Abstract
Muscle growth is associated with an activation of the mTOR signaling pathway and satellite cell regulators. The purpose of this study was to determine whether 17 selected genes associated with mTOR/muscle protein synthesis and the satellite cells/myogenic program are differentially expressed in young and older human skeletal muscle at rest and in response to a potent anabolic stimulus [resistance exercise + essential amino acid ingestion (RE+EAA)]. Twelve male subjects (6 young, 6 old) completed a bout of heavy resistance exercise. Muscle biopsies were obtained before and at 3 and 6 h post RE+EAA. Subjects ingested leucine-enriched essential amino acids at 1 h postexercise. mRNA expression was determined using qRT-PCR. At rest, hVps34 mRNA was elevated in the older subjects (P < 0.05) while there was a tendency for levels of myoD, myogenin, and TSC2 mRNA to be higher than young. The anabolic stimulus (RE+EAA) altered mRNAs associated with mTOR regulation. Notably, REDD2 decreased in both age groups (P < 0.05) but the expression of Rheb mRNA increased only in the young. Finally, cMyc mRNA was elevated (P < 0.05) in both young and old at 6 h post RE+EAA. Furthermore, RE+EAA also increased expression of several mRNAs associated with satellite function in the young (P < 0.05), while expression of these mRNAs did not change in the old. We conclude that several anabolic genes in muscle are more responsive in young men post RE+EAA. Our data provide new insights into the regulation of genes important for transcription and translation in young and old human skeletal muscle post RE+EAA.
Keywords: mTOR, REDD2, Rheb, resistance exercise, sarcopenia, aging, skeletal muscle
during the recovery period of resistance exercise, muscle protein synthesis exceeds that of protein breakdown resulting in an overall increase in net protein balance (44). Although effective, the muscle growth response following repeated bouts of resistance exercise in older individuals is less than their younger counterparts (36, 56). This may be due to an inability to fully stimulate muscle protein synthesis following an acute bout of resistance exercise (47). We recently showed that an acute anabolic stimulus (resistance exercise combined with essential amino acid ingestion) can stimulate muscle protein synthesis in older men; however, the response is delayed compared with young men (23). Undoubtedly, more information is needed at the molecular level to determine why skeletal muscle from older individuals is less responsive to an acute anabolic stimulus.
An important pathway controlling the synthesis of new proteins is the mammalian target of rapamycin (mTOR) (10). Contractile and nutritional signals converge on mTOR, making it a key regulator of muscle growth. mTOR has two well known downstream targets: ribosomal S6 kinase-1 (S6K1) and eukaryotic initiation factor 4E-binding protein-1 (4E-BP1). Recently, in drosophila, the transcription factor cMyc has also been identified as a target of TOR via regulation of ribosomal biogenesis (26, 50). Thus, the combined actions of mTOR on S6K1, 4E-BP1, and cMyc can contribute significantly to activation of protein synthesis in skeletal muscle.
In order for mTOR to activate its primary effectors, appropriate upstream signals must be received. mTOR has many positive and negative regulators that control its function. For instance, translationally controlled tumor protein (TCTP) is a positive regulator of the small GTPase, Rheb (Ras-homologue enriched in brain), therefore enhancing mTOR activity and cell growth (29). mTOR is also positively regulated by amino acids, and in particular by leucine (2, 3). To date, two molecules have been implicated in mediating signals from amino acids to mTOR: human vacuolar protein sorting-34 (hVps34) and mitogen-activated protein kinase kinase kinase kinase-3 (MAP4K3; 12, 25, 43). On the other hand, REDD1 and REDD2 have been identified as negative regulators of mTOR (17). REDD1 is altered in response to various cellular stresses such as hypoxia, dexamethasone, starvation, and muscle contraction (11, 17, 24, 48, 55). REDD1 has been shown to interact with 14–3-3 and modify the function of the tuberous sclerosis 1 (TSC1) and 2 (TSC2) complex (18) leading to inhibition of mTOR signaling. REDD2 has also been shown to inhibit mTOR (17) but the mechanisms are still not known. Another novel negative regulator of mTOR is PRAS40 (proline-rich Akt substrate-40). The function of PRAS40 remains elusive but it has been shown to bind mTOR, via raptor, which represses mTOR activity (51). Unfortunately much of these data are generated from rodents, drosophila, and cell lines. Therefore, few data, if any, are available on upstream regulators of mTOR within human skeletal muscle and how they respond to an anabolic stimulus.
Muscle growth can also be controlled by the regulation of satellite cells. Satellite cells respond to various injurious and anabolic stimuli. The sequence of satellite cell events involves activation, proliferation, and fusion to existing myofibers (19). Regulation of satellite cell function is controlled by many different factors, including growth factors (i.e., IGF-1, myostatin) and members of the myogenic regulatory family (i.e., myoD, myogenin), all of which are acutely responsive to a single bout of resistance exercise in humans (7, 8, 34, 39, 45, 57). However, no information is available in humans on the E3 ubiquitin ligase, mindbomb2 (mib2; also known as skeletrophin). Mindbomb2, an important player in Notch signaling, cleaves the intracellular region of the Notch ligand, which in turn allows the Notch receptor complex to translocate into the nucleus and upregulate genes important for myogenesis (35). Activation of Notch signaling is important for proliferation of satellite cells (15, 16). Interestingly, Notch signaling has been shown to be disregulated in aging skeletal muscle (13, 14).
Muscle protein synthesis is stimulated in both young and old men following an acute anabolic stimulus (resistance exercise followed by essential amino acid ingestion; RE+EAA), however, the response was delayed in the older men (23). The purpose of this study, in an effort to better understand the molecular mechanisms that may explain this delayed response, was to identify select and novel mRNAs associated with mTOR and satellite cell regulation after an acute anabolic stimulus (RE+EAA) in young and older men. We hypothesized that expression patterns of mTOR signaling-associated mRNAs (REDD1, REDD2, PRAS40, TSC1, TSC2, TCTP, Rheb, hVps34, MAP4K3, mTOR, S6K1, cMyc) and mRNAs associated with satellite cell function [insulin-like growth factor receptor (IGF-1R), myoD, myogenin, mib2, myostatin] would be differentially expressed in young and old human skeletal muscle both at rest and following RE+EAA.
METHODS
Subjects
We studied 6 young (29 ± 2 yr) and 6 older (70 ± 2 yr) men. These volunteers were studied previously and therefore the subject characteristics can be found elsewhere (23). The subjects were not engaged in any regular exercise training at the time of the enrollment, however, they were physically independent and overall healthy. Screening of subjects were performed with clinical history, physical exam, and laboratory tests including complete blood count with differential, liver and kidney function tests, coagulation profile, fasting blood glucose and oral glucose tolerance test, hepatitis B and C screening, HIV test, TSH, lipid profile, urinalysis, drug screening, electrocardiogram, and stress test on a treadmill or bicycle. On two separate occasions, subjects were tested for maximal strength by performing a one repetition maximum (1RM) on a leg extension machine (Cybex-VR2, Medway, MA). The starting weight used during the resistance exercise portion of this study was 70% of the subjects' predetermined 1RM. All subjects gave informed written consent before participating in the study, which was approved by the Institutional Review Board of the University of Texas Medical Branch (which is in compliance with the Declaration of Helsinki).
Experimental Design
All subjects were admitted to the General Clinical Research Center of the University of Texas Medical Branch the day prior to the exercise study, and a dual-energy X-ray absorptiometry (DEXA) scan (Hologic QDR 4500W, Bedford, MA) was performed to measure body composition and lean mass. The subjects were then fed a standard dinner and a snack at 2200. All subjects were studied following an overnight fast under basal conditions. They refrained from exercise for 24 h prior to study participation, and were studied at the exact same time (i.e., between 0500 and 1500).
A baseline muscle biopsy was obtained from the lateral portion of the vastus lateralis using a 5-mm Bergström biopsy needle under sterile procedure and local anesthesia (1% lidocaine). Subjects were escorted to the Cybex leg extension machine and then performed eight sets of ten repetitions of two-legged extension exercises set to 70% of their 1RM. The rest period between sets was 3 min. Following the last exercise set, the subjects were then transported back to their hospital bed for the remainder of the study. One hour following the completion of exercise, another muscle biopsy was taken. This was immediately followed with the ingestion of 20 g of a leucine-enriched essential amino acid mixture. The composition of the essential amino acid mixture was published previously (23). Subsequent muscle biopsies were sampled at 3 and 6 h postexercise. The baseline and 1 h muscle biopsy and the 3 and 6 h muscle biopsy were sampled from incision sites 1 and 2, respectively, with the incision sites ∼7 cm from each other. The needle for each muscle biopsy was angled in a different direction to prevent needle biopsy effects from repeated sampling. Muscle tissue was immediately blotted and frozen in liquid nitrogen and stored at −80°C until analysis. To answer our hypothesis, only the baseline and 3 and 6 h post RE+EAA muscle biopsies were used in this report to assess changes in our variables of interest.
RNA Extraction
Total RNA was isolated by homogenizing 30–40 mg tissue with a homogenizing dispenser (T10 Basic Ultra Turrax, IKA, Wilmington, NC) in a solution containing 1.0 ml of Tri reagent and 4 μl of polyacryl carrier (Molecular Research Center, Cincinnati, OH). The RNA was separated into an aqueous phase using 0.2 ml of chloroform and precipitated from the aqueous phase using 0.50 ml of isopropanol. Extracted RNA was washed with 1 ml of 75% ethanol, dried, and then suspended in a known amount (1.5 μl/mg tissue) of nuclease-free water. RNA was quantified spectrophotometrically (BioRad, SmartSpec, Hercules, CA) at a wavelength of 260 nm. RNA concentration was calculated on the basis of total RNA yield. RNA quality was assessed by RNA agarose gel electrophoresis followed by visulation of the 18 and 28S ribosomal RNA bands under ultraviolet light. RNA was DNase-treated using a commercially available kit (DNA-free, Ambion, Austin, TX).
cDNA Synthesis
One microgram of total RNA was reverse transcribed into cDNA according to the manufacturers' directions (iScript, BioRad). Briefly, a 20 μl reaction mixture consisting of 1 μg of total RNA, 4 μl of 5× iScript Reaction Mix, 1 μl of iScript Reverse Transcriptase, and a known amount of nuclease-free water and placed into the thermocycler (IQ5 Real-Time PCR cycler, BioRad) with the following temperature/time protocol: 25°C for 5 min, 42°C for 30 min, and 85°C for 5 min. All isolated RNA and cDNA samples were stored at −80°C until further analysis.
PCR Primers
Primer pairs were customized using Beacon Designer 5.0 software (Premier Biosoft, Palo Alto, CA) in which they were designed to avoid homology (BLAST analysis) and secondary structures. Primers were purchased from Invitrogen (Carlsbad, CA). Primer sequences are found in Table 1. Primers were considered optimal if they produced: 1) primer efficiencies between ∼90 and 100% and 2) a single DNA product of predicted size as identified with a melt analysis and DNA agarose gel.
Table 1.
Primer sequences (5′ to 3′) used for qRT-PCR
| Gene | General Description | Forward | Reverse | Accession No. |
|---|---|---|---|---|
| mRNAs Associated with mTOR | ||||
| REDD1 | negative regulator of mTOR | CTGGAGAGCTCGGACTGC | TCCAGGTAAGCCGTGTCTTC | NM_019058 |
| REDD2 | negative regulator of mTOR | CCCAGAGAGCCTGCTAAGTG | TTGCTTTGATTTGGACAGACA | NM_145244 |
| TSC1 | negative regulator of mTOR | TAGGCTGGAGGGACTGTGAG | CCTGGTGTCTTTCATGGTCA | NM_000368 |
| TSC2 | negative regulator of mTOR | CCGCAGCATCAGTGTGTC | CACTGGTGAGGGACGTCTG | NM_000548 |
| Rheb | positive regulator of mTOR | TTTTTGGAATCTTCTGCTAAAGAAA | AAGACTTGCCTTGTGAAGCTG | NM_005614 |
| TCTP | positive regulator of mTOR | GTCGTCGTCTCCCTTCAG | AGCGAGTCATCAATGTTACC | NM_003295 |
| *cMyc | downstream target of mTOR; positive regulator of ribosomal biogenesis | CGTCTCCACACATCAGCACAA | TGTTGGCAGCAGGATAGTCCTT | NM_002467 |
| PRAS40 | negative regulator of mTOR | GAACGGATAAGAGAAGAGG | CGGTGACTCAGAAATTCG | NM_032375 |
| hVps34 | nutrient regulator of mTOR | GAAGCAGATGGATCAGAACC | CCAGCCAATCTACTTTCACC | NM_002647 |
| MAP4K3 | nutrient regulator of mTOR | AGCCCAGCAAAGCCATCC | GCCATCTCGTTCACCATTTAAC | NM_003618 |
| *mTOR | positive regulator of protein synthesis | TCGTGCCTGTCTGATTCTC | GATTCATGCCCTTCTCTTTGG | NM_004958 |
| *S6K1 | downstream target of mTOR; positive regulator of protein synthesis | ACTCAATTTGCCTCCCTACC | AGAATGGATGAGCTTGAACTTC | NM_003161 |
| mRNAs Associated with Satellite Cell Function | ||||
| mib2 | E3-ligase for notch signaling | CTACGAGACCGCTCACTC | CCTTCGCTCCCTGGAAAATG | NM_080875 |
| *myostatin | negative regulator of satellite cell proliferation | CCGTCGAGACTCCTACAAC | ACACTGTCTTCACATCAATGC | AF019627 |
| *IGF-1R | promotes satellite cell activation and proliferation | ATGCGGTGTCCAATAACTAC | TTGTTGATGGTGGTCTTCTC | NM_000875 |
| *myoD | promotes satellite cell proliferation | GGTCCCTCGCGCCCAAAAGATT | CAGTTCTCCCGCCTCTCCTACCTCAA | NM_002478 |
| *myogenin | promotes satellite cell differentiation | TGGAGCTGTATGAGACATCC | CTCGTAGCCTGGTGGTTC | NM_002479 |
| Housekeeping Genes | ||||
| *GAPDH | housekeeping gene | AGGTGAAGGTCGGAGTCAAC | GCTCCTGGAAGATGGTGATG | J02642 |
| β2M | housekeeping gene | CCAAAGATTCAGGTTTACTCAC | TCAACTTCAATGTCGGATGG | NM_004048 |
Semi-Quantitative Real-Time PCR
Determination of relative mRNA expression was performed by real-time RT-PCR using the iQ5 Multicolor Real-Time PCR cycler (BioRad) or an ABI 7500 system (Applied Biosystems, Foster City, CA) dependent on the gene of interest. cDNA was analyzed using SYBR Green fluorescence (iQ SYBR Green Supermix, BioRad) when used in conjunction with the iQ5 cycler or with Power SYBR Green Master Mix (Applied Biosystems) when used in conjunction with the ABI PCR cycler system. Each reaction contained Sybr green, a mixture of forward and reverse primers, cDNA template, and a known amount of sterile water. The total volume of the reaction tube was 25 μl. All samples were run in duplicate. An initial cycle for 5 min at 95°C was used to denature the cDNA. This was followed with 40 PCR cycles consisting of denaturation at 95°C for 20 s and primer annealing and extension at 55°C for 30 s. For REDD1, REDD2, TSC1, TSC2, and Rheb, cycle conditions were the following: stage 1, 95°C for 10 min; stage 2, 40 cycles of 95°C for 15 s followed 60°C for 1 min. After all PCR runs, a melt analysis followed.
Housekeeping Gene Normalization and Calculations
The geometric means of GAPDH and β2M were used to normalize the genes of interest as recommended by Vandesompele et al. (53). As a result, we determined the slope of the housekeeping genes for young and old to be −0.027 and −0.031, respectively, with no differences occurring across time or between young and old. Relative fold changes in mRNA within and baseline differences between groups were determined from the Ct values using the 2−ΔΔCt as described by Livak and Schmittgen (38) where ΔΔCt = (Cttarget gene − Cthousekeeping gene)time x − (Ctmean target gene − Ctmean housekeeping gene)baseline. Time x would represent any point of interest. In the case of baseline values, the use of this method will produce a fold change of ∼1 because 20 = 1.
Immunoblot Analysis
Details to immunoblotting can be found elsewhere (22). Briefly, frozen tissue was homogenized, centrifuged for 10 min at 4°C, and supernatant was collected. Total protein concentrations were determined using the Bradford assay (Smartspec Plus, BioRad). The supernatant was diluted (1:1) in a 2× sample buffer mixture containing 125 mM Tris, pH 6.8; 25% glycerol; 2.5% SDS; 2.5% β-mercaptoethanol; and 0.002% bromophenol blue boiled for 3 min at 100°C. Equal amounts of total protein (50 μg) were loaded into each lane and the samples were separated by electrophoresis (150 V for 60 min) on a 7.5% or 15% polyacrylamide gel as determined by the size of the target protein (Criterion, BioRad). Each sample was loaded in duplicate and each gel contained an internal loading control (rodent skeletal muscle) and molecular weight ladder (Precision Plus, BioRad). Following electrophoresis, protein was transferred to a polyvinylidene difluoride membrane (BioRad) at 50 V for 60 min. Blots were incubated in primary antibody overnight at 4°C (antibody dilutions are described below). The next morning, blots were incubated in secondary antibody for 1 h at room temperature. Chemiluminescent solution (ECL plus, Amersham BioSciences, Piscataway, NJ) was applied to each blot. After a 5-min incubation, optical density measurements were obtained with a phosphoimager (ChemiDoc, BioRad) and densitometric analysis was performed using Quantity One 4.5.2 software (BioRad). Protein density values were normalized to an internal loading control (which was loaded on each respective blot) and the final protein data were expressed as arbitrary units (AU).
Antibodies
The antibodies used (1:1,000) were the following: REDD1 (Proteintech Group, Chicago, IL) TSC2 (Cell Signaling, Danvars, MA), hVps34 (Invitrogen), mTOR (Cell Signaling), S6K1 (Cell Signaling), cMyc (Santa Cruz Biotechnology, Santa Cruz, CA), myogenin (Santa Cruz Biotechnology), and myoD (Santa Cruz Biotechnology). Anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody was purchased from Amersham Bioscience (1:2,000).
Statistical Analysis
Data are reported as means ± SE. Between and within group differences were tested using repeated-measures ANOVA. When main effects existed, Bonferonni post hoc tests were conducted to assess interaction effects between particular time points and age groups. For baseline comparisons unpaired t-tests were conducted. Statistical significance was determined to be P < 0.05 unless otherwise indicated. All analyses were done with SigmaStat software (version 3.5).
RESULTS
Basal mRNA Expression
A total of 17 genes were examined in human skeletal muscle for each subject in both the young and old groups (Table 1). At baseline, hVps34 mRNA expression was significantly higher in the older men compared with the young (P < 0.05), while there was a tendency for myoD (P = 0.06), myogenin (P = 0.07), and TSC2 (P = 0.09) mRNAs to be higher in the older men (Table 2). There were no differences in mRNA expression between groups at baseline for the other 13 genes (P > 0.05; Table 2).
Table 2.
Basal mRNA expression between young and old subjects
| Young | Old | P Value | |
|---|---|---|---|
| REDD1 | 1.13±0.30 | 2.18±0.72 | 0.21 |
| REDD2 | 1.07±0.14 | 0.88±0.18 | 0.43 |
| TSC1 | 1.02±0.09 | 1.04±0.09 | 0.88 |
| TSC2 | 1.02±0.09 | 1.26±0.08 | 0.09† |
| Rheb | 1.01±0.05 | 0.93±0.03 | 0.25 |
| TCTP | 1.09±0.20 | 1.34±0.26 | 0.45 |
| cMyc | 1.13±0.23 | 2.12±0.53 | 0.12 |
| PRAS40 | 1.10±0.19 | 1.38±0.34 | 0.50 |
| hVps34 | 1.05±0.13 | 2.45±0.50 | 0.02* |
| MAP4K3 | 1.08±0.19 | 1.49±0.19 | 0.15 |
| mTOR | 1.05±0.14 | 1.18±0.23 | 0.64 |
| S6K1 | 1.02±0.09 | 1.31±0.21 | 0.24 |
| mib2 | 1.09±0.21 | 1.37±0.37 | 0.52 |
| myostatin | 1.17±0.27 | 1.08±0.34 | 0.84 |
| IGF-1R | 1.08±0.18 | 1.41±0.23 | 0.29 |
| myoD | 1.06±0.16 | 1.59±0.19 | 0.06† |
| myogenin | 1.09±0.21 | 2.04±0.42 | 0.07† |
Data expressed as fold change from young (mean ± SE).
Significantly different from young P < 0.05.
Significantly different from young P ≤ 0.09.
Values determined by the 2−ΔΔCt method.
Baseline expression of select proteins associated with muscle growth.
As a follow-up, we measured select proteins (based on the genes that tended to be elevated in the older men) at baseline to determine if differences exist between the age groups. Overall, all of the select proteins measured were not significantly different (P > 0.05) between young and old at baseline: REDD1 (young: 1.35 ± 0.11; old: 1.50 ± 0.25 AU), TSC2 (young: 5.52 ± 1.39; old: 5.59 ± 4.40 AU), cMyc (young: 0.62 ± 0.12; old: 0.65 ± 0.11 AU), mTOR (young: 0.66 ± 0.06; old: 0.66 ± 0.11 AU), S6K1 (young: 2.22 ± 0.28; old: 1.91 ± 0.40 AU), myoD (young: 0.82 ± 0.04; old: 0.71 ± 0.13 AU), and myogenin (young: 5.21 ± 0.98; old: 5.27 ± 0.36 AU). However, in agreement with our gene expression data, there was a tendency (P = 0.07) for hVps34 to be higher in the old compared with the young (0.55 ± 0.07 and 0.71 ± 0.09 AU, respectively).
mRNA Expression in Response to an Acute Anabolic Stimulus
mRNAs associated with negative mTOR signaling.
There was a main effect across time for REDD1 mRNA to decrease post RE+EAA compared with baseline (P < 0.05). Further evaluation of interactions identified REDD1 mRNA expression to be significantly lower at 6 h post RE+EAA in the old compared with baseline (P < 0.05; Fig. 1A). REDD2 mRNA expression was significantly lower (∼50%) at 3 and 6 h post RE+EAA in both young and older subjects compared with baseline (Fig. 1B; P < 0.05). TSC1 mRNA expression tended (P = 0.05) to be lower at 3 h and was significantly lower (∼25%; P < 0.05) at 6 h post RE+EAA in the older subjects compared with baseline (Fig. 1C) while TSC2 mRNA expression was significantly lower (∼25%; P < 0.05) at 3 and 6 h post RE+EAA in these same subjects compared with baseline (Fig. 1D). The expression level of TSC1 and TSC2 was different between the age groups at 6 h post RE+EAA (P < 0.05). PRAS40 mRNA expression was unchanged over time and between the age groups post RE+EAA (data not shown).
Fig. 1.
mRNAs associated with negative mTOR regulation. Data represent REDD1 (A), REDD2 (B), TSC1 (C), and TSC2 (D) mRNA expression between young and older subjects 3 and 6 h post resistance exercise + essential amino acid ingestion (RE+EAA). *Significantly different from basal (P < 0.05). #Significantly different from older subjects (P < 0.05).
mRNAs associated with positive mTOR signaling.
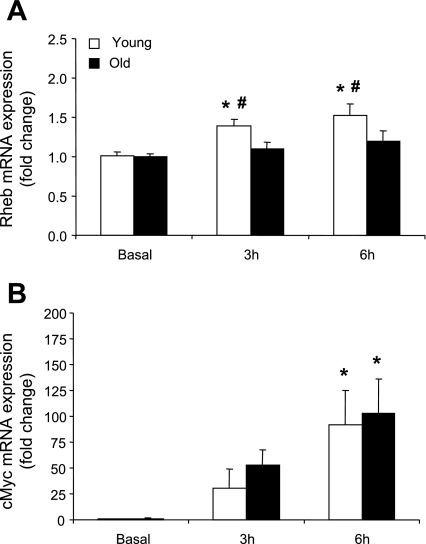
Rheb mRNA expression was significantly elevated (∼1.5-fold) at 3 and 6 h post RE+EAA only in the young subjects compared with baseline (P < 0.05; Fig. 2A) while cMyc mRNA expression was significantly elevated at 6 h post RE+EAA in both the young and older subjects compared with baseline (P < 0.05; Fig. 2B). TCTP, mTOR, and S6K1 mRNA expression as well as the nutrient regulators (hVPS34 and MAP4K3) were unchanged over time and between the age groups post RE+EAA (data not shown).
Fig. 2.
mRNAs associated with positive mTOR regulation. Data represent Rheb (A) and cMyc (B) mRNA expression between young and older subjects 3 and 6 h post RE+EAA. *Significantly different from basal (P < 0.05). #Significantly different from older subjects (P < 0.05).
mRNAs associated with satellite cell function.
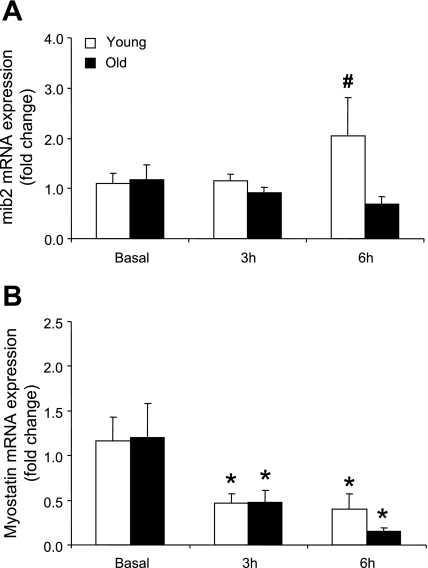
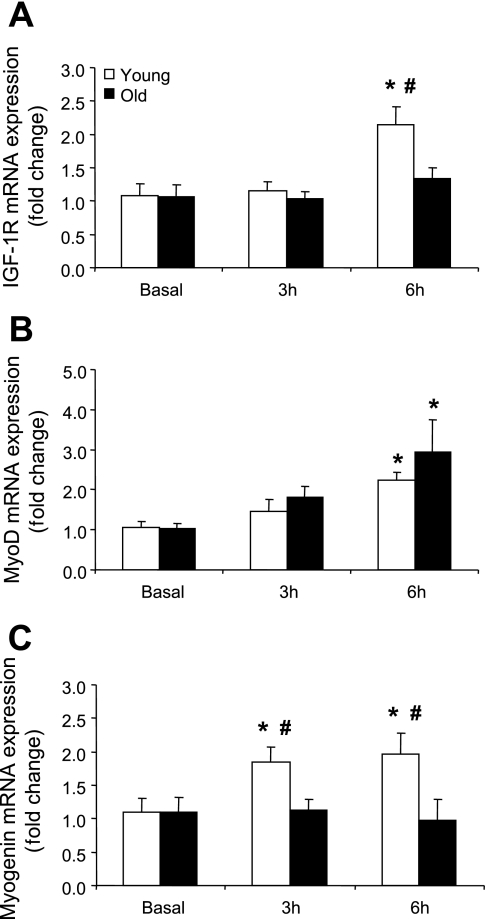
mib2 mRNA expression was significantly different between the age groups at 6 h post RE+EAA (P < 0.05; Fig. 3A) while myostatin mRNA expression was significantly lower (∼50%) compared with baseline at 3 and 6 h post RE+EAA in young and older men (P < 0.05; Fig. 3B). IGF-1R mRNA expression was significantly elevated in the young by ∼2.0-fold at 6 h post RE+EAA compared with baseline, and this was significantly greater than the older subjects at this same time point (P < 0.05; Fig. 4A). MyoD mRNA expression significantly increased (∼2.0-fold) at 6 h post RE+EAA in young and old subjects compared with baseline (P < 0.05; Fig. 4B). Myogenin mRNA expression was significantly higher (∼1.75-fold) in the young at 3 and 6 h post RE+EAA compared with baseline and this was significantly higher than the older men at these same time points (P < 0.05; Fig. 4C).
Fig. 3.
mRNAs associated with satellite cell regulation. Data represent mib2 (A) and myostatin (B) mRNA expression between young and older subjects 3 and 6 h post RE+EAA. *Significantly different from basal (P < 0.05). #Significantly different from older subjects (P < 0.05).
Fig. 4.
mRNAs associated with satellite cell regulation. Data represent IGF-1R (A), myoD (B), and myogenin (C) mRNA expression between young and older subjects 3 and 6 h post RE+EAA. *Significantly different from basal (P < 0.05). #Significantly different from older subjects (P < 0.05).
mRNA Expression Due to Repeated Muscle Sampling
A potential caveat to measuring mRNA expression with the current experimental design is a possible artificial change in mRNA expression due to repeated muscle biopsy sampling from a single incision. To test this possibility, we measured mRNA expression in the two basal muscle biopsy samples in the young men (i.e., biopsies taken from the first incision and separated by 1 h in time) for three of the genes in which we observed the largest treatment effect (i.e., cMyc, myoD, and myostatin). We found that there were no significant changes (P > 0.05) in mRNA expression between the two baseline muscle biopsy samples (i.e., the second biopsy obtained from the same incision had the same mRNA expression pattern as the original biopsy sample for all 3 genes). Although there have been mixed findings in the literature, changes in mRNA expression are dependent on the variable of interest and the biopsy timing, design, and technique (40, 54). Therefore, it appears that our findings are a result of the experimental treatment rather than a prior biopsy effect.
DISCUSSION
Noteworthy findings from this study were: 1) REDD2 mRNA expression, a negative regulator of mTOR signaling, was significantly reduced in skeletal muscle of both young and older men post RE+EAA; 2) Rheb mRNA expression, a positive regulator of mTOR signaling, increased post RE+EAA only in the young men; and 3) aging is associated with an elevated basal expression of hVps34 mRNA, a nutrient sensor linked to mTOR activation, and an inability to increase myogenin and IGF-1R mRNA expression in human skeletal muscle post RE+EAA. Together, our findings indicate that select genes associated with the regulation of protein synthesis and the myogenic program are differently expressed in muscle of young and older men at rest and post RE+EAA. In addition, these results are the first to report expression levels for many of these genes (e.g., REDD2, Rheb, TSC1, TSC2, hVps34, and mib2) in human skeletal muscle.
We found that REDD2 mRNA expression in skeletal muscle (Fig. 1B) was significantly decreased at several time points in both young and old individuals post RE+EAA. This is the first report of REDD2 mRNA expression in human skeletal muscle while REDD1 has shown to be reduced following low-intensity resistance exercise (24). REDD2 and its paralog REDD1 have been identified as negative regulators of mTOR (17), possibly through the control of the TSC1/2 complex (18). Thus a decrease in REDD2 mRNA expression is consistent with an activation of mTOR signaling following an anabolic stimulus (9, 21–23, 30). The decrease in REDD2 mRNA during the recovery from this potent anabolic stimulus is in agreement with an increase in mTOR phosphorylation and elevated rates of protein synthesis in these same subjects (23). One of the many important questions within the muscle hypertrophy field has been in understanding the prolonged activation of mTOR signaling for many hours postexercise. Previous collected data from these subjects suggest that amino acids may be providing the positive signal for mTOR activation since intracellular levels of leucine are still elevated at 6 h (see Table 3 in Ref. 23). Our results, however, provide preliminary evidence that decreased levels of REDD1/REDD2 may also contribute to the maintained increase in mTOR phosphorylation and protein synthesis post RE+EAA.
The current gene expression data support our previous work (21–23) indicating that mTOR activation during post-exercise recovery is playing an important role in regulating the acute muscle growth response. Of particular significance, TSC1 (Fig. 1C) and TSC2 (Fig. 1D) mRNA expression were decreased in the old, while Rheb (Fig. 2A) mRNA was modestly increased only in the young post RE+EAA. Although, TSC1 and TSC2 mRNAs were decreased only in the old post RE+EAA, this may be related to the higher expression levels at rest in the older subjects (Table 2). In drosophila, upregulated Rheb activity is associated with cell growth (46) and is a direct target of TSC2 GAP activity (58). It is intriguing to speculate that an enhanced Rheb mRNA expression in the young may translate into increased Rheb protein levels with repeated bouts of exercise. Therefore differential expression of Rheb may be one (of many) contributing factors to differences in muscle mass between young and old following resistance exercise training (36). Together, our data highlights that select mRNAs associated with mTOR are altered post RE+EAA in human skeletal muscle.
mTOR has also been identified as a sensor of nutrients particularly amino acids. Therefore, we evaluated the mRNA expression of the class 3 phosphatidylinositol kinase (hVps34) and the MAP kinase, MAP4K3, both of which have been associated with trafficking amino acids to mTOR (12, 25, 43). Interestingly, hVps34 mRNA at baseline was higher (2.4-fold) in the older compared with young subjects (Table 2). As a follow-up, we measured the protein expression of hVps34 and found a similar trend for the protein expression to be elevated in the older compared with the young subjects at baseline (P = 0.07). These findings are intriguing since older subjects appear to have a blunted protein synthesis response to small doses of essential amino acids (31, 32). Therefore, elevated basal levels of hVps34 may suggest a compensatory mechanism to restore nutrient sensitivity in the aging muscle cell. In contrast to our hypothesis, we did not identify any changes in mRNA expression of hVps34 and MAP4K3 within the time constraints of this experiment (data not shown). However, a recent review by MacKenzie et al. (41) indicates that mVps34 is activated in skeletal muscle 3 h after lengthening contractions. Furthermore, a recent publication indicated that influx of amino acids can cause intercellular calcium increases with subsequent activation of hVps34 and mTOR (27). Since muscle contraction releases calcium (4), resistance exercise and amino acids may synergistically regulate hVps34 activity but not alter mRNA levels. Less is known about the interaction of MAP4K3 and amino acid signaling but it will be interesting to evaluate its role following nutrient ingestion and exercise in human skeletal muscle.
Not surprisingly, the mRNAs associated with mTOR and its downstream target, S6K1, were unchanged throughout the time course of study in both younger and old subjects post RE+EAA (data not shown). This is in contrast to the rapid increase in phosphorylation status of these respective proteins (23). However, we note an elevation in cMyc mRNA in young and older subjects post RE+EAA (Fig. 2B). In support, previous data have indicated that c-Myc mRNA and protein are readily altered in young humans and animals after muscle contraction or stretch (1, 52). Not only can mTOR affect translation initiation and elongation, but it can also influence mRNA expression, particularly genes associated with ribosomal assembly (26, 42, 50). Using reporter gene activity, Teleman et al. (50) found that cMyc, a regulator of ribosome biosynthesis, was a downstream target of mTOR and is upregulated in the presence of insulin and downregulated in response to rapamycin. Furthermore, c-Myc has been identified as an important upregulator of the eIF4F translation initiation complex in cell lines (37). Thus these data suggest that RE+EAA influences protein translation initiation (through the phosphorylation of mTOR pathway proteins), but also alters the gene expression of the protein cMyc, which is associated with synthesis of new ribosomes and regulation of translation initiation complexes in young and old subjects.
Our data (Table 2 and Fig. 4) are consistent with the work of others that have shown elevations in myogenic regulatory factor mRNA in the old at rest and that resistance exercise alters the expression of human skeletal muscle mRNAs associated with satellite cell regulation in both young and older subjects (8, 28, 33, 34, 45). We also report for the first time in human skeletal muscle, an age-related differential expression pattern post RE+EAA of a novel gene known as mindbomb 2 (mib2; Fig. 3A). Specifically, mib2 cleaves the intracellular domain of the Notch ligand complex, thereby contributing to a cascade of events that eventually leads to expression of Notch regulated genes associated with satellite cell proliferation. The current data indicate that the lack of mib2 induction may be correlative (albeit weakly) with the blunted satellite cell response reported in older subjects following a bout of resistance exercise (20). Further investigation is warranted to evaluate mib2 in proliferating satellite cells post RE+EAA in human skeletal muscle.
We also found increases in IGF-1R (Fig. 4A) and myogenin (Fig. 4C) mRNA levels only in the young, while myoD (Fig. 4B) mRNA expression, although elevated, was not significantly different between the age groups post RE+EAA. This further suggests an acute dysregulation at the satellite cell level in the muscle of older subjects post RE+EAA. The role of IGF-1 receptor in contributing to muscle hypertrophy has been questioned in a recent study of synergist ablation induced hypertrophy in mice (49). But, a study by Bamman et al. (5) found an association between the magnitude of hypertrophy and upregulation of myogenin mRNA in young and older individuals following resistance exercise training. Interestingly, data from this group also show that older subjects have altered basal protein expression levels (6) and an attenuated myogenin mRNA response to resistance exercise compared with young subjects (34). Taken together, it remains to be determined whether the differential expression of myogenin and mib2 mRNA in older subjects post RE+EAA is a key factor responsible for the smaller increase in muscle hypertrophy following resistance exercise training (36, 56).
We previously showed that the young subjects from this study had a significant increase in muscle protein synthesis at 3 and 6 h post RE+EAA (23). Some of the mRNA expression data from the current study support this finding (i.e., reduced REDD2 and myostatin mRNA and increased Rheb mRNA expression at both 3 and 6 h post RE+EAA) in the young men. On the other hand, muscle protein synthesis was delayed in the older men (i.e., protein synthesis was unchanged at 3 h post RE+EAA and eventually increasing by 6 h post RE+EAA) and the link between mRNA expression and muscle protein synthesis is more complicated. For example, REDD2 and myostatin mRNA expression was reduced at 3 h post RE+EAA in the old men and yet their protein synthesis rate was unchanged. However, Rheb, IGF-1R, and myogenin mRNA expression was unchanged in the older men at all time points post RE+EAA. It remains to be determined whether an inability of particular genes to fully respond to RE+EAA is playing a role in the delayed activation of muscle protein synthesis in older men.
In summary, we report that following a treatment designed to maximally stimulate protein synthesis, REDD2 mRNA expression decreased in both the young and old human skeletal muscle. Furthermore, the expression of mRNAs associated with mTOR regulation (Rheb) and satellite cell function (mib2, IGF-1R, myogenin) were significantly altered in the young but unresponsive or delayed in the old. Finally, older subjects had an elevated expression level of hVps34 mRNA (and a tendency for higher hVps34 protein levels), which suggest altered nutrient sensitivity in these subjects. We conclude that following RE+EAA, mRNA expression of several genes associated with human skeletal muscle protein synthesis in both young and old are altered. However, select mRNAs were dysregulated in the old compared with their young counterparts. This differential expression of key genes associated with protein synthesis and the myogenic program may be associated with age-related changes in protein synthesis and the overall maintenance of skeletal muscle mass. Future studies are needed to determine whether long-term resistance exercise training in combination with essential amino acid supplementation can restore the acute anabolic response to youthful levels in older individuals.
GRANTS
This study was supported by grants from National Institute on Aging/National Institute of Arthritis and Musculoskeletal and Skin Diseases to B. B. Rasmussen (AR049877), E. Volpi (AG018311), and K. A. Esser (AR45617). Additional support came from National Institute on Aging Grant #P30 AG024832 and M01 RR00073 from the General Clinical Research Branch, National Center for Research Resources.
Acknowledgments
We thank the nurses and staff at the General Clinical Research Center for their assistance in screening, admitting, and assisting with the subjects during data collection.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Alway SE Overload-induced C-Myc oncoprotein is reduced in aged skeletal muscle. J Gerontol A Biol Sci Med Sci 52: B203–211, 1997. [DOI] [PubMed] [Google Scholar]
- 2.Anthony JC, Anthony TG, Kimball SR, Vary TC, Jefferson LS. Orally administered leucine stimulates protein synthesis in skeletal muscle of postabsorptive rats in association with increased eIF4F formation. J Nutr 130: 139–145, 2000. [DOI] [PubMed] [Google Scholar]
- 3.Anthony JC, Yoshizawa F, Anthony TG, Vary TC, Jefferson LS, Kimball SR. Leucine stimulates translation initiation in skeletal muscle of postabsorptive rats via a rapamycin-sensitive pathway. J Nutr 130: 2413–2419, 2000. [DOI] [PubMed] [Google Scholar]
- 4.Aydin J, Korhonen T, Tavi P, Allen DG, Westerblad H, Bruton JD. Activation of Ca(2+)-dependent protein kinase II during repeated contractions in single muscle fibres from mouse is dependent on the frequency of sarcoplasmic reticulum Ca(2+) release. Acta Physiol (Oxf) 191: 131–137, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Bamman MM, Petrella JK, Kim JS, Mayhew DL, Cross JM. Cluster analysis tests the importance of myogenic gene expression during myofiber hypertrophy in humans. J Appl Physiol 102: 2232–2239, 2007. [DOI] [PubMed] [Google Scholar]
- 6.Bamman MM, Ragan RC, Kim JS, Cross JM, Hill VJ, Tuggle SC, Allman RM. Myogenic protein expression before and after resistance loading in 26- and 64-yr-old men and women. J Appl Physiol 97: 1329–1337, 2004. [DOI] [PubMed] [Google Scholar]
- 7.Bamman MM, Shipp JR, Jiang J, Gower BA, Hunter GR, Goodman A, McLafferty CL Jr, Urban RJ. Mechanical load increases muscle IGF-I and androgen receptor mRNA concentrations in humans. Am J Physiol Endocrinol Metab 280: E383–E390, 2001. [DOI] [PubMed] [Google Scholar]
- 8.Bickel CS, Slade J, Mahoney E, Haddad F, Dudley GA, Adams GR. Time course of molecular responses of human skeletal muscle to acute bouts of resistance exercise. J Appl Physiol 98: 482–488, 2005. [DOI] [PubMed] [Google Scholar]
- 9.Blomstrand E, Eliasson J, Karlsson HK, Kohnke R. Branched-chain amino acids activate key enzymes in protein synthesis after physical exercise. J Nutr 136: 269S–273S, 2006. [DOI] [PubMed] [Google Scholar]
- 10.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3: 1014–1019, 2001. [DOI] [PubMed] [Google Scholar]
- 11.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG Jr. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 18: 2893–2904, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem 280: 33076–33082, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Carey KA, Farnfield MM, Tarquinio SD, Cameron-Smith D. Impaired expression of Notch signaling genes in aged human skeletal muscle. J Gerontol A Biol Sci Med Sci 62: 9–17, 2007. [DOI] [PubMed] [Google Scholar]
- 14.Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science 302: 1575–1577, 2003. [DOI] [PubMed] [Google Scholar]
- 15.Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433: 760–764, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev Cell 3: 397–409, 2002. [DOI] [PubMed] [Google Scholar]
- 17.Corradetti MN, Inoki K, Guan KL. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem 280: 9769–9772, 2005. [DOI] [PubMed] [Google Scholar]
- 18.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev 22: 239–251, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhawan J, Rando TA. Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol 15: 666–673, 2005. [DOI] [PubMed] [Google Scholar]
- 20.Dreyer HC, Blanco CE, Sattler FR, Schroeder ET, Wiswell RA. Satellite cell numbers in young and older men 24 hours after eccentric exercise. Muscle Nerve 33: 242–253, 2006. [DOI] [PubMed] [Google Scholar]
- 21.Dreyer HC, Drummond MJ, Pennings B, Fujita S, Glynn EL, Chinkes DL, Dhanani S, Volpi E, Rasmussen BB. Leucine-enriched essential amino acid and carbohydrate ingestion following resistance exercise enhances mTOR signaling and protein synthesis in human muscle. Am J Physiol Endocrinol Metab 294: E392–E400, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dreyer HC, Fujita S, Cadenas JG, Chinkes DL, Volpi E, Rasmussen BB. Resistance exercise increases AMPK activity and reduces 4E-BP1 phosphorylation and protein synthesis in human skeletal muscle. J Physiol 576: 613–624, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drummond MJ, Dreyer HC, Pennings B, Fry CS, Dhanani S, Dillon EL, Sheffield-Moore M, Volpi E, Rasmussen BB. Skeletal muscle protein anabolic response to resistance exercise and essential amino acids is delayed with aging. J Appl Physiol 104: 1452–1461, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drummond MJ, Fujita S, Takashi A, Dreyer HC, Volpi E, Rasmussen BB. Human muscle gene expression following resistance exercise and blood flow restriction. Med Sci Sports Exerc 40: 691–698, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J 403: 13–20, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guertin DA, Guntur KV, Bell GW, Thoreen CC, Sabatini DM. Functional genomics identifies TOR-regulated genes that control growth and division. Curr Biol 16: 958–970, 2006. [DOI] [PubMed] [Google Scholar]
- 27.Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab 7: 456–465, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hameed M, Orrell RW, Cobbold M, Goldspink G, Harridge SD. Expression of IGF-I splice variants in young and old human skeletal muscle after high resistance exercise. J Physiol 547: 247–254, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 445: 785–788, 2007. [DOI] [PubMed] [Google Scholar]
- 30.Karlsson HK, Nilsson PA, Nilsson J, Chibalin AV, Zierath JR, Blomstrand E. Branched-chain amino acids increase p70S6k phosphorylation in human skeletal muscle after resistance exercise. Am J Physiol Endocrinol Metab 287: E1–E7, 2004. [DOI] [PubMed] [Google Scholar]
- 31.Katsanos CS, Kobayashi H, Sheffield-Moore M, Aarsland A, Wolfe RR. Aging is associated with diminished accretion of muscle proteins after the ingestion of a small bolus of essential amino acids. Am J Clin Nutr 82: 1065–1073, 2005. [DOI] [PubMed] [Google Scholar]
- 32.Katsanos CS, Kobayashi H, Sheffield-Moore M, Aarsland A, Wolfe RR. A high proportion of leucine is required for optimal stimulation of the rate of muscle protein synthesis by essential amino acids in the elderly. Am J Physiol Endocrinol Metab 291: E381–E387, 2006. [DOI] [PubMed] [Google Scholar]
- 33.Kim JS, Cross JM, Bamman MM. Impact of resistance loading on myostatin expression and cell cycle regulation in young and older men and women. Am J Physiol Endocrinol Metab 288: E1110–E1119, 2005. [DOI] [PubMed] [Google Scholar]
- 34.Kim JS, Kosek DJ, Petrella JK, Cross JM, Bamman MM. Resting and load-induced levels of myogenic gene transcripts differ between older adults with demonstrable sarcopenia and young men and women. J Appl Physiol 99: 2149–2158, 2005. [DOI] [PubMed] [Google Scholar]
- 35.Koo BK, Yoon KJ, Yoo KW, Lim HS, Song R, So JH, Kim CH, Kong YY. Mind bomb-2 is an E3 ligase for Notch ligand. J Biol Chem 280: 22335–22342, 2005. [DOI] [PubMed] [Google Scholar]
- 36.Kosek DJ, Kim JS, Petrella JK, Cross JM, Bamman MM. Efficacy of 3 days/wk resistance training on myofiber hypertrophy and myogenic mechanisms in young vs. older adults. J Appl Physiol 101: 531–544, 2006. [DOI] [PubMed] [Google Scholar]
- 37.Lin CJ, Cencic R, Mills JR, Robert F, Pelletier J. c-Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res 68: 5326–5334, 2008. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] method. Methods 25: 402–408, 2001. [DOI] [PubMed] [Google Scholar]
- 39.Louis ES, Raue U, Yang Y, Jemiolo B, Trappe SW. Time course of proteolytic, cytokine, and myostatin gene expression after acute exercise in human skeletal muscle. J Appl Physiol 103: 1744–1751, 2007. [DOI] [PubMed] [Google Scholar]
- 40.Lundby C, Nordsborg N, Kusuhara K, Kristensen KM, Neufer PD, Pilegaard H. Gene expression in human skeletal muscle: alternative normalization method and effect of repeated biopsies. Eur J Appl Physiol 95: 351–360, 2005. [DOI] [PubMed] [Google Scholar]
- 41.Mackenzie MG, Hamilton DL, Murray JT, Baar K. mVps34 is activated by an acute bout of resistance exercise. Biochem Soc Trans 35: 1314–1316, 2007. [DOI] [PubMed] [Google Scholar]
- 42.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev 18: 423–434, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci USA 102: 14238–14243, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips SM, Tipton KD, Aarsland A, Wolf SE, Wolfe RR. Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol Endocrinol Metab 273: E99–E107, 1997. [DOI] [PubMed] [Google Scholar]
- 45.Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Myogenic gene expression at rest and after a bout of resistance exercise in young (18–30 yr) and old (80–89 yr) women. J Appl Physiol 101: 53–59, 2006. [DOI] [PubMed] [Google Scholar]
- 46.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 5: 566–571, 2003. [DOI] [PubMed] [Google Scholar]
- 47.Sheffield-Moore M, Paddon-Jones D, Sanford AP, Rosenblatt JI, Matlock AG, Cree MG, Wolfe RR. Mixed muscle and hepatic derived plasma protein metabolism is differentially regulated in older and younger men following resistance exercise. Am J Physiol Endocrinol Metab 288: E922–E929, 2005. [DOI] [PubMed] [Google Scholar]
- 48.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol 25: 5834–5845, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spangenburg EE, Le Roith D, Ward CW, Bodine SC. A functional insulin-like growth factor receptor is not necessary for load-induced skeletal muscle hypertrophy. J Physiol 586: 283–291, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7: 21–32, 2008. [DOI] [PubMed] [Google Scholar]
- 51.Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jeno P, Arrieumerlou C, Hall MN. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS ONE 2: e1217, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trenerry MK, Carey KA, Ward AC, Cameron-Smith D. STAT3 signaling is activated in human skeletal muscle following acute resistance exercise. J Appl Physiol 102: 1483–1489, 2007. [DOI] [PubMed] [Google Scholar]
- 53.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: 0034.0031–0034.0012, 2002. [DOI] [PMC free article] [PubMed]
- 54.Vissing K, Andersen JL, Schjerling P. Are exercise-induced genes induced by exercise? FASEB J 19: 94–96, 2005. [DOI] [PubMed] [Google Scholar]
- 55.Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. J Biol Chem 281: 39128–39134, 2006. [DOI] [PubMed] [Google Scholar]
- 56.Welle S, Totterman S, Thornton C. Effect of age on muscle hypertrophy induced by resistance training. J Gerontol A Biol Sci Med Sci 51: M270–275, 1996. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Creer A, Jemiolo B, Trappe S. Time course of myogenic and metabolic gene expression in response to acute exercise in human skeletal muscle. J Appl Physiol 98: 1745–1752, 2005. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 5: 578–581, 2003. [DOI] [PubMed] [Google Scholar]