Abstract
The alternative splicing of pre-mRNAs is a critical mechanism in genomic complexity, disease, and development. Studies of the receptor for advanced glycation end-products (RAGE) indicate that this gene undergoes a variety of splice events in humans. However, no studies have extensively analyzed the tissue distribution in other species or compared evolutionary differences of RAGE isoforms. Because the majority of studies probing RAGE function have been performed in murine models, we therefore performed studies to identify and characterize the splice variants of the murine RAGE gene, and we compared these to human isoforms. Here, using mouse tissues, we identified numerous splice variants including changes in the extracellular domain or the removal of the transmembrane and cytoplasmic domains, which produce soluble splice isoforms. Comparison of splice variants between humans and mice revealed homologous regions in the RAGE gene that undergo splicing as well as key species-specific mechanisms of splicing. Further analysis of tissue splice variant distribution in mice revealed major differences between lung, kidney, heart, and brain. To probe the potential impact of disease-like pathological states, we studied diabetic mice and report that RAGE splice variation changed dramatically, resulting in an increase in production of soluble RAGE (sRAGE) splice variants, which were not associated with detectable levels of sRAGE in murine plasma. In conclusion, we have determined that the murine RAGE gene undergoes extensive splicing with distinct splice isoforms being uniquely distributed in different tissues. These differences in RAGE splicing in both physiological and pathogenic states further expand our understanding of the biological repertoire of this receptor in health and disease.—Kalea, A. Z., Reiniger, N., Yang, H., Arriero, M., Schmidt, A. M., Hudson, B. I. Alternative splicing of the murine receptor for advanced glycation end-products (RAGE) gene.
Keywords: DNA cloning, mRNA, soluble receptor, species-specific splicing, protein evolution
Alternative splicing has emerged as a major mechanism to generate protein diversity from a single gene (1). Splicing of mRNA can have varied effects on the resulting protein produced, including the addition or removal of domains of a protein, the introduction of a stop codon to produce a truncated form of a protein, or even the production of a completely novel protein from the same gene (1). These changes can be cell- or tissue-specific and may occur at different developmental stages and under different pathophysiological states (1,2,3,4). Comparisons of alternative splicing between humans and mice have shown that although conservation of some major splice variants exists, a considerable fraction of splice events is species-specific (2, 5, 6). Thorough analysis of gene-specific splice variants across species is essential to lead to a better understanding of biological processes. In particular, the receptor for advanced glycation end-products (RAGE) gene has been demonstrated in humans to generate ∼20 alternative splice variants (7,8,9,10,11,12). RAGE is a cell surface receptor for a broad range of ligands, including the late products of nonenzymatic glycation of macromolecules, S100s, HMGB1, and amyloid fibrils (13,14,15,16). Splice variants of RAGE may affect the ligand-binding domain by insertion and removal of parts of the V domain and the removal of the transmembrane domain, thereby producing secreted isoforms (7, 9). However, information regarding the evolutionary nature of alternative splicing of RAGE from studies in species other than the human is limited (17). Because the majority of studies probing RAGE function in biology and disease to date have been based on experiments performed in murine models (14, 18,19,20,21,22), studies to improve the understanding of the RAGE gene in mice are essential, as the development of novel human therapeutic agents is modeled, at least in part, in this organism. In our present study, we report for the first time the extensive screening, identification, and definition of the murine RAGE gene and its splice variants.
MATERIALS AND METHODS
Materials
All animal studies were performed with the approval of the Institutional Animal Care and Use Committee of Columbia University and conform with the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health.
The Mouse I Multiple Tissue cDNA Panel (Clontech Laboratories Inc., Mountain View, CA, USA) was used for initial splice variant screening. The panel (lung, heart, kidney, and brain) is normalized, with first-strand cDNA pooled from >200 male/female BALB/C mice, aged 8–12 wk and disease-free.
For screening of splice variants in a RAGE-related pathogenic state, we purchased nondiabetic m/db mice from the Jackson Laboratory (Bar Harbor, ME, USA) and generated from these male homozygous (db/db) obese diabetic and heterozygous (m/db) lean nondiabetic mice. Animals were sacrificed at age 9 months, and the renal cortex was harvested and snap-frozen. Tissue was homogenized using QIAshredders (Qiagen, Valencia, CA, USA), and RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). A total of 3 μg of RNA was pooled from each group of mice (m/db vs. db/db, n=3 per group), and cDNA was generated using Superscript III (Invitrogen). Soluble RAGE (sRAGE) levels in plasma and urine from each mouse were measured (n=3 per group) using the Mouse RAGE DuoSet ELISA (R&D Systems, Minneapolis, MN, USA). This ELISA is designed to detect any murine sRAGE species and uses an eight-point standard curve with recombinant sRAGE in the range from 62.5 to 8000 pg/ml. The antibodies used in the ELISA for measurement are raised against the recombinant extracellular domain of RAGE and therefore would be predicted to detect sRAGE produced by either cleavage or alternative splicing.
Splice variant identification
Primers were designed to amplify within the 5′ and 3′ ends of the full length RAGE gene, and the resulting polymerase chain reaction (PCR) product sequence of the RAGE mRNA to be amplified was analyzed for restriction enzyme recognition sequences using the online tool NEBcutter V2.0 (http://tools.neb.com/NEBcutter2/index.php). Restriction enzyme sites were selected to cut the 1276-bp PCR product into a series of bands of differing size, ranging between 100 and 400 bp. The two restriction enzymes, BamHI and PstI, were chosen from the restriction map to give five bands of easily distinguishable size difference.
A primary PCR was performed on cDNA using the RAGE forward [mRAGE 5′-untranslated region (UTR) 1, 5′-CTGGAGCCTGGGAAGGAAG-3′] and reverse (mRAGE 3′-UTR1, 5′-CTGGTTGGAGAAGGAAGTGC-3′) primers with a Platinum Taq High Fidelity DNA polymerase system (Invitrogen). The PCR product was diluted 1:500, and a nested secondary PCR was performed with RAGE 5′-UTR2 (5′-AGGAAGCACCATGCCAGC-3′) and RAGE reverse nested primer (RAGE 3′-UTR2, 5′-GGATGGAATGTGGGGGAG-3′). The resulting PCR product was purified using the Ultra PCR Clean-Up kit (ABgene, Epsom, UK) and cloned into the TOPO TA vector. More than 100 bacterial colonies were selected as before (7), and PCR was performed using the mRAGE 5′-UTR2 forward primer and mRAGE 3′-UTR2 reverse primer. To verify product size, 5 μl of the PCR product was electrophoresed on a 1.5% agarose gel. The remaining PCR product was digested overnight at 37°C with 10 U of BamHI and PstI (Invitrogen) and electrophoresed for 2 h on a 3% Midi-Agarose gel (ABgene) at 100 V. For each tissue, 100 colonies were screened by this method, and a relative percentage of distribution was assigned. PCRs from colonies displaying different restriction patterns from the predicted full-length RAGE were repeated to verify the result. Plasmid DNA was purified from the respective colony as described above and the resulting DNA was sequenced using T7, M13R, and a RAGE-specific primer (RAGE 5′-AGCTGACAGTGATCCCCAC-3′).
Bioinformatics
Sequence data were viewed and annotated using the FinchTV program (Geospiza, Inc., Seattle, WA, USA) and aligned to mouse RAGE mRNA (NM_007425) and RAGE DNA (AF030001) using Clustalw (http://www.ebi.ac.uk/clustalw/). Protein translation of predicted proteins from splice variants was performed using ExPASy (http://us.expasy.org/tools/dna.html).
Nonsense-mediated decay (NMD) predictions were made by calculating the distance between the last splice site and the stop codon. When the final splice site was more than 50 bases from the stop codon, the variants were designated as a putative NMD target (23). Splice site prediction analysis was performed using NNSPLICE 0.9 (http://www.fruitfly.org/seq_tools/splice.html). The sequence data from this study have been submitted to GenBank as follows: mRAGE (EU520325), mRAGEv1 (EU570240), mRAGEv2 (EU570241), mRAGEv3 (EU570242), mRAGEv4 (EU570243), mRAGEv5 (EU570244), mRAGEv6 (EU570245), mRAGEv7 (EU570246), mRAGEv8 (EU570247), mRAGEv9 (EU906857), mRAGEv10 (EU906858), mRAGEv11 (EU906859), mRAGEv12 (EU906860), mRAGEv13 (EU906861), mRAGEv14 (EU906862), mRAGEv15 (EU906863), mRAGEv16 (EU906864), and mRAGEv17 (EU906865).
RESULTS
Identification of murine RAGE splice variants
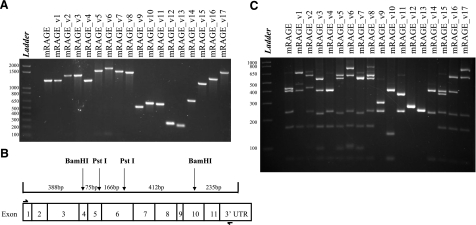
To identify RAGE splice variants in murine tissue, a combination of PCR, cloning, and restriction endonuclease digestion was used, as described previously (7). In brief, PCR primers were designed to amplify from upstream of the initiation codon to the 3′-UTR to capture any changes in all exons and introns. cDNA from murine tissue was then amplified by PCR, and the resulting mixture of splice variants from each tissue was cloned. One hundred clones were then selected, another round of PCR amplification was performed on each clone, and the resulting product was digested with restriction endonuclease enzymes to allow resolution of smaller bands to distinguish RAGE splice variants. As demonstrated in Fig. 1A, the canonical murine RAGE full-length isoform resulted in a PCR product of 1276 bp. Differences between splice variants could be seen for those products (mRAGE_v9–v14) with splice variants resulting from large exon and intron changes (Fig. 1A). A restriction digest was designed to give five DNA fragments of clearly differing sizes as depicted in the schematic diagram shown in Fig. 1B. Digestion products comprised RAGE from exon 1–4 (388 bp), exon 4–5 (75 bp), exon 5–6 (166 bp), exon 6–10 (412 bp), and exon 10–3′-UTR (235-bp). The digestion would reveal the previously identified RAGE splice variants. The digest of the major full-length RAGE isoform (mRAGE) and all variants detected in mouse lung, brain, kidney, and heart cDNA is shown in Fig. 1C. In total, 17 alternative splice variants were detected by PCR and restriction digest of more than 100 selected RAGE-positive clones for each tissue as demonstrated in Fig. 1C.
Figure 1.
Detection of mouse RAGE splice variants. A) Total PCR product for all different clones detected from the mouse lung, kidney, brain, and heart cDNA for RAGE. B) Restriction map of full-length RAGE cDNA with BamHI and PstI. Restriction sites are shown by arrows in the respective exon where they occur. Resulting DNA fragments are shown in base pairs; boxes represent exons. Primers used are indicated by half arrows. C) Restriction digest of the mouse RAGE cDNA PCR products with BamHI and PstI. The corresponding splice variant classification is shown above the digestion. DNA fragments were sized against a 1-kb DNA ladder as indicated on each gel.
Characterization of murine RAGE splice variants
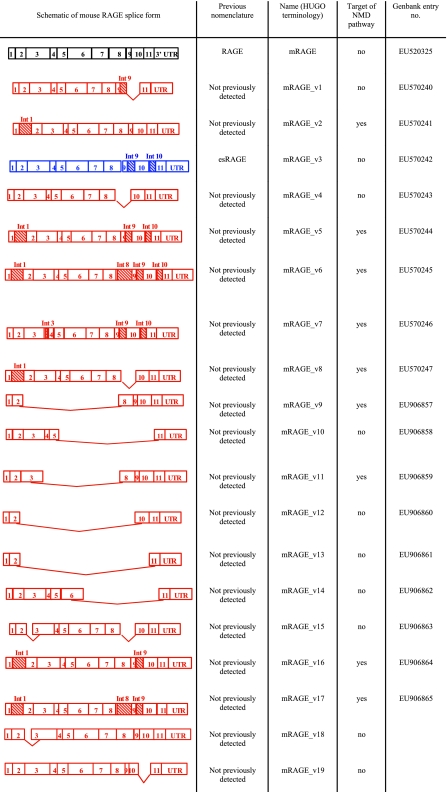
DNA sequencing of the different splice variants detected by PCR and restriction endonuclease digestion analysis revealed the splice changes occurring in the murine RAGE gene. These splice variants were then classified according to the consensus outlined by the Human Gene Nomenclature Committee (24), and the results are presented in Fig. 2. The majority of splice variants involved either inclusion of introns (introns 1, 8, 9, and 10), deletion of exon 9, or removal of large numbers of exons. Previously identified splice events from mice, which include the retention of intron 9 (mRAGE_v16) and the retention of intron 9 and intron 10 (mRAGE_v3), were detected (17) in addition to the canonical full-length form of RAGE (mRAGE). The majority of splice variants were novel and consisted of inclusion of intron 9 and deletion of exon 10 (mRAGE_v1), inclusion of intron 1 (mRAGE_v2) and in combination with other splice variation (mRAGE_v5, v6, v8, v16, and v7), deletion of exon 9 (mRAGE_v4), inclusion of intron 3 (mRAGE_v7), and removal of part of exon 3 (mRAGE_v15). In addition, a number of rare splice forms were detected, which resulted in deletion of large numbers of exons, including exons 2–8 (mRAGE_v9), exons 5–11 (mRAGE_v10), exons 3–8 (mRAGE_v11), exons 2–10 (mRAGE_v12), exons 2–11 (mRAGE_v13), and exons 6–11 (mRAGE_v14).
Figure 2.
Splice domain organization map of the different variants detected in mouse lung, kidney, brain, and heart cDNA for RAGE. Previously identified variants are shown in blue; novel variants in red.
Bioinformatic analysis of murine RAGE splice variants
After the characterization of variants, we performed bioinformatic analysis on murine RAGE splice variants to predict the changes in the protein sequence as a result of splice variation. Inclusion of intron 9 (mRAGE_v1 and mRAGE_v3) resulted in a reading frameshift in this intron at amino acid 330 and resulted in the loss of the transmembrane as well as the cytoplasmic domains, thereby creating a unique C-terminal sequence (EGLD). This change would result in a potential secreted isoform of mRAGE. Deletion of exon 9 (mRAGE_v4) resulted in the deletion of 9 amino acids (ETGDEGPAE) in the extracellular domain at amino acid 320, without altering the reading frame and retaining the transmembrane and cytoplasmic domains. Inclusion of intron 1 (mRAGE_v2, v5, v6, v8, v16, and v17) resulted in a reading frameshift at amino acid 19 and a resulting premature stop codon in intron 1.
Inclusion of intron 3 (mRAGE_v7) resulted in a reading frameshift at amino acid 118 and a premature stop codon 21 amino acids after this. Deletion of part of exon 3 resulted in the removal of 9 amino acids (NTGRTEAWK) of the V domain of the extracellular region of RAGE from amino acids 54–62.
Major protein changes were predicted for mRAGE_v9, v10, v11, v12, v13, and v14 due to removal of numerous exons. As detailed in Table 1, these would result in reading frameshifts producing short truncated proteins or deletion of major sections of the RAGE protein.
TABLE 1.
Splice variants of mouse RAGE resulting in large changes to the protein sequence
| RAGE splice variant | Amino acid frameshift | Amino acid termination | Amino acid deletion |
|---|---|---|---|
| mRAGE_v9 | 27 | 68 | |
| mRAGE_v10 | 154 | 155 | |
| mRAGE_v11 | 119 | 138 | |
| mRAGE_v12 | 28 | 30 | |
| mRAGE_v13 | 30 | 40 | |
| mRAGE_v14 | 189–383 |
For mRAGE_v9–v14, amino acid frameshift number and termination codon or amino acids deleted are shown.
To assess which variants would be likely candidates for degradation by the NMD pathway, which removes aberrant nontranslated mRNA species, we assessed transcripts according to the rule that a stop codon >50 nucleotides upstream of the penultimate exon would target a variant for degradation (23). As shown in Fig. 2, the splice variants that are candidates for NMD include mRAGE_v2, v5, v6, v7, v8, v9, v11, v16, and v17.
Tissue distribution of mouse RAGE splice variants
We next assessed the relative distribution of splice variants of mouse RAGE across different tissues. By assessing the digestion patterns of the 100 selected clones from lung, brain, kidney, and heart, an estimate of splice variation was established and reported as a percentage. These data are presented in Table 2. In these screened tissues, the canonical form of full-length RAGE was the most prevalent splice variant detected with the relative percentage compared to other forms being heart > kidney > lung > brain. After full-length RAGE, the next most prevalent form was mRAGE_v4, which is the result of the deletion of exon 9, occurring as 41% (lung), 13% (kidney), and 18% (heart) of the total RAGE splice variant percentage. Interestingly, this splice variant was not detected in brain. Other common variants included mRAGE_v2, which results from the inclusion of intron 1, occurring in brain and kidney (40 and 8%, respectively) and in brain, kidney, and heart in combination with other splice domain changes (mRAGE_v5, v6, v8, v16, and v17). In addition, variants predicted to produce secreted, soluble forms of RAGE were detected at relatively low levels (3% for mRAGE_v1 and 3% for mRAGE_v3) and only in kidney. All other variants from mRAGEv7–v17 were rare and detected only once in the respective tissue, as shown in Table 2.
TABLE 2.
Prevalence of mouse RAGE splice variants in lung, brain, kidney, and heart
| RAGE splice variant | Lung (%) | Brain (%) | Kidney (%) | Heart (%) |
|---|---|---|---|---|
| mRAGE | 53 | 44 | 57 | 74 |
| mRAGE_v1 | 3 | |||
| mRAGE_v2 | 39 | 8 | ||
| mRAGE_v3 | 5 | |||
| mRAGE_v4 | 41 | 12 | 21 | |
| mRAGE_v5 | 9 | 6 | 1 | |
| mRAGE_v6 | 7 | 1 | ||
| mRAGE_v7 | 2 | |||
| mRAGE_v8 | 6 | |||
| mRAGE_v9 | 2 | |||
| mRAGE_v10 | 1 | |||
| mRAGE_v11 | 1 | |||
| mRAGE_v12 | 1 | |||
| mRAGE_v13 | 1 | 1 | ||
| mRAGE_v14 | 1 | |||
| mRAGE_v15 | 1 | |||
| mRAGE_v16 | 1 | |||
| mRAGE_v17 | 1 |
Percentage expression frequency of each variant is a result obtained from the screening of cloned cDNA colonies. For all tissues, n = 100.
Conservation and species-specific splice variation of the mouse and human RAGE gene
Comparisons between the murine splice variants of RAGE with those previously detected by our and other groups in the human RAGE gene (7,8,9,10,11,12) demonstrated both conserved and distinct species specific variants.
As shown in Supplemental Fig. 1, alignment of the human and mouse DNA/RNA sequence reveals complete conservation of the 11 cassette exons of RAGE and an overall >60% conservation at the DNA level. Similarly, at the mRNA and protein levels, the canonical mouse and human RAGE are highly similar, being 79% similar at the mRNA (Supplemental Fig. 2) and 78% similar at the protein level (Supplemental Fig. 3).
In mice, the major mechanisms of generating splice variation of RAGE were the retention of introns (intron 1, 8, 9, and 10) or the deletion of cassette exons (exon 9 or multiple exons). In comparison, human splice variation of RAGE included all splice mechanisms including intron retention (intron 1 only) and deletion of cassette exons (exons 8 and 10). In addition, a number of splice variants in the human involved alternative acceptor and donor sites of exons and introns (7).
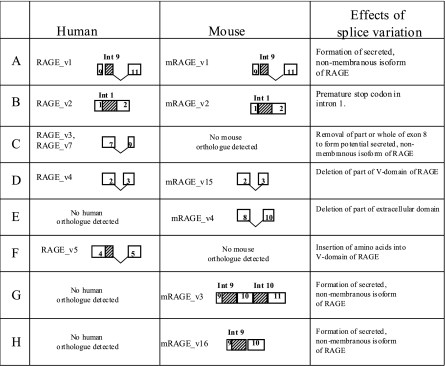
Species-specific isoforms were detected in mice and not previously detected in humans, as shown in Fig. 3 (7). The deletion of exon 9 (mRAGE_v4) was found to occur in most tissues screened, but a similar splice variant was not previously detected in humans (7).
Figure 3.
Conservation and species-specific orthologues of human and mouse RAGE gene. Exon and intron splice variations for selected splice variants detected for humans and mice. Species-specific splice variants are indicated.
In humans, two major common forms of splice variation were previously detected and resulted in the removal of the transmembrane domain, leading to a secreted form of RAGE and domain changes in the ligand-binding domain (7). In contrast, although mechanisms were detected to form soluble forms of RAGE in mice, these were found to be rare in this species. In addition, splice variation around the ligand-binding domain was also a rare event in mice.
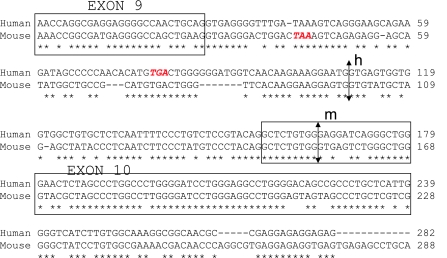
Further investigation of mouse splice variation to form soluble variants revealed that mRAGE_v1 was the orthologous form of human RAGE_v1 resulting from the inclusion of intron 9 and removal of exon 10 (Fig. 3). However, as detected previously, mRAGE_v3, although different in the exons/introns spliced (intron 9 and 10 inclusion), would form the same protein ending in the unique amino acid sequence, EGLD (amino acids 330–333) (17). Alignment and analysis of the genomic sequence of human and mouse RAGE revealed distinct differences in the formation of human and mouse RAGE_v1. First, because of intronic differences, the stop codon for mRAGE_v1 is earlier than that for RAGE_v1, resulting in a shorter truncated protein (Fig. 4). Whereas the murine RAGE_v1 protein ends with the amino acids EGLD as shown above, the human equivalent ends with the unique sequence EGFDKVREAEDSPQHM (amino acids 332–347). Second, analysis of the alternative donor sites in intron 9 reveals distinct differences in the splice sites leading to mouse and human RAGE_v1. For humans and mice, the alternative acceptor sites in intron 9 (human) and exon 10 (mouse) are not conserved between these species (indicated by arrows in Fig. 4).
Figure 4.
The human and mouse RAGE_v1 splice variant is formed by different splice sites. DNA sequence of human and mouse RAGE aligned by ClustalW. Stop codon for RAGE_v1 is shown in red; alternative donor splice sites are indicated by arrows for human (h) and mouse (m).
Distribution of mouse RAGE splice variants in the diabetic kidney
As the mice used in the tissue distribution study were relatively young and disease-free, we were interested in examining the effects of pathological settings on RAGE splice variant distribution. Using the type 2 diabetic mouse model (the db/db mouse), which has been used in numerous studies to probe the role for RAGE in the pathogenesis of diabetes complications (25,26,27), we investigated splice variant changes in age-matched m/db (nondiabetic) and db/db (diabetic) mice. With the same approach as with the screening of mouse tissue, 100 clones from m/db and db/db mice were selected and an estimate of splice variation was established and reported as a percentage. As shown in Table 3, there were obvious changes in RAGE splice variant distribution. First, we detected two rare novel variants in the diabetic mice: mRAGE_v18, which resulted from the deletion of part of exon 10, and mRAGE_v19, which resulted from deletion of part of exon 3, that were not seen in the nondiabetic m/db mouse (Table 2). Second, in particular and strikingly, relative mRAGEv1 levels increased from 3 to 9% in the diabetic state, suggesting an increase in the generation of the soluble splice variant of RAGE. To further explore these data, we performed experiments to determine whether this increase seen in mRAGE_v1 mRNA levels translated into an increase in the generation of sRAGE protein levels in the circulation of mice. For these experiments we used the murine-specific sRAGE ELISA, which is designed to measure any murine extracellular domain sRAGE species in the range from 62.5 to 8000 pg/ml. We were unable to detect sRAGE levels in murine plasma or urine from either diabetic or nondiabetic mice within this range.
TABLE 3.
Prevalence of mouse RAGE splice variants in nondiabetic and diabetic mouse kidney
| RAGE splice variant | Nondiabetic (%) | Diabetic (%) |
|---|---|---|
| mRAGE | 68 | 67 |
| mRAGE_v1 | 3 | 9 |
| mRAGE_v2 | 0 | 1 |
| mRAGE_v4 | 29 | 20 |
| mRAGE_v18 | 0 | 2 |
| mRAGE_v19 | 0 | 1 |
Percentage expression frequency of each variant is a result obtained from the screening of cloned cDNA colonies. For all conditions, n = 100.
DISCUSSION
RAGE is a multiligand member of the immunoglobulin superfamily (13,14,15,16). Studies have implicated RAGE in the pathogenesis of numerous disease states including cancer, vascular disease, and neurodegeneration, with the majority of supportive data obtained in model studies in mice (14, 18,19,20,21,22). Human studies have revealed that extensive splicing of the RAGE gene occurs to generate a repertoire of protein products with diverse function (7,8,9,10,11,12). However, there is limited information regarding the splice variation of the mouse RAGE gene (17). To fully understand the biology of RAGE in development and disease, a thorough evaluation of the splice variation that is conserved and species-specific is essential for the design and optimal testing of RAGE-directed therapeutic agents. To address this, we performed extensive analysis of splice variants of murine RAGE in tissues relevant to the biology of RAGE, including lung, kidney, brain, and heart. We finally performed studies in diabetic mice to test the role of disease pathogenesis on RAGE splice variant distribution. Although we found a relative increase in the mRAGE_v1 soluble splice variant at the mRNA level, we were unable to detect any sRAGE protein in mouse plasma in homeostasis or diabetes.
As hypothesized, the full-length form of RAGE occurred most frequently in all tissues studied, and this result was similar to that observed in human studies (7,8,9,10,11,12). In previous human studies, the most common alternative splice variants involved the production of secreted forms of RAGE resulting from frameshifts at the C terminus to remove the transmembrane and cytoplasmic domains (7). The only previous study of mouse RAGE splice variants identified two splice variants (here designated mRAGE_v3 and mRAGE_v16), which were demonstrated to be an orthologous protein form of RAGE_v1 (17) but distinctly different from RAGE_v1 in their exon/intron splice pattern. In our study, not only did we detect these splice variants but we also identified a novel murine splice ortholog of RAGE_v1 (mRAGE_v1) (Fig. 3, row A). However, apart from the kidney, we could only detect these splice variants in combination with the premature translation-terminating splice variant of intron 1 inclusion (mRAGE_v5, v6, v16, and v17). In addition, the occurrence of mRAGE_v1 and mRAGE_v3 in the kidney was found to be very rare. Therefore, in contrast with humans in whom RAGE_v1 is found in abundance and is perhaps the major mechanism for producing circulating sRAGE forms (7), in mice soluble splice variants of RAGE appeared to be rare. This difference in the biology of RAGE between humans and mice is an important distinction. In humans, clinical studies have suggested a relationship between the circulating sRAGE levels and a range of pathogenic disease states, including diabetes and its vascular complications, neuronal dysfunction, and cardiovascular disease (28,29,30,31,32,33). Therefore, to address potential changes in different physiological or pathogenic states, we performed additional splice screening in kidney from age-matched nondiabetic and diabetic mice and investigated potential changes in sRAGE levels in the murine plasma. At the mRNA level, in the diabetic mouse kidney a relative increase in mRAGE_v1 levels was observed compared with that in the nondiabetic mouse control kidney. Therefore, we concluded that in mice the diabetic state would result in increased production of sRAGE protein. To our surprise, measurement of both plasma and urine from both nondiabetic and diabetic mice using an ELISA that detects any sRAGE species, did not detect any sRAGE protein within the range of the standard curve used (lower limit of detection, 62.5 pg/ml). Of note, it has been demonstrated in vitro that sRAGE can be formed by multiple mechanisms in both mice and humans and may result from the cleavage or “shedding” of the extracellular domain from the full-length form at the cell surface (34, 35), as well as from alternative splicing as suggested here, and this result was also supported by Harashima et al. (17). Therefore, if sRAGE is produced readily in mice by either mechanism, one would expect that we would have detected this in the murine circulation. It is important to note, however, that our result does not preclude the existence of sRAGE in the mouse, as there are a number of factors to consider. First, it is possible that sRAGE is produced in mice at a much lower level beyond the sensitivity of the method used in this study (lower limit of detection is 62.5 pg/ml). If murine sRAGE is present in this protein range, this is still several orders of magnitude lower than that seen in humans (28,29,30,31,32,33). Second, it is remotely possible that this method may not be suitable for measuring murine plasma and/or urine. This scenario is highly unlikely as the antibodies used in this ELISA would certainly recognize the forms of “murine sRAGE” predicted by our studies and those of others. Further, similar studies on the human sRAGE protein have demonstrated that not only is this protein very stable and measurable in numerous ELISA dilution reagents (unpublished results) but it is also detectable and measurable in both serum and plasma (36). Finally, under other pathophysiological conditions or in other tissue or cell types not studied in this work, sRAGE may be produced to detectable levels. Evidence for this theory was also presented by Hanford et al. (37), who demonstrated that a “soluble” RAGE protein can be purified from mouse lung and that the level of this protein changes in the developing rat lung (38). Further studies are needed to investigate these mechanisms and to determine whether sRAGE is detectable at all in mice.
Therefore, the question arises, how might these different splice variants affect the biology of RAGE in humans and mice? It seems that in humans, one possibility is that splice variants affecting the V domain, for example, may change the ligand affinity of RAGE (7) and may lead to a larger evolved repertoire of RAGE-ligand interactions. Although splice variants of this domain are rarely seen in mice, the deletion of exon 9 (mRAGE_v4), which changes the sequence of the extracellular domain, may be an evolutionary equivalent of these human variants. mRAGE_v4 was predicted to form a mature protein only missing a 9-amino acid region in the extracellular domain, in the region proximal to the transmembrane domain, just after the last C-type domain. It is plausible that the effect of removal of this region of the extracellular domain in mice may affect ligand binding compared with that of the full-length RAGE form, by changing the tertiary structure of the extracellular domain. Interestingly, to date, in neither species have variants been identified that change the highly conserved protein sequence of the cytoplasmic domain (apart from its entire removal with the transmembrane domain). The absence of splice variants changing the cytoplasmic domain would support multiple studies in which this domain has been shown to be essential for RAGE function through its ability to transduce intracellular signaling (19, 39,40,41). Most intriguingly, the key distinction in the biology of splicing of RAGE between humans and mice is that under normal physiological states in mice, there is a relative lack of splice variants that produce an sRAGE (7). Although diabetic mice revealed a relative frequency of soluble splice variants similar to that seen in human tissue (7), this increase in soluble splice transcripts did not appear to translate to a similar change in sRAGE protein in the murine circulation. Therefore, a key difference between humans and mice is the relative deficiency of circulating sRAGE in mice. It is possible though that other mechanisms exist in mice to regulate the RAGE-ligand interaction to compensate for this relative deficiency of sRAGE. Further studies are required to elucidate why and how sRAGE is produced and regulated in humans as well as the biological effects of this evolved difference.
In conclusion, we have found that the murine RAGE gene undergoes extensive splicing, which is uniquely distributed in different tissues. Comparison with the human splice variants of RAGE reveals a number of conserved splice variants as well as species-specific isoforms in both mice and humans. Further definition and experimental validation of the unique and conserved splice variants will help us better understand the biology and regulation of the RAGE gene in both species.
Supplementary Material
Acknowledgments
This work was supported by grants from the U.S. Public Health Service and the Juvenile Diabetes Research Foundation. B.I.H. is a recipient of a Career Development Award from the Juvenile Diabetes Research Foundation International. A.M.S. is a recipient of a Scholar Award from the Juvenile Diabetes Research Foundation International. A.Z.K. is a recipient of a postdoctoral fellowship from the Juvenile Diabetes Research Foundation International. N.R. is a recipient of a National Research Service Award from the National Institutes of Health.
References
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj T A, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Lareau L F, Green R E, Bhatnagar R S, Brenner S E. The evolving roles of alternative splicing. Curr Opin Struct Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Xu Q, Lee C. Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 2003;31:5635–5643. doi: 10.1093/nar/gkg786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Xu Q, Lee C. Widespread production of novel soluble protein isoforms by alternative splicing removal of transmembrane anchoring domains. FEBS Lett. 2003;555:572–578. doi: 10.1016/s0014-5793(03)01354-1. [DOI] [PubMed] [Google Scholar]
- Nurtdinov R N, Neverov A D, Favorov A V, Mironov A A, Gelfand M S. Conserved and species-specific alternative splicing in mammalian genomes. BMC Evol Biol. 2007;7:249. doi: 10.1186/1471-2148-7-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurtdinov R N, Artamonova I I, Mironov A A, Gelfand M S. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum Mol Genet. 2003;12:1313–1320. doi: 10.1093/hmg/ddg137. [DOI] [PubMed] [Google Scholar]
- Hudson B I, Carter A M, Harja E, Kalea A Z, Arriero M, Yang H, Grant P J, Schmidt A M. Identification, classification, and expression of RAGE gene splice variants. FASEB J. 2007;22:1572–1580. doi: 10.1096/fj.07-9909com. [DOI] [PubMed] [Google Scholar]
- Malherbe P, Richards J G, Gaillard H, Thompson A, Diener C, Schuler A, Huber G. cDNA cloning of a novel secreted isoform of the human receptor for advanced glycation end products and characterization of cells co-expressing cell-surface scavenger receptors and Swedish mutant amyloid precursor protein. Brain Res Mol Brain Res. 1999;71:159–170. doi: 10.1016/s0169-328x(99)00174-6. [DOI] [PubMed] [Google Scholar]
- Yonekura H, Yamamoto Y, Sakurai S, Petrova R G, Abedin M J, Li H, Yasui K, Takeuchi M, Makita Z, Takasawa S, Okamoto H, Watanabe T, Yamamoto H. Novel splice variants of the receptor for advanced glycation end-products expressed in human vascular endothelial cells and pericytes, and their putative roles in diabetes-induced vascular injury. Biochem J. 2003;370:1097–1109. doi: 10.1042/BJ20021371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter C, Hauke S, Flohr A M, Rogalla P, Bullerdiek J. Tissue-specific expression patterns of the RAGE receptor and its soluble forms—a result of regulated alternative splicing? Biochim Biophys Acta. 2003;1630:1–6. doi: 10.1016/j.bbaexp.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Park I H, Yeon S I, Youn J H, Choi J E, Sasaki N, Choi I H, Shin J S. Expression of a novel secreted splice variant of the receptor for advanced glycation end products (RAGE) in human brain astrocytes and peripheral blood mononuclear cells. Mol Immunol. 2004;40:1203–1211. doi: 10.1016/j.molimm.2003.11.027. [DOI] [PubMed] [Google Scholar]
- Ding Q, Keller J N. Splice variants of the receptor for advanced glycosylation end products (RAGE) in human brain. Neurosci Lett. 2005;373:67–72. doi: 10.1016/j.neulet.2004.09.059. [DOI] [PubMed] [Google Scholar]
- Neeper M, Schmidt A M, Brett J, Yan S D, Wang F, Pan Y C P, Elliston K, Stern D, Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- Hofmann M A, Drury S, Fu C F, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, Neurath M F, Slattery T, Beach D, McClary J, Nagashima M, Morser J, Stern D, Schmidt A M. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for s100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen J X, Nagashima M, Lundh E R, Vijay S, Nitecki D, Morser J, Stern D, Schmidt A M. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin: mediation of neurite outgrowth and co-expression of RAGE and amphoterin in the developing nervous system. J Biol Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- Yan S D, Chen X, Fu J, Chen M, Zhu H, Roher A, Slattery T, Zhao L, Nagashima M, Morser J, Migheli A, Nawroth P, Stern D, Schmidt A M. RAGE and amyloid-β peptide neurotoxicity in Alzheimer’s disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- Harashima A, Yamamoto Y, Cheng C, Tsuneyama K, Myint K M, Takeuchi A, Yoshimura K, Li H, Watanabe T, Takasawa S, Okamoto H, Yonekura H, Yamamoto H. Identification of mouse orthologue of endogenous secretory receptor for advanced glycation end-products: structure, function and expression. Biochem J. 2006;396:109–115. doi: 10.1042/BJ20051573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park L, Raman K G, Lee K J, Lu Y, Ferran L J, Chow W S, Stern D, Schmidt A M. Suppression of accelerated diabetic atherosclerosis by the soluble receptor for advanced glycation endproducts. Nat Med. 1998;4:1025–1031. doi: 10.1038/2012. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Blood D C, del Toro G, Canet A, Lee D C, Qu W, Tanji N, Lu Y, Lalla E, Fu C, Hofmann M A, Kislinger T, Ingram M, Lu A, Tanaka H, Hori O, Ogawa S, Stern D, Schmidt A M. Blockage of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405:354–360. doi: 10.1038/35012626. [DOI] [PubMed] [Google Scholar]
- Kislinger T, Tanji N, Wendt T, Qu W, Lu Y, Ferran L J, Taguchi A, Olson K, Goova M T, Hofmann M A, Cataldegirmen G, D'Agati V, Pischetsrieder M, Stern D M, Schmidt A M. Receptor for advanced glycation end products mediates inflammation and enhanced expression of tissue factor in vasculature of diabetic apolipoprotein E-null mice. Arterioscler Thromb Vasc Biol. 2001;21:905–910. doi: 10.1161/01.atv.21.6.905. [DOI] [PubMed] [Google Scholar]
- Harja E, Bu D X, Hudson B I, Chang J S, Shen X, Hallam K, Kalea A Z, Lu Y, Rosario R H, Oruganti S, Nikolla Z, Belov D, Lalla E, Ramasamy R, Yan S F, Schmidt A M. Vascular and inflammatory stresses mediate atherosclerosis via RAGE and its ligands in apoE−/− mice. J Clin Invest. 2008;118:183–194. doi: 10.1172/JCI32703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson B I, Wendt T, Bucciarelli L G, Rong L L, Naka Y, Yan S F, Schmidt A M. Diabetic vascular disease: it’s all the RAGE. Antioxid Redox Signal. 2005;7:1588–1600. doi: 10.1089/ars.2005.7.1588. [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat L E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Wain H M, Bruford E A, Lovering R C, Lush M J, Wright M W, Povey S. Guidelines for human gene nomenclature. Genomics. 2002;79:464–470. doi: 10.1006/geno.2002.6748. [DOI] [PubMed] [Google Scholar]
- Wendt T, Harja E, Bucciarelli L, Qu W, Lu Y, Rong L L, Jenkins D G, Stein G, Schmidt A M, Yan S F. RAGE modulates vascular inflammation and atherosclerosis in a murine model of type 2 diabetes. Atherosclerosis. 2006;185:70–77. doi: 10.1016/j.atherosclerosis.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Goova M T, Li J, Kislinger T, Qu W, Lu Y, Bucciarelli L G, Nowygrod S, Wolf B M, Caliste X, Yan S F, Stern D M, Schmidt A M. Blockade of receptor for advanced glycation end-products restores effective wound healing in diabetic mice. Am J Pathol. 2001;159:513–525. doi: 10.1016/S0002-9440(10)61723-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T M, Tanji N, Guo J, Kislinger T R, Qu W, Lu Y, Bucciarelli L G, Rong L L, Moser B, Markowitz G S, Stein G, Bierhaus A, Liliensiek B, Arnold B, Nawroth P P, Stern D M, D'Agati V D, Schmidt A M. RAGE drives the development of glomerulosclerosis and implicates podocyte activation in the pathogenesis of diabetic nephropathy. Am J Pathol. 2003;162:1123–1137. doi: 10.1016/S0002-9440(10)63909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone C, Emanuele E, D'Angelo A, Buzzi M P, Belvito C, Cuccia M, Geroldi D. Plasma levels of soluble receptor for advanced glycation end products and coronary artery disease in nondiabetic men. Arterioscler Thromb Vasc Biol. 2005;25:1032–1037. doi: 10.1161/01.ATV.0000160342.20342.00. [DOI] [PubMed] [Google Scholar]
- Katakami N, Matsuhisa M, Kaneto H, Matsuoka T A, Sakamoto K, Nakatani Y, Ohtoshi K, Hayaishi-Okano R, Kosugi K, Hori M, Yamasaki Y. Decreased endogenous secretory advanced glycation end product receptor in type 1 diabetic patients: its possible association with diabetic vascular complications. Diabetes Care. 2005;28:2716–2721. doi: 10.2337/diacare.28.11.2716. [DOI] [PubMed] [Google Scholar]
- Basta G, Sironi A M, Lazzerini G, Del Turco S, Buzzigoli E, Casolaro A, Natali A, Ferrannini E, Gastaldelli A. Circulating soluble receptor for advanced glycation end products is inversely associated with glycemic control and S100A12 protein. J Clin Endocrinol Metab. 2006;91:4628–4634. doi: 10.1210/jc.2005-2559. [DOI] [PubMed] [Google Scholar]
- Koyama H, Shoji T, Yokoyama H, Motoyama K, Mori K, Fukumoto S, Emoto M, Shoji T, Tamei H, Matsuki H, Sakurai S, Yamamoto Y, Yonekura H, Watanabe T, Yamamoto H, Nishizawa Y. Plasma level of endogenous secretory RAGE is associated with components of the metabolic syndrome and atherosclerosis. Arterioscler Thromb Vasc Biol. 2005;25:2587–2593. doi: 10.1161/01.ATV.0000190660.32863.cd. [DOI] [PubMed] [Google Scholar]
- Geroldi D, Falcone C, Emanuele E, D'Angelo A, Calcagnino M, Buzzi M P, Scioli G A, Fogari R. Decreased plasma levels of soluble receptor for advanced glycation end-products in patients with essential hypertension. J Hypertens. 2005;23:1725–1729. doi: 10.1097/01.hjh.0000177535.45785.64. [DOI] [PubMed] [Google Scholar]
- Emanuele E, D'Angelo A, Tomaino C, Binetti G, Ghidoni R, Politi P, Bernardi L, Maletta R, Bruni A C, Geroldi D. Circulating levels of soluble receptor for advanced glycation end products in Alzheimer disease and vascular dementia. Arch Neurol. 2005;62:1734–1736. doi: 10.1001/archneur.62.11.1734. [DOI] [PubMed] [Google Scholar]
- Galichet A, Weibel M, Heizmann C W. Calcium-regulated intramembrane proteolysis of the RAGE receptor. Biochem Biophys Res Commun. 2008;370:1–5. doi: 10.1016/j.bbrc.2008.02.163. [DOI] [PubMed] [Google Scholar]
- Raucci A, Cugusi S, Antonelli A, Barabino S M, Monti L, Bierhaus A, Reiss K, Saftig P, Bianchi M E. A soluble form of the receptor for advanced glycation endproducts (RAGE) is produced by proteolytic cleavage of the membrane-bound form by the sheddase a disintegrin and metalloprotease 10 (ADAM10) FASEB J. 2008;22:3716–3727. doi: 10.1096/fj.08-109033. [DOI] [PubMed] [Google Scholar]
- Brown L F, Fraser C G. Assay validation and biological variation of serum receptor for advanced glycation end-products. Ann Clin Biochem. 2008;45:518–519. doi: 10.1258/acb.2008.008043. [DOI] [PubMed] [Google Scholar]
- Hanford L E, Enghild J J, Valnickova Z, Petersen S V, Schaefer L M, Schaefer T M, Reinhart T A, Oury T D. Purification and characterization of mouse soluble receptor for advanced glycation end products (sRAGE) J Biol Chem. 2004;279:50019–50024. doi: 10.1074/jbc.M409782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizotte P P, Hanford L E, Enghild J J, Nozik-Grayck E, Giles B L, Oury T D. Developmental expression of the receptor for advanced glycation end-products (RAGE) and its response to hyperoxia in the neonatal rat lung. BMC Dev Biol. 2007;7:15. doi: 10.1186/1471-213X-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson B I, Kalea A Z, Arriero M D, Harja E, Boulanger E, D'Agati V, Schmidt A M. Interaction of the RAGE cytoplasmic domain with diaphanous-1 is required for ligand-stimulated cellular migration through activation of Rac1 and Cdc42. J Biol Chem. 2008;283:34457–34468. doi: 10.1074/jbc.M801465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen H J, Fages C, Rauvala H. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-κB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J Biol Chem. 1999;274:19919–19924. doi: 10.1074/jbc.274.28.19919. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T, Yan S F, Yan S D, Belov D, Rong L L, Sousa M, Andrassy M, Marso S P, Duda S, Arnold B, Liliensiek B, Nawroth P P, Stern D M, Schmidt A M, Naka Y. Central role of RAGE-dependent neointimal expansion in arterial restenosis. J Clin Invest. 2003;111:959–972. doi: 10.1172/JCI17115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.