Abstract
Sexual dimorphisms are typically attributed to the hormonal differences arising once sex differentiation has occurred. However, in some sexually dimorphic diseases that differ in frequency but not severity, the differences cannot be logically connected to the sex hormones. Therefore, we asked whether any aspect of sexual dimorphism could be attributed to chromosomal rather than hormonal differences. Cells taken from mice at d 10.5 postconception (PC) before sexual differentiation, at d 17.5 PC after the first embryonic assertion of sexual hormones, and at postnatal day 17 (puberty) were cultured and exposed to 400 μM ethanol or 20 μM camptothecin or to infection with influenza A virus (multiplicity of infection of 5). The results showed that untreated male and female cells of the same age grew at similar rates and manifested similar morphology. However, they responded differently to the applied stressors, even before the production of fetal sex hormones. Furthermore, microarray and qPCR analyses of the whole 10.5 PC embryos also revealed differences in gene expression between male and female tissues. Likewise, the exposure of cells isolated from fetuses and adolescent mice to the stressors and/or sex hormones yielded expression patterns that reflected chromosomal sex, with ethanol feminizing male cells and masculinizing female cells. We conclude that cells differ innately according to sex irrespective of their history of exposure to sex hormones. These differences may have consequences in the course of sexually dimorphic diseases and their therapy.—Penaloza, C., Estevez, B., Orlanski, S., Sikorska, M., Walker, R., Smith, C., Smith, B., Lockshin R. A., Zakeri, Z. Sex of the cell dictates its response: differential gene expression and sensitivity to cell death inducing stress in male and female cells.
Keywords: apoptosis, estrogen, ethanol, gender, testosterone
Most animals manifest sex-related differences in nonsexual tissues and cells. In invertebrates they are generated in the absence of sex hormones and must be derived from chromosomal differences, whereas in vertebrates the differences are usually attributed to the effects of embryonic and postadolescent hormones. While many of the differences, particularly the most spectacular pubertal and postpubertal ones, are clearly dependent on hormones, many are less certain. For instance, hormonal ablation or supplementation does not completely eliminate or recreate sexual differences (1). Furthermore, sexually dimorphic diseases such as neurodegenerative, autoimmune, and cardiovascular diseases that differ in frequency, but often not severity, cannot be readily explained by the hypothesis that hormones alone create these differences (2,3,4,5,6,7,8,9). Reevaluation of this hypothesis is particularly germane in view of the use of hormonal supplementation in clinical practice (6, 10, 11).
Several researchers (12) have reported sexual dimorphism at the cell level. In vitro, male hippocampal neurons are more sensitive to and more readily killed by peroxynitrite than female neurons. Male primary rat hippocampal neurons survive better under normal (95%) oxygenation but are more sensitive to hypoxia than their female counterparts (13). We (14) have also shown that the amount of cell death can differ between the sexes in isolated embryonic cells exposed to similar conditions. However, the mechanisms responsible for these differences, other than hormonal influences, have not been explored. In particular, since prior exposure to hormones triggers a response that generates a positive feedback and locks a cell into a specific pathway, one must ask how male and female cells would respond if they had never been exposed to their own sex hormones. This question has not been previously asked.
Dimorphic gene expression has been studied in several situations. Amador-Noguez et al. (15) found a large subset of sex-dimorphic genes in adult liver of male and female Ames dwarf mice examined by gene array (15). In this study, when Ames dwarf mice mutated for deficiency in growth hormone, prolactin, and thyroid-stimulating hormone (prop1df/df), 110 originally sex-dimorphic genes changed their levels of expression in a sex-dependent manner, when compared with the parental strain (15). Although the focus of that investigation was not to study sex-dimorphic gene expression, and therefore the authors did not discuss their findings in that context, the study provides evidence for the existence of the dimorphic gene expression. Other studies (16, 17) have looked at differences in gene expression in embryos. Thus, Xu et al. (17) used the brains of embryos at embryonic day 13.5 (ED13.5) and adult mice and found general differences in Y-linked genes and X-chromosome homologs as in the case of Usp9y, Usp9x, Uty, and Utx. However they looked only at brain tissue, limiting their study to X- and Y-chromosomal genes after the initiation of sex differentiation. In another study, Dewing et al. (16) using the brain of ED10.5 embryos (before the onset of sex hormone production) found dimorphic expression of several genes in the absence of stress. They found an expression of autosomal Cyp7b1, a member of the cytochrome P-450 family of drug-metabolizing enzymes, and nuclear orphan receptor ROR-α4 (Rorα4) higher in males and other genes higher in females, as in the case of the X-chromosome gene Eif2s3x (16). Overall, they described 51 sex-dimorphic genes (16).
Several other genes display a sex-dimorphic expression under stress (18,19,20,21). Cyp1A1, a member of the cytochrome P-450 family responsible for metabolism of toxins, is more responsive to tobacco carcinogens in female human lung cells than in male cells (21). In response to electroshock, the female pituitary gland, mesenteric lymph nodes, and liver have an attenuated response of heat shock protein 72 (HSP72) compared with male organs (22). Active p38 mitogen-activated protein kinase is also increased in male rats compared with female rats in the presence of trauma-hemorrhage in splenic and peritoneal macrophages (23).
Thus, there is reason to hypothesize that mammalian cells can differ intrinsically by sex irrespective of past or current concentrations of sex hormones. Therefore, we tested the hypothesis that the chromosomal differences can by themselves generate differences in cell behavior. We found that male and female cells, independent of hormonal influences, respond differently to stressors such as ethanol, camptothecin, or influenza A virus infection. There were differences in basal gene expression between male and female embryos at stages where there were no hormonal influences. We conclude that male and female cells have intrinsic differences that are not dependent on hormones, which arise before full differentiation, allowing hormonal responsiveness. However, addition of exogenous sex hormones can alter the response of the cells to stress and lethal stress.
MATERIALS AND METHODS
Animals, cell isolation, culturing, and treatment
Female Swiss Webster mice were placed with males overnight for 12–16 h for mating, and detection of vaginal plugs was designated ED0.5 of gestation. Pregnant females were killed by CO2 and cervical dislocation at ED10.5 and ED17.5, and adolescent mice were likewise killed at postnatal days 4 and 17 (PN4 and PN17). Dissected embryos and/or tissues were placed in chilled sterile 1× PBS. Each embryo was removed from its embryonic sac and placed in chilled DMEM. For sexing ED10.5 embryos, a small piece of tail was placed in a sterile PCR tube and stored at −20°C for sexing by PCR as described below.
To isolate cells from whole embryos or embryonic and postnatal kidney, the tissues were sandwiched between sterile mesh (Sefar Nitex 03–150/38; Sefar Filtration, Inc, Depew, NY, USA). The cells were released by applied pressure to the sandwich, leaving the connective tissues trapped between the mesh. Suspended cells were collected by centrifugation at 3000 rpm and resuspended in media containing DMEM supplemented with 10% FBS, 1% 100 U/ml penicillin, and 100 μg/ml streptomycin; divided into several 35 mm plates; and incubated at 37°C in a humidified atmosphere with 5% CO2. Cells were cultured for ∼5 d or to 70% confluence before treatment. The medium was changed every 3 d. Before treatment with the stressors, the cells were seeded at 5 × 105 cells/plate for 24 h. The cultures represent mixed cell cultures of whole embryo or kidney tissues without further enrichment.
To assess cell response, cultured cells were exposed to 2% ethanol in 5% FBS. FBS was lowered to 5% to slow cell division even though lower FBS increases cell death. Amounts of cell death were therefore always directly compared with controls. Treated cells were placed at 37°C for 24 h in the presence of ethanol, before the assessment of cell death. Control samples contained the solvent [DMSO in the case of camptothecin (CPT) and virus-diluting medium for viruses] added to reduced serum (5% FBS) culture medium.
Cell viability by trypan blue exclusion
Dying cells, whether necrotic or apoptotic, become permeable, allowing dyes such as trypan blue to enter the cell (24). Since our results with trypan blue were completely consistent with other assays, such as Live/Dead and Hoechst 33258 (Hoechst, Frankfurt, Germany) staining (data not shown), we relied primarily on this simple and reliable assay. At the end of treatment, cells were scraped, and the medium containing attached and floating cells was pelleted; resuspended in an equal volume of 0.4% trypan blue (Sigma, St. Louis, MO, USA) in 1× PBS, pH 7.4; and incubated at room temperature for 3 min. At least 200 cells (dead or alive) were counted under a microscope, and each treatment was done at least three times on three different samples. Blue cells were scored as nonviable.
The percentage of cell death was calculated as 100 × the number of dead cells, divided by the total cell number. For each experiment, this number for the experimental sample was normalized by subtracting the basal level of cell death observed in the control (9–20%) for each treatment. Statistical significance of the results was calculated by standard t test; values of P < 0.05 were considered significant.
Staining of apoptotic cells with bis-benzimide
The nuclear fragmentation of apoptotic cells was assessed by staining the treated cells with the DNA-binding fluorochrome bis-benzimide (Hoechst 33258). Cells were scraped, pelleted, and washed with 1× PBS and resuspended in 3% paraformaldehyde in 1× PBS, fixed for 15 min, washed once in 1× PBS, and incubated with 16 μg/ml bis-benzimide in PBS at room temperature for 25 min. The microliters of cell suspension in bis-benzimide were placed on a slide, and cells were examined using a Leitz fluorescence microscope. Cells with condensed chromatin or fragmented nuclei were considered to be apoptotic. At least 200 cells were counted for each sample of three or more independent experiments.
DNA fragmentation assay by TUNEL
DNA fragmentation was detected by using TUNEL peroxidase (TUNEL POD) combined with nonisotopic digoxigenin-11-dUTP and terminal transferase according to our previously published method (25) and the manufacturer’s instructions (Roche Molecular Biochemicals, Mannheim, Germany). Briefly, slides were incubated with permeabilization solution (0.1% Triton X-100 in 0.1% sodium citrate) on ice for 2 min, and endogenous peroxidase activity was quenched with 0.3% hydrogen peroxide in methanol at room temperature for 30 min. After two rinses in PBS, the TUNEL reaction mixture (9 vol of TUNEL label and 1 vol of TUNEL enzyme) was applied to slides, which were incubated for 30 min at 37°C, followed by 3 washes with PBS. TUNEL POD was then applied to the slides to bind to the FITC-dUTP enzymatically added to the free end of the oligonucleotide and visualized with diaminobenzidine (Research Genetics, Huntsville, AL, USA). The slides were counterstained with methylene blue and mounted with Permount (Fisher Scientific, Burr Ridge, IL, USA).
Sex determination
For sexing ED10.5 embryos, which are anatomically undifferentiated, PCR was necessary. A small piece of tail was placed in a sterile PCR tube. DNA was isolated by digestion with 2 μl proteinase K (10 mg/ml in dH2O) and 50 μl PCR-D buffer and left overnight at 65°C. The enzyme was denatured at 95°C for 10 min, and 1 μl was transferred into a PCR tube to which was added 21 μl dH2O, 25 μl Master Mix (Sigma), 2 μl MgCl2, and 1 μl Primer Mix (25 pmol/μl of Zfy and Zfxya; 12.5 pmol/μl of Zfx primers). Primers for Zfy were 5′-CTCCTGATGGACAAACTTTACGTCTC and 3′-GCTGAGCCTCTTTGGTATCTGAGAAA; primers for Zfxya were 5′-GAGAGCATGGAGGGCCATG and 3′-GAGTACAGGTGTGCAGCTC; and Zfx primers were 5′-CTCTGAAGAAGAGACAAGTT and 3′-CTGTGTAGGATCTTCAATC. PCR was done for 40 total cycles (1 cycle=94°C for 45 s, 60°C for 25 s, and 72°C for 60 s) in the Eppendorf 2200 thermal cycler (Eppendorf North America, Westbury, NY, USA). The results of the DNA amplification were visualized on a 12% native polyacrylamide gel in tris-borate-EDTA buffer using positive controls (preamplified male and female samples) alongside and viewed by ultraviolet illumination. Male samples showed two bands, a 124-bp band for the Zfy gene, and a 134-bp band for the gene Zfx and females showed a single band at 134 bp for the Zfx gene (Fig. 1A).
Figure 1.
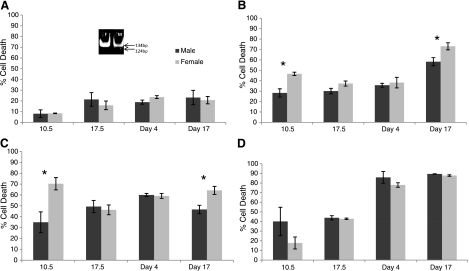
Male and female cells show differences in level of cell death induced by different insults. Cell death measured by trypan blue exclusion. Dark bars, male; light bars, females; 10.5, cells from whole embryos; 17.5, cells from ED17.5 kidney; day 4, kidney cells from PN4 mice; day 17, kidney cells from PN17 mice. A) Amount of cell death in control primary mixed cell cultures (controls) from ED10.5 whole embryos, ED17.5 kidney, and PN4 and PN17 kidney; male and female cells exposed to culture medium (DMEM supplemented with 10% FBS and 1% penicillin/streptomycin) only. Cells show no sex differences in basal level of cell death. Inset: native polyacrylamide gel with PCR amplification (see Materials and Methods) of Zfy and Zfx, indicative of X- and Y-chromosome-specific genes. Males (right) possess both bands; females (left) have only a single band. B) Cultured cells as in A exposed to 400 μM ethanol. Higher female cell sensitivity to ethanol is seen in cells from ED10.5 whole-embryo, ED17.5 kidney, and PN17 kidney mixed cell cultures, but not in mixed cells cultures from PN4 kidney. C) Cultured cells as in A exposed to 20 μM CPT show female-biased sensitivity at ED10.5 and PN17, but no differences are seen for ED17.5 and PN4 kidney mixed cell cultures. D) Cultured cells as in A infected with influenza A with a multiplicity of infection of 5 show no sex differences in amount of cell death sex, but older cells are more sensitive. Difference at ED10.5 for influenza A infection was not significant by Student’s t test in triplicate experiments. Data represent percentage total cell death for all conditions.
Sexing of other stages, i.e., ED17.5 and PN4 and PN17 mice was done by visualization of sex organs by dissection.
Microarray analysis
Total RNA was isolated using TRI reagent (Molecular Research Center, Cincinnati, OH, USA) and treated with DNase I (Qiagen, Valencia, CA, USA). For the pooled microarray experiments, pools of total RNA from either male or female embryo were arrayed against a control containing equal amounts of total RNA from the male and female pools. The individual embryo arrays were hybridized as male/female pairs. The RNA samples were reverse transcribed, and the cDNA was labeled with either Cy3 or Cy5 using the amino-allyl method adapted from the University Health Network Microarray Centre (Toronto, ON, Canada; http://www.microarrays. ca). Labeled cDNA was hybridized to mouse M7.4k1 and M15k4 array slides (University Health Network Microarray Centre,) at 37°C for 18 h. The slides were washed and then scanned using a ScanArray 5000 confocal scanner (Packard Bioscience, Waltham, MA, USA). Image files were quantitated using QuantArray v3 (Packard Bioscience).
Microarray data processing and analysis
Raw intensity data from QuantArray were subject to preprocessing in Microsoft Excel (Microsoft, Redmond, WA, USA) using an in-house Excel add-in written for the purpose. Background subtracted spot intensities were logged (base 2), and suspect data were filtered out before normalization. The following criteria were used for filtering: negative intensity values, signal/background <1.5, and intensity values in the lowest fifth percentile for either channel. In addition, spots identified manually as poor quality during quantitation, buffer controls, and positive controls were removed. The remaining data were normalized using methods that correct for intensity-dependent bias using a trimmed running average method and for spatial biases using a Euclidean distance-weighted Gaussian function based on the work of Workman et al. (27) and Colantuoni et al. (26). The logged (base 2) ratio of sample to control signals was then determined for each spot, and spot duplicates were averaged. The data were analyzed in Excel to select for spots that differed between sexes by >1.33-fold with P < 0.05 (using either paired or two-sample t test as appropriate). Annotation for spots of interest, identified by their clone ID, was obtained from the annotation database SOURCE (http://source.stanford.edu; ref. 28).
The Virtual Reality (VR) module of BioMiner, a data mining software package designed and built in-house, was used to generate three-dimensional virtual reality representations of the female vs. male microarrays. The VR module implements a simplified version of DIG (Hybrid Strategies Corp., Ottawa, ON, Canada). It is used for visualizing complex, high-dimensional data by mapping the data to three dimensions with minimal loss of the underlying structure of the original data. Subsets of the normalized intensity data from the female vs. male embryo microarrays were iterated in the VR algorithm until minimum absolute error was achieved. Technical details of the algorithm and its application to gene expression data have been published previously (29).
Quantitative (q)RT-PCR
Small PCR products (100–200 bp) were amplified in quadruple on a Roche LightCycler 2.0 real-time PCR machine, using universal PCR conditions [65°C to 59°C touchdown, followed by 40 cycles (15 min at 95°C, 10 min at 59°C, and 10 min at 72°C)]. cDNA (500 pg) was amplified in 20 μl reactions (0.3× Sybr-green, 3 mM MgCl2, 200 μM dNTPs, 200 μM primers, and 0.5 U Platinum TaqDNA polymerase; Roche). Primer dimers were assessed by amplifying primers without cDNA. Primers were retained if they produced no primer-dimers or nonspecific signal only after 40 cycles. Results were calculated as relative intensity compared with female expression. The last cycle was retained as baseline for comparison with “absent” genes. Data were plotted as cycle threshold (CT) values in which the CT value represents the cycle at which fluorescence is first detected. By this representation, a lower cycle number indicates a higher initial concentration of mRNA, and each decrease of one cycle indicates a doubling of initial concentration.
RESULTS
Developmental timing that reflects influence of endogenous hormones affects sex-dimorphic behavior of the cells
Individual mice from the same litter were genetically almost identical except for the sex chromosomes; hence, any dimorphism in cell behavior might be attributed to its sex. To study the effects of the sex on cell behavior, we developed an in vitro system. We used cells cultured from whole mouse embryos at ED10.5 and primary mixed kidney fibroblast cells derived from male and female mouse fetuses at ED17.5 and cells from mice at PN4 and PN17, which were maintained in culture for 4–14 d as described in Materials and Methods. At ED10.5–11.5, the gonads have not yet differentiated and the embryo has been exposed only to maternal hormones; at ED17.5, the fetuses have been exposed to the fetal hormones that led to their secondary sexual differentiation, while PN4 and PN17 mice exhibit prepubertal to adolescent levels of sex hormones.
For sex determination, the ED10.5 embryos were placed individually in a culture dish, and a piece of tail tissue was taken for PCR analyses using primers for Zfy, Zfxya, and Zfx (see Materials and Methods). Male embryos were identified by the appearance of two bands (124 and 134 bp) on a polyacrylamide gel, whereas female embryos contained only the 124-bp band (Fig. 1A, inset).
We found no significant differences between the sexes in cell morphology, overall size, number of divisions, or viability in the culture of cells obtained from ED10.5 embryos, ED17.5 fetuses, or PN4 or PN17 mice. However, there was an age-dependent difference in the rate of cell division between cells derived from the postnatal and embryonic tissues, as the latter divided faster (data not shown). The basal level of cell death also increased with the age of the donor, i.e., from ∼9% in ED10.5 to ∼17% in ED17.5 and ∼20% in PN17 cultures (Fig. 1A).
To assess the effect of development and endogenous hormones on cell response to different stressors (ethanol, camptothecin, and influenza virus infection), we used cells from ED10.5 whole-embryo and mixed kidney cells of ED17.5, PN4, and PN17 mice and measured the rate of cell death as an indicator of their sensitivity. Cells were seeded in equal numbers (105 cells/ml) and exposed to either 400 μM ethanol and 20 μM CPT (used as the toxin) for 24 h or influenza A virus infection with a multiplicity of infection of 5 for 48 h. The concentration and exposure time were selected based on for the kinetics of death and targeted ∼50% death in the more resistant sex. Cell death was evaluated by trypan blue dye exclusion (Fig. 1B). By this measure of cell death, cells from ED10.5 embryos showed the largest sexual dimorphism in response to both ethanol and CPT. Ethanol killed 47% of female (F) cells but only 28% of male (M) cells for a ratio F:M = 1.7:1 (Fig. 1B). Although the total rate of cell death was lower, the dimorphism was similar in mixed kidney cells of ED17.5, where 37% of female cells were killed by ethanol, compared with 30% of male cells (F:M=1.2:1; Fig. 1B). These differences were eliminated in the PN4 postnatal cultures (Fig. 1B) but reappeared at PN17. When cells from PN17 kidneys were examined, 73% of female cells but only 58% of male cells were killed by ethanol (F:M=1.3:1; Fig. 1B). These percentages were calculated after normalization against control levels of death and considered significant by Student’s t test values of P < 0.05.
In the case of CPT, cells from ED10.5 whole embryos maintained the highest level of dimorphism, reaching 70% death in female cells but only 35% death in male cells (F:M=2:1; Fig. 1C). ED17.5 kidney mixed cell cultures and PN4 cultures showed no dimorphism (Fig. 1C). The dimorphic response was recovered in PN17 mixed cell cultures in which 64% of the female cells, but only 47% of the male cells, died (F:M=1.4:1; Fig. 1C).
No dimorphic response was seen at any age in response to influenza A virus, although high levels of death were achieved (Fig. 1B–D). The apparent elevated level of cell death in 10.5 embryonic males cells subjected to virus in contrast with the female (Fig. 1D) did not achieve statistical significance in six independent experiments.
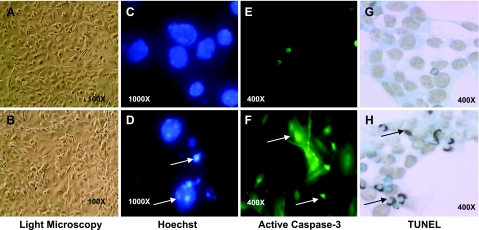
To confirm the validity of the trypan blue assay, we used other methods and also evaluated the type of cell death. We found, by general morphology, assay of nuclear condensation by Hoechst analysis, and estimation of DNA fragmentation using TUNEL, a level of cell death similar to that obtained by measurement of trypan blue exclusion (Fig. 2). Furthermore, caspase-3 activation assays, the hallmark of apoptosis, also indicated activation of caspase-3 by the three stressors, thus confirming apoptotic cell death.
Figure 2.
Cells from male and female ED10.5 whole embryos behave similarly in culture and die by apoptosis. Cells from experiments illustrated in Fig. 1 were used for this analysis as described in Material and Methods. A, B) Light microscopic images of male (A) and female (B) cells of ED10.5 whole embryo before stress possess similar morphology (shape and size) and rate of growth. C–H) Images are from female cells only, as male cells showed identical patterns under the same conditions (data not shown). C, D) Cells labeled with Hoechst stain for DNA condensation and fragmentation; control cells (C) show no chromatin condensation, indicative of apoptosis, before stress, but condensation (arrows) was seen in both female (D) and male cells exposed to 400 μM ethanol. Similar results were seen when cells were exposed to CPT or influenza virus (not shown). E, F) Cells labeled with anti-active caspase-3 antibody, indicative of apoptotic cell death; no caspase-3 activation is seen under control conditions (E) but caspase-3 is activated (arrows) in ethanol-treated cells (F). G, H) Similarly, cells labeled with TUNEL, indicative of DNA fragmentation. No DNA fragmentation is seen under control conditions (G), while DNA fragmentation (arrows) is present in ethanol treated cells (H). Original magnifications are indicated.
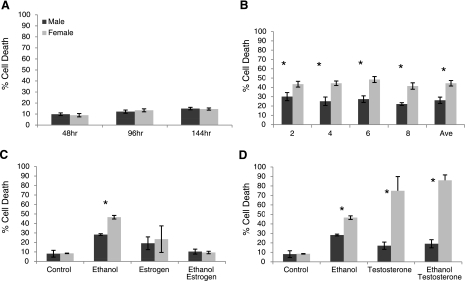
Since primary cell cultures may self-select over time, we analyzed cell behavior at different passages. Regardless of the number of passages, the basal level of cell death and ethanol-induced cell death remained the same, indicating that the effect is not dependent on the passage number (Fig. 3A, B). A similar pattern was also found with other stressors, such as CPT and influenza (data not shown).
Figure 3.
All cells are derived from ED10.5 whole embryos. Cell death is measured by trypan blue exclusion. Dark bars, male; light bars, females. A) ED10.5 mixed cells were maintained in culture medium for 48, 96, and 144 h. There were no differences between male and female control cells at basal level, although cell death increased with time of culture. B) ED10.5 whole-embryo cell cultures were passed and exposed after passages 2, 4, 6, and 8 to 400 μM ethanol and then averages of 3 experiments were plotted. Female sensitivity bias was maintained beyond the first few passages, and rates of death did not differ significantly from passage to passage. C) Influence of estradiol was analyzed by coexposing cells to the stressor ethanol and estradiol. ED10.5 mixed cell cultures exposed to control conditions (control), 400 μM ethanol (ethanol), 5 nM estradiol (estrogen), or both (ethanol estrogen). Female cells maintained the higher sensitivity to ethanol-induced cell death when compared with males, and estrogen did not affect cell death in either sex; however, estrogen completely protected cells of both sexes against ethanol (ethanol estrogen). D) ED10.5 mixed cell cultures were exposed to control culture conditions (control), 400 μM ethanol (ethanol), 5 nM testosterone (testosterone), or both (ethanol testosterone). Female cells maintained higher levels of death when exposed to ethanol alone. Testosterone alone had no effect on male cultures but was toxic to female cells. Testosterone was slightly protective against ethanol for male cells but not for female cells (ethanol testosterone). All treatments were done in triplicate. *P < 0.05; Student’s t test. Data represent percentage total cell death for all conditions.
Thus the data indicate that sex-differential cell behavior is dependent on the sex of the cell at the first level and that hormones during development do not fundamentally alter this difference. This suggests that the difference in cell behavior and sensitivity appears to be driven by the genetic sex of the cells to start with and that the effects of other factors such as hormones are superimposed on this difference. We therefore evaluated the influence of sex hormones on these cells.
Exogenous estrogen protects male and female cells against ethanol, while testosterone is toxic to female cells
Our data suggest that the exposure of the fetus to its own hormones is unlikely to dictate the dimorphic response found in ED10.5 cells. To more specifically examine the effect of hormones on the differential response of the cells, ED10.5 whole-embryo cell cultures were exposed to ethanol in the presence and absence of 5 nM 17β-estradiol. Estradiol alone is not toxic, and cell death remained at control level. More intriguing, estradiol protects cells from ethanol-induced cell death. In these situations, cell death is reduced by 18% in males and 37% in females in comparison to ethanol alone (Fig. 3C).
Testosterone on its own appears to exert a slight protective effect on male ED10.5 whole-embryo mixed cells exposed to ethanol (∼28% death reduced to ∼19%; Fig. 3D). In contrast, testosterone was toxic to female cells (∼9% cell death increased to ∼75%). This toxicity was not affected by further addition of ethanol (Fig. 3D).
Sex-dimorphic pattern of gene expression before the onset of gonadal hormone production
We performed microarray analysis of gene expression using equal amounts of RNA obtained from 6 individual male and female ED10.5 whole embryos, each representing an n = 1, which were hybridized as male/female pairs. Experiments were performed in duplicate with dye reversal on the UHN M15k and M7.4k microarrays (see Materials and Methods). These are double-spotted arrays containing 15,264 and 7407 nonredundant sequence-verified mouse expressed sequence tags. The clones originate from the National Institute of Aging (Bethesda, MD, USA).
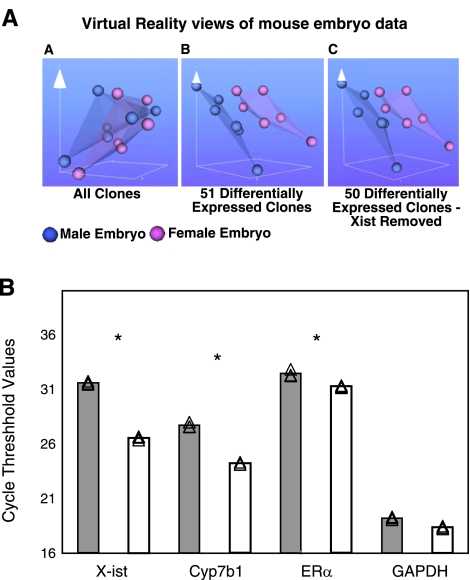
Data processing yielded 51 differentially expressed clones that were found on the M7.4k and M15k arrays (P<0.05; ratio>1.33-fold; see Table 1 for list). These sets of data were then iterated using the VR module of a data mining software package (BioMiner) designed and built in-house (National Research Council). The VR method was developed to visualize complex, high-dimensional data by mapping it to three dimensions with minimal loss of the underlying structure of the original data (29). This method allowed generation of 3-D VR representations of the female vs. male microarrays. The VR plots of intensity data of all clones showed that the separation of the samples was still maintained even after the exclusion of the most differentially expressed Xist gene (absolute error 0.027, 862 iterations), indicating that the separation of these data sets was based not solely on the expression of a single gene but resulted from the cumulative differences in the expression of the remaining 50 genes (Fig. 4A). The differential expression of Xist confirmed the validity of our gene array experiment, as this gene has been reported to be expressed more abundantly in females compared with males (see Table 1). Our results, in general, confirm findings of others, as indicated in Table 2. Some of the differences from others may be explained by the different sources of tissues [we used entire ED10.5 embryos, whereas Dewing et al. (16) used only the head region]. While the gene array differences appear low, they represent underestimates, which were amplified significantly when verified via qRT-PCR.
TABLE 1.
Differentially expressed clones in male/female paired microarrays
| Clone | Accession (3′) | Accession (5′) | Unigene | Gene symbol | Gene name | Fold change (F/M) |
|---|---|---|---|---|---|---|
| H4069B04 | BQ561239 | BQ561240 | Mm.274770 | Xist | Inactive X specific transcripts | 2.09 |
| H4007E02 | BQ551125 | BQ551126 | Mm.446163 | Vps4b | Vacuolar protein sorting 4b (yeast) | 1.56 |
| H3159E02 | BG076407 | BG088932 | Mm.25300 | Marveld2 | MARVEL (membrane-associating) domain containing 2 | 1.48 |
| H4001F10 | BQ550149 | BQ550150 | — | — | — | 1.48 |
| H3043D10 | BG066476 | — | Mm.394036 | C79296 | Expressed sequence C79296 | 1.47 |
| H4075C11 | BQ562320 | BQ562321 | Mm.270307 | Ncf2 | — | 1.43 |
| H4071H08 | BQ561704 | BQ561705 | Mm.31192 | Pomt1 | Protein-O-mannosyltransferase 1 | 1.42 |
| H4078D02 | BQ562852 | BQ562853 | — | — | — | 1.41 |
| H4078E04 | BQ562878 | BQ562879 | — | — | — | 1.39 |
| H4015A10 | BQ552323 | BQ552324 | Mm.293754 | Tmem65 | Transmembrane protein 65 | 1.38 |
| H4072A10 | BQ561741 | BQ561742 | Mm.391339 | ORF34 | Open reading frame 34 | 1.38 |
| H3116B04 | BG072884 | BG085668 | Mm.250599 | Pycrl | Pyrroline-5-carboxylate reductase-like | 1.38 |
| H4072G07 | BQ561856 | BQ561857 | Mm.442331 | — | — | 1.38 |
| H4021A10 | BQ553279 | BQ553280 | Mm.392098 | Atp6v0a2 | ATPase, H+ transporting, lysosomal V0 subunit A2 | 1.37 |
| H3048H12 | BG066958 | — | — | — | — | 1.37 |
| H3114G09 | — | — | — | — | — | 1.37 |
| H3071F12 | BG069006 | BG082030 | Mm.375388 | — | — | 1.37 |
| H4075D07 | BQ562335 | BQ562336 | Mm.234422 | Zfp719 | Zinc finger protein 719 | 1.36 |
| H3149C09 | BG075607 | BG088151 | Mm.271980 | Gprasp1 | G protein-coupled receptor associated sorting protein 1 | 1.36 |
| H3069A12 | BG068765 | BG081790 | — | — | — | 1.36 |
| H4063A08 | BQ560226 | BQ560227 | Mm.334193 | Mier2 | Mesoderm induction early response 1, family member 2 | 1.35 |
| H3155H12 | BG076126 | BG088653 | Mm.28189 | Cdc42se1 | CDC42 small effector 1 | 1.35 |
| H3047C10 | BG066820 | BG079985 | Mm.395650 | Slc6a6 | Solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 1.34 |
| H4053G03 | BQ558692 | BQ558693 | Mm.465307 | C330019G07Rik | RIKEN cDNA C330019G07 gene | 0.75 |
| H3060B09 | BG067961 | BG081019 | Mm.291936 | Map4k5 | Mitogen-activated protein kinase kinase kinase kinase 5 | 0.75 |
| H4003F04 | BQ550483 | BQ550484 | Mm.254997 | Zfp386 | Zinc finger protein 386 (Kruppel-like) | 0.74 |
| H4027E05 | BQ554333 | BQ554334 | Mm.233547 | Col15a1 | Procollagen, type XV | 0.74 |
| H3035G09 | BG065836 | BG079104 | Mm.403562 | — | — | 0.74 |
| H3013C07 | BG063914 | BG077266 | Mm.437275 | — | — | 0.74 |
| H3025C03 | BG064912 | — | Mm.24593 | 4833439L19Rik | RIKEN cDNA 4833439L19 gene | 0.74 |
| H3089G11 | BG070667 | BG083496 | — | — | — | 0.74 |
| H3102H10 | BG071763 | BG084608 | Mm.381359 | — | — | 0.74 |
| H4036C08 | BQ555807 | BQ555808 | Mm.221499 | Disp2 | Dispatched homolog 2 (Drosophila) | 0.73 |
| H3115D05 | BG072823 | — | Mm.332336 | Mrfap1 | Morf4 family associated protein 1 | 0.73 |
| H3148A04 | BG075500 | BG088046 | Mm.450686 | — | — | 0.72 |
| H3159A09 | BG076375 | BG088899 | Mm.216217 | 2900011O08Rik | RIKEN cDNA 2900011O08 gene | 0.72 |
| H3047G02 | BG066854 | BG080009 | Mm.284592 | Senp3 | SUMO/sentrin specific peptidase 3 | 0.72 |
| H3002E07 | BG062964 | BG076437 | Mm.41933 | Pitrm1 | Pitrilysin metallepetidase 1 | 0.71 |
| H3044C01 | — | CK334516 | Mm.334706 | LOC382044 | Liver carboxylesterase N-like | 0.71 |
| H4031A11 | BQ554920 | BQ554921 | Mm.398543 | Cugbp2 | CUG triplet repeat, RNA binding protein 2 | 0.71 |
| H3083F03 | BG070008 | BG083022 | Mm.259688 | Tmem87b | Transmembrane protein 87B | 0.70 |
| H3063F11 | BG068277 | BG081312 | — | — | — | 0.68 |
| H3094C02 | BG071056 | BG084050 | Mm.13433 | Dnmt3l | DNA (cytosine-5-)-methyltransferase 3-like | 0.68 |
| H4060A01 | BQ559726 | BQ559727 | Mm.28124 | Gfer | Growth factor, erv1 (S. cerevisiae)-like (augmenter of liver regeneration) | 0.68 |
| H4011C10 | — | BQ551744 | — | — | — | 0.67 |
| H4012H12 | BQ552024 | BQ552025 | Mm.309520 | Nploc4 | Nuclear protein localization 4 homolog (S. cerevisiae) | 0.66 |
| H3151D06 | — | — | — | — | — | 0.66 |
| H4071F08 | — | BQ561661 | Mm.277217 | Dgkd | Diacylglycerol kinase, delta | 0.65 |
| H3151B08 | BG075745 | BG088288 | Mm.410587 | — | — | 0.64 |
| H4071H10 | BQ561708 | BQ561709 | — | — | — | 0.63 |
| H4079B12 | BQ562999 | BQ563000 | Mm.451145 | — | — | 0.52 |
Table shows the clones and accession numbers as well as fold difference [female/male (F/M)] for genes found to be sex dimorphic by our gene arrays. Calculation of fold differences is a safe underestimate, as validation of select genes by RT-PCR documented significantly higher fold differences than indicated by the arrays.
Figure 4.
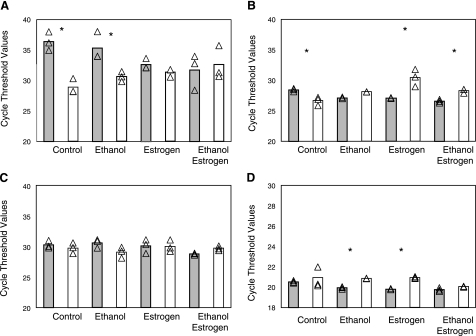
Sex differences in gene expression in unstressed ED10.5 whole embryos. A) Virtual reality views of paired male/female embryo microarray intensity data created using the BioMiner VT module. Each sphere represents intensity data from one channel of paired male/female microarray. Blue spheres represent male data, pink represent female data. VR representation of all clones (AA), 51 differentially expressed clones (AB), and the 50 differentially expressed clones with the clone for X-ist removed (AC). B) qRT-PCR verification of sample genes from the microarray. Source of cDNA was whole male or female embryo homogenate at ED10.5. Samples were normalized at the RNA and cDNA levels for equal cDNA loading. Gray bars, male; white bars, female. Experiments were done in triplicate. Individual CT points are shown together with mean values. *P < 0.05; Student’s t test. Ordinate is average CT of fluorescence detection for real-time PCR. Each increment in CT indicates a starting transcript concentration ∼0.5× that of CT-1. X-ist was expressed significantly more in female embryos, while ERα and Cyp7b1 were higher, and GAPDH was not sexually dimorphic.
TABLE 2.
Comparison of qPCR with published data
| Gene | Fold change (F/M)
|
||
|---|---|---|---|
| qPCRa | Dewing et al.(16) | Xu et al.(17)b | |
| Cyp7b1 | 1.49 | 0.53 | Not performed |
| Eif2s3x | 1.62 | 1.90 | ∼2 |
| Jarid1c (Smcx) | 4.29 | Not performed | ∼2 |
| Utx | 4.42 | Not performed | ∼2 |
| Uty | Male only | Not performed | Male only |
Table represents a comparison of genes present in our arrays together with data obtained by others. Not all the genes were assayed by other groups and conflicts may derive from differences in choice of tissue or other differences. See Discussion.
Normalized to GAPDH.
Northern blot.
Subsequently, we selected a few genes for further verification by qRT-PCR using RNA from the same ED10.5 tissues as in the gene arrays. These were X-ist and Cyp7b1, the expression of which was higher in female than in male embryos (the difference in microarrays was >2-fold), estrogen receptor-α (ERα; the female level was equivalent to 150% of the male level), and the housekeeping gene GAPDH. Thus, the ED10.5 female embryos had 35.5-fold higher transcript level of X-ist than males. There was a 10.9-fold higher level of Cyp7b1 and a 2-fold higher level of ERα, whereas there was no difference between the sexes in the GAPDH transcript (Fig. 4B). The X-ist gene is known to be expressed in a sex dimorphic manner, with females having higher transcript levels than males (30); however, the published study (16) indicates that Cyp7b1 is higher in male brain tissues than in females. The reason for this difference is presently not known.
Stress affects the expression of sex-dimorphic genes
We also asked whether stress influences the expression of the sex-dimorphic genes. We cultured cells from whole ED10.5 embryos and exposed them to the ethanol-induced stress. The expressions of X-ist, Cyp7b1, ERα, and GAPDH were measured by qRT-PCR. After ethanol exposure, the female cells still contained more X-ist transcripts than their male counterparts (26-fold), albeit much less than in female controls. Exposure of cells to estrogen (5 nM 17β-estradiol), on the other hand, eliminated the dimorphic expression of X-ist, regardless of the presence or absence of the ethanol cotreatment (Fig. 5A), suggesting that X-ist is an estrogen-responsive gene.
Figure 5.
Gene activity is sexually dimorphic in standard cell culture but can be altered by additives. Cells exposed to 400 μM ethanol and or in combination with estradiol as in Fig. 3 were used for analysis of gene expression. Gray bars, male; white bars, female. A) X-ist sex differences were maintained during cell culture for ED10.5 whole-embryo control cultures (control). Ethanol treatment (400 μM) did not alter gene expression, while 5 nM estradiol with or without ethanol suppressed sex differences in the expression of X-ist, as a result of a loss in transcript in female cells and an up-regulation of transcription in male cells. B) Differences in Cyp7b1 expression (female higher) were maintained in basal culture (control). Estradiol with or without ethanol reversed gene expression, whereby male transcription was up-regulated, while female transcription was down-regulated. Ethanol alone eliminated sex dimorphism in Cyp7b1 expression. C) ERα expression was independent of sex regardless of culture conditions used. D) Housekeeping gene GAPDH transcript levels were independent of sex under control culture conditions (control) and in the presence of both estrogen and ethanol, while individually estradiol and ethanol stimulated significantly more transcript in male cells. Scale in D differs from other panels because of high transcript levels for the gene. *P < 0.05, male vs. female in each condition.
One of the genes found to be transcribed more by female than male cells was the steroid metabolism gene Cyp7b1, which was expressed 10.9-fold higher in female whole-embryo cell extracts than in male extracts. This trend was maintained in cultured cells, with female cells expressing 3.43-fold more Cyp7b1 than male cells. However, treatment with ethanol decreased the female preferential expression, reducing it to 0.5-fold of male expression; estradiol had a similar effect, but the combination of estradiol and estrogen was not additive (Fig. 5B). Thus, our data suggest a sexually dimorphic gene regulatory mechanism. The mechanism of regulation is unknown; it could be different levels of transcription factors, or ligands to such transcription factors, as in the case of estrogen and estrogen receptors. However, we find that ERα is not sex dimorphic under any of the tested cell culture conditions (Fig. 5C).
The basal expression of the housekeeping gene GAPDH was not sexually dimorphic in the control cells or as determined by RNA from whole tissue (Fig. 5D). However, exposure to either ethanol or estrogen increased transcription in males, resulting in a sex dimorphism. The combination of estrogen and ethanol increased transcription in female cells as well, attenuating the dimorphism (Fig. 5D). Thus, even housekeeping genes may respond differentially according to sex.
DISCUSSION
Here we report a previously unrecognized differential cell sensitivity and sex-dependent gene regulation in isolated cells in the presence and absence of sex hormones and in response to stress. We show that cultured cells of different sexes but from similar tissues and developmental timing behave similarly in the absence of stress. Under stress such as induction of cell death, however, these cells respond in a manner that is modulated by sex and developmental stage. We assume that cells of the same developmental stage from female or male fetuses are at similar stages of development and in controlled levels of hormones in culture, and thus any differences observed should reflect only the sex dimorphisms. This potential confounding factor requires further study but does not undercut the basic argument that the cells differ intrinsically.
Male and female ED10.5 whole-embryo mixed cell cultures, ED17.5 mixed kidney cell cultures, and PN4 and PN17 mixed kidney cultures behave similarly with respect to cell morphology, rate of proliferation, and basal levels of cell death in the absence of stress. Ethanol, however, elicits a sexually dimorphic response, with female cells being significantly more sensitive than male cells. ED10.5 embryos have not initiated gonadal development and therefore, by extension, production of sex steroids. Thus, any molecular or behavioral differences can be attributed to sex differences and not hormonal influences. By ED 17.5, the situation is more complex, but cells still respond differently. Female cells are significantly more sensitive than males to ethanol-induced cell death. These sex differences in stress responses cannot be attributed to sex hormones alone.
We followed this pattern throughout development by exposing kidney mixed cell cultures from embryos at ED17.5 and mice at PN17 to ethanol. At both posthormonal production stages of development, at ED17.5 and at PN17 (puberty for the young mouse), the differences are maintained. We therefore conclude that a predisposition for sex dimorphism before sex hormone production is partially maintained and later altered by hormones. Other stressors such as camptothecin also induce a sex-dimorphic cell response, in a manner similar to ethanol, documenting that the sex-dimorphic response is a more general phenomenon. Differences at the cell level potentially can affect the overall physiology of the organism.
We attempted to mimic hormonal influences by introducing exogenous estrogen or testosterone at the higher physiological range. Estrogen exerted a protective effect in the presence of ethanol, but on its own has little to no effect on the death of the cells. The protection against ethanol could derive from any of several effects, whereby the hormone acts as antioxidant or proliferative agent. Testosterone on the other hand increased the death of female cells, while male cells were unaffected. The high level of death induced by testosterone was not further increased by the addition of ethanol, indicating that in this case male cells were unresponsive to testosterone but that the hormone was detrimental to female cells. In summary, estradiol exerts a protective effect against alcohol in both male and female cells, while testosterone is detrimental to female cells only and is detrimental even in the absence of stress.
These differences are reflected in genetic activity. A previous study of Dewing et al. (16) described 51 sex-dimorphic genes in ED10.5 mouse brains, 7 of which were confirmed by PCR. We likewise found 51 dimorphic genes of a total of 22,000 (0.2%), although not all the same genes. Approximately half of our dimorphic genes were upregulated in females and half upregulated in males. Twenty-seven of Dewing et al.’s dimorphic genes were represented on our arrays. However, in our hands, only the expression of Xist was consistent with their data. This discrepancy perhaps derives from the fact that different regions of embryos were used for the different arrays. Dewing et al. (16) used the brain region of the ED10.5 embryos, while we used whole embryos. Subsequently, we sought to examine by qPCR the expression of 5 genes described to be sex dimorphic by others (16, 17): cytochrome P-450, family 7, subfamily b, polypeptide 1 (Cyp7b1); eukaryotic translation initiation factor 2, subunit 3, structural gene X-linked (Eif2s3x); and the X- and Y-linked partners of ubiquitously transcribed tetratricopeptide repeat gene (Utx and Uty), reported previously to be sexually dimorphic (16, 17), as well as Jumonji, AT rich interactive domain 1c (Jarid1c), reported by to be sex dimorphic (17). We confirmed a higher expression in females of all X-chromosome genes [Eif2s3x: 1.6-, 1.9-, and 2-fold in our data and those of Dewing et al. (16) and Xu et al. (17), respectively]; Utx: 4.4- and 2-fold in our data and those of Xu et al., respectively; and Jarid1c: 4.3- and 2-fold in our data and those of Xu et al., respectively. We also confirmed the presence of the Y-linked gene Uty in males only (see Table 2). However, our results differed from those of Dewing et al. in the direction of dimorphism of Cyp7b1. Our data showed 1.5-fold higher expression in females than males compared with Dewing et al.’s 1.9-fold higher in males. This difference may result from tissue specificity as well as strain specificity, as Dewing et al. used the head region of the embryos, while we used whole embryos.
The sex differences in basal expression were maintained in response to stimuli. In cells derived from ED10.5 in the presence or absence of ethanol or estrogen, gene expression frequently mimicked the dimorphic cell response we observed in our cell death analysis. Cyp7bl gene expression followed our previous results in which control female cells expressed significantly higher transcript levels than male cells, while addition of ethanol increased male transcription and decreased female transcription. The activity of Cyp7b1, which catalyzes the synthesis of 7α-hydroxy dehydroepiandrosterone and 7α-hydroxypregnenolone (31), directly correlates to its transcript levels and responsiveness to estrogen (18, 32), indicating that these differences are physiologically significant. Similarly, the dimorphism of X-ist transcripts (expressed at significantly higher levels in females in control cells) is reduced by ethanol. The reduction in difference of transcription of X-ist results both from an increase in male X-ist transcription and a reduction in female transcription of X-ist. X-ist functions in the absence of translation in that the transcript itself silences genes on the extra X-chromosome in females, therefore, presumptively in our case feminizing our male cells in culture and masculinizing the female cells in response to ethanol. The ethanol thus tends to reverse sex-dimorphic gene regulation, a result with potential wide-ranging consequences, including social behaviors and, more cogently, sexually dimorphic response to disease. Overall, we find a possible sex-dependent gene regulatory mechanism, whereby stress and hormones affect gene expression differently between the sexes. This mechanism may explain the sex-dimorphic cellular response observed and may further explain sex dimorphism in physiology and pathology.
Acknowledgments
This work was supported by funding from National Institutes of Health Grant RO3 to Z.Z.; Minority Access to Research Careers Undergraduate Student Training for Academic Research (MARC U-STAR) to Z.Z.; and the National Science Foundation New York City Louis Stokes Alliance for Minority Participation (LSAMP) and the City University of New York Alliance for Graduate Education and the Professoriate (CUNY AGEP). We thank Ms. Nikezic-Ardolic for technical impact on the original idea and methodologies for this manuscript. We also thank Dr. Tahereh Entezari-Zaher, Dr. Mohammad Javdan, and Tamjeed Sikder for technical and intellectual assistance.
References
- Ceribelli A, Pino M S, Cecere F L. Gender differences: implications for clinical trials and practice. J Thorac Oncol. 2007;2:15–18. doi: 10.1097/01.JTO.0000268635.25579.7e. [DOI] [PubMed] [Google Scholar]
- Bain C F D, Speizer F E, Thun M, Hertzmark E, Rosner B A, Colditz G A. Lung cancer rates in men and women with comparable histories of smoking. J Natl Cancer Inst. 2004;96:826–834. doi: 10.1093/jnci/djh143. [DOI] [PubMed] [Google Scholar]
- Beeson P. Age and sex association of 40 autoimmune diseases. Am J Med. 1994;96:457–462. doi: 10.1016/0002-9343(94)90173-2. [DOI] [PubMed] [Google Scholar]
- Cutolo M, Capellino S, Sulli A, Serioli B, Secchi M E, Villaggio B, Straub R H. Estrogens and autoimmune diseases. Ann N Y Acad Sci. 2006;1089:538–547. doi: 10.1196/annals.1386.043. [DOI] [PubMed] [Google Scholar]
- Lockshin M D. Invited review: sex ratio and rheumatic disease. J Appl Physiol. 2001;91:2366–2373. doi: 10.1152/jappl.2001.91.5.2366. [DOI] [PubMed] [Google Scholar]
- Lockshin M D. Sex differences in autoimmune disease. Orthop Clin North Am. 2006;37:629–633. doi: 10.1016/j.ocl.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Shulman L M, Bhat V. Gender disparities in Parkinson’s disease. Expert Rev Neurother. 2006;6:407–416. doi: 10.1586/14737175.6.3.407. [DOI] [PubMed] [Google Scholar]
- Vaccarino V, Parsons L, Every N R, Barron H V, Krumholz H M. Sex-based differences in early mortality after myocardial infarction. N Engl J Med. 1999;341:217–225. doi: 10.1056/NEJM199907223410401. [DOI] [PubMed] [Google Scholar]
- Zorgdrager A, Keyser J D. The premenstrual period and exacerbations in multiple sclerosis. Eur Neurol. 2002;48:204–206. doi: 10.1159/000066166. [DOI] [PubMed] [Google Scholar]
- Gompel A, Piette J C. Systemic lupus erythematosus and hormone replacement therapy. Menopause Int. 2007;13:65–70. doi: 10.1258/175404507780796433. [DOI] [PubMed] [Google Scholar]
- Lockshin M D, Buyon J P. Estrogens and lupus: Bubbling cauldron or another overrated witches’ brew? Arthritis Rheum. 2007;56:1048–1050. doi: 10.1002/art.22631. [DOI] [PubMed] [Google Scholar]
- Du L, Bayir H, Lai Y, Zhang X, Kochanek P M, Watkins S C, Graham S H, Clark R S B. Innate gender-based proclivity in response to cytotoxicity and programmed cell death pathway. J Biol Chem. 2004;279:38563–38570. doi: 10.1074/jbc.M405461200. [DOI] [PubMed] [Google Scholar]
- Heyer A, Hasselblatt M, von Ahsen N, Hafner H, Siren A-L, Ehrenreich H. In vitro gender differences in neuronal survival on hypoxia and in 17β-estradiol-mediated neuroprotection. J Cereb Blood Flow Metab. 2005;25:427–430. doi: 10.1038/sj.jcbfm.9600056. [DOI] [PubMed] [Google Scholar]
- Nikezic-Ardolic M, Lin L, Milcevic M, Zakeri Z. Gender differences in cellular response. Lupus. 1999;8:375–379. doi: 10.1177/096120339900800509. [DOI] [PubMed] [Google Scholar]
- Amador-Noguez D, Zimmerman J, Venable S, Darlington G. Gender-specific alterations in gene expression and loss of liver sexual dimorphism in the long-lived Ames dwarf mice. Biochem Biophys Res Commun. 2005;332:1086–1100. doi: 10.1016/j.bbrc.2005.05.063. [DOI] [PubMed] [Google Scholar]
- Dewing P, Shi T, Horvath S, Vilain E. Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Mol Brain Res. 2003;118:82–90. doi: 10.1016/s0169-328x(03)00339-5. [DOI] [PubMed] [Google Scholar]
- Xu J, Burgoyne P S, Arnold A P. Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet. 2002;11:1409–1419. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- Dulos J, van der Vleuten M A, Kavelaars A, Heijnen C J, Boots A M. CYP7B expression and activity in fibroblast-like synoviocytes from patients with rheumatoid arthritis: regulation by proinflammatory cytokines. Arthritis Rheum. 2005;52:770–778. doi: 10.1002/art.20950. [DOI] [PubMed] [Google Scholar]
- Higashi E, Fukami T, Itoh M, Kyo S, Inoue M, Yokoi T, Nakajima M. Human CYP2A6 is induced by estrogen via estrogen receptor. Drug Metab Dispos. 2007;35:1935–1941. doi: 10.1124/dmd.107.016568. [DOI] [PubMed] [Google Scholar]
- Isensee J, Witt H, Pregla R, Hetzer R, Regitz-Zagrosek V, Ruiz Noppinger P. Sexually dimorphic gene expression in the heart of mice and men. J Molec Med. 2008;86:61–74. doi: 10.1007/s00109-007-0240-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollerup S, Ryberg D, Hewer A, Phillips D H, Haugen A. Sex differences in lung CYP1A1 expression and DNA adduct levels among lung cancer patients. Cancer Res. 1999;59:3317–3320. [PubMed] [Google Scholar]
- Nickerson M, Kennedy S L, Johnson J D, Fleshner M. Sexual dimorphism of the intracellular heat shock protein 72 response. J Appl Physiol. 2006;101:566–575. doi: 10.1152/japplphysiol.00259.2006. [DOI] [PubMed] [Google Scholar]
- Angele M K, Nitsch S, Knoferl M W, Ayala A, Angele P, Schildberg F W, Jauch K W, Chaudry I H. Sex-specific p38 MAP kinase activation following trauma-hemorrhage: involvement of testosterone and estradiol. Am J Physiol Endocrinol Metab. 2003;285:E189–E196. doi: 10.1152/ajpendo.00035.2003. [DOI] [PubMed] [Google Scholar]
- Karasavvas N E R, Bittman R, Lockshin R, Hockenbery D, Zakeri Z. BCL-2 suppresses ceramide-induced cell killing. Cell Death Differ. 1996;3:149–151. [PubMed] [Google Scholar]
- Ahuja H S, James W, Zakeri Z. Rescue of the limb deformity in Hammertoe mutant mice by retinoic acid-induced cell death. Dev Dyn. 1997;208:466–481. doi: 10.1002/(SICI)1097-0177(199704)208:4<466::AID-AJA3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Henry G, Zeger S, Pevsner J. SNOMAD (standardization and normalization of microarray data): web-accessible gene expression data analysis. Bioinformatics. 2002;18:1540–1541. doi: 10.1093/bioinformatics/18.11.1540. [DOI] [PubMed] [Google Scholar]
- Workman C, Jensen L J, Jarmer H, Berka R, Gautier L, Nielser H B, Saxild H-H, Nielsen C, Brunak S, Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3:1–16. doi: 10.1186/gb-2002-3-9-research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Sherlock G, Binkley G, Jin H, Matese J C, Hernandez-Boussard T, Rees C A, Cherry J M, Botstein D, Brown P O, Alizadeh A A. SOURCE: a unified genomic resource of functional annotations, ontologies, and gene expression data. Nucleic Acids Res. 2003;31:219–223. doi: 10.1093/nar/gkg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R, Smith B, Liu Q Y, Fazel F, Valdes J J, Liu Z, Lach B. Data mining of gene expression changes in Alzheimer brain. Artif Intell Med. 2004;31:137–154. doi: 10.1016/j.artmed.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Peippo J, Farazmand A, Kurkilahti M, Markkula M, Basrur P K, King W A. Sex-chromosome linked gene expression in in-vitro produced bovine embryos. Mol Hum Reprod. 2002;8:923–929. doi: 10.1093/molehr/8.10.923. [DOI] [PubMed] [Google Scholar]
- Rose K A, Stapleton G, Dott K, Kieny M P, Best R, Schwarz M, Russell D W, Björkhem L, Seckl J, Lathe R. Cyp7b, a novel brain cytochrome P450, catalyzes the synthesis of neurosteroids 7a-hydroxy dehydroepiandrosterone and 7a-hydroxy pregnenolone. Proc Natl Acad Sci USA. 1997;94:4925–4930. doi: 10.1073/pnas.94.10.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Norlin M. Regulation of steroid hydroxylase CYP7B1 by androgens and estrogens in prostate cancer LNCaP cells. Biochem Biophys Res Commun. 2006;344:540–546. doi: 10.1016/j.bbrc.2006.03.175. [DOI] [PubMed] [Google Scholar]