Abstract
Discovery and characterization of novel secreted enzymes of Mycobacterium tuberculosis are important for understanding the pathogenesis of one of the most important human bacterial pathogens. The proteome of M. tuberculosis contains over 400 potentially secreted proteins, the majority of which are uncharacterized. A family of seven cutinase-like proteins (CULPs) was identified by bioinformatic analysis, expressed and purified from Escherichia coli, and characterized in terms of their enzymatic activities. These studies revealed a functional diversity of enzyme classes based on differential preferences for substrate chain length. One member, Culp1, exhibited strong esterase activity, 40-fold higher than that of Culp6, which had strong activity as a lipase. Another, Culp4, performed moderately as an esterase and weakly as a lipase. Culp6 lipase activity was optimal above pH 7.0, and fully maintained to pH 8.5. None of the CULP members exhibited cutinase activity. Site-directed mutagenesis of each residue of the putative catalytic triad in Culp6 confirmed that each was essential for activity toward all fatty acid chain lengths of nitrophenyl esters and lipolytic function. Culp1 and Culp2 were present only in culture supernatants of M. tuberculosis, while Culp6, which is putatively essential for mycobacterial growth, was retained in the cell wall, suggesting the proteins play distinct roles in mycobacterial biology.—West, N. P., Chow, F. M. E., Randall, E. J., Wu, J., Chen, J., Ribeiro, J. M. C., Britton, W. J. Cutinase-like proteins of Mycobacterium tuberculosis: characterization of their variable enzymatic functions and active site identification.
Keywords: secreted enzymes, activity
Mycobacterium tuberculosis continues to be one of the leading causes of death from an infectious disease, claiming 5500 lives each day (1). One-third of the world’s population is estimated to be infected with the pathogen, and resistance to multiple front-line drugs has emerged. The current vaccine against tuberculosis (TB), Mycobacterium bovis bacille Calmette Guerin (BCG), is failing to stop its spread, and new, more effective treatments are urgently required. As a result, TB has been declared a public health emergency. Secreted proteins of M. tuberculosis have attracted much attention in attempts to understand the molecular mechanisms of pathogenesis. Reasons for this include the fact that a large number of proteins are secreted from M. tuberculosis during in vitro culture and that many of these proteins are important for virulence, as they are secreted at key stages of infection, such as the macrophage phagosome (2), where live bacilli effectively arrest maturation and phagolysosomal fusion (3, 4). Several of these proteins perform important enzymatic functions, as membrane-bound or secreted proteins, and they can enhance mycobacterial survival and virulence. Among the most well known groups of secreted proteins to perform a vital role in the cell wall are the members of the Antigen 85 complex, which are essential mycolyl transferases required for the terminal transfer of mycolic acid during cell wall biosynthesis (5). Other secreted enzymes such as SapM and PtpA inhibit phagolysosomal fusion through their action on host molecules required for this process (6, 7). A further group of secreted enzymes known to perform important functions are the serine hydrolase/lipases, which may act as cell wall-associated virulence factors (8) or in triacylglycerol utilization under nutrient-limiting conditions (9), inferring a role that may occur during latent infection. Mining of the M. tuberculosis genome sequence (10) has highlighted the importance of lipid metabolism for mycobacteria with lipases being over-represented compared to other bacteria. For example, Escherichia coli possesses 50 enzymes involved in lipid metabolism, while the M. tuberculosis genome contains 5 times this number (11).
Seven cutinase-like proteins (CULPs), 6 of which have putative secretion signals, were selected for further functional studies, based on the outcome of a bioinformatic analysis. This family of putative enzymes are so-named because they share a similar predicted functional site as the well-characterized cutinase from Fusarium solani, a fungi pathogenic to plants (12), facilitating fungal invasion as a secreted virulence factor (13). Cutinases are hydrolytic enzymes with specificity for cutin, a complex glycolipid polymer consisting of hydroxy and epoxy fatty acids (14). Few examples of bacterial cutinases exist in the literature, with those identified to date being largely confined to saprophytic species and those pathogenic to plants, such as the pseudomonads (15, 16). Recently we have demonstrated different immune responses to CULP members, with three conferring protection as DNA vaccines against M. tuberculosis challenge in mice (17). In this study, the seven M. tuberculosis CULPs were expressed as recombinant E. coli proteins and purified for enzymatic characterization. There was functional diversity and variation in location within the CULP family, with none of the member proteins able to hydrolyze cutin, the natural substrate of a cutinase. The active site of Culp6, which is a putatively essential enzyme (18), was identified and the subcellular location of Culp6 was established for the first time. This novel secreted enzyme may represent a future drug target to control mycobacterial infections.
MATERIALS AND METHODS
Reagents
E. coli DH5α was used for plasmid preparations during cloning experiments and grown in Luria-Bertani (LB) broth or on LB agar. Ampicillin (100 μg/ml) or kanamycin (50 μg/ml) was supplemented as necessary. H37Rv genomic DNA and M. tuberculosis cellular fractions were obtained from Colorado State University (Fort Collins, CO, USA).
Bioinformatic procedures
Bioinformatic analyses was performed as described previously (17). Briefly, the proteome of M. tuberculosis H37Rv was batch submitted to the SignalP server (19) (http://www.cbs.dtu.dk/services/SignalP/) to identify putative signal peptides indicative of secretion using both the neural network and hidden Markov models of prediction applied to gram-positive bacteria. The proteome was further compared to the COG and CDD databases (20, 21) using the program rpsblast (22) and also to the nonredundant protein database of the National Center for Biotechnology Information (NCBI), where blastp was utilized. Initial sequence alignment was performed with ClustalW (23), whereas final alignment of catalytic residues was done manually. Mega3 was used to obtain the phylogram using the neighbor-joining algorithm, bootstrapped 10,000 times (24).
Protein expression and purification of inclusion bodies
CULPs were cloned without signal sequences into an E. coli expression vector (pET19b; Merck, San Diego, CA, USA) following amplification of each from H37Rv genomic DNA. This resulted in overexpression of N-terminally His-tagged cytoplasmic proteins. Cytoplasmic inclusion bodies consisting of the recombinant proteins accumulated following IPTG (0.5 mM) induction and were purified as described previously (17).
Protein refolding
Refolding potential of each protein was screened in a set of 15 buffers, prepared as described in the Quick Fold Protein Refolding kit manual (AthenaES, Baltimore, MD, USA). A buffer representing optimal refolding conditions composed of 50 mM Tris (pH 8.5), 9.6 mM NaCl, 0.4 mM KCl, 0.4 M sucrose, 0.05% PEG 3550, 0.5% Triton X-100, 2 mM MgCl2, 2 mM CaCl2, with 1 mM GSH and 0.1 mM GSSH was selected to fold all proteins. Proteins were rapidly diluted (20×) into refolding buffer and were allowed to fold overnight at 4°C with gentle agitation. Correct protein folding was analyzed by FPLC on a Sephadex-200 gel filtration column (GE Healthcare, Piscataway, NJ, USA). Refolded protein (100 μl) was loaded to the column, and chromatography was performed at a flow rate of 0.5 ml/min. Eluted protein peaks were detected by UV absorption, and peak retention times were compared to a standard curve giving approximate molecular masses.
Enzyme assays
The esterase and lipase potential of the CULPs were determined as described by Winkler and Stuckmann (25) with minor adaptations. The stock solutions of the substrates, p-nitrophenyl butyrate (pNPB), p-nitrophenyl laurate (pNPL), p-nitrophenyl myristate (pNPM), p-nitrophenyl palmitate (pNPP), and p-nitrophenyl stearate (pNPS) were obtained from Sigma (St. Louis, MO, USA) or MP Biomedicals (Irvine, CA, USA) and prepared in isopropanol to 10 mM. These substrate solutions were mixed 1:9 with a solution containing 50 mM sodium phosphate (pH 8.0), 2.3 mg/ml sodium deoxycholate, and 1 mg/ml gum arabic. To 10 μl of enzyme solution (100 μg/ml), 240 μl of the above reaction mixture was added in a 96-well microwell plate, mixed, and incubated at 37°C for 30 min. The accumulation of p-nitrophenol was measured spectrophotometrically at 405 nm, and concentrations were calculated by comparisons to p-nitrophenol standards. Lipolytic activity as determined by Tween cleavage was performed as previously reported (26). Briefly, a reaction mix was prepared with 33 mM CaCl2 and 0.33% Tween 20 in buffer (50 mM Tris for pH 7.0 and pH 8.0; 50 mM MES for pH 6.0). Fifty microliters of enzyme was added prior to 1 h incubation at 37°C. Two hundred fifty microliters was transferred to a 96-well microwell plate, and turbidity was assessed at 405 nm. Activity in this assay is reported as U/μg of protein, with 1 U defined as the amount of enzyme required to effect a change in OD (405 nm) of 0.01 under assay conditions.
Cutinase activity was determined as previously reported (27) with the following modification. Apple cutin, prepared from mature apples (28), served as the substrate. For a typical assay, 1 mg of enzyme and 100 mg of apple cutin were added into buffer (50 mM Tris–HCl, pH 7.5) in a final volume of 10 ml. The tube was shaken (125 rpm) for 18 h at 37°C for Clp1–7 and 60°C for Thermobifida fusca cutinase. At the end of reaction, the remaining cutin was removed by centrifugation. The resulting supernatant was acidified with acetic acid, and the released cutin monomers were extracted with chloroform. The organic soluble material was collected and dried under a stream of nitrogen. The dried cutin monomers were converted to their corresponding methyl esters and then silylated with bis-(trimethylsilyl) trifluoroacetamide (BSTFA). The silylated methyl esters were dissolved in hexane and analyzed by Finnigan GC/MS 4610B on a 122-7032 DB-WAX 30 M capillary column (Thermo Finnegan, San Jose, CA, USA) with temperature programmed as following: 125°C for 5 min, 4°C/min to 250°C, 250°C for 15 min.
Enzyme inhibition assays
Diethyl 4-nitrophenyl phosphate (paraoxon; Sigma) was solubilized in acetonitrile to 100 mg/ml. A working stock was prepared by dilution into 100% methanol to 10 mg/ml. Assay concentrations were then prepared by aqueous dilution. Equivalent methanol concentrations were tested for inhibition of enzyme activity and were found to have no effect (data not shown). Inhibitor was mixed 1:1 with 100 μg/ml enzyme solution and incubated at RT for 30 min. p-Nitrophenyl esterase assays were then performed as described previously: pNPB was used as the substrate for Culp1 and 4, while pNPL was used as the substrate for Culp6. Assays were carried out for 60 min at 37°C.
Site-directed mutagenesis
Site-directed mutagenesis of Ser-175, Asp268 and His299 of Culp6 was carried out using the Stratagene QuikChange II kit (Stratagene, La Jolla, CA, USA) according to manufacturer’s instructions. Briefly, template plasmid from Dam+ E. coli DH5α was amplified using a pair of complementary primers containing the desired mutations by a non-strand-displacing, high-fidelity polymerase in a linear amplification. A methylation-sensitive restriction endonuclease, DpnI, was then added, degrading the methylated template but leaving the nicked, unmethylated copies containing the mutation. The nonlinearized plasmids were then transformed into supercompetent E. coli XL1-Blue and screened by sequencing for the presence of the desired mutation.
PAGE, antibody production, and Western blot analysis
Protein samples were separated by two-phase 4%/12% SDS-polyacrylamide gel electrophoresis and stained with Coomassie blue. Mouse polyclonal anti-Culp1, 2 and 6 antibodies were raised for subcellular location experiments. Mice were immunized 3×, at 2-wk intervals, with 10 μg of antigen injected subcutaneously on the back. Antigen was delivered with 250 μg of dimethyl-dioctadecylammonium (DDA) and 25 μg monophosphoryl lipid (MPL) as adjuvant (29). Blood was collected 4 wk after final immunization and allowed to clot before serum was clarified by centrifugation. For Western blot, equivalent amounts of the protein samples visualized by Coomassie staining were loaded along with prestained molecular mass standards (Bio-Rad, Richmond, CA, USA) and transferred to PVDF membrane. Blots were probed for polyhistidine tag with HisDetector Nickel-HRP (KPL, Gaithersburg, MD, USA) or anti-CULP (1:250) antibodies as required. Anti-mouse HRP-conjugated secondary antibodies were used; blots were developed with SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL, USA) and visualized on a Kodak 4000MM Image Station (Eastman Kodak, Rochester, NY, USA).
RESULTS
CULP identification
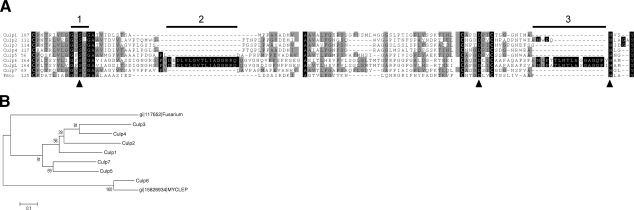
The signalP server (19) was queried to identify the presence of putative secretion signals in the M. tuberculosis proteome. Proteins identified in initial analysis with possible functional similarities were grouped, revealing six proteins (Table 1) that clustered with homologies ranging from 17 to 63% at an amino acid level. Another protein, Culp5 (Rv1758), was added to the group based on sequence homologies, although the lack of a classic signal sequence indicates that it may be cytoplasmic in its subcellular location. A previous study (30) identified two of these proteins on silver-stained 2-dimensional electrophoretic gels of M. tuberculosis culture filtrates, named Cfp21 (Rv1984c) and Cfp25 (Rv2301), which are reported here as Culp1 and Culp2, respectively. These proteins, like other members of the family, share homology to the fungal cutinase, but until now, cutinase activity had not been empirically determined. Therefore, we have referred to these and the other members of the family as CULPs. The full-length CULPs of M. tuberculosis align with varying amino acid homology with the well-characterized cutinase of the fungi, Fusarium solani, and the sole Mycobacterium leprae homologue, ML0099, referred to here as CulpL. Of note is the conservation of the active catalytic triad between species. Each CULP contains the serine, aspartic acid, and histidine residues that comprise the active site, critical for functional F. solani cutinase (Fig. 1A). A comprehensive analysis of the protein sequences reveals significant homology about the catalytic triad of each. CULPs are α/β hydrolases (31), possessing the conventional catalytic triad comprised of Ser, Asp, and His, with the serine residue located within the conserved pentapeptide, G-X-S-X-G (Fig. 1A, bar 1). Within all of the sequences shown in Fig. 1, the pentapeptide motif describing all of the sequences is G-[YF]-S-[QL]-G. The Tyr residue is the most common in position 2, phenylalanine being observed in Culp5, Culp6, and CulpL of M. leprae, while only Culp2 contains the leucine residue in position 4, all of the others containing asparagine. Uniquely, Culp5 and Culp6 have the pentapeptide sequence of a family VI-type lipase, i.e., G-F-S-Q-G (32), as does the CulpL ortholog. The spatial organization of the active site region is also conserved, except that Culp6 and CulpL of M. leprae possess two extra amino acid blocks of ∼15 aa each, one after the catalytic Ser and the other between the key Asp and His residues (Fig. 1A, bars 2 and 3). The first extra block is flanked by Gly, while the second is flanked by Pro, suggesting they may form extra loops in relation to the general enzyme framework. This may lead to an alternative conformation around the active site of Culp6, which further suggests that this enzyme has an alternative substrate to the other CULPs. The phylogram shows strong bootstrap support for orthology between Culp6 and the M. leprae CulpL, and suggests that the remaining M. tuberculosis are products of gene duplication, in particular the pairs Culp3/Culp4 and Culp7/Culp5 (Fig. 1B).
TABLE 1.
Proteomic characteristics of the family of CULPs of M. tuberculosis
| CULP | Rv number | Signal peptide | Cleavage | Length (aa) | M (kDa) | Mat M (kDa) | pI |
|---|---|---|---|---|---|---|---|
| Culp1 | Rv1984c | + | 32−33 | 217 | 21.8 | 18.6 | 5.52 |
| Culp2 | Rv2301 | + | 32−33 | 219 | 22.6 | 19.6 | 4.96 |
| Culp3 | Rv3451 | + | 26−27 | 247 | 24.9 | 22.3 | 5.79 |
| Culp4 | Rv3452 | + | 45−46 | 226 | 23.7 | 18.9 | 9.61 |
| Culp5 | Rv1758 | − | − | 174 | 17.9 | 17.9 | 4.7 |
| Culp6 | Rv3802c | + | 26−27 | 336 | 35.5 | 32.5 | 6.5 |
| Culp7 | Rv3724 | + | 30−31 | 187 | 21.2 | 17.8 | 6.93 |
M, molecular mass; Mat M, mature molecular mass; pI, isoelectric point.
Figure 1.
Alignment and phylogram of the CULPs of M. tuberculosis, M. leprae, and F. solani. A) Alignment of residues of the catalytic triad (indicated by arrowheads) and surrounding regions. Residues conserved in all proteins are indicated with black background; those shaded gray are identical in ≥50% of the proteins. Conserved cysteines are boldface in black background. Bar 1 indicates the conserved pentapeptide motif G-X-S-X-G containing the catalytic serine. Bars 2 and 3 indicate insertion of amino acids found in Culp6 and in the M. leprae ortholog, respectively. B) Bootstrapped phylogram from alignments of whole sequences. Numbers represent percentage bootstrap support. Scale bar indicates 10% amino acid sequence divergence. Symbol gi| indicates the NCBI accession number for the F. solani and M. leprae (MYCLEP) proteins.
Functional analysis
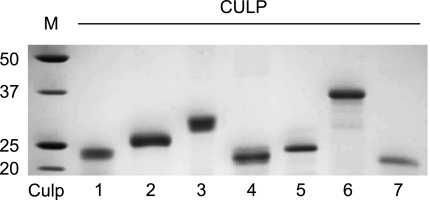
All seven CULPs were cloned and expressed as N-terminally His tagged recombinant E. coli proteins. Following overexpression, Co2+-charged immobilized metal affinity chromatography (IMAC) was utilized to purify the chemically solubilized CULPs. The CULPs were then refolded in a buffer series, which varied in their components in order to address critical and often unique physiochemical refolding conditions for the proteins. These included pH, ionic strength, oxidation state, and the presence of chaotropic and polar agents. The resultant proteins were pure, refolded, and soluble proteins by SDS-PAGE (Fig. 2), allowing further functional characterization of the individual proteins. Paradoxically, Culp5 migrates slower than Culp4, even though the predicted molecular masses are 17.9 and 18.9 kDa, respectively. This pattern of migration was consistently observed and may be attributed to the two proteins possibly binding different molar amounts of SDS.
Figure 2.
Purified, refolded, recombinant CULPs. Coomassie blue-stained CULPs shown following E. coli expression and HIS-tag IMAC purification and separation by SDS-PAGE. Molecular mass standards (M) with sizes (kDa) are at left.
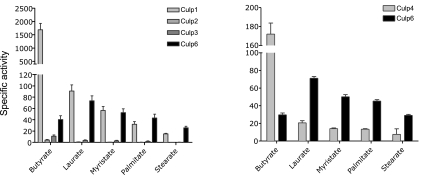
To establish whether the M. tuberculosis CULPs function as enzymes, and if so to classify them to a specific enzymatic class, a series of functional assays were performed. Functional and structural properties of F. solani cutinase have been comprehensively examined, and the enzyme is well established as a serine hydrolase (12). Comparison of the protein sequences of the two species suggested that the enzymes might share functional characteristics. Therefore, the CULPs of M. tuberculosis were assessed for their esterase, lipase, and cutinase activities. To delineate the activities of the CULP specificity for substrate fatty acid chain length, and thereby allow a more definitive description in terms of enzymatic class, the enzymes were assessed for their capacity to cleave a range of fatty acid substrates. The substrates used were esters of p-nitrophenyl and butyric (C4), lauric (C12), myristic (C14), palmitic (C16), and stearic (C18) acids. Cleavage of these esters liberates p-nitrophenol, which can be measured spectrophotometrically (25, 33, 34). The CULPs demonstrated a divergence in optimal fatty acid chain length specificity, with Culp1 proving an effective esterase owing to its ability to cleave pNPB efficiently (Fig. 3). In fact, Culp1 had an average specific activity (1688 pmol·min−1·mg−1) ∼40-fold that of Culp6 for the same substrate. Further, the activity of Culp1 decreased sharply as substrate chain length increased, with its specific activity reduced 20-fold for the substrate with which it displayed the next highest activity, i.e., pNPL. The activity of Culp1 continued to decrease as fatty acid chain length increased. In comparison, although the overall activity of Culp6 declined after a peak for medium-length fatty acids (lauric acid), its activity was greater than Culp1 when the chain length was C16 or more. These results infer that Culp6 possesses lipolytic activity in addition to esterase activity. Culp4, whose homologue in Mycobacterium smegmatis had activity as a phospholipase A in a recent study (35), was able to effectively cleave butyric acid from the nitrophenyl ester with a mean specific activity of 182 pmol · min−1 · mg−1, ranking it only behind Culp1 as the most active esterase of the family. Cleavage of pNPP and pNPS was most efficiently achieved by Culp6, again indicating lipolytic activity, while Culp1 and Culp4 were also able to cleave the longer-chain length fatty acids, albeit to a lesser degree than Culp6. Culp2 and Culp3 performed weakly as esterases for pNPB (Fig. 3), while Culp5 and Culp7 demonstrated activities at or near background levels for all substrates tested, at pH ranging from 5.0–8.0 (results not shown). Also, performing these assays in reducing or oxidizing buffers did not appreciably alter their activities (results not shown).
Figure 3.
CULPs of M. tuberculosis possess divergent enzymatic function. Specific activity of CULPs demonstrates substrate specificity with fatty acid substrates of different lengths. Specific activity represents pmol p-nitrophenol liberated · min−1 · mg−1 enzyme. Results are the means ± se of triplicate experiments performed at pH 8.0. Culp5 and Culp7 showed no activity in assays tested (not shown).
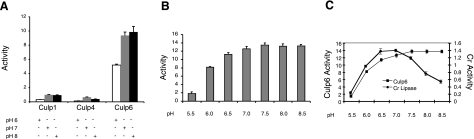
To define further the lipase activities of Culp1, Culp4, and Culp6, the cleavage of polyoxyethylene sorbitans (Tweens) (26, 36) was measured. This assay is reported to be up to 36-fold more sensitive than the titrimetric assay and at least 4-fold more sensitive than the colorimetric assay of pNPP (37). We also extended this assay by varying the buffer system in order to examine enzyme activity in either basic or acidic conditions. The activities for these three CULPs were, therefore, determined at pH 6, 7, and 8 (Fig. 4). When the CULPs were tested for the cleavage of polyoxyethylene sorbitan monolaurate (Tween 20), Culp6 demonstrated lipolytic activity up to 10-fold higher than either Culp1 or Culp4, and this activity was optimum at pH 8.0 (Fig. 4A). Culp1 was able to hydrolyze substrate to a maximum of 1 U/μg at a pH of 7.0 and 8.0, while Culp4 demonstrated a similarly low level of activity, but only at pH 7.0. No other CULP demonstrated lipolytic activity as measured by this assay at any pH, including Culp5, despite sharing a family VI-type lipase serine pentapeptide with that of Culp6 (results not shown).
Figure 4.
Lipolytic activity demonstrated by cleavage of polyoxyethylene sorbitan monolaurate (Tween 20). A) CULPs with activity for pNPP were tested at varying pH. B) Culp6 activity for Tween 20 performed from pH 5.5–8.5. Bars represent means ± se of triplicate experiments. C) Comparison of Culp6 activity (U/μg protein) to that of a purified lipase of Candida rugosa (Cr). Data points are means ± se of triplicate experiments.
To establish a pH profile of Culp6 activity, the cleavage of Tween 20 was examined over a broad pH range. Activity was highest at neutral to alkaline conditions, with performance dropping markedly below pH 6 (Fig. 4B). Interestingly, Culp6 lipase activity was maintained above pH 7.0, whereas a genuine lipase of Candida rugosa, in addition to having ∼10-fold lower activity, was far less active above pH 7.0 (Fig. 4C).
To establish whether the proteins were folded correctly or merely soluble aggregates, gel filtration was conducted by FPLC. Analysis of the elution chromatograms indicated that all of the proteins were soluble and eluted at expected molecular masses consistent with them being correctly folded, with the exception of Culp2, which demonstrated a majority of incorrectly folded protein (results not shown). It is possible that Culp2 aggregated under the conditions of gel filtration, yet we cannot rule out similar aggregation during enzyme analysis, resulting in apparent inactivity.
Cutinase activity
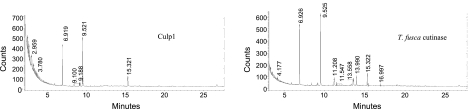
Because of the presence of a cutinase motif comprising the putative active site of these proteins, a number of them have been named as cutinases in the annotated M. tuberculosis genome. As an absence of experimental evidence for this activity exists, we have defined them as CULPs. We sought to establish whether they do indeed possess cutinase activity, and thus we performed assays of cutin hydrolysis (Fig. 5). None of the 7 CULPs exhibited cutinase activity, as C16 and C18 hydroxyl fatty acids, the characteristic cutin components, were absent in the enzymatic reaction products (Table 2). These products were clearly present following hydrolysis of cutin by T. fusca cutinase, which served as the positive control (Fig. 5). The present nomenclature is, therefore, more appropriate.
Figure 5.
Enzymatic reaction products from cutinase activity analysis. Example mass chromatograms for Culp1 and positive control, T. fusca cutinase, illustrating peak retention times (min).
TABLE 2.
Enzymatic reaction products detected in assays for cutinase activity of M. tuberculosis CULPs
| Cutin hydrolysis product | Reaction time (min) | Hydrolysis area (%)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| T. fusca cutinase | Culp1 | Culp2 | Culp3 | Culp4 | Culp5 | Culp6 | Culp7 | ||
| Hexadecanoic acid | 6.926 | 30.94 | 24.25 | 37.92 | 25.11 | 40.26 | 32.44 | 27.09 | 23.18 |
| 9-Octadecenoic acid | 9.1 | 0.912 | 1.55 | 5.1 | 0.512 | 0.571 | 1.757 | 0.349 | 0.492 |
| 9,12-Octadecadienoic acid | 9.188 | 1.696 | 1.825 | 0.345 | 0.316 | 2.166 | 2.143 | 0.913 | 0.554 |
| Octadecanoic acid | 9.525 | 36 | 32.16 | 39.06 | 31.08 | 44.36 | 41.73 | 32.59 | 27.68 |
| 16-Hydroxyhexadecanoic acid | 11.547 | 0.843 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| 18-Hydroxyoctadeca-9-enoic acid | 13.017 | 1.97 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| 18-Hydroxyoctadeca-9,12-dienoic acid | 13.558 | 7.18 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| 10,16-Dihydroxyhexadecanoic acid | 13.99 | 5.8 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
N.D, not determined.
Inhibition of enzyme activity
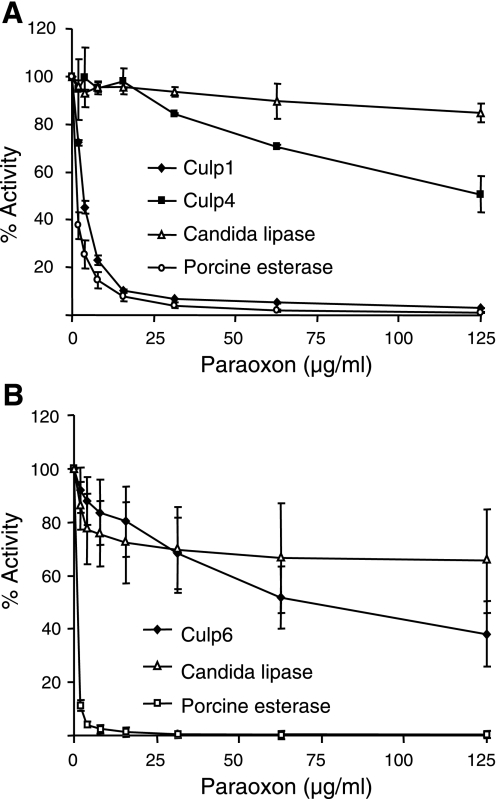
Esterases can be classified by the interaction with organophosphates, such as paraoxon. In this context, if an esterase can hydrolyze organophosphate esters, it is referred to as class A. If it is inhibited by organophosphate esters, it falls into class B and is classified as a carboxylesterase (EC 3.1.1). Paraoxon acts as an inhibitor by phosphorylating the hydrozyl group of the catalytic serine residue (38). F. solani cutinase is rapidly inhibited by low concentrations of paraoxon (39, 40), as are other esterases, such as porcine liver esterase (41). Lipases, such as that of C. rugosa used as a control in this study, are also inhibited by paraoxon, but this inhibition is reduced because of the protection of the catalytic serine by the lid domain of the enzyme (42). We therefore tested the sensitivity of the M. tuberculosis CULPs to inhibition by paraoxon in order to establish their classification as A or B esterases and the level of exposure of their catalytic serine to the solvent. Inhibition of CULP activity by paraoxon was varied. Culp1 was almost completely inhibited (>99%) in its ability to cleave pNPB at high inhibitor concentrations, with an IC50 ∼ 1.9 μg/ml (Fig. 6A). Culp4 was less susceptible to inhibition, with ∼65 times the inhibitor concentration needed to quench activity to the same degree (IC50∼125 μg/ml); a similar pattern of resistance to inhibition was seen for Culp6, where an IC50 ∼ 65 μg/ml was observed for activity to pNPL (Fig. 6B). The rapid and complete inhibition of Culp1 by paraoxon firmly classifies this enzyme as a carboxylesterase with an active site readily accessible to the inhibitor. Culp4 and Culp6 can also be classified as carboxylesterases; however, the high concentrations of inhibitor required may indicate a shielding of the enzyme active site, as seen with the C. rugosa lipase.
Figure 6.
Dose-related paraoxon inhibition of CULPs. Graphs demonstrate dose-dependent inhibition of Culp1 and Culp4 (A) and Culp6 (B) to hydrolyze pNPB and pNPL, respectively. Candida lipase and porcine esterase are included as inhibition-resistant and inhibition-sensitive controls, respectively.
Active site identification of Culp6
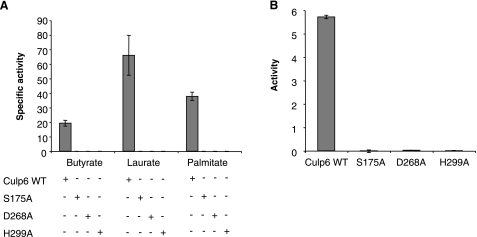
The active site of Culp6 was defined by site-directed mutagenesis. Three mutant proteins were created, each with a single residue of the putative catalytic triad (i.e., Ser-175, Asp268, and His299) being replaced with alanine. For each of the altered proteins, all activity was lost for both short- and long-chain substrates (Fig. 7A). Further, lipase activity was eliminated, as measured by the cleavage of Tween 20 (Fig. 7B).
Figure 7.
Site-directed mutagenesis of active site results in loss of function. A) Specific activity of mutated proteins for different chain length substrates. B) Lipase activity as measured by cleavage of Tween 20. Results are means ± se of triplicate experiments performed at pH 8.0.
Subcellular location of CULPs
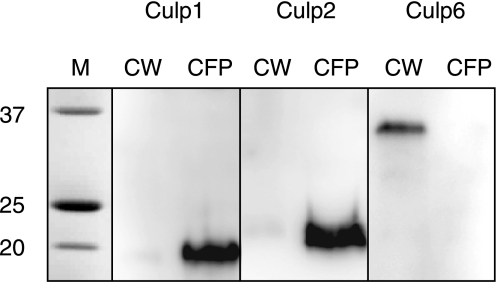
Subcellular location and secretion potential are known for few CULPs. Culp1 and Culp2 have been identified in M. tuberculosis culture filtrates (30), and Culp4 has been reported to be located in the cell wall of M. tuberculosis but not to be secreted into the culture supernatant (35). To establish the secretion status of Culp6 and to further characterize that for Culp1 and Culp2, high-titer antibodies were raised against the respective recombinant proteins and used to probe subcellular fractions of M. tuberculosis via immunoblotting. We confirmed the presence of Culp1 and Culp2 in culture filtrate and failed to detect appreciable levels either in the cell wall (Fig. 8) or the cytoplasm (not shown), illustrating that the majority of protein is destined for immediate secretion. By contrast Culp6 was present in the cell wall fraction alone (Fig. 8), with no detectable levels in the culture filtrate protein, despite possessing a typical signal sequence, as predicted by both the neural network and hidden Markov models. This finding strongly suggests a role for this enzyme in cell wall biosynthesis and/or maintenance.
Figure 8.
Western blot analysis of M. tuberculosis CULPs in culture filtrate protein (CFP) and cell wall (CW) fractions. CULP-specific antibodies revealed that Culp1 and Culp2 were found only in CFPs, whereas Culp6 was entirely restricted to the CW fraction. Anti-CULP antibodies were raised against purified proteins.
DISCUSSION
This study aimed to characterize the CULPs of M. tuberculosis, a little-known family of putatively secreted proteins, some of which are emerging to perform important functions. Three members have strong enzyme activity as either esterases or lipases and are all carboxylesterases, as defined by enzyme inhibition. The study defined the active site of the enzyme family by site-directed mutagenesis of Culp6, which has been reported as essential for replication of M. tuberculosis (18), as well as its operational pH optimum and cellular location. This work also demonstrates that these proteins do not possess cutinase activity, as would be inferred by the annotation as “cutinases” in the genome.
Secreted gene products of M. tuberculosis are important for virulence and survival of the bacillus and pathogenesis of tuberculosis. For example, the mycolyl transferases of the Antigen 85 complex are critical for cell wall development (5) and are major targets of the immune responses in animal and human mycobacterial infections (43, 44). The first secreted CULPs of M. tuberculosis to be identified were Culp1 and Culp2, which were present in short-term culture filtrate (30). These proteins were found to be potent inducers of IFN-γ from memory T lymphocytes from M. tuberculosis-infected mice, and they elicited strong delayed type hypersensitivity responses in M. tuberculosis-sensitized guinea pigs. Subsequent studies revealed that one or both of these antigens were also recognized by T cells from tuberculosis patients (45, 46). Recently, we have demonstrated that Culp1 and Culp2 as individual antigens can confer protective immunity in mice against M. tuberculosis challenge (17). In addition to the potential use of Culp1 as a protective vaccine antigen, this study defines it as an esterase with high specific activity, rapidly hydrolyzing pNPB. As an enzyme secreted into the extracellular milieu and largely absent from the bacterial cytoplasm or cell wall, we suggest that Culp1 may interact with host molecules and contribute to the virulence and intracellular survival of M. tuberculosis, as demonstrated for other secreted enzymes. Culp1 is unlikely to be essential for the viability of M. tuberculosis, as it is absent from the majority of M. bovis (BCG) vaccine strains, which lack the region of deletion 2 (RD2) containing the culp1 homologue of M. bovis (47).
The secreted phosphatases, SapM and PtpA, and the serine/threonine protein kinase G, PknG (48), influence the survival of M. tuberculosis within macrophages by inhibiting phagosome maturation and acidification and host vacuolar protein sorting. This inhibition of the normal macrophage responses to phagocytosed bacteria has important consequences to the outcome of infection. Interestingly, although Culp2 displayed minimal enzyme activity in the assays performed here, its homologue was found in a genetic screen of M. bovis BCG to inhibit phagosome acidification (49), widening the functional responsibilities of this family. To date, Culp2 is the only member of the CULP family that has been attributed an intracellular function within host macrophages, but it is possible that other members of the group may also contribute to intracellular survival.
Although all but one of the CULPs have putative secretion signals, only two, Culp1 and Culp2, have been proven to be secreted, while two, Culp4 and Culp6, are cell wall associated. The homologue of Culp4 in M. smegmatis, which has activity as a mycobacterial phospholipase A, was secreted by M. smegmatis. Similar phospholipase activity was present only in the cell wall fraction of M. tuberculosis, suggesting that the M. tuberculosis Culp4 is retained in the cell wall (35). The gene encoding Culp6, Rv3802c, was one of ∼800 genes identified by a genomic screen to be essential for growth of M. tuberculosis in vitro (18). We have demonstrated that Culp6 is not exported fully from the cell but is also retained in the cell wall. This variation in cellular location suggests divergent roles for these homologous enzymes, and localization within the cell wall may indicate a role for Culp6 in cell wall biosynthesis and/or remodeling. Furthermore, the chromosomal location of culp6 is directly downstream of the mycolyl transferases, fbpA and fbpC1 (antigens 85A and 85C, respectively), and immediately upstream of fadD32, a probable fatty acid Co-A synthase, and of other key enzymes in lipid metabolism. This genomic orientation may suggest a direct role in cell wall modification or lipid metabolism for this lipase. Another possible role for CULP members with lipase activity is as scavenger enzymes using host lipids to provide a carbon source for the mycobacterium, a process that is important for mycobacterial replication within host cells (50, 51).
Analysis of the enzymatic function of the protein family revealed an unexpected diversity between them. The strong esterase activity of Culp1 distinguishes it as having a preference for short-chain fatty acids. Culp6 has a peak activity for medium-chain length fatty acids, such as myristic acid, but possesses stronger activity than Culp1 for substrates with chain lengths longer than C14, categorizing Culp6 as a lipase. It is possible that the extra amino acids found between the components of the catalytic triad in Culp6 (Fig. 1A) confer this specificity toward larger substrates. Although there is overlap in activities of these two proteins, this does not necessarily indicate enzymatic redundancy. Culp6 has been identified as a potentially essential gene from genomic mutational studies (18). In addition, Culp6 is conserved within all mycobacterial genomes examined, and its M. leprae ortholog, CulpL, is the only intact cutinase present in the drastically restricted genome of M. leprae (Fig. 1). This further supports the notion that culp6 is essential for mycobacterial survival. Although the active site residues identified in Culp6 are conserved in other CULP members, Culp2, Culp3, Culp5, and Culp7 had little or no activity with any of the tested substrates, indicating possible divergence of substrate specificity. FLPC analysis indicated that all proteins but Culp2 were predominantly folded correctly. With Culp2 aggregating during gel filtration, we cannot rule out that the same has occurred under activity assay conditions and is one reason for perceived inactivity.
The catalytic triad of two of the CULPs, Culp2 and Culp6, differ from the other members in having additional amino acids separating the second and third residues of the triad, the majority of which were not shared between them. Culp2, which is essential for the inhibition of phagosome maturation, has an extra spacing of 6 aa compared to the others, while the extension is 19 aa in length for Culp6. Although this does not in itself point to a particular substrate, it is consistent with the notion that they have unique substrates.
All of the CULPs that displayed activity in assays for p-nitrophenyl ester hydrolysis were inhibited with paraoxon, suggesting they are class B esterases. The divergent sensitivity to the inhibitor between Culp1, Culp4, and Culp6 may be informative of the degree of exposure of the catalytic serine to the solvent. Our data show that Culp1 has similar inhibitory sensitivity to both porcine liver esterase and published data for F. solani cutinase (40), strongly suggesting that its catalytic site is readily accessible. Our data also suggest that the active sites of Culp4 and Culp6 are not as readily accessible to a small-molecule inhibitor. This possible contrast in structure may be supported by the additional residues in the region of the catalytic site in both Culp4 and Culp6 when compared to Culp1 (Fig. 1).
This pattern of sequence homology, yet with functional diversity, has been attributed to gene duplication followed by eventual functional drift in other homologous/divergent classes of enzymes. One explanation states that when two paralogs of a bifunctional gene arise from duplication, each copy may be driven by selective pressures to specialize in one of its original functions, resulting in the creation of two essential genes (52). A more conventional view of gene duplication is that the process produces two redundant proteins, thereby relaxing selective pressures on each and so allowing the accumulation of mutations, which can eventuate in functional divergence (53). Either of these theories is consistent with the characteristics of the CULP family and may explain why M. tuberculosis maintains multiple CULPs without obvious redundancy. By contrast, M. leprae has maintained within its reduced genome only one CULP member, namely CulpL, which shows 75% amino acid homology with Culp6. This further suggests an essential role for Culp6 in survival of mycobacteria. This fact may also reflect the restriction of environmental niches and hosts permissive for each pathogen, which for M. leprae are primarily humans.
The CULPs are emerging as a fascinating group of mycobacterial proteins, important in numerous events during host infection. We have previously demonstrated that members of the CULPs are protective in a murine model of tuberculosis (17). We now report diversity of function and cellular location within the group and characterized the components of the active site of Culp6, which is retained within the cell wall, suggesting Culp6 performs a vital role in its biosynthesis and maintenance. Further study of these proteins may contribute to understanding the pathogenic process and potentially yield novel targets for effective antimycobacterial therapies.
Acknowledgments
This study was supported by the National Health and Medical Research Council of Australia and the New South Wales Government through its infrastructure grant to the Centenary Institute, and by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S.A. M. tuberculosis materials, including subcellular protein fractions and DNA, were obtained through the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases (NIAID) contract HHSN266200400091C, “Tuberculosis Vaccine Testing and Research Materials,” awarded to Dr. J. Belisle, Colorado State University (Fort Collins, CO, USA). The authors thank Dr. A. Sher (NIAID, NIH) for material support, N. Field and V. Roknic for their technical assistance, and M. Jormakka, A. Guilfoyle, and K. Vincent (Structural Biology, Centenary Institute) for protein fold analysis.
References
- Dye C. Global epidemiology of tuberculosis. Lancet. 2006;367:938–940. doi: 10.1016/S0140-6736(06)68384-0. [DOI] [PubMed] [Google Scholar]
- Beatty W L, Russell D G. Identification of mycobacterial surface proteins released into subcellular compartments of infected macrophages. Infect Immun. 2000;68:6997–7002. doi: 10.1128/iai.68.12.6997-7002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J A, Hart P D. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J Exp Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D G. Mycobacterium tuberculosis: here today, and here tomorrow. Nat Rev Mol Cell Biol. 2001;2:569–577. doi: 10.1038/35085034. [DOI] [PubMed] [Google Scholar]
- Belisle J T, Vissa V D, Sievert T, Takayama K, Brennan P J, Besra G S. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997;276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- Vergne I, Chua J, Lee H H, Lucas M, Belisle J, Deretic V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2005;102:4033–4038. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach H, Papavinasasundaram K G, Wong D, Hmama Z, Av-Gay Y. Mycobacterium tuberculosis virulence is mediated by PtpA dephosphorylation of human vacuolar protein sorting 33B. Cell Host Microbe. 2008;3:316–322. doi: 10.1016/j.chom.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Lun S, Bishai W R. Characterization of a novel cell wall-anchored protein with carboxylesterase activity required for virulence in Mycobacterium tuberculosis. J Biol Chem. 2007;282:18348–18356. doi: 10.1074/jbc.M700035200. [DOI] [PubMed] [Google Scholar]
- Deb C, Daniel J, Sirakova T D, Abomoelak B, Dubey V S, Kolattukudy P E. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J Biol Chem. 2006;281:3866–3875. doi: 10.1074/jbc.M505556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Canaan S, Maurin D, Chahinian H, Pouilly B, Durousseau C, Frassinetti F, Scappuccini-Calvo L, Cambillau C, Bourne Y. Expression and characterization of the protein Rv1399c from Mycobacterium tuberculosis. A novel carboxyl esterase structurally related to the HSL family. Eur J Biochem. 2004;271:3953–3961. doi: 10.1111/j.1432-1033.2004.04335.x. [DOI] [PubMed] [Google Scholar]
- Egmond M R, de Vlieg J. Fusarium solani pisi cutinase. Biochimie (Paris) 2000;82:1015–1021. doi: 10.1016/s0300-9084(00)01183-4. [DOI] [PubMed] [Google Scholar]
- Schafer W. The role of cutinase in fungal pathogenicity. Trends Microbiol. 1993;1:69–71. doi: 10.1016/0966-842x(93)90037-r. [DOI] [PubMed] [Google Scholar]
- Purdy R E, Kolattukudy P E. Hydrolysis of plant cuticle by plant pathogens. Purification, amino acid composition, and molecular weight of two isozymes of cutinase and a nonspecific esterase from Fusarium solani f. pisi. Biochemistry. 1975;14:2824–2831. doi: 10.1021/bi00684a006. [DOI] [PubMed] [Google Scholar]
- Fett W F, Gerard H C, Moreau R A, Osman S F, Jones L E. Screening of nonfilamentous bacteria for production of cutin-degrading enzymes. Appl Environ Microbiol. 1992;58:2123–2130. doi: 10.1128/aem.58.7.2123-2130.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett W F, Wijey C, Moreau R A, Osman S F. Production of cutinolytic esterase by filamentous bacteria. Lett Appl Microbiol. 2000;31:25–29. doi: 10.1046/j.1472-765x.2000.00752.x. [DOI] [PubMed] [Google Scholar]
- West N P, Wozniak T M, Valenzuela J, Feng C G, Sher A, Ribeiro J M, Britton W J. Immunological diversity within a family of cutinase-like proteins of Mycobacterium tuberculosis. Vaccine. 2008;26:3853–3859. doi: 10.1016/j.vaccine.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C M, Boyd D H, Rubin E J. Genes required for mycobacterial growth defined by high-density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Tatusov R L, Fedorova N D, Jackson J D, Jacobs A R, Kiryutin B, Koonin E V, Krylov D M, Mazumder R, Mekhedov S L, Nikolskaya A N, Rao B S, Smirnov S, Sverdlov A V, Vasudevan S, Wolf Y I, Yin J J, Natale D A. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko A R, Shoemaker B A, Thiessen P A, Geer L Y, Bryant S H. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Winkler U K, Stuckmann M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J Bacteriol. 1979;138:663–670. doi: 10.1128/jb.138.3.663-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt J, Cooley J D, Purdy C W, Straus D C. Lipase activity from strains of Pasteurella multocida. Curr Microbiol. 2000;40:306–309. doi: 10.1007/s002849910061. [DOI] [PubMed] [Google Scholar]
- Chen S, Tong X, Woodard R W, Du G, Wu J, Chen J. Identification and characterization of bacterial cutinase. J Biol Chem. 2008;283:25854–25862. doi: 10.1074/jbc.M800848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton T J, Kolattukudy P E. Determination of the structures of cutin monomers by a novel depolymerization procedure and combined gas chromatography and mass spectrometry. Biochemistry. 1972;11:1885–1896. doi: 10.1021/bi00760a025. [DOI] [PubMed] [Google Scholar]
- Hovav A H, Fishman Y, Bercovier H. Gamma interferon and monophosphoryl lipid A-trehalose dicorynomycolate are efficient adjuvants for Mycobacterium tuberculosis multivalent acellular vaccine. Infect Immun. 2005;73:250–257. doi: 10.1128/IAI.73.1.250-257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldingh K, Rosenkrands I, Jacobsen S, Rasmussen P B, Elhay M J, Andersen P. Two-dimensional electrophoresis for analysis of Mycobacterium tuberculosis culture filtrate and purification and characterization of six novel proteins. Infect Immun. 1998;66:3492–3500. doi: 10.1128/iai.66.8.3492-3500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D L, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken S M, Harel M, Remington S J, Silman I, Schrag J, Sussman J L, Verschueren K H G, Goldman A. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- Arpigny J L, Jaeger K E. Bacterial lipolytic enzymes: classification and properties. Biochem J. 1999;343:177–183. [PMC free article] [PubMed] [Google Scholar]
- Lesuisse E, Schanck K, Colson C. Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. Eur J Biochem. 1993;216:155–160. doi: 10.1111/j.1432-1033.1993.tb18127.x. [DOI] [PubMed] [Google Scholar]
- Kordel M, Hofmann B, Schomburg D, Schmid R D. Extracellular lipase of Pseudomonas sp. strain ATCC 21808: purification, characterization, crystallization, and preliminary X-ray diffraction data. J Bacteriol. 1991;173:4836–4841. doi: 10.1128/jb.173.15.4836-4841.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker S K, Curtin K M, Vasil M L. Purification and characterization of mycobacterial phospholipase A: an activity associated with mycobacterial cutinase. J Bacteriol. 2007;189:4153–4160. doi: 10.1128/JB.01909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Tigerstrom R G, Stelmaschuk S. The use of Tween 20 in a sensitive turbidimetric assay of lipolytic enzymes. Can J Microbiol. 1989;35:511–514. doi: 10.1139/m89-079. [DOI] [PubMed] [Google Scholar]
- Gupta R, Rathi P, Gupta N, Bradoo S. Lipase assays for conventional and molecular screening: an overview. Biotechnol Appl Biochem. 2003;37:63–71. doi: 10.1042/ba20020059. [DOI] [PubMed] [Google Scholar]
- Testa B, Mayer J M. Classification, localization and some physiological roles of hydrolytic enzymes. Verlag Helvetica Chimica Acta, Zurich: Wiley-VCH, Weinheim and Cambridge; Hydrolysis in Drug and Prodrug MetabolismChemistry, Biochemistry, and Enzymology. 2003:25–39. [Google Scholar]
- Martinez C, Nicolas A, van Tilbeurgh H, Egloff M P, Cudrey C, Verger R, Cambillau C. Cutinase, a lipolytic enzyme with a preformed oxyanion hole. Biochemistry. 1994;33:83–89. doi: 10.1021/bi00167a011. [DOI] [PubMed] [Google Scholar]
- Walz I, Schwack W. Cutinase inhibition by means of insecticidal organophosphates and carbamates. Part 1: Basics in development of a new enzyme assay. Eur Food Res Technol. 2007;225:593–601. [Google Scholar]
- Quistad G B, Casida J E. Sensitivity of blood-clotting factors and digestive enzymes to inhibition by organophosphorus pesticides. J Biochem Mol Toxicol. 2000;14:51–56. doi: 10.1002/(sici)1099-0461(2000)14:1<51::aid-jbt7>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Pernas M A, Lopez C, Rua M L, Hermoso J. Influence of the conformational flexibility on the kinetics and dimerisation process of two Candida rugosa lipase isoenzymes. FEBS Lett. 2001;501:87–91. doi: 10.1016/s0014-5793(01)02630-8. [DOI] [PubMed] [Google Scholar]
- Huygen K, Lozes E, Gilles B, Drowart A, Palfliet K, Jurion F, Roland I, Art M, Dufaux M, Nyabenda J, De Bruyn J, Van Vooren J, DeLeys R. Mapping of TH1 helper T-cell epitopes on major secreted mycobacterial antigen 85A in mice infected with live Mycobacterium bovis BCG. Infect Immun. 1994;62:363–370. doi: 10.1128/iai.62.2.363-370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launois P, DeLeys R, Niang M N, Drowart A, Andrien M, Dierckx P, Cartel J L, Sarthou J L, Van Vooren J P, Huygen K. T-cell-epitope mapping of the major secreted mycobacterial antigen Ag85A in tuberculosis and leprosy. Infect Immun. 1994;62:3679–3687. doi: 10.1128/iai.62.9.3679-3687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sable S B, Kumar R, Kalra M, Verma I, Khuller G K, Dobos K, Belisle J T. Peripheral blood and pleural fluid mononuclear cell responses to low-molecular-mass secretory polypeptides of Mycobacterium tuberculosis in human models of immunity to tuberculosis. Infect Immun. 2005;73:3547–3558. doi: 10.1128/IAI.73.6.3547-3558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldingh K, Andersen P. Immunological evaluation of novel Mycobacterium tuberculosis culture filtrate proteins. FEMS Immunol Med Microbiol. 1999;23:159–164. doi: 10.1111/j.1574-695X.1999.tb01235.x. [DOI] [PubMed] [Google Scholar]
- Mahairas G G, Sabo P J, Hickey M J, Singh D C, Stover C K. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, Klebl B, Thompson C, Bacher G, Pieters J. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 2004;304:1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- Stewart G R, Patel J, Robertson B D, Rae A, Young D B. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 2005;1:269–278. doi: 10.1371/journal.ppat.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler P R, Bulmer K, Ratledge C. Enzymes for biosynthesis de novo and elongation of fatty acids in mycobacteria grown in host cells: is Mycobacterium leprae competent in fatty acid biosynthesis? J Gen Microbiol. 1990;136:211–217. doi: 10.1099/00221287-136-1-211. [DOI] [PubMed] [Google Scholar]
- Wheeler P R, Ratledge C. Use of carbon sources for lipid biosynthesis in Mycobacterium leprae: a comparison with other pathogenic mycobacteria. J Gen Microbiol. 1988;134:2111–2121. doi: 10.1099/00221287-134-8-2111. [DOI] [PubMed] [Google Scholar]
- Hughes A L. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Ohno S. New York: Springer-Verlag; Evolution by Gene Duplication. 1970 [Google Scholar]