Abstract
Vaccinia virus A26 protein is an envelope protein of the intracellular mature virus (IMV) of vaccinia virus. A mutant A26 protein with a truncation of the 74 C-terminal amino acids was expressed in infected cells but failed to be incorporated into IMV (W. L. Chiu, C. L. Lin, M. H. Yang, D. L. Tzou, and W. Chang, J. Virol 81:2149-2157, 2007). Here, we demonstrate that A27 protein formed a protein complex with the full-length form but not with the truncated form of A26 protein in infected cells as well as in IMV. The formation of the A26-A27 protein complex occurred prior to virion assembly and did not require another A27-binding protein, A17 protein, in the infected cells. A26 protein contains six cysteine residues, and in vitro mutagenesis showed that Cys441 and Cys442 mediated intermolecular disulfide bonds with Cys71 and Cys72 of viral A27 protein, whereas Cys43 and Cys342 mediated intramolecular disulfide bonds. A26 and A27 proteins formed disulfide-linked complexes in transfected 293T cells, showing that the intermolecular disulfide bond formation did not depend on viral redox pathways. Finally, using cell fusion from within and fusion from without, we demonstrate that cell surface glycosaminoglycan is important for virus-cell fusion and that A26 protein, by forming complexes with A27 protein, partially suppresses fusion.
Vaccinia virus, the prototype of the Orthopoxvirus genus of the family Poxviridae, infects many cell lines and animals (13) and produces several forms of infectious particles, among which the intracellular mature virus (IMV) is the most abundant form inside cells. The IMV can be wrapped with additional Golgi membrane, transported through microtubules, and released from cells as extracellular enveloped viruses (10). The IMV has evolved to enter host cells through plasma membrane fusion (1, 3, 12, 29, 47) or endocytosis (11, 48). Recently, Mercer et al. reported that IMV entered HeLa cells through apoptotic mimicry and macropinocytosis (32), and Huang et al. reported that IMV enters into HeLa cells through a dynamin-dependent fluid-phase endocytosis that required the cellular protein VPEF (22).
The IMV contains more than 75 viral proteins. Of these, more than 10 viral envelope proteins are known to be involved in vaccinia virus entry into cells (6, 34, 55). Vaccinia virus contains at least five attachment proteins, with H3, A27, and D8 binding to cell surface glycosaminoglycans (GAGs) (7, 21, 28), A26 protein binding to the extracellular matrix protein laminin (5), and L1 protein binding to unidentified cell surface molecules (14). A27 protein also binds to the viral A17 protein through its C-terminal region (35, 50), and it was recently shown that the coexpression of A17 and A27 proteins resulted in cell fusion in transiently transfected 293T cells (27). In this study, we demonstrate the formation through disulfide bonds of complexes between two viral attachment proteins, A26 and A27, and we determine the cysteine residues that are critical for these disulfide bonds. We also address the biological role of the A26-A27 protein complex formation in cell fusion regulation.
MATERIALS AND METHODS
Cells, viruses, and reagents.
BSC40, 293T, L, and sog9 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (from Biological Industries, Israel). sog cells were derived from L cells and expressed no GAGs on the cell surface (2, 17). The Western Reserve (WR) strain of the vaccinia virus was used as the wild type (Wt-VV) in this study. A mutant virus, WR32-7/Ind14K, derived from the WR strain, was obtained from G. L. Smith (38); in this study, for simplicity, it is referred to as IA27L. IA27L virus contains an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible A27L open reading frame (ORF) and an accidental mutation of A26L resulting in the expression of a truncated A26 protein of 426 amino acids (aa) (5). IA27L-A26WR was a recombinant virus generated from IA27L virus and constitutively expressed a full-length A26L (WR) protein of 500 aa, as previously described (5). Recombinant viruses vG4Li (53), vE10Ri (42), and vA17LΔ5 (54) expressed viral G4, E10, or A17 protein induced by IPTG, respectively, and were provided by Bernard Moss. All viruses were grown on BSC40 cells, and IMVs were purified by two successive centrifugations through 36% and 25 to 40% sucrose gradient layers as described previously (25). Rabbit antibodies (Abs) for vaccinia virus A26, A27, H3, and L1 proteins were described previously (5, 8, 21, 28). Rabbit anti-L1 Ab (A7) recognized both reduced and oxidized forms of L1 protein in immunoblots, whereas mouse monoclonal Ab (MAb) 2D5, provided by Yasuo Ichihashi, recognized only the oxidized form of L1 protein (23, 24). An anti-hemagglutinin (HA) tag, MAb HA.11, was purchased from Covance Research Products Inc. A reducing agent, tris-(2-carboxyethyl) phosphine (TCEP), was purchased from Pierce Inc. An alkylating agent, N-ethylmaleimide (NEM), was purchased from Sigma Inc.
Construction of IA27L-A26KO virus.
The IA27L-A26KO virus was generated based on previously established procedures (5). To delete the mutated A26L ORF in IA27L, 293T cells were infected with IA27L virus and subsequently transfected with 2 μg of the plasmid pA25/lacZ/A28-KS(−), containing a late promoter (p11K)-driven lacZ expression cassette flanked by A25L and A28L ORF sequences. These cells were cultured in growth medium supplemented with 5 mM IPTG and harvested at 2 days postinfection (p.i.) Recombinant IA27L-A26KO virus was isolated by three rounds of plaque purification in agar containing 150 μg/ml of 5-bromo-4-chloro-3-indolyl-ß-d-galactopyranoside (X-Gal) as described previously (4).
Alkylation of free thiols in samples with NEM for immunoprecipitation or immunoblot analyses.
To avoid artificial reduction and disulfide bond rearrangement during cell rupture, cell lysates were prepared in the presence of an alkylating agent, NEM, based on two established procedures, with slight modifications (26, 30, 43, 52). Both methods gave identical results in immunoblots analyses, as described below. For the NEM lysis method, cells (mock infected, virus infected at 24 h p.i., or plasmid transfected) were placed on ice and washed twice with freshly prepared cold phosphate-buffered saline (PBS) containing 20 mM NEM (Sigma) and subsequently solubilized in 50 mM Tris (pH 8.0)-1% sodium dodecyl sulfate (SDS) containing 40 mM NEM. The lysates were used for immunoprecipitation (described below) or mixed with an equal volume of SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer (150 mM Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 0.04% bromophenol blue), with or without 40 mM TCEP (Pierce), boiled for 10 min, and loaded on NuPAGE 4 to 12% Bis-Tris denaturing gels (Invitrogen). For the trichloroacetic acid (TCA)-NEM method, cells were washed once with cold PBS and treated with PBS containing 10% TCA. After centrifugation at 10,000 × g for 15 min at 4°C, protein precipitates were washed three times with cold acetone. The pellets were dissolved in 50 mM Tris (pH 8.0), 1% SDS, 40 mM NEM (with or without 40 mM TCEP) (Pierce). The samples subsequently were mixed with an equal volume of SDS-PAGE loading buffer, boiled for 10 min, and loaded on NuPAGE 4 to 12% Bis-Tris denaturing gels. For preparing the alkylated form of vaccinia virus IMV (WR strain), infected BSC40 cell monolayers were washed with PBS containing 20 mM NEM for 10 min, and cell pellets were collected by centrifugation. The cell pellets were dounced in TM buffer (10 mM Tris-HCl, pH 7.5, 5 mM MgCl2) containing 20 mM NEM, and IMV preparations were performed using 36% and 25 to 40% sucrose gradient as described above (25).
Immunoprecipitation.
Cells (mock infected, virus infected at 24 h p.i., or plasmid transfected) were lysed in lysis buffer (1% Triton X-100-1% deoxycholate-0.1% SDS-150 mM NaCl-1 mM EDTA-50 mM Tris [pH 7.4]), with or without 20 mM NEM, and insoluble materials were removed by centrifugation (10,000 × g, 15 min, 4°C). Cell lysates were further precleared by incubation with 50 μl of a 50% protein A-Sepharose bead slurry with gentle agitation for 1 h at 4°C in a tube rotator. After brief centrifugation, the precleared supernatant was incubated with an anti-A26 (1:50) or anti-A27 (1:100) antibody at 4°C for 2 h to overnight for protein immunoprecipitation and subsequently incubated with 50 μl of a 50% protein A-Sepharose bead slurry (GE Healthcare Bio-Sciences AB) for another 2 h of incubation at 4°C. The immunoprecipitates were washed five times with lysis buffer and saved for immunoblot analyses.
Immunoblot analysis.
For samples not treated with NEM, the following procedures were performed. Purified IMV virions, whole-cell lysates, or immunoprecipitates from virus-infected cells were dissolved in SDS-PAGE loading buffer (150 mM Tris [pH 6.8], 20% glycerol, 4% SDS, 0.04% bromophenol blue). When proteins needed to be reduced, β-mercaptoethanol (or TCEP) was added to samples at a final concentration of 10% (20 mM). The samples then were boiled for 10 min, separated on NuPAGE 4 to 12% Bis-Tris denaturing gels (Invitrogen Inc.), and transferred to nitrocellulose membranes for immunoblot analysis with anti-A26 (1:6,000 dilution), anti-A27 (1:1,500), anti-H3 (1:2,000), anti-A17 (1:1,000), anti-L1 (1:1,000 for 2D5 and A7), and anti-HA (1:1,000) Abs with either secondary Ab (alkaline phosphatase-conjugated goat anti-rabbit or goat anti-mouse immunoglobulin [Ig]) or a One-Step Complete IP-Western kit (GenScript Inc.) and developed according to the manufacturer's instructions.
Complementation assays for studying complex formation. (i) Plasmid construction and site-directed mutagenesis.
The plasmids pA25/A26(WR)-KS(−) and A27L-TOPO, containing A26L and A27L ORFs, were generated and described previously (5). Other A26L and A27L mutants, A26L-C43A, A26L-C128A, A26L-C162A, A26L-C342A, A26L-441A, A26L-C442A, A26L-C441/442A, A27L-C71A, A27L-C72A, and A27L-C71/72A, were generated from pA25/A26(WR)-KS(−) and A27L-TOPO by site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene) using the following complementary oligonucleotides: A26L-C43A (5′-AAA ATC GAA GCA AAT ACA GCC ATC AGT AGA AAA CAT AGA-3′), A26L-C128A (5′-ATT TTA TAT ATA GTA TTT GCT ATA ATA TCT GGT AAG AAT-3′), A26L-C162A (5′-GAG CGT ATC AAG TAT GCA GCT AAG CAA ATA TTA CAC GGT-3′), A26L-C342A (5′-GTA ATA ATA GAC AAT GAA GCC GCT AAT ATT CAG TCG AGT-3′), A26L-C441A (5′-AAT TTC ATC GGA CAC TAT GCT TGT GAT ACA GCG GCA GTT-3′), A26L-C442A (5′-TTC ATC GGA CAC TAT TGT GCT GAT ACA GCG GCA GTT GAT-3′), A26L-C441/442A (5′-AAT TTC ATC GGA CAC TAT GCT GCT GAT ACA GCG GCA GTT GAT-3′), A27L-C71A (5′-TTT GAA CAA ATA GAA AAG GCT TGT AAA CGC AAC GAT GAA-3′), A27L-C72A (5′-GAA CAA ATA GAA AAG TGT GCT AAA CGC AAC GAT GAA GTT-3′), A27L-C71/72A (5′-TTT GAA CAA ATA GAA AAG GCT GCT AAA CGC AAC GAT GAA GTT-3′). The underlined bases represent the mutated codons, and only the sense primers are shown. All of the plasmid clones were confirmed by DNA sequencing.
(ii) Trans-complementation assays.
The trans-complementation assays were performed essentially as described previously (5). Confluent 293T cells in 60-mm dishes were infected with IA27L-A26KO or IA27L-A26WR at a multiplicity of infection (MOI) of 5 PFU/cell for 1 h in the absence of IPTG. After infection, cells were washed with PBS, and 1 μg of individual plasmids (A26L-C43A, A26L-C128A, A26L-C162A, A26L-C342A, A26L-C441A, A26L-C442A, A26L-C441/442A, A27L-C71A, A27L-C72A, and A27L-C71/72A) was transfected into cells using 10 μl Lipofectamine (Invitrogen). After a 5-h incubation, the mixtures were removed and fresh complete medium was overlaid. Cells were harvested at 24 h p.i. for immunoprecipitation and immunoblot analyses as described above.
Expression of vaccinia virus A26 and A27 protein in 293T cells.
Vaccinia virus A26 (WR) and A27 (WR) ORF sequences were codon optimized (GenScript Inc.) and cloned into the mammalian expression vector pLKO-AS3.1-EGFP3′ (National RNAi Core, Academia Sinica, Taiwan). 293T cells were cotransfected with plasmids expressing A26 and A27 proteins using calcium phosphate transfection procedures, and the cells were harvested at 1 day posttransfection for immunoprecipitation (anti-A27 Abs at 1:40) and immunoblot analyses (anti-A26 at 1:1,000 and anti-A27 at 1:200) as described above.
Cell fusion assays. (i) Cell fusion from within.
BSC40 cells were infected with wild-type WR vaccinia virus (Wt-VV), IA27L virus, or IA27L-A26WR virus at an MOI of 5 PFU per cell and incubated in culture medium with or without 5 mM IPTG to regulate A27 protein expression (5, 37, 38). Cell fusion was monitored up to 24 h p.i., and cells were photographed using phase-contrast optics on a Nikon inverted microscope.
(ii) Live cell imaging.
Alternatively, live cell imaging was performed to monitor cell fusion from within as described below. BSC40 cells were seeded onto coverslips in 60-mm dishes (7.5 × 105 per dish) and subsequently infected with Wt-VV, IA27L, and IA27L-A26WR at an MOI of 5 PFU per cell at 37°C for 1 h. The infected cells were washed with PBS, transferred to the microscope stage, and maintained in growth medium, and the samples were kept in a warm 37°C chamber with 5% CO2 throughout the recording time up to 34 h p.i. Images were taken initially from 30 min p.i. to 10 h p.i. at 1 frame per 30 min and then switched to 1 frame per 10 min from 10 to 34 h p.i. Three microscopes, a Carl Zeiss confocal LSM 510 Pascal (20× Plan-Neofluar lens), an LSM 510 Meta NLO (0.8-fold; 20× Plan Apochromat), and a wide-field Zeiss Axiovert (1.6×; 10× Plan Neofluar lens), were used to acquire individual images of cells infected with Wt-VV, IA27L, and IA27L-A26WR at the same time. Images were analyzed using AxioVision release 4.6. At the end of recording, the infected cells were fixed in 4% paraformaldehyde, permeabilized in PBS-0.2% saponin, and stained with anti-vaccinia virus Abs (1:5,000), fluorescein isothiocyanate-conjugated goat anti-rabbit IgG Abs (diluted 1:1,000), and 0.5 μg/ml of 4′,6′-diamidino-2-phenylindole (DAPI) to ensure that all cells were infected. Images were acquired by LSM 510 Meta NLO confocal microscopy (20× Plan Apochromat).
(iii) Cell fusion from without.
Cell fusion from without was performed as described previously (16). Briefly, L cells and sog9 cells were seeded on coverslips in 24-well plates and, after overnight incubation, were incubated with 6 μg vaccinia virus (∼4,000 particles/cell) in PBS-AM (PBS, 0.05% bovine serum albumin, 10 mM MgCl2) for 1 h, washed, and then treated with PBS (pH 4.7) at 37°C for 2 min. The cells were incubated at 37°C for 1 h with complete medium containing 40 μg/ml cordycepin (Sigma) and fixed for 15 min with 4% paraformaldehyde in PBS at 25°C, and then the plasma membrane was stained with PKH26 (Sigma) and nuclei were stained with Hoechst 33258 (Invitrogen Inc.). Images were collected with an LSM 510 Meta confocal laser scanning microscope (Carl Zeiss) using a 40× objective lens and confocal microscopy software (release 2.8; Carl Zeiss), and nuclei numbers were counted from multiple images (>300 cells). To quantify cell fusion, we calculated the percentage of fused cells (i.e., cells containing more than one nucleus) using the following equation: percentage of cell fusion = (sum of nuclei added from all fused cells/sum of nuclei added from total seeded cells) × 100. When we subdivide the above-described fused cell population into small-sized (2 to 5 nuclei/cell), medium-sized (6 to 10 nuclei/cell), and large-sized (>10 nuclei/cell) fused cell populations, the quantification was calculated as follows: percentage of small-sized fused cells = 100 × (sum of nuclei added from all the small-sized fused cells/sum of nuclei added from total seeded cells); percentage of medium-size fused cells = 100 × (sum of nuclei added from all the medium-sized fused cells/sum of nuclei added from total seeded cells); and percentage of large-size fused cells = 100 × (sum of nuclei added from all the large-sized fused cells/sum of nuclei added from total seeded cells). The experiments were repeated twice, and in each experiment more than 300 cells were counted for each virus.
RESULTS
A26 and A27 proteins form 70-/90-kDa protein complexes through disulfide bond formation in the infected cells, and the complexes are packaged into IMV particles.
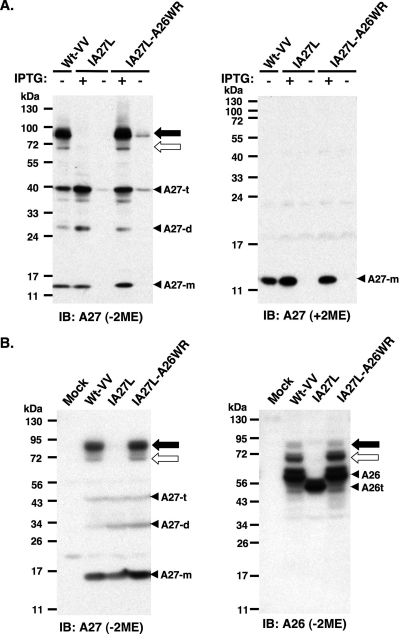
The immunoblot analyses of cell lysates harvested from HeLa cells infected with Wt-VV, IA27L (37, 38), and IA27L-A26WR (5) were performed in reducing and nonreducing conditions using anti-A27 Ab (Fig. 1A). The expression of A27 protein in Wt-VV was constitutive, whereas in IA27L and IA27L-A26WR it was induced by IPTG, as previously described (5, 37, 38). In reducing SDS-PAGE (with 2-mercaptoethanol [2ME]), A27 protein migrated as a 14-kDa protein in the infected cells; however, it migrated as multiple species in nonreducing conditions (without 2ME), suggesting that disulfide bonds mediated the formation of these complexes. A27 protein monomers of 14 kDa, dimers of 28 kDa, and trimers of 42 kDa were detected, which is consistent with previous reports (36). More interestingly, A27 protein also formed two high-molecular-mass complexes of 70 kDa (Fig. 1A) and 90 kDa (Fig. 1A) in cells infected with Wt-VV and IA27L-A26WR but not IA27L (Fig. 1A). Because IA27L-A26WR (5) was derived from IA27L virus by the replacement of a truncated A26 protein (aa 1 to 426) with a full-length A26 protein (1 to 500 aa), this implied that A26 (aa 427 to 500) was required for the formation of the 70-/90-kDa protein complexes. The samples were boiled and denatured before gel separation, so the 70-/90-kDa protein complex detected on nonreducing gels most likely is formed through disulfide bonds. As expected, in the absence of IPTG, all of the monomer, dimer, and trimer forms of A27 protein, as well as the 70- and 90-kDa complexes, were barely detected in cells infected with IA27L and IA27L-A26WR.
FIG. 1.
A26 and A27 proteins formed protein complexes through disulfide bond formation in BSC40 cells. (A) Immunoblot (IB) analysis of A27 protein in virus-infected cell lysates following boiling and separation by reducing (with 2ME) and nonreducing (without 2ME) SDS-PAGE. The expression of the A27L ORF in wild-type vaccinia virus (Wt-VV) was constitutive, whereas in IA27L and IA27L-A26WR viruses it was induced by 5 mM IPTG. (B) Immunoblot analysis of A27 and A26 proteins in virus-infected cells cultured in medium containing IPTG. Lysates were boiled, separated on nonreducing (without 2ME) SDS-PAGE, and probed with anti-A26 (1:1,000) and anti-A27 (1:1,000) Abs. The 90- and 70-kDa protein complexes are indicated by black and white arrows, respectively. Three protein bands representing A27 monomers (A27-m), dimers (A27-d), and trimers (A27-t) also were detected. A26t, truncated A26 protein (aa 1 to 426).
The above data suggested the possibility that A26 protein interacts with A27 protein to form the 70-/90-kDa protein complexes. We therefore tested whether 70-/90-kDa protein complexes also contain A26 protein in the infected cells. In immunoblots performed under nonreducing conditions, not only anti-A27 but also anti-A26 Ab detected two protein complexes of 70 and 90 kDa in size (Fig. 1B). It is worth noting that we consistently observed that anti-A26 Ab reacted better with the 70-kDa protein complex than with the 90-kDa form on gels, whereas anti-A27 Ab exhibited the opposite preference (Fig. 1B).
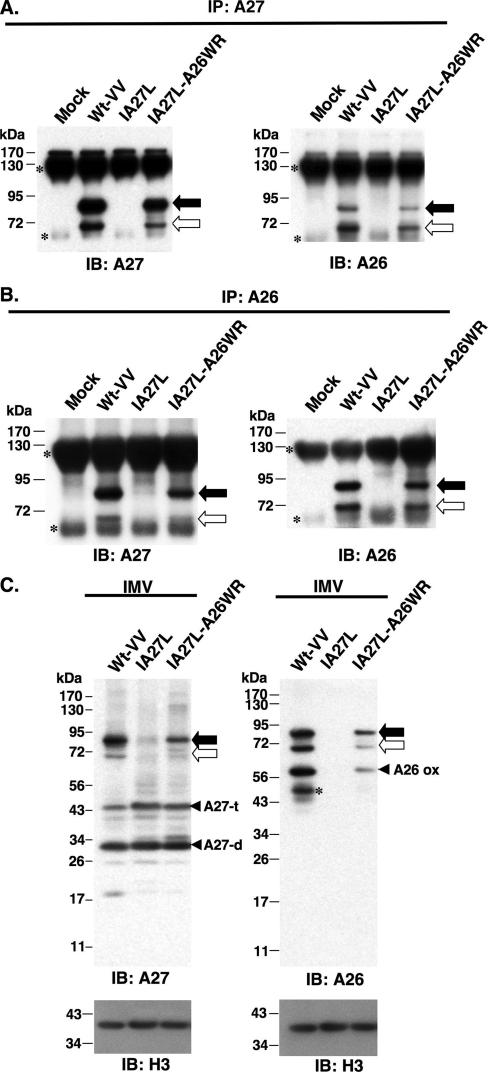
To provide direct evidence that these 70-/90-kDa protein complexes contained A26 and A27 proteins linked by disulfide bonds, we examined cell lysates harvested at 24 h p.i. by immunoprecipitation. Anti-A27 Ab brought down the 70- and 90-kDa protein complexes from cells infected with Wt-VV or IA27L-A26WR but not IA27L (Fig. 2A). More importantly, these immunoprecipitated 70-/90-kDa protein complexes contained A26 protein (Fig. 2A). Reciprocal immunoprecipitations performed using anti-A26 Ab also immunoprecipitated A26 protein in the 70-/90-kDa protein complex that contained A27 protein (Fig. 2B). To avoid artificial disulfide bond rearrangement during cell rupture, cell lysates were prepared using 10% TCA precipitation and an alkylating agent, N-ethylmaleimide (26, 30, 43, 52), as described in Materials and Methods for immunoblot analyses. The results revealed that, regardless of NEM treatment, 70-/90-kDa protein complexes remained intact in the infected cells (data not shown). Taken together, these results showed that A26 and A27 proteins formed 70- and 90-kDa protein complexes through disulfide bonds. The sizes of the 70-/90-kDa complexes differed by the molecular size of an A27 monomer, implying a stoichiometry for the 70-/90-kDa complex of an A26 monomer in complex with either an A27 dimer or an A27 trimer.
FIG. 2.
Immunoprecipitation (IP) of A26-A27 protein complexes from cell lysates. BSC40 cells were infected with viruses as shown at the top at an MOI of 10 PFU/cell and were harvested at 24 h p.i. Cell lysates were immunoprecipitated with anti-A27 Ab (1:100) (A) or anti-A26 Ab (1:50) (B). The immunoprecipitates were washed, boiled, and separated on SDS-PAGE and analyzed by immunoblotting using anti-A26 and anti-A27 Abs as described for panel A. The black and white arrows indicate the 90- and 70-kDa protein complexes, while small asterisks indicate Ab heavy and light chains. (C) A26-A27 protein complexes were stably incorporated in IMV particles. Equivalent amounts of each IMV were separated by SDS-PAGE and analyzed by immunoblotting with anti-A26 (1:1,000), anti-A27 (1:1,000), and anti-H3 (1:1,000) Abs. The black and white arrows indicate the 90- and 70-kDa protein complexes, respectively. The asterisk represents the degraded form of A26 protein. IB, immunoblot.
To test whether the A26-A27 protein complexes are packaged in IMVs, we purified IMV particles of Wt-VV, IA27L, and IA27L-A26WR viruses and loaded equal amounts on gels for immunoblots, as shown by the anti-H3 Ab blot (Fig. 2C). The anti-A27 Ab blot revealed that A26-A27 protein complexes were incorporated into IMV particles. Additionally, some dimer and trimer forms of A27 protein also were detected (Fig. 2C). Anti-A26 Ab also detected A26-A27 protein complexes as well as A26 monomer and its degraded form in gels (Fig. 2C). These data suggest that, on IMV particles, not all of the A26 and A27 proteins are part of covalently linked A26-A27 complexes. When IMVs were purified with reagents containing NEM to avoid disulfide oxidation during cell rupture, anti-A27 Ab still detected the 70-/90-kDa complexes on IMV particles (data not shown).
Cysteines 71 and 72 of A27 protein are important for A26-A27 protein complex formation.
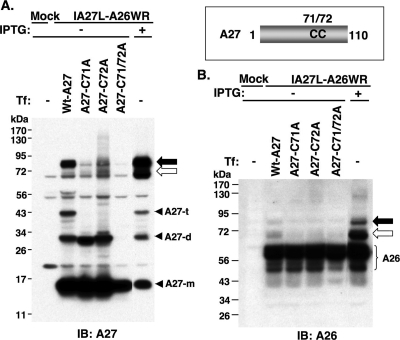
A27 protein forms covalently linked trimers in infected cells through disulfide bonds (36), although the homotrimer formation is not dependent on disulfide bonds formed between cysteine residues 71 and 72 (50). To identify how A26 and A27 proteins form intermolecular disulfide bonds, we used in vitro mutagenesis to generate mutant A27 proteins with either single or double cysteine-to-alanine mutations at cysteines 71 and 72 (Fig. 3). The plasmids were immediately transfected into 293T cells that were infected with IA27L-A26WR in the absence of IPTG, so that A26 protein was expressed from the viral genome and A27 protein was provided in trans from the transfected plasmids. Wild-type A27 protein provided in trans formed the 70-/90-kDa protein complexes with the A26 protein expressed from the viral genome (Fig. 3A and B). Mutant A27 C71A and C72A proteins partially reduced A26-A27 protein complex formation, whereas the C71/72A double mutant A27 protein completely lost the ability to form 70-/90-kDa protein complexes (Fig. 3A). Anti-A26 Ab had a weaker reactivity to the complexes on blots than anti-A27 Abs and detected the 70-/90-kDa complexes only in cells expressing the wild-type A27 protein (Fig. 3B). It is known that C71 and C72 of A27 protein mediated A27 protein trimerization (36), and we noticed that C71A and C72A single-mutant A27 proteins existed as dimers and C71/72A double-mutant protein as monomers in the transfected cells (Fig. 3A), suggesting that A27 dimers contain disulfide bonds formed between C71-C71 or C72-C72 and A27 trimers must contain an additional disulfide bond between C71 and C72. Taken together, the data here showed that C71 and C72 also are involved in the formation of the intermolecular disulfide bonds between A26 and A27 proteins.
FIG. 3.
Cysteines 71 and 72 of A27 protein are required for the formation of A26-A27 protein complexes. Schematic representation of the cysteine residues 71 and 72 on A27 protein. BSC40 cells were mock infected or infected with IA27L-A26WR and subsequently transfected with individual plasmids encoding wild-type A27 or C71A, C72A, or C71/72A double mutant proteins. Cells were harvested at 24 h p.i., boiled, and separated on nonreducing SDS-PAGE for immunoblot (IB) analyses using anti-A27 Ab (A) or anti-A26 Ab (B). The black and white arrows indicate the 90- and 70-kDa protein complexes, respectively. A27-m, A27-d, and A27-t represent monomers, dimers, and trimers of A27 protein, respectively. Tf, transfection.
Cysteines 441 and 442 of A26 protein are necessary for A26-A27 protein complex formation, whereas C43 and C342 form an intramolecular disulfide bond.
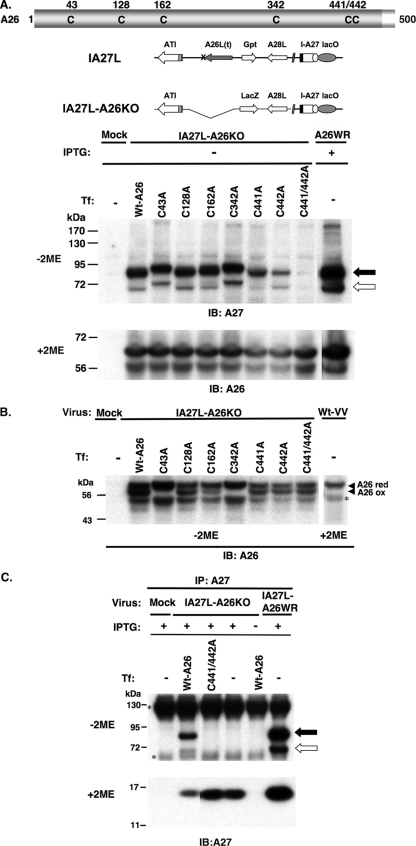
The A26 protein contains six cysteines at residues 43, 128, 162, 342, 441, and 442 (Fig. 4A). We generated a series of mutant A26 proteins that contained either single or double cysteine-to-alanine mutations. To perform complementation analyses, we generated an IA27L-A26KO virus (Fig. 4A) to remove the truncated A26 (aa 1 to 426) protein from IA27L virus (5, 38). BSC40 cells were infected with IA27L-A26KO virus in the presence of IPTG so that the A27 protein was expressed from the viral genome. These cells then were transfected with plasmids expressing either wild-type or mutant A26 protein to determine which cysteine is important for disulfide bond formation between A26 and A27 proteins (Fig. 4A). The transfected wild-type A26 protein and the C43A, C128A, C162A, and C342A mutants were able to form the 70-/90-kDa complexes in the infected cells in the presence of IPTG, although we noticed that the C43A and C342A mutants caused a slightly slower migration of the complexes (Fig. 4A). The C441A and C442A mutations reduced the amount of the 70-/90-kDa protein complexes formed, while the C441/442A double mutant almost completely eliminated complex formation (Fig. 4A). All of the transfected A26 proteins, including the C441/442 mutant, were well expressed (Fig. 4A), so the inability to form 70-/90-kDa complexes was not due to a lack of A26 protein expression in cells. Interestingly, besides the 70-/90-kDa protein complexes, anti-A26 Ab also detected multiple forms of A26 monomer protein in the transfected cells (Fig. 4B). The top band migrated similarly to the A26 protein band in the reducing gel (with 2ME), suggesting that it is the reduced form of A26 protein. The middle band was present in wild-type A26 protein and C128A, C162A, C441A, C442A, and C441/442A mutants but was absent in C43A and C342A mutant proteins, suggesting that the middle band represented the form of A26 protein containing an intramolecular disulfide bond between C43 and C342. The bottom band was present in all of the constructs with various intensities, and we suspected that it is the degraded form of A26 protein. Finally, to be sure that A27 protein did not form disulfide-linked protein complexes with the A26 C441/442 mutant protein, we performed immunoprecipitation using anti-A27 Ab. The wild-type A27 protein brought down wild-type A26 protein, but not the mutant C441/442 protein, in the infected cells (Fig. 4C). The same complexes were formed in cells infected with IA27L-A26WR, which served as a control.
FIG. 4.
(A) Cysteines 441 and 442 of A26 protein are required for the intermolecular disulfide bond formation of A26-A27 protein complexes. All of the six cysteine residues of A26 protein as well as the genome representations of IA27L and IA27L-A26KO viruses are shown at the top. BSC40 cells were mock infected or infected with IA27L-A26KO or IA27L-A26WR, subsequently transfected with individual plasmids encoding wild-type A26 or C43A, C128A, C162A, C342A, C441A, C442A, and C441/442A double mutant proteins, harvested at 24 h p.i., and then boiled to denature proteins for immunoblot analyses (IB) on nonreducing (without 2ME) gel using anti-A27 Ab (1:1,000) or on reducing (with 2ME) SDS-PAGE using anti-A26 (1:1,000) Ab. The black and white arrows indicate the 90- and 70-kDa protein complexes, respectively. (B) C43 and C342 form an intramolecular disulfide bond in A26 protein. The cell lysates shown in panel A were boiled and separated by nonreducing (without 2ME) SDS-PAGE and stained with anti-A26 (1:1,000) Ab. The top migrating band is the reduced form of full-length A26 protein (A26 red). The middle band is the oxidized form of A26 protein (A26 ox), while the asterisk indicates the degraded product of A26 protein. (C) A26 C441/442A mutant protein failed to form disulfide bonds with A27 protein. The immunoprecipitation (IP) of cell lysates described in panel A was performed using anti-A27 Ab (1:100) and was analyzed by SDS-PAGE on nonreducing (without 2ME) or reducing (with 2ME) gels using anti-A27 Ab (1:1,000). The asterisks (left) indicate Ig heavy and light chains. Tf, transfection.
A26-A27 complex formation in the infected cells does not require virion morphogenesis, A17 protein, or the viral redox system.
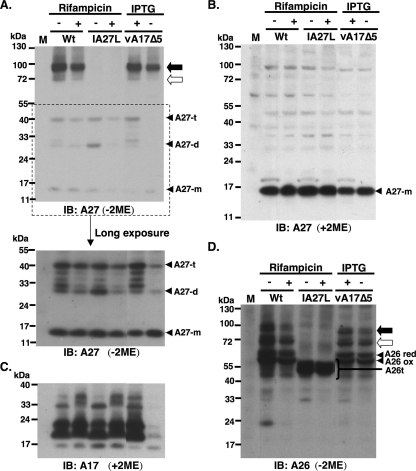
We next investigated whether viral A26 and A27 proteins form disulfide bonds during virion morphogenesis in the infected cells. Rifampin blocked viral core protein processing (data not shown) without significantly affecting A26 and A27 protein levels in cells (Fig. 5B and D); however, 70- and 90-kDa complexes still formed in these drug-treated cells, as detected by anti-A27 Ab (Fig. 5A) and anti-A26 Ab (Fig. 5D), showing that intermolecular bond formation between A26 and A27 proteins occurred prior to virion morphogenesis. Because the A27 protein has been shown to bind to A17 protein (35), we tested whether A17 protein is involved in A26-A27 complex formation. BSC40 cells were infected with a recombinant virus, vA17Δ5, expressing A17 protein in the presence of IPTG (54). The infected cells were harvested at 24 h p.i. for immunoblot analyses (Fig. 5A and D). The results showed that the association between A17 and A27 protein is not a prerequisite for the formation of A26-A27 complexes, since the 70-/90-kDa complexes still formed in the infected cells when A17 expression was repressed in the absence of IPTG (Fig. 5C).
FIG. 5.
A26-A27 protein complexes formed prior to virion assembly and did not require A17 protein. (A) Immunoblot analyses (IB) of 70-/90-kDa complex in the infected cells using anti-A27 Ab. BSC40 cells were infected with individual vaccinia viruses as shown at the top at an MOI of 10 PFU per cell. Wt, wild-type vaccinia virus (WR). Cell lysates were boiled and separated by SDS-PAGE on nonreducing (without 2ME) gels and probed with anti-A27 Abs. The bottom portion shows a longer exposure of a portion of the blot (marked by the dotted line). The black and white arrows indicate the 90- and 70-kDa protein complexes, respectively. A27-m, A27-d, and A27-t represent monomers, dimers, and trimers of A27 protein, respectively. (B) Samples were treated with reducing condition (with 2ME) and loaded in the same order as that used for panel A and were probed with anti-A27 Ab. (C) Samples were loaded in the same order as that for panel A and were treated with reducing conditions (with 2ME) and probed with anti-A17 Ab. (D) Samples were loaded in the same order as that used for panel A and were probed with anti-A26 Ab. A26 red, reduced form of A26 protein; A26 ox, oxidized form of A26 protein; and A26t, truncated A26 protein (aa 1 to 426) (5); M, mock-infected cells; Wt, wild type virus.
Vaccinia virus encodes a unique redox pathway comprising viral G4 (18, 53), E10 (42), and A2.5 proteins (45), each of which is essential for virus morphogenesis and for catalyzing the intramolecular disulfide bond formation of viral L1 and F9 proteins (43, 52). To determine whether disulfide bond formation between A26 and A27 proteins is mediated by the virus-encoded redox pathway, we tested whether 70- and 90-kDa complex formation depends on viral G4 and E10 proteins by using two recombinant viruses, vG4Li (53) and vE10Ri (42), expressing G4 and E10 protein, respectively, under IPTG induction. BSC40 cells were infected with vG4Li or vE10Ri at an MOI of 5 PFU per cell, were cultured in medium with or without 50 μM IPTG for 24 h, and were harvested with NEM-containing reagents, as described in Materials and Methods, to avoid disulfide exchange during cell rupture. These cell lysates subsequently were boiled and analyzed by immunoblots under nonreducing (without TCEP) or reducing (with TCEP) conditions. As shown in Fig. 6, 70- and 90-kDa protein complexes were readily detected by anti-A27 Ab (Fig. 6A) and anti-A26 Ab (Fig. 6B) in cells infected with vG4Li in the absence or presence of IPTG, suggesting that the formation of the disulfide bond between A26 and A27 protein did not require G4 protein. The control experiments showed that HA-tagged G4 protein expression (Fig. 6C) as well as virus growth in cultures were dependent on IPTG addition (Fig. 6D), as expected. When cell lysates infected with vE10Ri were analyzed similarly by immunoblot analyses using anti-A27 Ab (Fig. 7A) and anti-A26 Ab (Fig. 7B), the formation of 70- and 90-kDa protein complexes was detected equally well with or without IPTG, supporting the notion that E10 protein is not required for 70-/90-kDa complex formation. The growth of vE10Ri was tightly regulated by IPTG (Fig. 7C). We did not have anti-E10 Ab, so immunoblot analyses were performed to detect L1 protein, the oxidation of which was shown to depend on E10 protein activity (42). As shown in Fig. 7D, oxidized L1 protein migrated faster than the reduced form of L1 protein on gels. In the absence of IPTG, the reduced form of L1 protein was detected in cells infected with vE10Ri, although some large amount of the oxidized form of L1 protein (detected by 2D5 MAb [23, 24]) still remained.
FIG. 6.
A26-A27 complex formation in cells infected with vG4Li. (A) BSC40 cells were infected with vG4Li and harvested at 24 h p.i. with 10% TCA precipitation (TCA ppt) and NEM as described in Materials and Methods. Protein samples were added with or without TCEP, boiled, and separated on gels using anti-A27 Ab. (B) Samples were loaded in the same order as that used for panel A and were probed with anti-A26 Ab. The asterisk represents the degraded A26 protein. (C) The expression of HA-tagged G4 protein (HA-G4) was induced by IPTG. (D) The growth of vG4Li virus was regulated by IPTG. IB, immunoblot.
FIG. 7.
A26-A27 complex formation in cells infected with vE10Ri. (A) BSC40 cells were infected with vE10Ri, and cells were harvested at 24 h p.i. with 10% TCA precipitation (TCA ppt) and NEM as described in Materials and Methods. Protein samples were added with or without TCEP, boiled, and separated on gels using anti-A27 Ab. (B) Samples were loaded in the same order as that used for panel A and were probed with anti-A26 Ab. The asterisk represents the degraded A26 protein. (C) The growth of vE10Ri virus was regulated by IPTG. (D) Immunoblot analyses (IB) of total L1 protein (top) and the oxidized form of L1 protein (L1 ox). L1 red, reduced form of L1 protein. 2D5 MAb only recognized the oxidized form of L1R on nonreducing gels (24).
Finally, to provide direct evidence that A26-A27 disulfide bonding does not require a viral redox system, A26 and A27 ORFs were coexpressed in 293T cells by transient transfections. Cells subsequently were harvested with or without NEM for immunoprecipitation using anti-A27 Abs and analyzed by immunoblot analyses with anti-A27 (Fig. 8A) and anti-A26 (Fig. 8B) Abs. Both 70- and 90-kDa protein complexes were readily detected in the immunoprecipitates, migrating similarly to the 70-/90-kDa complexes identified in the virus-infected cells (Fig. 8). When the transfected cells were harvested in the presence of NEM to avoid disulfide exchange, the 70-/90-kDa complexes remained detected (data not shown). Taking these results together, we concluded that the disulfide-linked complexes containing A26 and A27 proteins could form in host cells without the aid of viral redox enzymes.
FIG. 8.
A26 and A27 protein formed protein complexes in transfected 293T cells. (A) 293T cells were either mock transfected or cotransfected with plasmids expressing A26 and A27 protein, and cell lysates were harvested for immunoprecipitation (IP) using anti-A27 Ab (1:100). The immunoprecipitates were washed, boiled, and separated on SDS-PAGE and analyzed by immunoblotting (IB) using anti-A27 Abs (1:1,000) as described for Fig. 2A. (B) Samples were loaded in the same order as that used for panel A and were analyzed by anti-A26 Ab (1:1,000). The black and white arrows indicate the 90- and 70-kDa protein complexes, respectively.
A26 protein partially suppresses cell fusion induced by A27 protein expression.
Since A26 protein formed complexes with A27 protein, it may modulate A27 protein functions in cells. A27 protein was required for the formation of enveloped virions; however, the expression of A26 protein did not alter the resulting extracellular enveloped virus titers (5). Previous experiments using IA27L to study the fusion of virus-infected cells showed that the cell-to-cell fusion of the infected cells (cell fusion from within) was dependent on A27 protein expression induced by IPTG and that A27 protein may participate in the cell fusion process, since anti-A27 MAb C3 (36) and soluble A27 protein (20) containing the GAG-binding domain blocked A27-dependent cell fusion. Although A27 protein is unlikely to be the fusion protein per se (40), these results suggested a regulatory role of GAG-A27 protein interaction in the cell fusion process. Moreover, Kochan et al. recently showed that A17-A27 protein induced cell-cell fusion, and they also hypothesized that A27 protein binding to GAGs pulls membranes in close proximity for A17 protein to exert cell fusion (27).
Since A26 forms a complex with A27 protein through the C-terminal region, it is conceivable that the large size (∼500 aa) of A26 protein (5) easily masks the GAG-binding region (∼14 aa) of A27 protein (20), making the latter topologically difficult to bind to GAGs on cells. If this is true, we would expect that the expression of A26 protein suppresses A27-dependent cell fusion. To provide an unbiased view of cell-to-cell fusion in cells infected with Wt-VV, IA27L, and IA27L-A26WR, we performed live cell imaging and monitored cell fusion from within from 10 to 34 h p.i. (Fig. 9A; also see movie S1 in the supplemental material). From the kinetics analyses of cell fusion and the size of fused cells, we concluded that the expression of A26 protein delayed the cell fusion kinetics and suppressed A27-dependent cell fusion from within. We also repeated the cell fusion from within on the mouse L cell line that is the parental cell line used to generate the GAG-deficient mutant cell line sog9 (2). The comparison of cell fusion from without between L and sog9 cells would allow us to validate whether cell surface GAGs are indeed important for cell-to-cell fusion. Experiments were performed, and the results are shown in movie S2 in the supplemental material. The data are consistent with Fig. 9A and, more interestingly, very little cell-to-cell fusion was seen in virus-infected sog9 cells that were stained with strongly anti-VV Abs, showing that GAGs are required for cell fusion.
FIG. 9.
A27-dependent cell fusion was partially inhibited by A26 protein. (A) Live imaging recording of cell fusion from within of virus-infected BSC40 cells by time-lapse immunofluorescence microscopy. BSC40 cells were infected with Wt-VV, IA27L, and IA27L-A26WR at an MOI of 5 PFU per cell and incubated at 37°C in the presence of 5 mM IPTG for 34 h, and cell images showing cell fusion development were collected at 30 min, 10 h, 20 h, 27 h, and 34 h p.i. The bottom color figures show confocal images of the infected cells fixed at 34 h p.i. and stained with anti-VV (1:5,000) Ab (green), fluorescein isothiocyanate-conjugated goat anti-rabbit Igs, and DAPI (blue). (B) Parental L or sog9 cells were infected with purified IMV virions for 1 h at 37°C, treated with PBS (pH 4.7) at 37°C for 2 min, incubated in normal medium for 1 h to develop cell fusion from without, and subsequently fixed. Plasma membrane was stained with PKH26 (red) and nuclei were stained with Hoechst 33258 (blue), and cells were visualized by confocal microscopy. White arrows refer to fused cells. (C) Quantification of cell fusion from without of L and sog9 cells in medium containing 5 mM IPTG as shown in panel B. The percentage of cell fusion was quantified based on equations described in Materials and Methods. (D) The fused cell population of L cells in panel C were subdivided into three subpopulation for quantification as described in Materials and Methods: small fused cells (2 to 5 nuclei per cell), medium fused cells (6 to 10 nuclei per cell), and large fused cells (>10 nuclei per cell). A total of >300 cells were counted for each virus, and the experiments were independently repeated twice.
We then tested whether IMV particles of Wt-VV, IA27L, and IA27L-A26WR mediate cell fusion from without with different efficiencies under acidic pH conditions. IMV particles of Wt-VV, IA27L, or IA27L-A26WR virus were purified and incubated with L and sog9 cells, and cell fusion from without was triggered by a brief acid treatment, as described in Materials and Methods (Fig. 9B). In L cells, IA27L virus was more fusogenic than Wt-VV or IA27L-A26WR and triggered cell fusion (≥2 nuclei per cell) in about 80% of L cells; however, Wt-VV and IA27L-A26WR particles triggered cell fusion in ∼50% L cells (Fig. 9C). When these fused cells (≥2 nuclei per cell) were further divided into small (2 to 5 nuclei per cell), medium (6 to 10 nuclei per cell), and large (>10 nuclei per cell) fused cell populations (Fig. 9D), it became clear that IA27L triggered more robust cell fusion, resulting in an increase in the number as well as the size of the fused cells. The incorporation of A26WR protein into IA27L virus, i.e., IA27L-A26WR, suppressed cell fusion, resulting in fewer and smaller fused cells, similarly to those triggered by Wt-VV (Fig. 9D), suggesting that virion A26 protein partially suppresses cell fusion. Interestingly, none of these three viruses triggered obvious cell fusion from without on sog9 cells (Fig. 9B and C), demonstrating that virus-to-cell fusion also required cell surface GAGs.
DISCUSSION
Vaccinia virus envelope A27 protein is known to be anchored onto the IMV through interaction with A17 protein via the C-terminal helix (35). In this study, we demonstrated that vaccinia virus A27 and A26 proteins form covalently linked protein complexes in the infected cells and are incorporated into IMV. This conclusion is consistent with an independent study done by Howard et al. (19). Moreover, we showed that intermolecular disulfide bond formation between A26 and A27 protein required cysteines 441 and 442 on A26 (Fig. 10A), consistently with our previous report that a truncated A26 protein missing the C-terminal aa 427 to 500 was not packaged into IMV particles (5). A26-A27 disulfide bonds also required cysteines 71 and 72 of A27 protein. Interestingly, these neighboring cysteine residues are situated in the C-terminal region with a significant amino acid homology between A26 and A27 proteins (Fig. 10B). It was previously found that the C-terminal region of the A26 protein from aa 443 to 472 showed 44% identity (31) to a coiled-coil region of A27 protein. Moreover, we found another homologous region of aa 422 to 440 that was predicted to contain an α-helical structure as well (Fig. 10B). That may allow the two cysteines of A26 to be positioned in an analogous way to those of the A27 protein, implying that the helical structure of the A26 C-terminal region mimics that of the A27 protein, providing a structural basis for A26 and A27 hetero-oligomerization that is further stabilized by intermolecular disulfide bonds. The A27 protein has been shown to exist on IMV mainly as homotrimers and some dimers, but there was no indication of the presence of an intermolecular disulfide bond between A26-A27 protein complexes (36). In our experiments, A26-A27 complexes could be detected with or without NEM in lysis buffer, although in the former case the complexes seemed to be more abundant in the infected cells. The formation of the intermolecular disulfide bond between A26 and A27 proteins apparently occurred in the infected cells without the need for virion morphogenesis or viral A17 protein. At this point, we do not know the stoichiometry of each protein in 70- and 90-kDa complexes. Most likely, it is that the size difference between 70- and 90-kDa complexes is due to an extra A27 molecule, although we have not completely ruled out the presence of other viral or even cellular proteins in the 90-kDa complex. Further experiments will be needed to verify the biochemical nature of these two complexes.
FIG. 10.
(A) Schematic drawing of A26 and A27 proteins showing all of the cysteine residues (blue) and the positions of the disulfide bonds. (B) Alignment of amino acids in the C-terminal regions of A26 and A27 proteins (red lines). The conserved amino acids are in red, and brown triangles mark the two cysteines on each protein forming intermolecular bonds. The peptide regions predicted to be α-helixes are underlined. The secondary structure prediction was performed using the HNN secondary structure prediction method at the website http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_nn.html (9).
Besides the two cysteines forming intermolecular disulfide bonds, cysteines 43 and 342 of A26 protein formed an intramolecular disulfide bond, i.e., an oxidized form of A26. Since these two cysteines lie on either side of a hydrophobic region of aa 113 to 130 (5), the presence of a disulfide bond between C43 and C342 must occur on the surface of the membrane, which argues that it is topologically impossible for this hydrophobic region to act as a transmembrane region that anchors A26 protein on the IMV envelope. Finally, we have no evidence of C128 and C162 of A26 protein forming disulfide bonds, but whether these two cysteine are embedded deep inside A26 protein and remained reduced currently is not known.
In eukaryotic yeasts and mammalian cells, the redox regulation is highly compartmentalized, and accumulative knowledge revealed that each of the major compartments have unique redox characteristics (15). Among the different cellular compartments, cytosol is highly reduced, whereas the endoplasmic reticulum (ER) and the secretory pathway contain proteins with oxidative functions, including Ero-1 and PDI, that introduce disulfides into proteins during refolding and export processes (15, 46). On the other hand, it has been suggested that some vaccinia virus membrane proteins formed disulfide bonds in the cytosol of virus-infected cells (30) and that vaccinia virus contains a complete redox pathway, including E10, G4, and A2.5 proteins for forming cytoplasmic disulfide bonds on viral proteins (43, 45). Based on these published results, we originally expected that A26 and A27 proteins would be similar to L1 protein in the need of viral redox enzymes for disulfide formation. In contrast, the 70-/90-kDa protein complexes and some oxidized forms of L1 protein still formed in the infected cells when no G4 or E10 protein was expressed. The data thus suggested that, while the repression of viral redox enzymes significantly affects virus growth, these enzymes are not required for the disulfide bond between A26 and A27 proteins. This conclusion is supported by our data using transiently transfected 293T cells in which A26 and A27 proteins readily formed 70-/90-kDa complexes, showing that the viral redox system is dispensable for A26-A27 disulfide bond formation. Interestingly, Rodriguez et al. had constructed a chimeric molecule of vaccinia virus A27 fused with human immunodeficiency virus Env and found that this molecule was partially glycosylated and could be expressed on the cell surface, suggesting that the chimera molecule has passed through the ER lumen for limited glycosylation (39). The chimera A27-Env molecule contains the N-terminal 110 aa of A27 sequences, implying that the A27 N-terminal region could serve as a signal peptide for targeting the chimera molecule to the ER/secretory pathway. Besides, A17 and A27 proteins, when transfected into 293T cells, formed a complex and transported to the cell surface in 293T cells (27), providing another example in which viral membrane protein complexes were formed and exported, most likely using the cellular ER/secretory pathway. Alternatively, although the eukaryotic cytosolic environment often is considered highly reduced, the slow oxidation of newly synthesized cytosolic proteins occurred and was correlated with the oxidized glutathione/glutathione ratio in the cytosol (33). Therefore, we cannot totally rule out a cytoplasmic contribution to A26-A27 protein disulfide bond formation.
In virus-infected cells, the 70-/90-kDa complexes need to bind to A17 and perhaps other viral proteins on viral membranes in order to be incorporated into IMV. Since A26, A27, and A17 proteins all are envelope proteins on the surface of IMVs, one might wonder whether A26 protein regulates any biological functions of the A27 protein during virus entry. Using cell fusion-from-within and cell fusion-from-without assays, we demonstrated that A26 protein partially suppresses virus-induced cell fusion, based on the results obtained from the kinetics and the extent of cell-to-cell fusion. Currently, our results are consistent with the model that A26 protein suppressed A27-dependent cell fusion by interfering with cell surface GAG interactions, which otherwise led to membrane fusion executed by either A17 fusion protein (27) or the multicomponent viral fusion complex (41). However, cell fusion mediated by vaccinia virus is very complex and mostly likely requires multiple protein complexes. It is worth noting that other viral proteins, such as viral A56/K2 proteins, have been shown to suppress cell fusion (49, 51). It therefore is possible that our data have other explanations. Further investigation is required to unravel the molecular details of the cell fusion of vaccinia virus.
Supplementary Material
Acknowledgments
We thank Sue-Ping Lee and Yu-Xun Chang for their technical assistance in microscopy work.
This work was supported by grants from the Academia Sinica and the National Science Council of the Republic of China (NSC97-2320-B-001-001MY3).
Footnotes
Published ahead of print on 15 April 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Armstrong, J. A., D. H. Metz, and M. R. Young. 1973. The mode of entry of vaccinia virus into L cells. J. Gen. Virol. 21533-537. [DOI] [PubMed] [Google Scholar]
- 2.Banfield, B. W., Y. Leduc, L. Esford, K. Schubert, and F. Tufaro. 1995. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J. Virol. 693290-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, A., and D. H. Metz. 1976. Further investigations on the mode of entry of vaccinia virus into cells. J. Gen. Virol. 32275-282. [DOI] [PubMed] [Google Scholar]
- 4.Chiu, W. L., and W. Chang. 2002. Vaccinia virus J1R protein: a viral membrane protein that is essential for virion morphogenesis. J. Virol. 769575-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu, W. L., C. L. Lin, M. H. Yang, D. L. Tzou, and W. Chang. 2007. Vaccinia virus 4c (A26L) protein on intracellular mature virus binds to the extracellular cellular matrix laminin. J. Virol. 812149-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, C. S., C. H. Chen, M. Y. Ho, C. Y. Huang, C. L. Liao, and W. Chang. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 802127-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, C. S., J. C. Hsiao, Y. S. Chang, and W. Chang. 1998. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 721577-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, C. S., C. Y. Huang, and W. Chang. 2005. Vaccinia virus penetration requires cholesterol and results in specific viral envelope proteins associated with lipid rafts. J. Virol. 791623-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25147-150. [DOI] [PubMed] [Google Scholar]
- 10.Condit, R. C., N. Moussatche, and P. Traktman. 2006. In a nutshell: structure and assembly of the vaccinia virion. Adv. Virus Res. 6631-124. [DOI] [PubMed] [Google Scholar]
- 11.Dales, S., and R. Kajioka. 1964. The cycle of multiplication of vaccinia virus in Earle's strain L cells. I. Uptake and penetration. Virology 24278-294. [DOI] [PubMed] [Google Scholar]
- 12.Doms, R. W., R. Blumenthal, and B. Moss. 1990. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J. Virol. 644884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenner, F. 1990. Poxviruses, p. 2113-2133. In B. Fields and D. M. Knipe (ed.), Virology. Raven Press, New York, NY.
- 14.Foo, C. H., H. Lou, J. C. Whitbeck, M. Ponce-de-LeÛn, D. Atanasiu, R. J. Eisenberg, and G. H. Cohen. Vaccinia virus L1 binds to cell surfaces and blocks virus entry independently of glycosaminoglycans. Virology 385368-382. [DOI] [PMC free article] [PubMed]
- 15.Go, Y. M., and D. P. Jones. 2008. Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta 17801273-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong, S. C., C. F. Lai, and M. Esteban. 1990. Vaccinia virus induces cell fusion at acid pH and this activity is mediated by the N-terminus of the 14-kDa virus envelope protein. Virology 17881-91. [DOI] [PubMed] [Google Scholar]
- 17.Gruenheid, S., L. Gatzke, H. Meadows, and F. Tufaro. 1993. Herpes simplex virus infection and propagation in a mouse L cell mutant lacking heparan sulfate proteoglycans. J. Virol. 6793-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gvakharia, B. O., E. K. Koonin, and C. K. Mathews. 1996. Vaccinia virus G4L gene encodes a second glutaredoxin. Virology 226408-411. [DOI] [PubMed] [Google Scholar]
- 19.Howard, A. R., T. G. Senkevich, and B. Moss. 2008. Vaccinia virus A26 and A27 proteins form a stable complex tethered to mature virions by association with the A17 transmembrane protein. J. Virol. 8212384-12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiao, J.-C., C.-S. Chung, and W. Chang. 1998. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J. Virol. 728374-8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiao, J. C., C. S. Chung, and W. Chang. 1999. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 738750-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, C. Y., T. Y. Lu, C. H. Bair, Y. S. Chang, J. K. Jwo, and W. Chang. 2008. A novel cellular protein, VPEF, facilitates vaccinia virus penetration into HeLa cells through fluid phase endocytosis. J. Virol. 827988-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ichihashi, Y., and M. Oie. 1996. Neutralizing epitope on penetration protein of vaccinia virus. Virology 220491-494. [DOI] [PubMed] [Google Scholar]
- 24.Ichihashi, Y., T. Takahashi, and M. Oie. 1994. Identification of a vaccinia virus penetration protein. Virology 202834-843. [DOI] [PubMed] [Google Scholar]
- 25.Joklik, W. K. 1962. The purification of four strains of poxvirus. Virology 189-18. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, T., S. Kishigami, M. Sone, H. Inokuchi, T. Mogi, and K. Ito. 1997. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc. Natl. Acad. Sci. USA 9411857-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kochan, G., D. Escors, J. M. Gonzalez, J. M. Casasnovas, and M. Esteban. 2008. Membrane cell fusion activity of the vaccinia virus A17-A27 protein complex. Cell Microbiol. 10149-164. [DOI] [PubMed] [Google Scholar]
- 28.Lin, C. L., C. S. Chung, H. G. Heine, and W. Chang. 2000. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 743353-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Locker, J., A. Kuehn, S. Schleich, G. Rutter, H. Hohenberg, R. Wepf, and G. Griffiths. 2000. Entry of the two infectious forms of vaccinia virus at the plasma membrane is signaling-dependent for the IMV but not the EEV. Mol. Biol. Cell 112497-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Locker, J. K., and G. Griffiths. 1999. An unconventional role for cytoplasmic disulfide bonds in vaccinia virus proteins. J. Cell Biol. 144267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKelvey, T. A., S. C. Andrews, S. E. Miller, C. A. Ray, and D. J. Pickup. 2002. Identification of the orthopoxvirus p4c gene, which encodes a structural protein that directs intracellular mature virus particles into A-type inclusions. J. Virol. 7611216-11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercer, J., and A. Helenius. 2008. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 320531-535. [DOI] [PubMed] [Google Scholar]
- 33.Østergaard, H., C. Tachibana, and J. R. Winther. 2004. Monitoring disulfide bond formation in the eukaryotic cytosol. J. Cell Biol. 166337-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Resch, W., K. K. Hixson, R. J. Moore, M. S. Lipton, and B. Moss. 2007. Protein composition of the vaccinia virus mature virion. Virology 358233-247. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez, D., J. R. Rodriguez, and M. Esteban. 1993. The vaccinia virus 14-kilodalton fusion protein forms a stable complex with the processed protein encoded by the vaccinia virus A17L gene. J. Virol. 673435-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez, J. F., E. Paez, and M. Esteban. 1987. A 14,000-Mr envelope protein of vaccinia virus is involved in cell fusion and forms covalently linked trimers. J. Virol. 61395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez, J. F., and G. L. Smith. 1990. Inducible gene expression from vaccinia virus vectors. Virology 177239-250. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez, J. F., and G. L. Smith. 1990. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 185347-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez, J. R., D. Rodriguez, and M. Esteban. 1991. Structural properties of HIV-1 Env fused with the 14-kDa vaccinia virus envelope protein. Virology 181742-748. [DOI] [PubMed] [Google Scholar]
- 40.Senkevich, T. G., and B. Moss. 2005. Vaccinia virus H2 protein is an essential component of a complex involved in virus entry and cell-cell fusion. J. Virol. 794744-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Senkevich, T. G., S. Ojeda, A. Townsley, G. E. Nelson, and B. Moss. 2005. Poxvirus multiprotein entry-fusion complex. Proc. Natl. Acad. Sci. USA 10218572-18577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senkevich, T. G., A. S. Weisberg, and B. Moss. 2000. Vaccinia virus E10R protein is associated with the membranes of intracellular mature virions and has a role in morphogenesis. Virology 278244-252. [DOI] [PubMed] [Google Scholar]
- 43.Senkevich, T. G., C. L. White, E. V. Koonin, and B. Moss. 2000. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl. Acad. Sci. USA 9712068-12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senkevich, T. G., C. L. White, E. V. Koonin, and B. Moss. 2002. Complete pathway for protein disulfide bond formation encoded by poxviruses. Proc. Natl. Acad. Sci. USA 996667-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Senkevich, T. G., C. L. White, A. Weisberg, J. A. Granek, E. J. Wolffe, E. V. Koonin, and B. Moss. 2002. Expression of the vaccinia virus A2.5L redox protein is required for virion morphogenesis. Virology 300296-303. [DOI] [PubMed] [Google Scholar]
- 46.Sevier, C. S., and C. A. Kaiser. 2008. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim. Biophys. Acta 1783549-556. [DOI] [PubMed] [Google Scholar]
- 47.Smith, G. L., and A. Vanderplasschen. 1998. Extracellular enveloped vaccinia virus. Entry, egress, and evasion. Adv. Exp. Med. Biol. 440395-414. [PubMed] [Google Scholar]
- 48.Townsley, A. C., A. S. Weisberg, T. R. Wagenaar, and B. Moss. 2006. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J. Virol. 808899-8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner, P. C., and R. W. Moyer. 2008. The vaccinia virus fusion inhibitor proteins SPI-3 (K2) and HA (A56) expressed by infected cells reduce the entry of superinfecting virus. Virology 380226-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vázquez, M.-I., G. Rivas, D. Cregut, L. Serrano, and M. Esteban. 1998. The vaccinia virus 14-kilodalton (A27L) fusion protein forms a triple coiled-coil structure and interacts with the 21-kilodalton (A17L) virus membrane protein through a C-terminal α-helix. J. Virol. 7210126-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagenaar, T. R., and B. Moss. 2009. Expression of the A56 and K2 proteins is sufficient to inhibit vaccinia virus entry and cell fusion. J. Virol. 831546-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White, C. L., T. G. Senkevich, and B. Moss. 2002. Vaccinia virus G4L glutaredoxin is an essential intermediate of a cytoplasmic disulfide bond pathway required for virion assembly. J. Virol. 76467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White, C. L., A. S. Weisberg, and B. Moss. 2000. A glutaredoxin, encoded by the G4L gene of vaccinia virus, is essential for virion morphogenesis. J. Virol. 749175-9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolffe, E. J., D. M. Moore, P. J. Peters, and B. Moss. 1996. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J. Virol. 702797-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoder, J. D., T. S. Chen, C. R. Gagnier, S. Vemulapalli, C. S. Maier, and D. E. Hruby. 2006. Pox proteomics: mass spectrometry analysis and identification of Vaccinia virion proteins. Virol. J. 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.