Abstract
A study of potential mycobacterial regulatory genes led to the isolation of the Mycobacterium smegmatis whmD gene, which encodes a homologue of WhiB, a Streptomyces coelicolor protein required for sporulation. Unlike its Streptomyces homologue, WhmD is essential in M. smegmatis. The whmD gene could be disrupted only in the presence of a plasmid supplying whmD in trans. A plasmid that allowed chemically regulated expression of the WhmD protein was used to generate a conditional whmD mutant. On withdrawal of the inducer, the conditional whmD mutant exhibited irreversible, filamentous, branched growth with diminished septum formation and aberrant septal placement, whereas WhmD overexpression resulted in growth retardation and hyperseptation. Nucleic acid synthesis and levels of the essential cell division protein FtsZ were unaltered by WhmD deficiency. Together, these phenotypes indicate a role for WhmD in mycobacterial septum formation and cell division.
The slow growth rate of mycobacteria is significant to the pathogenesis and treatment of mycobacterial infections; yet little is known about cell division and its regulation in these bacteria. This slow growth rate necessitates protracted chemotherapeutic regimens for tuberculosis and leprosy, which in turn leads to poor patient compliance and the emergence of drug-resistant organisms.
Recently, much progress has been made toward an understanding of the temporal and spatial regulation of prokaryotic cell division (1–4). Doubling times for mycobacteria are significantly longer than those of better characterized model prokaryotes: 18–24 h in Mycobacterium tuberculosis, the cause of tuberculosis, 14 days in Mycobacterium leprae, the cause of leprosy, and 3 h in Mycobacterium smegmatis, a saprophyte. The basis for these prolonged cell division times is unknown, although the limited number of rRNA operons (one in M. tuberculosis, two in M. smegmatis, as compared with seven in Escherichia coli) and the metabolic costs of maintaining the complex cell wall that characterizes this genus are often cited as possible contributing factors (5–7).
Here we show that the M. smegmatis whmD gene is essential for cell division. whmD was identified in a search for homologues of Streptomyces coelicolor proteins known to function in postexponential phase adaptation. The WhmD protein is homologous to WhiB, an 87 aa S. coelicolor protein that is dispensable for normal vegetative growth but is required for the maturation of aerial hyphae during sporulation (8, 9). S. coelicolor whiB mutants fail to assemble FtsZ rings in their aerial hyphae, leading to the arrest of hyphal development before crosswall formation (10). WhiB is postulated to function as a transcription factor, although the exact mechanism by which it regulates spore formation is not yet established. Recently a family of whiB-like genes has been identified in Streptomyces and Mycobacteria (11), and functional assessments of some of these genes have been reported (12, 13). Our data show that WhmD, like WhiB, is important for the formation and maturation of the division septum.
Methods
Strains and Media.
M. smegmatis mc26 1-2c was grown in Middlebrook 7H9 liquid medium and on Middlebrook 7H10 agar. These media were supplemented with albumin dextrose complex (ADC; 5 g/liter BSA, 2 g/liter dextrose, 0.85 g/liter NaCl), 0.2% glycerol, and antibiotics as appropriate (50 μg/ml hygromycin, 25 μg/ml kanamycin, 30 μg/ml apramycin). Acetamide at either 0.2% or 0.02% was used to induce expression of whmD from pJG1012, and withdrawal of acetamide was achieved by washing cells in M7H9 twice. E. coli DH5α was used for all subcloning procedures and was grown in LB medium/agar.
Identification and Cloning of M. smegmatis whmD.
The degenerate primers WB1AL (5′-GGGAATTCTGGCARSWVCRIGSICTSTG-3′) and WB4AR (5′-GCGCAAGCTTCGYTCSSWYTCSSWSAGICCICCCCA-3′) were used to amplify a 200-bp fragment of the M. smegmatis whmD gene by PCR. This DNA fragment was used to probe a Lambda ZapII library of EcoRI-digested M. smegmatis genomic DNA obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Of approximately 4,000 plaques examined in the primary screen, one gave a positive signal after secondary screening. A phagemid with a 783-bp insert containing the whmD ORF was isolated and sequenced, and the size and origin of this fragment were confirmed by Southern blotting of EcoRI-digested M. smegmatis genomic DNA. The sequence of this DNA fragment has been entered into GenBank under accession number AF164439. To obtain a larger DNA fragment carrying the whmD locus, a pUC19-based plasmid library of 2.5- to 3.5-kb PmlI genomic DNA fragments was generated and probed with the 783-bp EcoRI restriction fragment. Two clones, pJGH1 and PJGI2, were isolated and found to contain the regions upstream and downstream of whmD, respectively.
Plasmid Constructs for whmD Gene Replacement.
The targeting vector pJG1007 contains a 2.0-kb PvuII fragment (containing approximately 1.9 kb of upstream sequences and the first 37 codons of whmD) and a 2.2-kb XbaI fragment (containing the whmD stop codon and downstream sequences) flanking the hyg gene from Streptomyces hygroscopicus. The hygromycin resistance cassette replaces the final 91 codons of the whmD coding region. The vector backbone of pJG1007 contains a kanamycin resistance cassette (aph) and sacB gene but does not contain a mycobacterial origin of replication and therefore can be maintained only by integration into the host chromosome (Fig. 1A). The complementing plasmid pJG1010 consists of a 1.9-kb BamHI-XbaI whmD-containing fragment cloned into the apramycin resistance-conferring shuttle vector pPE207 (14), kindly provided by Julian Davies (Univ. of British Columbia, Vancouver, BC, Canada). The acetamide-regulated complementing plasmid pJG1012 expresses whmD from the inducible acetamidase promoter of M. smegmatis (15). Its parental vector pJG1011, a pPE207 derivative, contains only the acetamidase regulatory region.
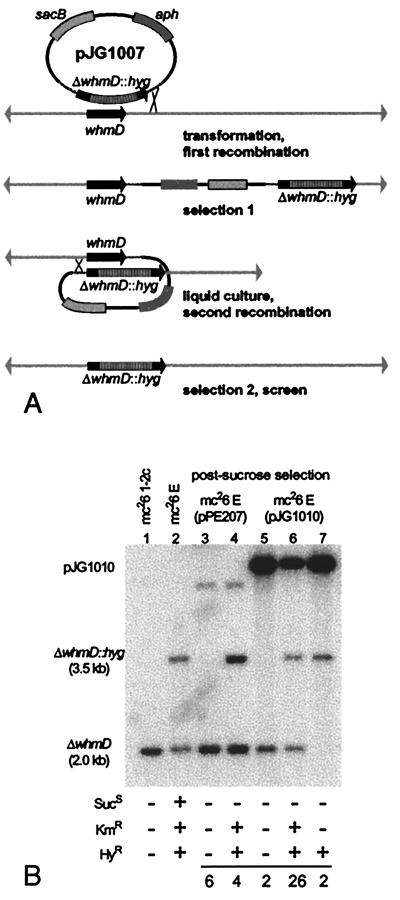
Figure 1.
(A) Two-step strategy for disruption of the M. smegmatis whmD gene. pJG1007 carries a partially deleted whmD gene with a cassette conferring HyR replacing the final 93 codons of whmD, a sacB marker conferring SucS, and an aph gene conferring KmR. Selection 1 results in KmR,HyR merodiploid integrants. Selection 2 (on sucrose) produces either the desired knockout (SucR, HyR, KmS) as shown, the wild-type (SucR, HyS, KmS), or sacB mutant merodiploids (SucR, HyR, KmR). (B) Southern blot of total DNA from representative sucrose-resistant clones and their parental strains. A 2-kb BamHI fragment containing whmD was used to probe BamHI-digested genomic DNA run on a 0.8% agarose gel. The last three lanes show the three types of postsucrose selection isolates obtained when the trans-complementing plasmid pJG1010 was present. The final lane shows the loss of the wild-type 2-kb BamHI fragment and the presence of the insertionally inactivated allele. A summary of the resistance phenotypes of analyzed clones is shown in Table 1, with each corresponding genotypic pattern as determined by Southern blotting. mc26 E, Merodiploid strain obtained after selection 1. After sucrose selection, 10 clones from mc26 E (pPE207) and 30 clones from mc26 E (pJG1010) were scored phenotypically. The numbers at the bottom indicate the frequency of each phenotype.
Replacement and Complementation of the M. smegmatis whmD Gene.
After electroporation into M. smegmatis, pJG1007 integrants were selected on hygromycin plus kanamycin. Southern blotting of PmlI-digested DNA isolated from M. smegmatis at this stage revealed that, in generating the merodiploid intermediate, homologous recombination between pJG1007 and the chromosomal whmD locus occurred with approximately equal efficiency in the upstream (18 of 37 clones) and downstream (19 of 37 clones) sequences flanking the hygromycin resistance (HyR) marker in pJG1007. After 24 h of growth in liquid culture in the absence of antibiotics, clones were subjected to a second round of selection on 10% sucrose with or without hygromycin, and sucrose-resistant clones were screened for kanamycin sensitivity. The complementing plasmids pJG1010 and pJG1012 were introduced into the merodiploid intermediate strain by electroporation. After sucrose selection, the presence of the aph gene, which cannot be scored phenotypically because of the apramycin resistance marker on the pPE207 derivatives, was scored by PCR.
Microscopy.
Differential interference contrast (DIC) microscopy and fluorescence microscopy were used to examine M. smegmatis morphology. Live bacteria were stained for 5 min at 37°C with the nucleic acid binding dye SYTO 11 (Molecular Probes) at a concentration of 5 μg/ml in Middlebrook 7H9 medium. SYTO 11, which fluoresces when bound to either RNA or DNA, was chosen for our analyses because of its permeance relative to 4′,6-diamidino-2-phenylindole (DAPI). Digital photography was performed by using a Photometrics (Tucson, AZ) PXL-1400 charge-coupled device (CCD) camera, and images were manipulated (background subtraction) by using the iplab program. Fluorescence images were artificially colored by using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Electron microscopy was performed on Zeiss EM10 or Phillips (Eindhoven, the Netherlands) CM120 instruments at the Johns Hopkins Medical Institutions Microscopy Facility. Three different fixation and staining protocols were used to maximize cell wall and nucleoid visualization and to orient the bacteria in a planar arrangement for sectioning. The first set of samples was fixed in 2% glutaraldehyde, 2% formaldehyde in Dulbecco's PBS (DPBS) (pH 7.4) plus 3 mM MgCl2 for 1 h and then washed three times for 5 min each in 0.1 M cacodylate. Specimens were then microwaved for 5 min in 1% OsO4 in 0.1 M cacodylate, washed three times for 5 min each in dH2O, and treated with 2% uranyl acetate for 30 min before rinsing with 50% ethanol. Samples were dehydrated by sequential 10-min rinses in ethanol (50%, 70%, 90%, and 100%, three times each). After 1 h in SPURRS (a low viscosity resin) diluted 1:1 in ethanol, samples were transferred to complete SPURRS overnight. The following day, the resin was changed three times before baking at 60°C overnight. A second set of samples was subjected to the above protocol with minor modifications. In an attempt to obtain a more planar arrangement of the mycobacteria before sectioning, after dehydration the bacteria were embedded in eponate on a poly-l-lysine-coated coverslip. The third set of samples was handled as in Takede et al. (16).
Western and Northern Blot Analyses.
Recombinant histidine-tagged WhmD was expressed in E. coli BL21(DE3) from pETD1, a derivative of pET15b containing the whmD ORF. The His-tagged WhmD protein was purified by using a nickel column (Invitrogen) and was used to immunize a New Zealand white (NZW) rabbit at Covance Laboratories (Denver, PA). WhmD antiserum was used at a 1:100 dilution in Western blots of M. smegmatis proteins that had been electrophoresed on a 12% tricine gel and transferred to nitrocellulose. Horseradish peroxidase-conjugated goat anti-rabbit IgG at a 1:3500 dilution and chemiluminescent substrate (Amersham) were used to detect the presence of WhmD. Detection of FtsZ by Western blot was achieved by using the polyclonal rabbit antiserum UK25A, raised against the E. coli FtsZ protein, generously provided by Joe Lutkenhaus (Univ. of Kansas Medical Center, Kansas City, KS). M. smegmatis proteins were electrophoresed on a 10% SDS/PAGE gel before transfer, and UK25A serum was used at a 1:2000 dilution to detect the M. smegmatis FtsZ protein.
Results
Identification of the M. smegmatis whmD Gene.
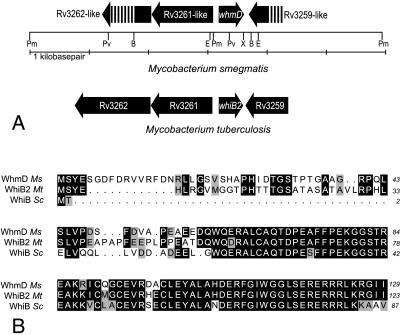
Southern blot surveys suggested the existence of a mycobacterial gene closely related to whiB of S. coelicolor (17). A partial M. smegmatis ORF was amplified by degenerate PCR using primers based on the sequence of the S. coelicolor whiB gene, and a full-length clone was subsequently isolated from a genomic library of M. smegmatis using the PCR product as a probe. We designated this gene whmD, because it was the fourth mycobacterial whiB homolog we had identified (11). A highly similar M. tuberculosis gene later appeared in the published genome sequence of the H37Rv strain and was annotated as whiB2 (18). The genomic context of the M. tuberculosis and M. smegmatis whmD genes is conserved (Fig. 2A). Additionally, whereas the C termini of the 129-aa M. smegmatis WhmD (WhmDMs) protein and the 123-aa WhmDMt (WhiB2) are virtually identical to the 87-aa S. coelicolor WhiB, the WhmD proteins contain a unique 38- to 48-aa N-terminal extension not found in WhiB (Fig. 2B). Although the M. tuberculosis whiB2 gene is annotated as an 89 codon ORF based on homology to whiB, the use of the more upstream start codon is supported by both the relative mobility of the WhmD protein seen in immunoblots (15–16 kDa) and the presence of a likely Shine-Dalgarno sequence centered 10 bp upstream of the proximal start codon. A total of seven WhiB-homologues are encoded in the M. tuberculosis genome and at least six are present in S. coelicolor; however, the gene family has not been identified in bacteria outside the order Actinomycetales (11).
Figure 2.
(A) Map of the M. smegmatis whmD locus. The restriction sites in this region are shown as abbreviations (B = BamHI, E = EcoRI, Pm = PmlI, Pv = PvuII, and X = XbaI). The local arrangement of predicted genes is shown by arrows. The arrangement of genes in M. tuberculosis H37Rv is shown below to illustrate the similarity of the gene arrangement in the two species. (B) Alignment of the M. smegmatis (Ms) WhmD and M. tuberculosis (Mt) WhiB2 (WhmD) proteins with the WhiB protein of S. coelicolor (Sc). Identities are shown in black and similarities in gray.
whmD Is an Essential Gene.
We attempted to disrupt the M. smegmatis whmD locus by a two-step gene replacement strategy by using a suicide vector in which a hygromycin-resistance cassette replaced the final 91 codons of the whmD ORF (Fig. 1A). The vector also contained the counterselectable sacB gene and the aph gene, which confer sucrose sensitivity (SucS) (19, 20) and kanamycin resistance (KmR), respectively. After electroporation, numerous HyR, KmR merodiploid clones were obtained, and the integration of the targeting vector at the whmD locus was confirmed by Southern blotting. These merodiploids were subjected to the second-step selection on medium containing sucrose. Clones capable of growth on sucrose were anticipated in three categories: ΔwhmD∷hyg (HyR, SucR, and KmS), wild-type (HyS, SucR, and KmS), or sacB mutant merodiploids (HyR, SucR, and KmR). Unexpectedly, no HyR, SucR, and KmS colonies (i.e., ΔwhmD∷hyg recombinants) were obtained after multiple attempts. As shown in Table 1, a detailed phenotypic evaluation of 742 SucR colonies obtained by second-step selection at either 30°C or 37°C failed to reveal a single whmD-disrupted mutant. However, SucR clones resulting from recombination on the same flank as the original integration, restoring an intact whmD locus, were readily obtained when hygromycin selection was omitted. Because the initial integration of the plasmid occurred with equal frequency on either flank, the failure to obtain knockouts could not be explained by recombinational bias toward one flank of the whmD gene. Instead, these results strongly suggested that disruption of whmD is deleterious for bacterial survival.
Table 1.
Results of second-step selection on 10% sucrose
| Temperature | Selection medium* | No. of colonies (%)
|
|||
|---|---|---|---|---|---|
| HyRKanS ΔwhmD | HySKanS wt whmD | HyRKanR sacB mutant | Total | ||
| 37°C | M7H10 + 10% sucrose | 0 (0%) | 114 (70%) | 50 (30%) | 164 |
| M7H10 + 10% sucrose, hygromycin B | 0 (0%) | 0 (0%) | 275 (100%) | 275 | |
| 30°C | M7H10 + 10% sucrose | 0 (0%) | 95 (40%) | 144 (60%) | 239 |
| M7H10 + 10% sucrose, hygromycin B | 0 (0%) | 0 (0%) | 64 (100%) | 64 | |
M7H10, Middlebrook 7H10 medium supplemented with albumin dextrose complex (ADC).
To evaluate whether whmD is in fact an essential gene, we introduced a whmD-containing plasmid conferring apramycin resistance (AmR) into the HyR,KmR merodiploid before sucrose counterselection. The complemented merodiploid strain gave rise to 2 of 30 (6.7%) HyS, KmS wild-type revertants, 26 of 30 (87%) HyR, KmR sacB mutants, and 2 of 30 (6.7%) HyR, KmS chromosomal whmD-disrupted mutants. Although the majority of SucR clones arose because of sacB mutation, plasmid excision events leading to the loss of the KmR marker occurred at an equal frequency on either flank. Thus, the presence of an episomal whmD gene permitted the disruption of the chromosomal whmD locus. Transformation with the parental vector pPE207 had no effect on our ability to disrupt the chromosomal whmD gene. Because the complementing plasmid contained no ORFs 3′ of whmD, successful disruption of the chromosomal allele only in the presence of extrachromosomal complementation indicates that whmD, itself, rather than a downstream gene, is essential in M. smegmatis.
Conditional Complementation Reveals a Role for WhmD in Septation and Cell Division.
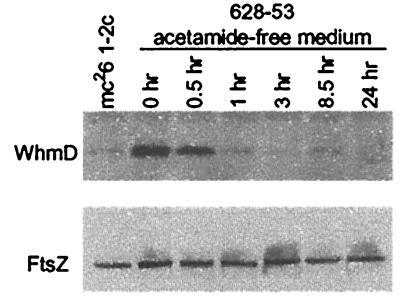
To assess the phenotypic consequences of whmD loss, we developed a strain in which the chromosomal whmD disruption could be conditionally complemented by whmD expression from an inducible promoter. We replaced the native whmD promoter with an acetamide-inducible promoter and regulatory gene cassette from the M. smegmatis acetamidase gene locus. Recently, this 3-kb M. smegmatis DNA cassette, which contains several putative accessory genes and a strong promoter (Pace) preceding the highly inducible acetamidase gene, has been shown to confer acetamide inducibility on downstream genes (15). After a plasmid, pJG1012, harboring the Pace∷whmD allele was introduced into a HyR, KmR whmD merodiploid strain, sucrose selection in the presence of 0.2% acetamide yielded a HyR,KmS strain, designated 628-53, that on Southern blot analysis contained the desired chromosomal ΔwhmD mutation. Immunoblots of lysates of M. smegmatis 628-53 showed that, in the presence of acetamide, WhmD was produced at levels greater than those seen in wild-type M. smegmatis, but WhmD levels declined rapidly after acetamide withdrawal (see Fig. 5). The addition of 0.2% acetamide to wild-type M. smegmatis did not alter WhmD expression levels.
Figure 5.
WhmD withdrawal does not influence FtsZ accumulation. Western blots to detect WhmD and FtsZ in lysates of M. smegmatis 628-53 at various times after transfer of the bacteria to acetamide free medium. Lysates containing matched amounts of protein were separated on a 12% tricine gel for detection of the 14.4-kDa WhmD protein, or on a 10% SDS/PAGE gel for detection of the ≈39-kDa FtsZ protein.
When strain 628-53 was transferred from medium containing glycerol and acetamide to medium containing only glycerol, the WhmD-underexpressing cells became defective in cell division, forming long, branched filaments over the course of 6 to 12 h (6 h equates to slightly more than two typical generations) (Figs. 3 and 4). During this time, the culture continued to accumulate biomass as assessed by turbidity measurements, and increasing filamentation resulted in macroscopic aggregation of the bacilli. Qualitative analyses showed that bacterial length increased rapidly after acetamide withdrawal, suggesting that the effect of whmD loss was immediate. Addition of acetamide to the culture medium, after filamentation had become obvious, did not lead to restoration of binary fission. The WhmD-deficient filaments retained their acid-fast staining properties.
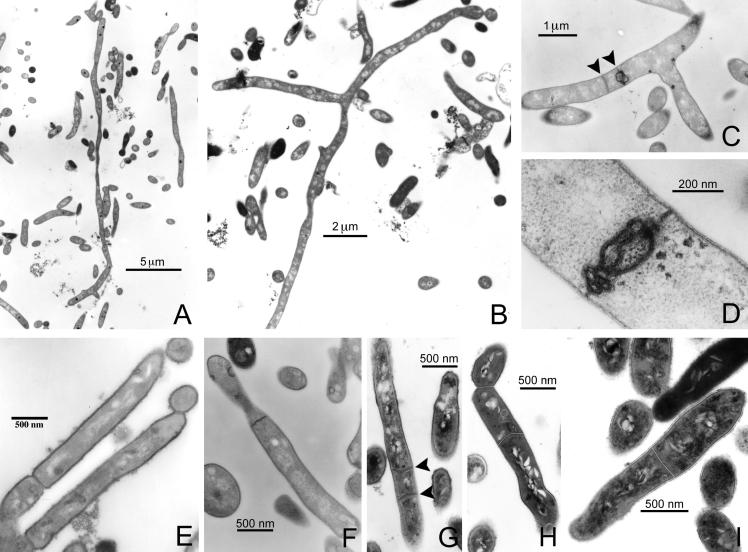
Figure 3.
(A–D) Transmission electron micrographs of ultrathin sections of M. smegmatis 628-53, which carries the plasmid pJG1012 (Pace∷whmD), allowing for the acetamide-dependent expression of WhmD. After growth for 13 h in the absence of acetamide, bacteria became filamentous, as can be seen in A and B. The longest bacterium in A is at least 26 μm in length, and no evidence of septa can be detected throughout its length. In B, a branched, aseptate bacterium is shown. The length of this bacterium from the upper right corner to the bottom center of the panel is 16 μm. (C and D) Defective septal structures seen in the WhmD-underexpressing mutant; (C) closely spaced septa (arrows) present in sections of single filaments. (E and F) The complemented mutant strain grown in 0.2% acetamide. Although the morphology of septa appears to be occasionally abnormal, as seen in F, these bacteria are capable of septum maturation and fission. (G–I) M. smegmatis-carrying plasmids pJG1012 (Pace∷whmD; G and H) or PJG1011 (Pace, empty vector; I). Bacteria in G–I were grown 0.2% acetamide and were fixed and stained by using tannic acid. Noteworthy are the multiple septa within a single bacterium (G, arrows) and asymmetric positioning of septa close to previous division sites (H). No unusual septation was seen in bacteria carrying the control plasmid pJG1011 (I).
Figure 4.
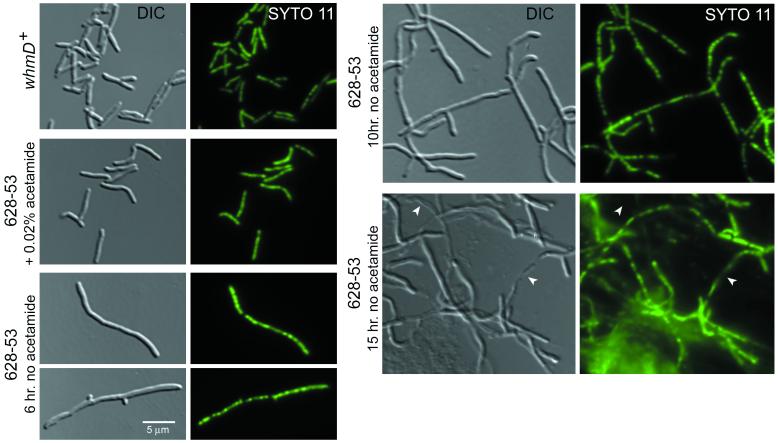
DIC and fluorescence microscopy analysis. The nucleic acid stain SYTO 11 was used to localize the nucleoids in wild-type and whmD mutant M. smegmatis grown in M7H9 with and without 0.2% acetamide. Parallel digital micrographs of the same field were captured by using DIC (Left) or fluorescence (Right) microscopy (×1000). After two washes in acetamide-free M7H9, M. smegmatis 628-53 was grown at 37°C in acetamide-free M7H9 (with 0.2% glycerol as a carbon source). The formation of filaments and some branching is obvious by 6 h (doubling time of M. smegmatis is 2.5 to 3 h). By 10 h, bacteria have become highly branched and filamentous, and larger, more obvious gaps in the nucleic acid staining pattern are visible. Significant lysis is seen by 15 h (arrows). Wild-type M. smegmatis mc26 1-2c (whmD+) and the complemented mutant (628-53 + 0.2% acetamide) are shown for comparison.
We studied the filaments for the presence of septa by examining thin sections of fixed samples by transmission electron microscopy (Fig. 3) (16). Although septa were observed in strain 628-53 grown in both acetamide-free and acetamide-containing media, the frequency, positioning, and character of the septa were altered when WhmD levels were depleted by acetamide withdrawal. Whereas wild-type M. smegmatis grown in the presence of acetamide were limited to a single, medially placed septum, the filamentous WhmD-deficient organisms often had multiple septa distributed unevenly throughout their length. In the absence of acetamide, the placement of completed septa was not medial; septa were commonly observed near branch points and the tips of filaments. Some WhmD-deficient filaments were completely devoid of septa; one such filament, from a culture grown for 13 h without acetamide, was 26 microns long. When septa were present in the mutant, their maturation appeared to be arrested because no evidence of cytokinesis was seen in mutant cells. In the presence of acetamide, M. smegmatis 628-53 was competent for cytokinesis (Fig. 3E), although the structures of some septa observed by transmission electron microscopy appeared to be abnormal, as illustrated in Fig. 3F.
Having observed that WhmD withdrawal reduced the frequency of septum formation, we investigated the effect of WhmD overexpression. Wild-type M. smegmatis, harboring the Pace∷whmD fusion plasmid (pJG1012) grown in the presence of acetamide, produced excess WhmD relative to similarly grown M. smegmatis carrying a plasmid bearing only Pace (pJG1011). On M7H10 agar, the M. smegmatis colony size was greatly reduced when whmD expression was induced by the presence of acetamide in the medium, but the WhmD-overexpressing strain showed only slightly slowed growth kinetics in liquid medium. Although liquid-grown, acetamide-induced M. smegmatis carrying pJG1012 appeared normal by DIC microscopy, we observed several examples of multiply septate bacteria when these cultures were examined by transmission electron microscopy (Fig. 3G). These observations suggest that WhmD levels affect the localization or efficiency of septum formation. Interestingly, whereas excess WhmD in a wild-type background led to misplaced septa, greater than wild-type levels of WhmD were required in trans to prevent filamentation in a ΔwhmD∷hyg background.
We evaluated the state of the nucleoids in the filamentous, WhmD-deficient M. smegmatis 628-53 by staining with SYTO 11, a highly permeant dye that fluoresces when bound to nucleic acids (Fig. 4). SYTO 11-stained filaments fluoresced in a beaded pattern throughout their length, suggesting that they contained multiple chromosomes and that nucleic acid synthesis had continued after WhmD withdrawal. After 10 h of growth in the absence of acetamide, larger gaps between nucleoids could be observed, and a significant amount of lysis was seen. By 15 h, we observed many instances of partially lysed filaments by DIC microscopy. These partial ghosts failed to stain with SYTO 11 in the lysed segments of their length but continued to stain in their unlysed segments, indicating that the septa present in these filaments can completely separate adjacent cytoplasmic compartments. These experiments suggest that the WhmD-deficient filaments are polyploid with respect to DNA content.
Withdrawal of WhmD Does Not Affect FtsZ Accumulation.
The polymerization of FtsZ in a ring at midcell is necessary for initiation of septum assembly. To determine whether WhmD deficiency reduced the expression of FtsZ, we performed Western blots of M. smegmatis 628-53 underexpressing WhmD during various stages of filamentation (Fig. 5). Despite the extensive filamentation by 6 h, FtsZ levels remained constant from 1 to 24 h whereas WhmD levels declined rapidly, reaching minimal levels after 3 h. Hence, the septal defects we observed after WhmD withdrawal are not due to insufficient levels of FtsZ.
Discussion
The cell division defects associated with WhmD deficiency provide an explanation for the essential nature of the whmD gene. Although loss of whmD has more severe consequences for M. smegmatis than does loss of whiB for S. coelicolor, the two mutant phenotypes are related. Both WhiB and WhmD appear to be important for septum formation and fragmentation, WhiB governing postexponential phase spore septum formation and WhmD binary fission. Whereas whiB expression is tightly linked to growth phase (21), both whmD mRNA and WhmD protein are present throughout exponential growth and into early stationary phase but are undetectable in 5-day-old cultures (data not shown). This expression pattern is consistent with a requirement for WhmD during periods of active cell division. The fact that WhiB is dispensable in S. coelicolor likely reflects the mycelial nature of this organism; Streptomycetes are unique in their ability to remain viable after deletion of the usually essential cell division genes ftsZ and ftsQ (10, 22, 23). The extensive and immediate filamentation we observed in a conditional whmD mutant, in conjunction with an apparent defect in septum formation and positioning, suggests a role for WhmD in the early stages of mycobacterial cell division, perhaps in FtsZ localization or polymerization or as a regulator of cell division genes other than ftsZ.
Several other proteins closely related to WhmD and WhiB are present in M. tuberculosis, S. coelicolor, and other high-GC Gram-positive bacteria. These proteins are apparently incapable of functionally substituting for WhmD, because loss of whmD alone is lethal. A homologue of whmD, whmB (also described as whiB3), is present in M. smegmatis, M. tuberculosis, and M. leprae. Although this gene is nonessential in M. smegmatis (13), the S. coelicolor orthologue of WhmB, WhiD, has been shown to be essential for spore formation (12).
It should be noted that the filamentous, largely aseptate morphology of the WhmD-deficient M. smegmatis 628-53 is similar to that seen when Mycobacterium avium is treated with low levels of the β-lactam antibiotic ampicillin (24). Several penicillin-binding proteins have been shown to be required for cell division; examples include the E. coli FtsI (PBP3) (25) and its Bacillus subtilis counterpart, PBP2B (26), which are septum-specific peptidoglycan synthetic enzymes. ponA mutants of B. subtilis, which are defective in PBP1 function, also show sporadic division defects (27). Along these lines, the septal defects of 628-53 may be due to a WhmD requirement for function of one or more cell division-associated murein synthetic enzymes. A better understanding of WhmD function may reveal additional components of the actinomycete cell division machinery and provide new potential targets for chemotherapeutic intervention against mycobacterial diseases.
Acknowledgments
We thank Gary Ketner, Michael Sayre, James Dick, and Arthur Dannenberg for helpful suggestions, Joseph Lutkenhaus for FtsZ antiserum, Tanya Parish for plasmids carrying the acetamidase regulatory region, Carol Cooke and Douglas Murphy for assistance with microscopy, and Naomi Gauchet for manuscript preparation. This work was supported by National Institutes of Health Grants AI36973 and AI37856.
Abbreviations
- Suc
sucrose
- Hy
hygromycin
- Km
kanamycin
- Am
apramycin
- superscript S
sensitivity
- superscript R
resistance
- DIC
differential interference contrast
Footnotes
The sequence reported in this paper has been deposited in the GenBank database (accession no. AF164439).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140225297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140225297
References
- 1.Bi E F, Lutkenhaus J. Nature (London) 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 2.Bramhill D. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 3.Lutkenhaus J, Addinall S G. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs C, Shapiro L. Proc Natl Acad Sci USA. 1999;96:5891–5893. doi: 10.1073/pnas.96.11.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bercovier H, Kafri O, Sela S. Biochem Biophys Res Commun. 1986;136:1136–1141. doi: 10.1016/0006-291x(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 6.Hiriyanna K T, Ramakrishnan T. Arch Microbiol. 1986;144:105–109. doi: 10.1007/BF00414718. [DOI] [PubMed] [Google Scholar]
- 7.Wheeler P R, Ratledge C. In: Tuberculosis: Pathogenesis, Protection and Control. Bloom B R, editor. Washington: Am. Soc. Microbiol.; 1994. pp. 353–385. [Google Scholar]
- 8.Chater K F. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 9.Davis N K, Chater K F. Mol Gen Genet. 1992;232:351–358. doi: 10.1007/BF00266237. [DOI] [PubMed] [Google Scholar]
- 10.Schwedock J, McCormick J R, Angert E R, Nodwell J R, Losick R. Mol Microbiol. 1997;25:847–858. doi: 10.1111/j.1365-2958.1997.mmi507.x. [DOI] [PubMed] [Google Scholar]
- 11.Soliveri J A, Gomez J E, Bishai W R, Chater K F. Microbiology. 2000;146:333–346. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- 12.Molle V, Palframan W J, Findlay K C, Buttner M J. J Bacteriol. 2000;182:1286–1295. doi: 10.1128/jb.182.5.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutter B, Dick T. Res Microbiol. 1999;150:295–301. doi: 10.1016/s0923-2508(99)80055-2. [DOI] [PubMed] [Google Scholar]
- 14.Paget E, Davies J. J Bacteriol. 1996;178:6357–6360. doi: 10.1128/jb.178.21.6357-6360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parish T, Mahenthiralingam E, Draper P, Davis E O, Colston M J. Microbiology. 1997;143:2267–2276. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- 16.Takade A, Takeya K, Taniguchi H, Mizuguchi Y. J Gen Microbiol. 1983;129:2315–2320. doi: 10.1099/00221287-129-7-2315. [DOI] [PubMed] [Google Scholar]
- 17.Soliveri J, Vijgenboom E, Granozzi C, Plaskitt K A, Chater K F. J Gen Microbiol. 1993;139:2569–2578. doi: 10.1099/00221287-139-11-2569. [DOI] [PubMed] [Google Scholar]
- 18.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 19.Gay P, Le Coq D, Steinmetz M, Ferrari E, Hoch J A. J Bacteriol. 1983;153:1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelicic V, Reyrat J M, Gicquel B. J Bacteriol. 1996;178:1197–1199. doi: 10.1128/jb.178.4.1197-1199.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soliveri J, Brown K L, Buttner M J, Chater K F. J Bacteriol. 1991;174:6215–6220. doi: 10.1128/jb.174.19.6215-6220.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick J R, Su E P, Driks A, Losick R. Mol Microbiol. 1994;14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 23.McCormick J R, Losick R. J Bacteriol. 1996;178:5295–5301. doi: 10.1128/jb.178.17.5295-5301.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizuguchi Y, Ogawa M, Udou T. Antimicrob Agents Chemother. 1985;27:541–547. doi: 10.1128/aac.27.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss D S, Pogliano K, Carson M, Guzman L M, Fraipont C, Nguyen-Disteche M, Losick R, Beckwith J. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- 26.Daniel R A, Williams A M, Errington J. J Bacteriol. 1996;178:2343–2350. doi: 10.1128/jb.178.8.2343-2350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen L B, Angert E R, Setlow P. J Bacteriol. 1999;181:3201–3211. doi: 10.1128/jb.181.10.3201-3211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]