Abstract
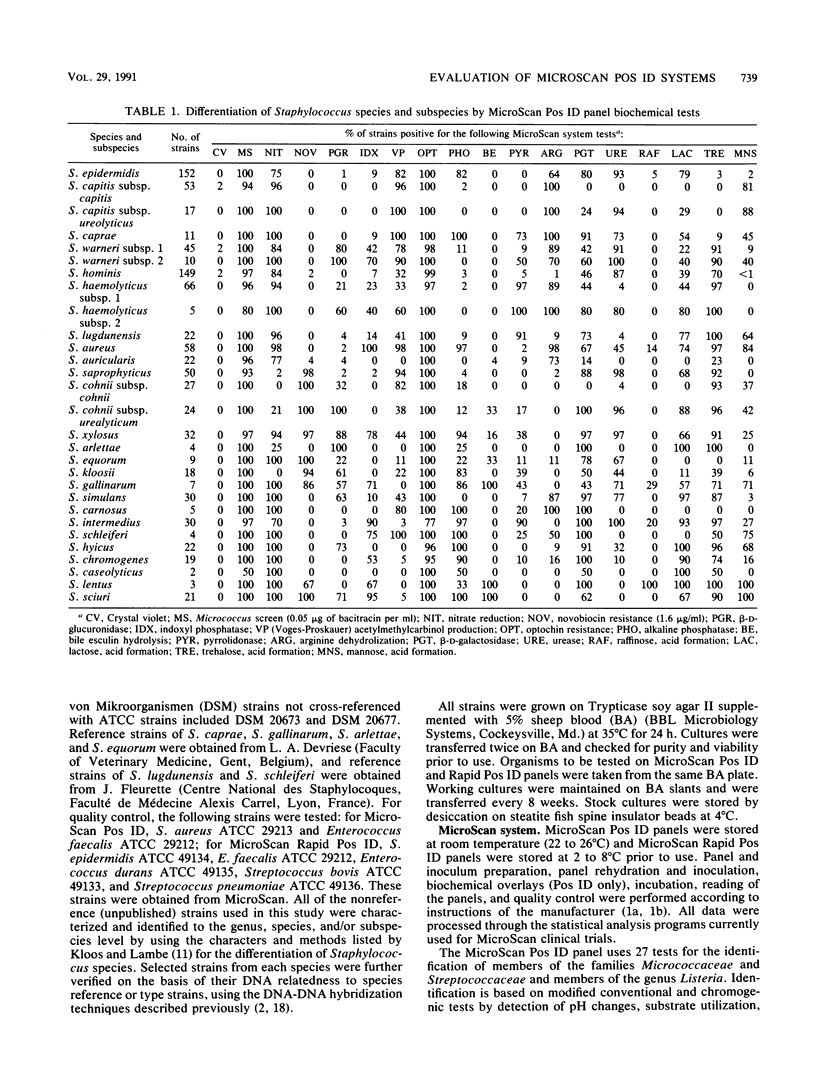
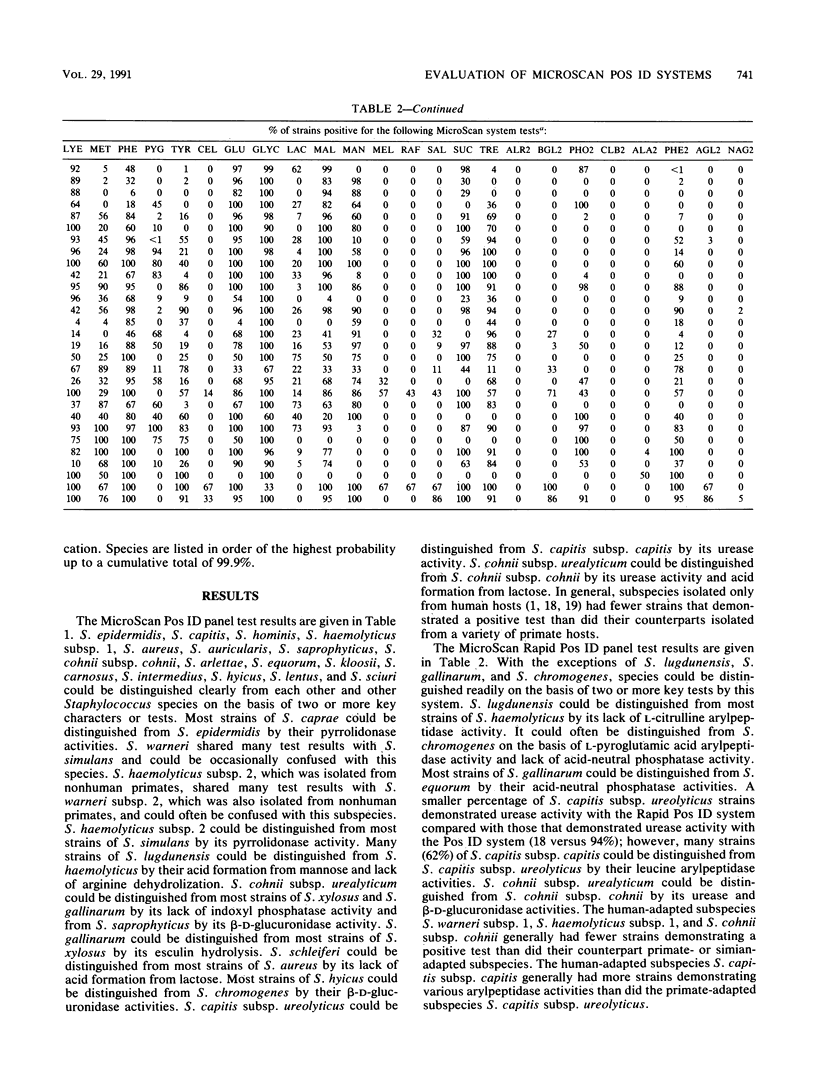
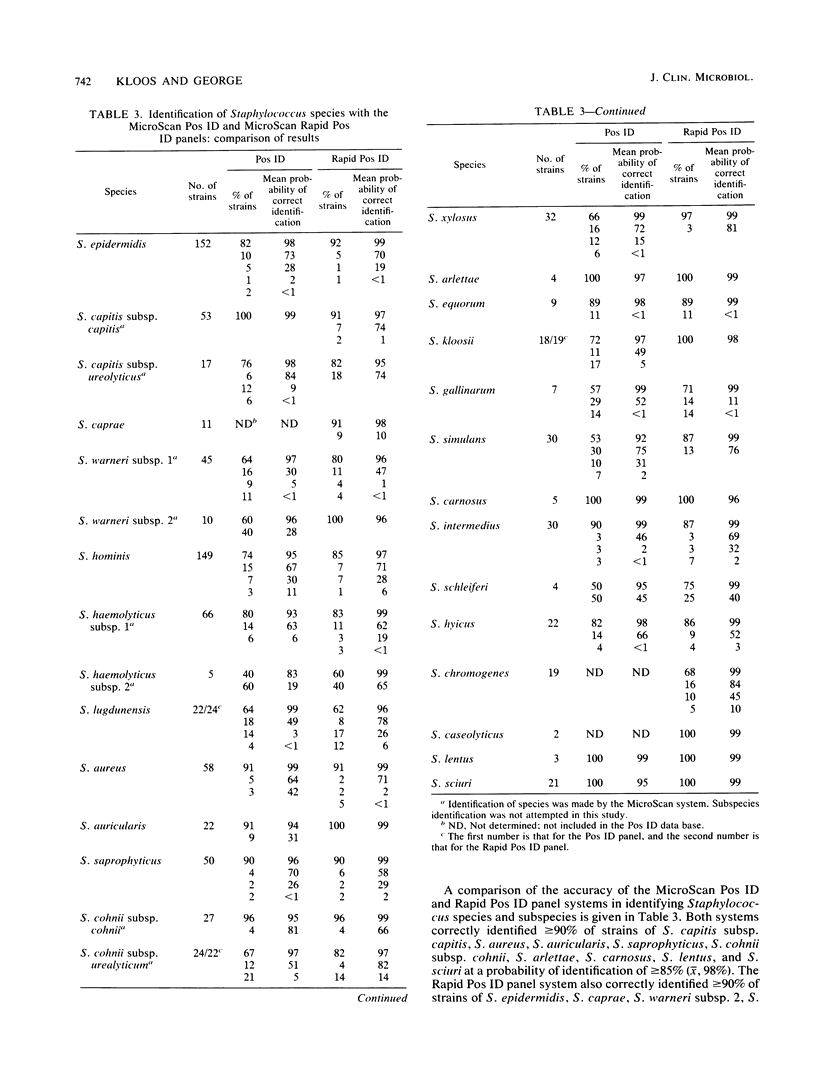
The accuracies of the MicroScan Pos ID and Rapid Pos ID panel systems (Baxter Diagnostic Inc., MicroScan Division, West Sacramento, Calif.) were compared with each other and with the accuracies of conventional methods for the identification of 25 Staphylococcus species and 4 subspecies. Conventional methods included those used in the original descriptions of species and subspecies and DNA-DNA hybridization. The Pos ID panel uses a battery of 18 tests, and the Rapid Pos ID panel uses a battery of 42 tests for the identification of Staphylococcus species. The Pos ID panel has modified conventional and chromogenic tests that can be read after 15 to 48 h of incubation; the Rapid Pos ID panel has tests that use fluorogenic substrates or fluorometric indicators, and test results can be read after 2 h of incubation in the autoSCAN-W/A. Results indicated that both MicroScan systems had a high degree of congruence (greater than or equal to 90%) with conventional methods for the species S. capitis, S. aureus, S. auricularis, S. saprophyticus, S. cohnii, S. arlettae, S. carnosus, S. lentus, and S. sciuri and, in particular, the subspecies S. capitis subsp. capitis and S. cohnii subsp. cohnii. The Rapid Pos ID panel system also had greater than or equal to 90% congruence with conventional methods for S. epidermidis, S. caprae, S. warneri subsp. 2, S. xylosus, S. kloosii, and S. caseolyticus. For both MicroScan systems, congruence with conventional methods was 80 to 90% for S. haemolyticus subsp. 1, S. equorum, S. intermedius, and S. hyicus; and in addition, with the Rapid Pos ID panel system congruence was 80 to 89% for S. capitis subsp. ureolyticus, S. warneri subsp. 1, S. hominis, S. cohnii subsp. urealyticum, and S. simulans. The MicroScan systems identified a lower percentage (50 to 75%) of strains of S. lugdunensis, S. gallinarum, S. schleiferi, and S. chromogenes, although the addition of specific tests to the systems might increase the accuracy of identification significantly.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bannerman T. L., Kloos W. E. Staphylococcus capitis subsp. ureolyticus subsp. nov. from human skin. Int J Syst Bacteriol. 1991 Jan;41(1):144–147. doi: 10.1099/00207713-41-1-144. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Rake A. V., Johnson K. E. Batch procedure for thermal elution of DNA from hydroxyapatite. Anal Biochem. 1969 Apr 4;28(1):447–459. doi: 10.1016/0003-2697(69)90199-7. [DOI] [PubMed] [Google Scholar]
- Hussain Z., Stoakes L., Stevens D. L., Schieven B. C., Lannigan R., Jones C. Comparison of the MicroScan system with the API Staph-Ident system for species identification of coagulase-negative staphylococci. J Clin Microbiol. 1986 Jan;23(1):126–128. doi: 10.1128/jcm.23.1.126-128.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos W. E., Schleifer K. H. Simplified scheme for routine identification of human Staphylococcus species. J Clin Microbiol. 1975 Jan;1(1):82–88. doi: 10.1128/jcm.1.1.82-88.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi J. T. Coagulase-negative staphylococci and the epidemiological typing of Staphylococcus epidermidis. Microbiol Rev. 1985 Jun;49(2):126–139. doi: 10.1128/mr.49.2.126-139.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Herwaldt L. A. Laboratory, clinical, and epidemiological aspects of coagulase-negative staphylococci. Clin Microbiol Rev. 1988 Jul;1(3):281–299. doi: 10.1128/cmr.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]



