blocking phosphorylation, it was reasonable to determine if PLP similarly targeted the NF-κB pathway. Therefore, the NF-κB reporter plasmid was transfected into 293T cells with SV40/Renilla luciferase control plasmid, and the cells were then treated with either SeV (MOI of 5), poly(I-C) (2 μg), or tumor necrosis factor alpha (TNF-α) (10 ng). As expected, these stimuli induced robust induction of the NF-κB/luciferase construct (Fig. 5A, B, and C). In contrast to earlier reports (13), cotransfection with HA-tagged PLP plasmid reduced luciferase expression following SeV, poly(I-C), or TNF-α treatment. We tested the V5-tagged PLP plasmid (named PLP Sol) used by Devaraj et al. (13) and demonstrated a similar block in NF-κB signaling (Fig. 5A, B, and C, PLP Sol lanes). Equal amounts of lysates were also used for Western blotting analysis to ensure equal expression for each transfection (see Fig. S3A, B, and C in the supplemental material).
FIG. 5.
PLP inhibits the NF-κB signaling pathway. (A) 293T cells were transfected with a 3xκB/luciferase reporter plasmid that reports NF-κB-mediated gene induction. The reporter was transfected with PLP, PLPΔUBL, or PLP-Sol-expressing plasmids (13). At 24 h after transfection, cells were treated wither either SeV (MOI of 5) (A), poly(I-C) (2 μg) (B), or TNF-α (10 ng) (C). Note that both PLP and PLP-Sol block NF-κB-mediated gene induction, while PLPΔUBL does not. (D) The effect of PLP on NF-κB signaling was assayed by Western blotting. 293T cells were transfected with GFP, PLP, PLPΔUBL, or UBL alone for 24 h. At 24 h posttransfection, cells were treated with 10 ng of TNF-α for 0, 15, 30, or 45 min and proteins were extracted at those time points. Western blots were analyzed for either total IKb or phospho-IKb. Below the blot is the numerical ratio of total IKb to phospho-IKb as quantified by IPLAB.
As both the IRF3 and NF-κB pathways are dependent on IKK kinase phosphorylation-based activation (26), PLP could be inhibiting the IKK kinase family, which mediates Ikbα phosphorylation. To test this hypothesis, 293T cells were transfected with a GFP plasmid or SARS-CoV PLP and treated with 10 ng of TNF-α per well for the specified time points (Fig. 5D). Proteins were isolated and assayed by Western blotting for Iκbα phosphorylation levels, using an Iκbα phospho-specific antibody. Iκbα phosphorylation decreased across the time points in the control lanes transfected with a plasmid encoding GFP (1-4), consonant with the expected proteasome degradation that occurs after phosphorylation (50). Under identical conditions, PLP coexpression stabilized the levels of Iκbα phosphorylation. We find that SARS-CoV PLP expression does not block Iκbα phosphorylation but in fact stabilizes IκBa protein levels by inhibiting its degradation.
PLP does not inhibit IκBα kinase function in vitro.
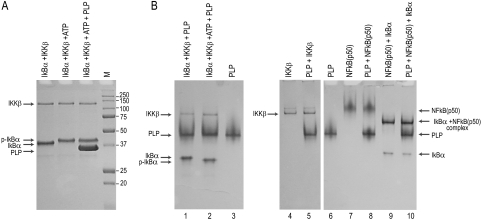
To examine if PLP interferes with IκBα phosphorylation in vitro, purified IκBα was incubated with purified IKKβ and ATP in both the absence and presence of PLP and then analyzed by SDS-PAGE and native PAGE. IκBα was successfully phosphorylated, but the presence of PLP did not reduce the kinase activity of IKKβ with its IκBα target (Fig. 6A). PLP does not appear to physically interact with either IκBα or phosphorylated IκBα as judged by the native gel analysis (Fig. 6B, lanes 1 and 2).
FIG. 6.
Analysis of the effect of PLP on IκBα kinase activity and of PLP interactions with various NF-κB pathway proteins. (A) IκBα was incubated with IKKβ and ATP to induce phosphorylation and then analyzed by SDS-PAGE. The same reactions were carried out in the presence of PLP to determine if PLP interferes with kinase activity. Locations of nonphosphorylated and phosphorylated IκBα, PLP, and IKKβ are indicated by arrows. Molecular mass marker (M) sizes are shown to the right of the gel in kDa. (B) Native gel analysis of PLP interactions with NF-κB pathway proteins. Proteins were mixed, as indicated above the gels, and incubated in 50 mM HEPES (pH 7.5) for 10 min at 4°C and then diluted with 2× sample buffer before being loaded onto the gels. Locations of the individual proteins are indicated by arrows to the left and right of the gels.
To determine if PLP interacts with other components of the NF-κB pathway, PLP was incubated with either individual proteins or specific protein complexes associated with the pathway. Samples were then analyzed by native PAGE (Fig. 6B). PLP did not bind the NF-κB(p50) subunit (lane 8), nor the NF-κB(p50)/IκBα complex (lane 10), under conditions in which a strong and expected association was noted between NF-κB(p50) and IκBα (Fig. 6B, lanes 7 and 9). These data indicate that the conditions in native PAGE do not interfere with the formation of an established complex. No association between PLP and the IKKβ kinase was observed either (Fig. 6B, lane 5). These in vitro data indicated that PLP does not likely block NF-κB signaling by inhibiting IKKβ kinase function.
NL63 PLP2, but not MHV PLP2, acts as an IFN antagonist.
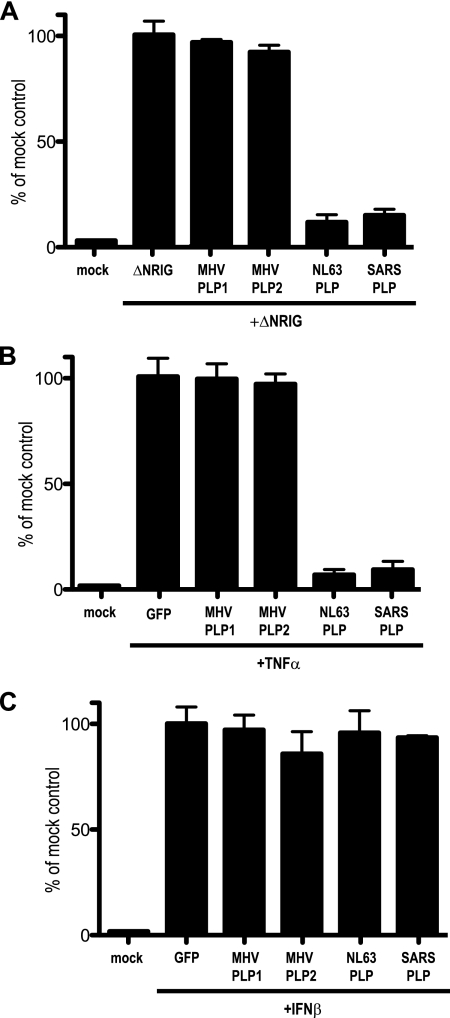
We next evaluated whether the IFN antagonism properties of the SARS-CoV PLP was conserved across other CoVs. Both MHV and HCoV-NL63 encode two PLPs, PLP1 and PLP2, the latter of each being the protein most homologous to SARS-CoV PLP2 and conserved across CoVs (24). To evaluate IFN antagonist activity, 293T cells were transfected with the IFN-β reporter plasmid, SV40/Renilla luciferase control plasmid, the constitutively active N-RIG construct, and either empty plasmid, MHV PLP1, MHV PLP2, or HCoV-NL63 PLP plasmid. At 24 h posttransfection, cells were lysed and assayed for levels of luciferase expression (Fig. 7). As shown in Fig. 2D, N-RIG-transfected cells expressed large amounts of luciferase, starkly contrasting the inhibition in expression noted following coexpression of N-RIG with SARS-CoV PLP. In contrast, cultures cotransfected with N-RIG and either MHV PLP1 or PLP2 did not block IFN-β induction, nor did they block NF-κB or STAT1 signaling (Fig. 7A to C). Importantly, NL63 PLP was able to block IFN-β induction as well as NF-κB signaling, similar to results reported with the SARS-CoV PLP. Equal amounts of lysates were also used for Western blotting analysis to ensure equal expression for each transfection (see Fig. S4A to C in the supplemental material). It is noted that MHV PLP1 expresses at a slightly lower level than to MHV PLP2 and SARS PLP. These data suggest that the CoV PLPs differentially antagonize the IFN pathway, although it is possible that MHV PLP antagonism might require an appropriate conformational presentation as part of NSP3.
FIG. 7.
PLP from NL63 but not MHV is an IFN antagonist. PLPs from NL63 and MHV were assayed for their ability to inhibit IFN-β, NF-κB, and STAT1 signaling in 293T cells. (A) In this assay, 293T cells were transfected with N-RIG and either GFP-, MHV PLP1-, MHV PLP2-, or NL63 PLP2-expressing plasmids and an IFN-β/luciferase reporter plasmid. At 24 h posttransfection, cells were assayed for luciferase expression. The value of induction in GFP plus N-RIG was set at 100%, with all other values in relation to it. Notice that MHV PLP1 and -2 do not block IFN-β induction, while NL63 and SARS-CoV PLP do. (B) 293T cells were transfected with GFP-, MHV PLP1-, MHV PLP2-, or NL63 PLP2-expressing plasmids and an NF-κB/luciferase reporter plasmid. At 24 h posttransfection, cells were treated with 100 ng of TNF-α; 6 h later, cells were assayed for luciferase expression. The value of induction in GFP plus TNF-α was set at 100%, with all other values in relation to it. Notice that MHV PLP1 and -2 do not block IFN-β induction, while NL63 and SARS-CoV PLP do. (C) 293T cells were transfected with either GFP-, MHV PLP1-, MHV PLP2-, or NL63 PLP2-expressing plasmids and an ISRE/luciferase reporter plasmid. At 24 h posttransfection, cells were treated with 100 ng of IFN-β; 6 h later, cells were assayed for luciferase expression. The value of induction in GFP plus IFNβ was set at 100%, with all other values in relation to it. Notice that none of the expressed proteins inhibits ISRE induction.
UBL domain of PLP is necessary for the IFN antagonism activity of PLP.
The crystal structure of the SARS-CoV PLP demonstrated two major domains: an N-terminal region containing a UBL domain and a C-terminal region containing the protease active site (42). To determine whether the UBL domain functioned in innate immune antagonism, we constructed a UBL-deleted PLP mutant (PLPΔUBL) that lacked the N-terminal 60 amino acids (aa) of PLP (aa 683 to 783 of NSP3 or aa 1 to 60 of the PLP expressed in this work), based largely on the domain segmentation represented in the crystal structure. The PLPΔUBL mutant, but not wild-type PLP, failed to block IFN-β induction by overexpression of RIG, MAVS, IKKi, and IRF3 (Fig. 2D). Moreover, PLPΔUBL was also unable to block NF-κB/luciferase induction (Fig. 5A, B, and C). Under identical conditions, wild-type PLP efficiently blocked IFN-β and NF-κB induction (Fig. 2D and 5A, B, and C).
To determine if the lone UBL domain was an inhibitor of IRF3, we repeated these assays expressing the 60-aa UBL domain and found that it was unable to block RIG-, MAVS-, IKKi-, or IRF3-mediated induction (Fig. 2E). One caveat to these experiments is that when expressed alone, the UBL domain may not fold correctly. We do not think this is an issue since UBL domains are globular structurally stable protein folds, and this domain resembles wild-type ubiquitin by crystal structure. However, the potential for misfolding is noted. These data suggested that the UBL domain of PLP is necessary for blocking IRF3 and NF-κB signaling yet was not sufficient to inhibit that signaling.
The UBL domain is not required for PLP protease activity.
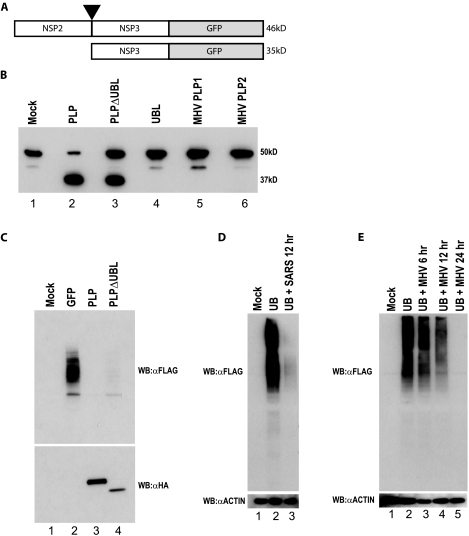
As previous studies have implicated the PLP protease function in IFN antagonism activity (13), we next determined whether the UBL-deleted PLP (PLPΔUBL), retained functional protease activity. To test for protease function of PLPΔUBL, we amplified a region of the SARS-CoV genome containing the C-terminal 80 aa of NSP2 and the N-terminal 100 aa of NSP3, spanning the NSP2/3 cleavage site that is normally proteolytically cleaved by PLP (Fig. 8A). The NSP2/3 peptide was fused to GFP in frame and expressed from a constitutive promoter in 293T cells. The uncleaved NSP2/NSP3/GFP fusion protein had the expected molecular mass of ∼46 kDa (Fig. 8B, lane 1) and was processed by PLP into an ∼35-kDa product (with a C-terminally tagged GFP) and an N-terminal ∼10-kDa product, with the former product detectable by GFP antibodies. Importantly, PLPΔUBL is capable of processing the precursor into the expected 35-kDa protein product, although the presence of the residual 46-kDa precursor protein suggested a slight reduction in proteolytic activity. Thus, the PLPΔUBL retains its ability to proteolytically process the target polyprotein. Importantly, both the SARS-CoV UBL domain and the MHV PLP1 and PLP2 proteins were also unable to cleave the precursor protein, demonstrating the specificity of the reaction for the substrate (Fig. 8B).
FIG. 8.
PLPΔUBL retains its protease and DUB functions. (A) A schematic of the SARS-CoV polyprotein cleavage assay is shown. The C terminal 100 aa of NSP2 through the N-terminal 80 aa of NSP3 were fused to GFP in an expression plasmid. PLP should cleave between NSP2 and -3 at the black arrowhead if the PLP protease is functional. The uncleaved product should be 46 kDa (46kD), and the cleaved product should be 35 kDa (35kD) when assayed on an SDS-PAGE gel. (B) 293T cells were transfected with either the cleavage reporter alone (mock) or the reporter and each plasmid noted above the Western blot. Proteins were extracted 24 h posttransfection and analyzed by Western blotting with an anti-GFP antibody to identify the cleaved or uncleaved cleavage reporter. The mock lane (lane 1) shows the full NSP2/3/GFP reporter. The PLP lane shows the cleaved product at ∼35 kDa. (C) PLPΔUBL retains its DUB activity. 293T cells were transfected with Flag-tagged ubiquitin and either GFP, PLP, or PLPΔUBL. Proteins were extracted 24 h posttransfection and analyzed by Western blotting (WB) with either an anti-HA antibody (αHA) (bottom panel) to visualize the PLP expression or anti-Flag antibody (αFLAG) to visualize Flag-tagged ubiquitin (top panel). (D) Vero cells were transfected with Flag-tagged ubiquitin and infected with icSARS-CoV at an MOI of 5 24 h posttransfection. Cells were lysed at 12 h postinfection and assayed for anti-Flag and antiactin (αACTIN) staining by Western blotting. (E) BHK cells were transfected with Flag-tagged ubiquitin and infected with MHV-A59 at an MOI of 5 24 h posttransfection. Cells were lysed at 6, 12, and 24 h postinfection and assayed for anti-Flag and antiactin staining by Western blotting.
PLPΔUBL retains DUB activity.
SARS-CoV PLP has been shown to have a significant amount of DUB activity (5). One explanation for PLPΔUBL losing its IFN antagonist activity is that the deletion of the UBL may specifically disrupt the DUB activity of PLP, which may be responsible for the IFN antagonism. To test this hypothesis, 293T cells were transfected with a Flag-tagged ubiquitin plasmid and either a plasmid expressing GFP, HA-tagged wild-type PLP, or HA-tagged PLPΔUBL. At 24 h posttransfection, cells were lysed and assayed by Western blotting with an anti-Flag antibody for the levels of Flag-tagged ubiquitin attached to proteins in the lysate (Fig. 8C, top panel). Flag-tagged ubiquitin is efficiently utilized by the cellular machinery and is conjugated to proteins in the cell, resulting in a smear of ubiquitinated proteins (Fig. 8C, lane 2). Following cotransfection with wild-type PLP plasmid and the tagged ubiquitin construct, PLP efficiently cleaves ubiquitin from normally ubiquitinated proteins. Importantly, PLPΔUBL also efficiently deubiquitinated proteins in the cell (Fig. 8C, lane 4). Thus, DUB activity was also uncoupled from the mechanism by which PLPΔUBL failed to antagonize IFN activity.
DUB activity of PLP could be due to an artifact of plasmid overexpression in cell culture. To exclude that possibility, we analyzed the DUB activity of SARS-CoV and MHV during virus infection. SARS-CoV DUB activity was analyzed by transfection of Vero E6 cells with the same Flag-tagged ubiquitin plasmid as before (Fig. 8D). At 24 h posttransfection, wild-type SARS-CoV (Urbani) was used to infect the cultures. At 12 h after infection, the cells were lysed and used for analysis by Western blotting. Similar experiments were performed using MHV-A59 for the infection: however, in those experiments, DBT cells were transfected with the Flag-tagged ubiquitin plasmid (Fig. 8E). In both cases, we found that ubiquitinated proteins are prevalent throughout uninfected cells. However, virus infection is characterized by an almost complete DUB of the cellular protein milieu. These data support the hypothesis that the DUB activity seen from plasmid-based expression of SARS-PLP is not an artifact of the expression system. Additionally, they demonstrates that MHV-A59 deubiquitinates during the course of its infection in cell culture, while its PLP2 does not seem to act as an antagonist in transfection studies. This may suggest that DUB is not a critical component of the IFN antagonism seen during infection or that MHV PLP requires additional flanking sequences for in vitro activity.
SARS-CoV PLP active site mutants show various degrees of IFN antagonism.
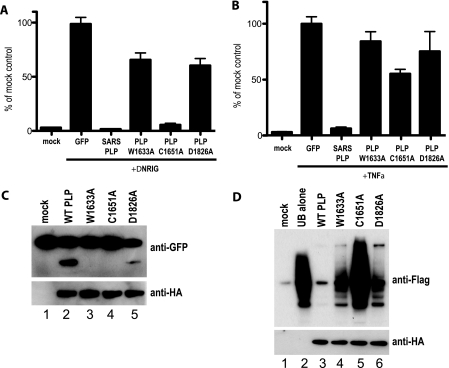
Since the PLPΔUBL mutant no longer blocked IRF3 or NF-κB signaling, we next determined whether various protease active site mutations had an effect on PLP's IFN antagonism activity. The SARS-CoV PLP active site is dependent upon a catalytic triad of amino acids, C1651, H1812, and D1826 (42). We mutated the C1651 and D1826 catalytic residues to alanine and also mutated amino acid W1633 of NSP3 to alanine, as this latter site likely blocks proteolytic activity by altering the dimensions of the protease pocket (42). When assayed in the presence of constitutive RIG-I expression, the W1633A and D1826A mutations were unable to block IFN-β induction, whereas the C1651A mutant retained near-wild-type activity in its IFN-β inhibition activity (Fig. 9A). To confirm levels of expression of the PLPs, Western blot analysis was performed on these lysates and we found that all PLPs are expressed at reasonable levels in the transfections (see Fig. S4D and E in the supplemental material).
FIG. 9.
PLP catalytic mutants play a role in IFN antagonism. (A) 293T cells were transfected with an IFN-β/luciferase plasmid that reports IRF3-mediated gene induction. The reporter was cotransfected with N-RIG and GFP, PLP, or the PLP W1633A, C1651A, or D1826A mutants. At 24 h after transfection, cells were lysed and assayed for luciferase expression. (B) 293T cells were transfected with a 3xκB/luciferase plasmid that reports NF-κB-mediated gene induction. The reporter was cotransfected with GFP, PLP, or the PLP W1633A, C1651A, or D1826A mutants. At 24 h after transfection, cells were treated with 10 ng of TNF-α, and 6 h after treatment, the cells were lysed and assayed for luciferase expression. (C) PLP mutants were tested for their cleavage activity using the construct and experimental design described in the legend to Fig. 8. The top panel is a Western blot for anti-GFP showing cleavage of the reporter protein, and the lower panel is a Western blot of the same extract probed with anti-HA antibody showing expression of the PLP mutants. (D) Deubiquitinase activities of the PLP mutants were assayed using the Flag-tagged ubiquitin assay described in the legend to Fig. 8. The top panel is a Western blot probed with anti-Flag antibody showing Flag-ubiquitin conjugation to cellular proteins. WT, wild type. The bottom panel is a Western blot with anti-HA antibody showing expression of the PLP variants.
When assayed for their ability to block NF-κB signaling, none of the three mutants showed the ability to block TNF-α-based induction of luciferase expression from the 3xκB/luciferase reporter plasmid (Fig. 9B). Interestingly, using the Flag-tagged ubiquitin assay, the W1633A and D1826A mutations allowed for robust DUB activity, while DUB activity was greatly reduced with the C1651A mutation (Fig. 9D). In our protease reporter assay, the W1633A and C1651A mutants lost the ability to cleave the GFP reporter substrate while the D1826A showed a reduction in proteolytic activity (Fig. 9C). These data demonstrate that both the protease activity and the UBL domain of PLP may play roles in IFN antagonism.
DISCUSSION
SARS-CoV is a highly pathogenic respiratory virus that causes 50% mortality rates in aged populations. Although the molecular basis for this highly virulent phenotype is unknown, our findings build upon earlier reports that implicate the SARS-CoV PLP as an IFN antagonist. Moreover, our novel IFN antagonist assay also identified the SARS-CoV NSP1, NSP7, NSP15, ORF6, and N protein as potential robust antagonists of host innate immune signaling pathways. Previous studies by our group and others have shown that NSP1 and ORF6 block IFN induction and IFN signaling by diverse mechanisms (18, 29, 53). Here we demonstrate that SARS-CoV PLP blocks the activation of IRF3 and NF-κB and that the UBL domain and catalytic site of PLP are necessary, but not sufficient, to block IFN-β and NF-κB promoter induction.
IFN antagonist screen.
VEE virus is a highly pathogenic alphavirus that has been genetically manipulated as a vaccine vector for the prevention of human and animal diseases (40). The VEE replicon system produces VRPs which are replication defective, meaning they can infect a cell but do not produce progeny viruses from that cell. VRPs are high-titer (<108 to 1010), single-hit particles that infect cells, replicate, and produce large amounts of the foreign protein without spreading beyond the initial target cell (11). Importantly VRPs induce high levels of type I IFN from all cell types we have tested, allowing for the detection of both IFN-β mRNA and type I IFN by bioassay. VRPs will efficiently infect (>95% of cells infected at an MOI of 5) mammalian, avian, and invertebrate cells, providing a screening platform for foreign genes that encode IFN antagonism activities in diverse species. Some advantages of the VRP screen include: (i) a large induction of type I IFN, allowing for a high signal/noise ratio in the assays; (ii) infection of virtually every cell in the culture; (iii) the capablity of infecting cell types that are highly resistant to transfection methodologies (e.g., MEFs derived from wild-type and gene knockout mice); and (iv) expression of the query gene in the context of virus infection rather than plasmid-based overexpression. Importantly, expression of antagonists from defective VRP vectors is safe since they do not spread in vitro or in vivo. One caveat to using VRP vectors is that transgene expression may potentially underrepresent or overrepresent the expression levels seen in wild-type SARS-CoV-infected cells. This potential problem is recognized and is why further follow-up with plasmid- and wild-type virus-based expression is used.
Wathelet et al. have evaluated the SARS ORF1ab NSPs for IFN antagonist activity and identified NSP1 and NSP3, using an SeV screen that coupled a plasmid-based antagonism expression with a luciferase-based readout for IFN induction (53). In addition to NSP1 and NSP3, the VRP approach also identified NSP7 and NSP15 as robust IFN antagonists. Again, the differences may reflect inherent differences between plasmid-based and VRP-based screens and/or differences in the innate levels of IFN induction associated with paramyxovirus and alphavirus infection. The most comprehensive interpretation of these data suggests that (i) combinations of screens may represent a better approach to identify potential candidates for downstream testing and (ii) more detailed studies of antagonism mechanisms in different viral genome backbones may be warranted.
IFN antagonists of SARS.
Many viruses encode proteins that block the innate immune response, and a detailed mechanistic understanding of the process may lead to the identification of therapeutic targets that can be used to attenuate replication and pathogenesis. For example, influenza viruses containing deletions of NS1, an IFN antagonist, are attenuated in vivo, suggesting a precise antiviral target for therapeutics (41). We have previously identified three SARS-CoV proteins, nucleocapsid, ORF3b, and ORF6, which antagonize host innate immune responses and demonstrated that ORF6 binds nuclear import factors to antagonize import of activated STAT1 (18, 29). Our finding, coupled with earlier reports, indicate that several replicase proteins of SARS-CoV encode important functions outside of a role in virus replication, suggesting an important role for ORF1 proteins in virulence and pathogenic disease outcomes that are uncoupled from virus load. Intriguingly, mutations in NSP1 (36, 53), NSP2 (22), and NSP14 (16) have been shown to affect various aspects of in vitro and in vivo infections and pathogenesis, although the exact mechanisms of their function is still under active investigation. Our findings support earlier work by Wathelet et al. (53), who demonstrated that NSP1 blocks several kinase, transcription, and translation steps that are normally induced during viral infection. In parallel, Narayanan et al. showed that NSP1 affects IFN-β gene transcript stability which augments NSP1's IFN antagonism activity (36). While the mechanisms of action of NSP1 and PLP have been well studied, the molecular mechanisms by which NSP7 and NSP15 block various aspects of the antiviral response are fertile ground for future investigations.
It is intriguing that SARS-CoV encodes at least seven IFN antagonist genes. Although the basis for encoding such a large repertoire of antagonists is unknown, poxviruses, paramyxoviruses, and filoviruses encode multiple IFN antagonism genes as well (28, 37, 44). Consistent with an important role for IFN in regulating the severity of CoV infection, MHV infection of IFN-α/β and IFN-γ receptor knockout mice enhances disease severity and rapidly progresses to fatal outcomes (8, 46). It has also been shown that SARS-CoV infection of either cell culture (18) or of macaques (12) results in a block in STAT1 translocation to the nucleus, suggesting that control of the IFN response is an important component in virus pathogenesis. It is likely that viruses encode multiple IFN antagonists to retard the induction of the innate immune responses during the early phases of infection to allow for efficient replication and spread. Alternatively, different antagonists may function more efficiently in different tissues and host cells that are targeted during infection.
PLP IFN antagonism.
Devaraj et al. (13) identified the SARS-CoV PLP as an IFN antagonist, implicating a direct interaction with IRF3 as a mechanism that blocked phosphorylation and nuclear import. In contrast, Wathelet et al. (53) identified NSP3 as an IFN antagonist but did not attempt to show an interaction directly with IRF3. In agreement with both laboratories, we show that PLP is an IFN antagonist that blocks the phosphorylation of IRF3 by a yet-to-be-identified upstream mechanism. However, we do not detect a direct IRF3-PLP interaction by immunoprecipitation assays using either cell lysates or purified protein components in in vitro biochemical assays. Additionally, a recent paper by Zheng et al. identified MHV PLP2 as a IFN antagonist and showed a direct interaction between IRF3 and PLP, both of which we cannot repeat in our experiments (58). Zheng et al. propose that the DUB activity of MHV PLP2 blocks IRF3 function, leading to the block in IFN antagonism. One caveat to their work is that PLP appears to nonspecifically deubiquitinate all proteins in the cell, including IRF3 (Fig. 8C, D, and E). They do not show this result in their paper, but we have demonstrated this fact here. Thus the interpretation that IRF3 DUB is solely critical for the IFN block is premature and not substantiated by the existing data. Devaraj et al. (13) also presented data showing that the catalytic activity of PLP was not essential for the IFN antagonist activity, although one mutant (C1651A) showed reduced activity compared with controls. Using a broader panel of active site mutants, we present data demonstrating that some of the mutants involving residues that participate directly in catalysis, or residues within the catalytic region of the active site, disrupt the IFN antagonism activity.
We also demonstrated a significant block in NF-κB signaling by PLP following stimulation with TNF-α, SeV, or dsRNA, in contrast to previous studies that assayed for antagonism of NF-κB signaling pathways by RT-PCR of the A20 message, an NF-κB-induced gene in some cell lines (13). The differences between our observations may reflect the sensitivity of our luciferase assay, the use of different cell types, or the differential induction of this particular A20 message via alternative signaling pathways. Importantly, we demonstrate a block in the NF-κB signaling pathway using three different inducers of NF-κB, including SeV, the dsRNA mimic poly(I-C), and TNF-α. Regardless of the inducer, we find that PLP is able to block NF-κB-mediated signaling. We do note that these contradictory findings certainly support the argument that multiple biochemical assays and inducers should be used to record NF-κB and IFN antagonism activities, coupled with well-defined mutants that display loss or gain of function.
Role of the UBL domain in IFN antagonism.
We found that the UBL domain of PLP was necessary but not sufficient for PLP's IFN antagonist activity. When the UBL was deleted from PLP, the protease and DUB functions of PLP were retained (Fig. 6). However, this form of PLP did not inhibit IRF3 phosphorylation or NF-κB signaling. Interestingly, the UBL domain alone was not able to inhibit these pathways either. Although the mechanism is unclear, an interaction between the UBL domain and the upstream effector proteins involved in activation of the pathway is a strong possibility (27). TBK1, IKKi, and IKKβ each contain UBL domains that are critical for their enzymatic activity, and the removal of these UBL domains has been shown to impair their function (27, 33). Interestingly, enzymatic activity can be restored with UBL domains derived from the reciprocal kinase, IKKi or TBK1. We are intrigued that the kinases and PLP both contain UBL domains, although we cannot show any direct involvement or direct protein-protein interactions in vitro or in vivo.
Role of the catalytic domain in IFN antagonism.
The catalytic region of PLP plays a role in IFN antagonism as well. Mutagenesis of the catalytic triad in the active site differentially affected IFN antagonism (Fig. 9). We found that mutagenesis of the active site at two different residues, which have been shown to knock out catalytic activity (6), also affects the antagonism activity. These data not only suggest that the catalytic site activity may be directly involved in the IFN and NF-κB antagonism phenotypes, but alternatively, the mutations may alter the PLP structure and, via long-range interactions, disrupt IFN antagonism activities. This possibility is strengthened by findings with the W1633A mutant, which loses IFN and NF-κB antagonism and protease activity but retains some DUB activity. Based on analysis of the X-ray structure, the W1633 residue is thought to be necessary for retention of the correct active site architecture. Mutation of this site is predicted to lead to a potential collapse of the active site pocket into a smaller pocket volume resulting from increased flexibility in the region created by the loss of a hydrogen bond. The decreased size would lead to the inability of the pocket to accommodate the larger leaving groups of the polyprotein but would perhaps still allow for the smaller isopeptide bonds of ubiquitin conjugates to bind, as observed in our assays. Although speculative, the catalytic site of PLP may bind and/or proteolytically degrade key cellular proteins that promote IFN expression. Alternatively, the mutations in the catalytic site may be affecting protein structure such that the cellular proteins interacting with wild-type PLP can no longer bind to the mutant form. Further X-ray structural analysis of the interactions with different substrates is warranted in order to tease out any structural effects on molecular recognition and catalysis.
In conclusion, we have used a novel IFN antagonist assay to identify NSP1, PLP, NSP7, NSP15, ORF6, and N as IFN antagonists. These findings, coupled with the identification of differential IFN antagonism activities encoded within different PLPs, suggest that Coronaviridae genomes likely encode multiple and different antagonists of innate immunity. The availability of a robust mouse model for studying the pathogenesis of SARS-CoV enables us to evaluate the role of these genes in pathogenesis. This allows for the ability to identify novel antagonists both in vitro and in vivo and to understand their mechanisms for evading the host innate immune response. Understanding the mechanism of action of each antagonist may also direct us to novel therapeutic targets on both the virus and the host.
Supplementary Material
Acknowledgments
We are grateful for the support of NIH grant P01AI060915 to A.D.M., NIH grant P01AI059443 to R.S.B., and NIH grant F32AI066542 to M.B.F.
We also thank Martha Collier for production of VRPs, John Hiscott for phospho-specific IRF3 antibodies, Luis Enjuanes and Jose Luis Nieto for SARS-CoV anti-E antibody, and Susan Baker for the V5-PLP plasmid and helpful discussions.
Footnotes
Published ahead of print on 15 April 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aguilar, P. V., A. P. Adams, E. Wang, W. Kang, A.-S. Carrara, M. Anishchenko, I. Frolov, and S. C. Weaver. 2008. Structural and nonstructural protein genome regions of eastern equine encephalitis virus are determinants of interferon sensitivity and murine virulence. J. Virol. 824920-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira, S., and H. Hemmi. 2003. Recognition of pathogen-associated molecular patterns by TLR family. Immunol. Lett. 8585-95. [DOI] [PubMed] [Google Scholar]
- 3.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasuriya, U. B. R., H. W. Heidner, J. F. Hedges, J. C. Williams, N. L. Davis, R. E. Johnston, and N. J. MacLachlan. 2000. Expression of the two major envelope proteins of equine arteritis virus as a heterodimer is necessary for induction of neutralizing antibodies in mice immunized with recombinant Venezuelan equine encephalitis virus replicon particles. J. Virol. 7410623-10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2006. Deubiquitinating activity of the SARS-CoV papain-like protease. Adv. Exp. Med. Biol. 58137-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 7915189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 9712289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cervantes-Barragan, L., R. Zust, F. Weber, M. Spiegel, K. S. Lang, S. Akira, V. Thiel, and B. Ludewig. 2006. Control of coronavirus infection through plasmacytoid dendritic cell-derived type I interferon. Blood 1091131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornell, C. T., W. B. Kiosses, S. Harkins, and J. L. Whitton. 2006. Inhibition of protein trafficking by coxsackievirus B3: multiple viral proteins target a single organelle. J. Virol. 806637-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz, C. D., H. Palosaari, J.-P. Parisien, P. Devaux, R. Cattaneo, T. Ouchi, and C. M. Horvath. 2006. Measles virus V protein inhibits p53 family member p73. J. Virol. 805644-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, N. L., I. J. Caley, K. W. Brown, M. R. Betts, D. M. Irlbeck, K. M. McGrath, M. J. Connell, D. C. Montefiori, J. A. Frelinger, R. Swanstrom, P. R. Johnson, and R. E. Johnston. 2000. Vaccination of macaques against pathogenic simian immunodeficiency virus with Venezuelan equine encephalitis virus replicon particles. J. Virol. 74371-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lang, A., T. Baas, T. Teal, L. M. Leijten, B. Rain, A. D. Osterhaus, B. L. Haagmans, and M. G. Katze. 2007. Functional genomics highlights differential induction of antiviral pathways in the lungs of SARS-CoV-infected macaques. PLoS Pathog. 3e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devaraj, S. G., N. Wang, Z. Chen, Z. Chen, M. Tseng, N. Barretto, R. Lin, C. J. Peters, C. T. Tseng, S. C. Baker, and K. Li. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 28232208-32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowell, S. F., and M. S. Ho. 2004. Seasonality of infectious diseases and severe acute respiratory syndrome—what we don't know can hurt us. Lancet Infect. Dis. 4704-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 3481967-1976. [DOI] [PubMed] [Google Scholar]
- 16.Eckerle, L. D., X. Lu, S. M. Sperry, L. Choi, and M. R. Denison. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 8112135-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4491-496. [DOI] [PubMed] [Google Scholar]
- 18.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 819812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230217-227. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252324-330. [DOI] [PubMed] [Google Scholar]
- 21.Gorbalenya, A. E., E. J. Snijder, and W. J. M. Spaan. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 787863-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham, R. L., A. C. Sims, S. M. Brockway, R. S. Baric, and M. R. Denison. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 7913399-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han, Y., H. Geng, W. Feng, X. Tang, A. Ou, Y. Lao, Y. Xu, H. Lin, H. Liu, and Y. Li. 2003. A follow-up study of 69 discharged SARS patients. J. Tradit. Chin. Med. 23214-217. [PubMed] [Google Scholar]
- 24.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 7813600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hengel, H., U. H. Koszinowski, and K. K. Conzelmann. 2005. Viruses know it all: new insights into IFN networks. Trends Immunol. 26396-401. [DOI] [PubMed] [Google Scholar]
- 26.Hiscott, J., N. Grandvaux, S. Sharma, B. R. Tenoever, M. J. Servant, and R. Lin. 2003. Convergence of the NF-kappaB and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. N. Y. Acad. Sci. 1010237-248. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda, F., C. M. Hecker, A. Rozenknop, R. D. Nordmeier, V. Rogov, K. Hofmann, S. Akira, V. Dotsch, and I. Dikic. 2007. Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes. EMBO J. 263451-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kash, J. C., E. Mühlberger, V. Carter, M. Grosch, O. Perwitasari, S. C. Proll, M. J. Thomas, F. Weber, H.-D. Klenk, and M. G. Katze. 2006. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J. Virol. 803009-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 3481953-1966. [DOI] [PubMed] [Google Scholar]
- 31.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 10214040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12143-156. [DOI] [PubMed] [Google Scholar]
- 33.May, M. J., S. E. Larsen, J. H. Shim, L. A. Madge, and S. Ghosh. 2004. A novel ubiquitin-like domain in IkappaB kinase beta is required for functional activity of the kinase. J. Biol. Chem. 27945528-45539. [DOI] [PubMed] [Google Scholar]
- 34.McWhirter, S. M., B. R. Tenoever, and T. Maniatis. 2005. Connecting mitochondria and innate immunity. Cell 122645-647. [DOI] [PubMed] [Google Scholar]
- 35.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 4371167-1172. [DOI] [PubMed] [Google Scholar]
- 36.Narayanan, K., C. Huang, K. Lokugamage, W. Kamitani, T. Ikegami, C.-T. K. Tseng, and S. Makino. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 824471-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283230-239. [DOI] [PubMed] [Google Scholar]
- 38.Park, M.-S., M. L. Shaw, J. Muñoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 771501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poon, L. L. M., D. K. W. Chu, K. H. Chan, O. K. Wong, T. M. Ellis, Y. H. C. Leung, S. K. P. Lau, P. C. Y. Woo, K. Y. Suen, K. Y. Yuen, Y. Guan, and J. S. Peiris. 2005. Identification of a novel coronavirus in bats. J. Virol. 792001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pushko, P., M. Parker, G. V. Ludwig, N. L. Davis, R. E. Johnston, and J. F. Smith. 1997. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 239389-401. [DOI] [PubMed] [Google Scholar]
- 41.Qian, X.-Y., F. Alonso-Caplen, and R. M. Krug. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 682433-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 1035717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 805156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivas, C., J. Gil, Z. Melkova, M. Esteban, and M. Diaz-Guerra. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology 243406-414. [DOI] [PubMed] [Google Scholar]
- 45.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schijns, V. E., C. M. Wierda, M. van Hoeij, and M. C. Horzinek. 1996. Exacerbated viral hepatitis in IFN-gamma receptor-deficient mice is not suppressed by IL-12. J. Immunol. 157815-821. [PubMed] [Google Scholar]
- 47.Schoneboom, B. A., J. S. Lee, and F. B. Grieder. 2000. Early expression of IFN-alpha/beta and iNOS in the brains of Venezuelan equine encephalitis virus-infected mice. J. Interferon Cytokine Res. 20205-215. [DOI] [PubMed] [Google Scholar]
- 48.Shabman, R. S., T. E. Morrison, C. Moore, L. White, M. S. Suthar, L. Hueston, N. Rulli, B. Lidbury, J. P.-Y. Ting, S. Mahalingam, and M. T. Heise. 2007. Differential induction of type I interferon responses in myeloid dendritic cells by mosquito and mammalian-cell-derived alphaviruses. J. Virol. 81237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Symons, J. A., A. Alcami, and G. L. Smith. 1995. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell 81551-560. [DOI] [PubMed] [Google Scholar]
- 50.Terzic, J., I. Marinovic-Terzic, F. Ikeda, and I. Dikic. 2007. Ubiquitin signals in the NF-kappaB pathway. Biochem. Soc. Trans. 35942-945. [DOI] [PubMed] [Google Scholar]
- 51.Theofilopoulos, A. N., R. Baccala, B. Beutler, and D. H. Kono. 2005. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 23307-336. [DOI] [PubMed] [Google Scholar]
- 52.Warfield, K. L., C. M. Bosio, B. C. Welcher, E. M. Deal, M. Mohamadzadeh, A. Schmaljohn, M. J. Aman, and S. Bavari. 2003. Ebola virus-like particles protect from lethal Ebola virus infection. Proc. Natl. Acad. Sci. USA 10015889-15894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wathelet, M. G., M. Orr, M. B. Frieman, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 8111620-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Webster, R. G. 2004. Wet markets—a continuing source of severe acute respiratory syndrome and influenza? Lancet 363234-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. G. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 765251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye, Y., K. Hauns, J. O. Langland, B. L. Jacobs, and B. G. Hogue. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 812554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 10012995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng, D., G. Chen, B. Guo, G. Cheng, and H. Tang. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 181105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou, Z., K. Hoebe, X. Du, Z. Jiang, L. Shamel, and B. Beutler. 2005. Antagonism between MyD88- and TRIF-dependent signals in B7RP-1 up-regulation. Eur. J. Immunol. 351918-1927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.