Abstract
ErbB4, a type I transmembrane receptor tyrosine kinase, is a member of the epidermal growth factor receptor family. Its cleavage releases an intracellular C-terminal domain (ICD), which can be either degraded following ubiqitination or translocated to the nucleus and regulate gene expression. There are 2 ErbB4 ICD isoforms: CYT-1 and CYT-2. We and others have previously reported that following cleavage, CYT-2 selectively translocates to the nucleus. In the current study we found that following cleavage, the intracellular levels of CYT-1 ICD decreased rapidly, while levels of CYT-2 ICD remained relatively stable. CYT-1 ICD degradation could be prevented by administration of either the proteasome inhibitor lactacystin or the lysosome inhibitor chloroquine, indicating both proteasomal and lysosomal degradation. Further studies implicated Nedd4, an E3 ubiquitin ligase, as a mediator of CYT-1 ubiquitination and degradation. The interaction of Nedd4 with CYT-1 was shown by coimmnunoprecipitation, an in vitro direct binding assay, and an in vitro ubiquitination assay. Three PPxY or PY motifs present in the CYT-1 C terminus are necessary for binding by Nedd4 WW domains, because impaired interactions are seen in mutation of any of the PY motifs. Nedd4-CYT-1 binding was associated with increased CYT-1 ubiquitination following proteasome inhibitor treatment. Impaired Nedd4 binding to CYT-1 by PY motif mutations led to increased CYT-1 ICD stability, whereas only one of the PY motif mutations (Y1056A), which disrupts the binding sites for both a WW domain and an SH2 domain of PI3 kinase, demonstrated enhanced nuclear translocation following HB-EGF treatment. These studies indicate that Nedd4 mediates ErbB4 CYT-1 ICD ubiquitination and degradation, and the prevention of both WW binding and PI3 kinase activity are required for ErbB4 nuclear translocation.—Zeng, F., Xu, J., Harris, R. C. Nedd4 mediates ErbB4 JM-a/CYT-1 ICD ubiquitination and degradation in MDCK II cells.
Keywords: E3 ubiquitin ligase, PY motif mutation, PI3 kinase, nuclear translocation
Erbb4, a type I transmembrane receptor tyrosine kinase, belongs to the epidermal growth factor (EGF) receptor family, which includes EGFR (ErbB1), ErbB2 (c-Neu), ErbB3, and ErbB4 (1). On ligand binding, these receptors trigger a variety of cellular responses by activating their kinase domains. Unlike the other receptors in this family, ErbB4 is expressed as alternatively spliced isoforms characterized by variant extracellular juxtamembrane (JM) domains and intracellular cytoplasmic (CYT) domains. The ErbB4 JM type a (JM-a) isoform is cleaved on ligand activation or activation of protein kinase C by phorbol ester administration. Subsequent regulated intramembrane proteolysis (RIP) leads to release of ErbB4 ICD into the cytoplasm, which then either is degraded or is translocated to the nucleus to regulate gene expression and cellular function (2,3,4).
Unlike EGFR, which has been shown to undergo efficient endocytosis and degradation on EGF stimulation (5), little is known about ErbB4 degradation. Although earlier studies showed that ErbB4 is endocytosis impaired, it can be ubiquitinated (6,7,8,9). Ubiquitination, the process by which ubiquitin is conjugated to lysine side chains of proteins, is now recognized to be the mechanism regulating degradation of most intracellular proteins. The transfer of ubiquitin is a multistep process that involves at least 3 classes of enzymes: ubiquitin-activating enzymes, generally called E1 enzymes; ubiquitin-conjugating enzymes or E2; and ubiquitin ligases, E3. E3 ubiquitin ligases are of particular importance because they confer substrate specificity to the system by interacting directly with substrate proteins and thereby directing the transfer of ubiquitin (10). Among the many E3 ubiquitin ligases, the family of “homologous to E6-AP C terminus” (HECT)-E3 ubiquitin ligases representing the yeast Rsp5p and its mammalian homologues Nedd4, AIP4/Itch, and Smurf has been shown to ubiquitinate membrane proteins and, in some instances, to induce their degradation (11).
Neuronal precursor cell-expressed, developmentally down-regulated protein 4 (Nedd4) is the prototypical protein in this family of E3 ubiquitin ligases that share common domain architectures. They are comprised of an N-terminal C2 domain, 3 or 4 tandem WW domains, and a C-terminal ubiquitin ligase HECT domain (12). WW domains represent globular domains consisting of 40 amino acids, 2 of which are highly conserved tryptophans (WW) (13). They function as protein interaction modules through binding to proline-rich peptide sequences (14). The WW domains of Nedd4 have been shown to bind to a PPxY or PY motif, where x represents any amino acid. In one well-characterized example, the WW domains of Nedd4 bind to PY motifs present in the cytosolic tails of the 3 subunits of the epithelial amiloride-sensitive sodium channel (ENaC). Mutations in the PY motifs disrupt the interaction between Nedd4-2 (also noted as Nedd4L: Nedd4-like protein) and the distal nephron epithelial sodium channel (ENaC), giving rise to a genetic form of hypertension termed Liddle’s syndrome (12, 15). Interestingly, like ENaC, ErbB4 is also expressed in the collecting duct in the rat kidney (4), with PY motifs in its C terminus, which makes it a candidate substrate for Nedd4 and/or Nedd4-2.
The 2 CYT isoforms of ErbB-4 represent alternative splice isoforms and differ by the presence (CYT-1) or absence (CYT-2) of a 16-aa peptide fragment containing 2 overlapping protein recognition motifs (PPAYTPM), PPxY and YxxM. The first motif is for WW domains of class I and requires tyrosine (Y) to be unphosphorylated for binding. The second motif will serve as a binding site for the phosphatidylinositol 3-OH kinase (PI3K) SH2-domain if this tyrosine residue is phosphorylated (16). In our previous studies we found that compared to CYT-1, CYT-2 selectively translocates to the nucleus after heparin-binding EGF-like growth factor (HB-EGF) binding to ErbB4 (4). Because those studies also indicated that HB-EGF induced MDCK II cell tubulogenesis selectively in cells expressing the CYT-2 isoform of ErbB4, the current studies were designed to investigate the underlying mechanisms mediating this differential processing of CYT-1 and CYT-2. We now report that compared to CYT-2 ICD, CYT-1 ICD is more rapidly degraded by both proteasomal and lysosomal pathways. Nedd4, an E3 ubiquitin ligase that can bind to ErbB4 CYT-1 through its PY motifs, plays an important role in ErbB4 CYT-1 ICD ubiquitination and degradation. Furthermore, the nuclear translocation of CYT-2 is the result of the absence of both the Nedd4 WW binding domain and PI3K SH2 binding domain.
MATERIALS AND METHODS
Cell culture and transfection
MDCK II cells were cultured at 37°C in a humidified air atmosphere with 5% CO2. and grown in DMEM/F12 (Gibco BRL; Carlsbad, CA, USA), supplemented with 10% FBS, streptomycin (100 μg/ml), and penicillin (100 U/ml). The transfected MDCK II cells stably expressing human full-length ErbB4 JM-a/CYT-1 (CYT-1) or JM-a/CYT-2 (CYT-2) isoforms that were cloned into the pcDNA3.1 vector were described in our previous studies (4). Cells were cultured in the presence of 1 mg/ml G418 (Research Products International Corp., Mt. Prospect, IL, USA). Cell transfection with the plasmid with CYT-1 PY motif mutations was performed as described previously (4).
Antibodies and reagents
Chloroquine, cycloheximide (CHX), lactacystin, N-ethylmaleimide (NEM), phorbol 13-myristate 12-acetate (PMA), and reduced glutathione were obtained from Sigma-Aldrich (St. Louis, MO, USA). Anti-ErbB4 antibody (c-18) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-ErbB4 antibody (Ab-2) was obtained from NeoMarkers (Fremont, CA, USA). Both ErbB4 antibodies (c-18 and Ab2) were raised against the ErbB4 C terminus and can recognize all splice variants of ErbB4. The β tubulin antibody and anti-ubiquitin antibody (P4D1) were from Cell Signaling Technology (Danvers, MA, USA). P4D1 can detect ubiquitin, polyubiquitin, and ubiquitinated proteins. Recombinant protein G agarose was obtained from Invitrogen Corporation (Carlsbad, CA, USA). Rabbit polyclonal anti-Nedd4 antibody was obtained from Abcam (Cambridge, MA, USA). Ubiquitin activating enzyme (E1, human recombinant), UbcH5b (human recombinant), ubiquitin, and the ubiquitin conjugation reaction buffer kit were obtained from BostonBiochem (Cambridge, MA, USA). Amicon Ultra-4 (10 K) was obtained from Millipore (Billerica, MA, USA).
ErbB4 ICD protein stability
To test ErbB4 ICD stability after PMA cleavage, MDCK II cells expressing either CYT-1 or CYT-2 were treated with PMA or vehicle (DMSO) for the indicated times. After washing 3 times with ice-cold Ca2+- and Mg2+-free phosphate-buffered saline, cells were lysed with ice-cold TGH buffer (1% Triton X-100; 10% glycerol; 20 mM Hepes, pH 7.2; 100 mM NaCl; 1 mM phenylmethylsulfonyl fluoride; 10 μg/ml leupeptin; 10 μg/ml aprotinin; and 1 mM Na3VO4). The lysates were incubated for 20 min on ice with intermittent vortexing. Insoluble material was removed by centrifugation (14,000 g, 15 min) at 4°C, and the supernatant was collected. Protein concentrations were assayed by Bradford’s method, using bovine serum albumin as the standard. Following SDS-PAGE, the proteins were transferred to PVDF membranes for Western blot. Immunoblots were blocked with TBST buffer (0.05% Tween 20, 150 mM NaCl, 50 mM Tris, pH 7.4) containing 5% nonfat milk powder (blocking buffer) at room temperature for 1 h. The membranes were probed overnight at 4°C with anti-ErbB4 antibody in the blocking buffer, washed 3 times with TBST buffer, and incubated with peroxidase-conjugated secondary antibody. After thorough washing, the chemiluminescence reaction was performed, and the membranes were exposed to ECL hyperfilm according to the manufacturer’s instructions (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
Proteasome and lysosome inhibition experiments
To study the effects of proteasome or lysosome inhibitors on ErbB4 protein stability, ErbB4-transfected cells were incubated with 5 μM lactacystin or 100 mM chloroquine for the indicated times before lysis. The control group was incubated with vehicle (DMSO) for the same period of time. Cells were lysed by the methods indicated above. Protein lysates were loaded onto 6% SDS-PAGE and immunoblotted with an anti-ErbB4 antibody.
Coimmunoprecipitation (co-IP) and ubiquitination assays
In preliminary studies, we determined that MDCK II cells express endogenous Nedd4 and Nedd4-2 mRNA (by RT-PCR, data not shown) and protein (by Western blot). Therefore, MDCK II cells transfected with ErbB4 are an ideal model for studying the interaction between endogenous Nedd4 and ErbB4. For this purpose the above cells were incubated with lactacystin for 16 h to block protein degradation and lysed in the lysis buffer plus 10 mM NEM to inhibit deubiquitination enzymes. For co-IP, 1 μg of anti-ErbB4 antibody (c-18) or anti-Nedd4 antibody was incubated for 2 h at 4°C with 500 μg of cellular protein followed by overnight incubation with protein G agarose. Immune complexes were washed 3 times with TGH lysis buffer, suspended in 2 × Laemmli buffer, and boiled for 5 min before loading onto 6% SDS-PAGE. The membranes were probed overnight at 4°C with either anti-ErbB4 antibody (immunoprecipitated with anti-Nedd4 antibody) or anti-Nedd4 antibody (immunoprecipitated with anti-ErbB4 antibody). The membranes containing proteins immunoprecipitated by anti-ErbB4 antibody were also used to detect protein ubiquitination level by immunoblotting with the anti-ubiquitin antibody. In addition, the same membrane was reprobed with either anti-Nedd4 or anti-ErbB4 antibody.
Glutathione S-transferase (GST) fusion proteins and pulldown assays
The plasmid encoding GST-Nedd4 (aa 52–777), which includes the C2 domain and 3 WW domains (17) (see Fig. 3A), was from Addgene.com (Addgene plasmid 11466). The GST-Nedd4 fusion protein was expressed in Escherichia coli BL-21 (DE3) and purified on glutathione-sepharose beads, as described by the manufacturer (GE Healthcare, Piscataway, NJ, USA).
Figure 1.
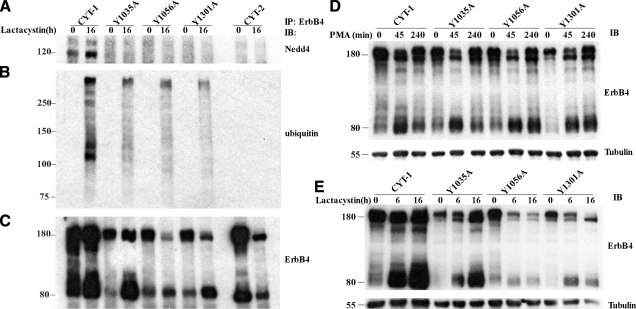
CYT-1 was degraded through both proteasomal and lysosomal pathways. A) Protein stability of full-length ErbB4 (180 kDa) and the 80-kDa ICD was measured by immunoblotting (IB) with anti-ErbB4 antibody at different time points after PMA administration. CYT-1 ICD was rapidly degraded 45 min after PMA-induced cleavage, while CYT-2 ICD levels remained relatively stable. B) The proteasome inhibitor, lactacystin, inhibited CYT-1 ICD degradation but had minimal effects on CYT-2 ICD level. C) The lysosome inhibitor, chloroquine, inhibited CYT-1 ICD degradation only at relatively late time points. D) Administration of the protein synthesis inhibitor, cytoheximide (CHX), decreased CYT-1 expression level. E) Simultaneous administration of lactacystin with CHX inhibited CYT-1 degradation, especially the 80-kDa ICD fragment.
Figure 2.
Increased ErbB4 CYT-1 ubiqintination was associated with enhanced Nedd4-CYT-1 binding. A) ErbB4 ubiquitination. To test the possible ubiquitination of ErbB4, especially the 80-kDa ICD, MDCK II cells were treated with lactacystin to block proteasomal-mediated protein degradation. Cell lysates were immunoprecipitated with anti-ErbB4 antibody, and immunoblotted with anti-ubiquitin antibody. There was prominent and selective ubiquitination of the CYT-1 isoform. B) Nedd4-ErbB4 interactions were detected by immunoprecipitation (IP) with an anti-ErbB4 antibody, and IB with an anti-ubiquitin antibody. Both Nedd4 (bottom band) and Nedd4-2 (top band) can interact with ErbB4 CYT-1. C) The above membrane was reprobed with anti-ErbB4 antibody to indicate the relative levels of ErbB4 protein used in IP. +, with lactacystin; −, DMSO (vehicle); vector, pcDNA3.1.
Figure 3.
Nedd4 directly bound to ErbB4 CYT-1 ICD both in vivo and in vitro. A) Illustration of GST-Nedd4 fusion protein. GST was fused with Nedd4, which has 3 WW domains and a C-terminal HECT domain. B) Co-IP assay. Cells were treated with lactacystin for 16 h, cell lysates were immunoprecipitated with anti-Nedd4 antibody, and the membrane was probed using anti-ErbB4 antibody. Results indicated that Nedd4 predominately bound to CYT-1 ICD, with very little binding to CYT-2. C) GST-Nedd4 pulldown assay. Equal amounts of protein lysates were loaded onto a GST-Nedd4 column. GST-Nedd4 exclusively bound to CYT-1 (both full-length and 80-kDa ICD), but not with CYT-2. D) CYT-1 and CYT-2 expression levels used for IP and binding assay.
For pulldown assays, equal amounts of isolated GST alone or GST-Nedd4 fusion proteins were incubated with 1 mg protein lysate and incubated for 4 h at 4°C. The column was then washed 3 times with TGH lysis buffer, and bound proteins were eluted in 2× SDS protein loading buffer (100 mM Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 0.2% bromphenol blue, and 200 mM DTT) and resolved by 6% SDS-PAGE. Immunoblots were probed with an anti-ErbB4 antibody and reprobed with an anti-Nedd4 antibody. Levels of the ErbB4 bound to Nedd4 were quantitated with an IS-1000 digital imaging system (Alpha Innotech, San Leandro, CA, USA).
Site-directed mutagenesis
ErbB4 CYT-1 PY motif mutants were generated by QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). We used pcDNA3-ErbB4 JM-a/CYT-1 plasmid as a PCR template. The mutagenic primers, each complementary to opposite strands of the plasmid DNA carrying codons for the required substitutions, were synthesized by Integrated DNA Technologies (Coralville, IA, USA). PCR amplification was performed using Pfu DNA polymerase according to the manufacturer’s protocol. The PCR product was treated with DpnI to digest the parental DNA template, leaving the newly synthesized mutant PCR product, which was then transformed into XL1-Blue supercompetent cells. Plasmid DNA was isolated and sequenced. Selected clones verified for the required mutations (see Fig. 4A) were used for transfection.
Figure 4.
Mutation of CYT-1 ICD PY motifs inhibited in vitro Nedd4 binding A) Illustration of PY motifs in CYT-1 and CYT-2 ICD. There are 3 PY motifs in CYT-1 ICD: PY1, PY2, and PY3. Subscript number indicates tyrosine location in CYT-1 isoform. PY2 motif is absent in CYT-2. PY motif mutations were made by changing Y or P to A in CYT-1 ICD. B) CYT-1 PY motif mutations affected Nedd4-ErbB4 binding in the in vitro direct binding assay. Equal amounts of protein lysates were loaded onto a GST-Nedd4 column. After elution, proteins were separated by SDS-PAGE and immunoblotted with anti-ErbB4 antibody (top panel). Membrane was stripped and reprobed with anti-Nedd4 antibody (bottom panel). C) The above membrane was reprobed with anti-ErbB4 antibody to indicate the ErbB4 protein expression level. D) Relative amounts of full-length ErbB4 bound to the Nedd4 column were calculated by correction for their basal expression level. E) Relative amounts of 80-kDa ICD bound to the Nedd4 column were calculated by correction for their basal expression level. *P < 0.05.
In vitro ubiqination assays
For in vitro ubiquitination assays, a GST-Nedd4 fusion protein was eluted in the elution buffer (50 mM Tris-HCl, 10 mM reduced glutathione, pH 8.0). Glutathione was removed by centrifuging through an Amicon Ultra-4 (10 K), and the concentrated GST-Nedd4 fusion protein was used in the in vitro ubiquitination assay, which was performed according to the manufacturer’s instructions (BostonBiochem). In a 30 μl reaction volume, 50 nM E1, 500 nM E2 (UbcH5b), 500 μM ubiquitin, 3 μl 10 × ubiquitination reaction buffer, 3 μl 10 × Mg-ATP (final 1mM), 1 μg GST-Nedd4 fusion protein, and 0.5 mg pooled proteins from cell lysis of MDCK II cells overexpressing ErbB4 were combined and incubated at 37°C for 1 h. The reaction was stopped by adding 70 μl TGH lysis buffer. Next, 10 μl was probed with anti-Nedd4 antibody, and the other 90 μl was used for ErbB4 immunoprecipitation, followed by immunoblotting with anti-ubiquitin antibody and anti-ErbB4 antibody.
Immunofluorescence (IF)
IF staining was performed as described previously (4). Cells plated on glass coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline plus 0.5 mM MgCl2 and 0.9 mM CaCl2 (PBS2+) and permeabilized with 0.2% Triton X-100 in PBS2+. After blocking with 5% normal goat serum and 1% BSA in PBS2+, samples were incubated with ErbB4 antibody (Ab2) overnight at 4°C in a moist chamber. Samples were then washed 3 times with PBS2+, followed by incubation for 30 min with the respective conjugated second antibody (Jackson ImmunoResearch Laboratories, West Grove, PA, USA). After 3 washes, cells were counterstained with TO-PRO- 3 in PBS for 10 min and mounted with VectaShield mounting media (Vector Laboratories, Burlingame, CA, USA). The images were examined using a Zeiss-410 confocal microscope (Carl Zeiss, Oberkochen, Germany) at the Vanderbilt Imaging Core facility. Cells with nuclear staining for ErbB4 were counted from at least 100 ErbB4 expressing cells, and the percentage of positive cells was calculated and compared to the level without HB-EGF treatment). Data represesent means ± se from 3 independent experiments.
Statistics
Graphic data are presented as means ± se. Student’s t test was used for comparing different treatments and cell lines. Differences with P < 0.05 were considered statistically significant.
RESULTS
ErbB4 JM-a/CYT-1 ICD is rapidly degraded following PMA cleavage
Previous studies have found that PMA, a protein kinase C activator, induces rapid and extensive proteolytic cleavage of the ErbB4 JM-a isoform (18). To determine the stability of ErbB4 ICD, MDCK II cells transfected with either CYT-1 or CYT-2 were treated with PMA for 45 min to achieve maximal ErbB4 cleavage. ErbB4 protein levels were determined by immunoblotting. Similar to previous reports (4, 18), PMA treatment for 45 min induced cleavage of a significant proportion of CYT-1 and CYT-2 isoforms. CYT-1 ICD (80 kDa) protein level decreased dramatically in the subsequent 80–120 min following cleavage, with gradual recovery of the full-length CYT-1 level. In contrast, CYT-2 ICD protein levels did not show significant decrement (Fig. 1A). These results suggested that CYT-1 ICD was rapidly degraded after ErbB4 cleavage, while CYT-2 ICD was relatively stable.
ErbB4 JM-a/CYT-1 ICD degradation is through both proteasomal and lysosomal pathways
As proteasomes and lysosomes are the 2 major organelles mediating intracellular protein degradation, we examined their possible involvement in the degradation of CYT-1 ICD and determined protein stability after either proteasome or lysosome inhibition. We have found that there is a significant amount of basal cleavage of ErbB4 in the transfected MDCK II cells grown in regular growth medium. For these longer-duration studies, we did not induce cleavage with PMA in these studies because phorbol esters have pleiotropic cellular effects in addition to their stimulation of metalloproteinase-mediated ErbB4 cleavage. As shown in Fig. 1B, CYT-1 protein levels, both full length and ICD, were dramatically increased after 8 h treatment with the proteasomal inhibitor, lactacystin. The increased full-length CYT-1 was predominantly the smaller molecular mass (Fig. 1B, SM) form, which may represent newly synthesized and not fully glycosylated ErbB4. Compared to the effects of lactacystin, the lysosomal inhibitor, chloroquine, exhibited a delayed time course to block CYT-1 degradation. The significant elevation of CYT-1 ICD level was seen only after 16 h treatment, compared to 8 h of lactacystin treatment (Fig. 1B, left side). In contrast, the same treatments had minimal effects on CYT-2 ICD protein expression levels (Fig. 1B, 1C, right side). At 4 and 8 h of lactacystin treatment, the level of the 80-kDa CYT-2 ICD exhibited a slightly increase, companied by the decreased full-length CYT-2 level, which may be a result of basal cleavage. The 80-kDa CYT-2 ICD level decreased gradually from 8 to 16 h after lactacystin treatment, which suggested that CYT-2 ICD might be degradated through a mechanism other than the proteasomal pathway.
To exclude the possibility that the accumulation of CYT-1 was the result of newly synthesized protein, cells were pretreated for 1 h with CHX, a protein synthesis inhibitor, followed by administration of lactacystin for different times. Cells treated with CHX alone were used as a protein synthesis inhibition control. As shown in Fig. 1D, levels of both the full-length and 80-kDa ICD of CYT-1 gradually decreased after CHX treatment. Simultaneous treatment with CHX and lactacystin led to CYT-1 ICD accumulation, whereas expression of the full-length ErbB4 level decreased (Fig. 1E). We presume that the decreased full-length ErbB4 level is secondary to decreased protein synthesis as well as the natural cleavage of this protein (18).
Ubiquitination of ErbB4 JM-a/CYT-1 is involved in its proteasomal degradation
We next investigated whether ubiquitination is involved in ErbB4 degradation. After treatment with lactacystin for 16 h, cell extracts were immunoprecipitated with anti-ErbB4 antibody and immunoblotted with anti-ubiquitin antibody. Elevated ubiquitin levels were specifically associated with the CYT-1 isoform (Fig. 2A). The results also suggested that CYT-1 and its cleaved ICD were polyubiquitinated, as suggested by detection of multiple bands with different molecular masses over 32 kDa (the sum of 4 ubiquitin molecules). Under the same conditions, we did not detect any increased ubiquitination associated with the CYT-2 isoform. These results suggested that compared to CYT-2, the CYT-1 isoform is readily ubiquitinated.
Nedd4-CYT-1 binding is associated with increased CYT-1 ubiquitination
It is well established that the E3 ubiquitin ligase, Nedd4, mediates specific protein ubiquitination through interaction between its WW domains and PY motifs in the substrates. ErbB4, which contains 2 or 3 PY motifs in its C terminus, is a candidate substrate for Nedd4. Protein lysates were immunoprecipitated with anti-ErbB4 antibody and immunoblotted with anti-Nedd4 antibody. The results showed that Nedd4 bound to ErbB4, with isoform discrimination, as only the CYT-1 isoform was coimmunoprecipitated with Nedd4 (Fig. 2B). Anti-Nedd4 antibody immunoblotting resulted in 2 bands. As the anti-Nedd4 antibody utilized recognizes both Nedd4 (120 kDa, the lower band) and Nedd4-2 (the upper, higher molecular band) isoforms, we assume that both Nedd4 and Nedd4-2 can recognize and interact with ErbB4. The presence of both Nedd4 and Nedd4-2 in MDCK II cells was also confirmed by reverse transcription PCR (data not shown), with Nedd4 being the predominant species.
Association of Nedd4 with ErbB4 was also documented by anti-Nedd4 antibody immunoprecipitation, followed by anti-ErbB4 immunoblotting. The results indicated that Nedd4 predominately bound to CYT-1 ICD, with weaker binding to full-length CYT-1 (Fig. 3B). To ascertain whether the observed interaction was direct, Nedd4-ErbB4 interactions were further assessed using an in vitro GST-Nedd4 pulldown assay. A GST-Nedd4 fusion protein directly and specifically interacted with CYT-1 protein but not with CYT-2 (Fig. 3C). In contrast to the co-IP studies in Fig. 3B, full-length CYT-1 also readily bound to the GST-Nedd4 column in addition to CYT-1 ICD. We propose that this may be due to conformational changes of CYT-1 protein caused by the lysis buffer. There was minimal Nedd4-CYT-2 interaction detected in either the Nedd4 immunoprecipitation or GST pulldown assay.
PY motif mutations in ErbB4 JM-a/CYT-1 ICD decreased Nedd4-ErbB4 binding
Given the differential binding of Nedd4 to CYT-1 and CYT-2 isoforms, we investigated the potential role of each of the PY motifs in the ErbB4 ICD. Since mutation of either proline (P) or the tyrosine (Y) in the PPxY motif will inhibit its ability to bind Nedd4 (19), we mutated all 3 PY motifs in CYT-1 ICD separately by changing Y to alanine (A), namely, Y1035A (PY1 motif mutation), Y1056A (PY2 motif mutation), and Y1301A (PY3 motif mutation) (illustrated in Fig. 4A). In the in vitro binding assay, we found that all 3 mutations affected Nedd4-CYT-1 binding, with Y1035A decreasing the binding, and either Y1056A or Y1302A mutations almost completely blocking the binding in both full-length CYT-1 and CYT-1 ICD (Fig. 4B, D, E).
The impaired Nedd4/PY mutation binding was also confirmed by a Nedd4-ErbB4 co-IP assay (Fig. 5A), which showed very weak Nedd4-CYT-1 binding in all 3 PY mutations and was comparable to the negative Nedd4-CYT-2 binding. Meanwhile, the decreased Nedd4-CYT-1 binding in the PY motif mutations also affected the level of CYT-1 ubiquitination (Fig. 5B).
Figure 5.
Mutation of CYT-1 ICD PY motifs inhibited in vivo Nedd4 binding and ubiquitination. A–C) CYT-1 PY motif mutations decrease Nedd4 binding and subsequent ubiquitination. Equal amounts of cell lysates were immunoprecipitated with anti-ErbB4 antibody, immunoblotted with anti-Nedd4 antibody (A), and then reprobed with either anti-ubiquintin antibody (B) or anti-ErbB4 antibody(C). Compared to CYT-1, CYT-2 and all 3 PY motif mutations showed decreased interaction with Nedd4 and decreased ubiquitination. ErbB4 protein expression level shown in C. D) Compared to mutation of the 1st WW binding domain (Y1035A), mutations of the 2nd or 3rd WW binding domain (Y1056A and Y1301A) exhibited a slower rate of degradation, as shown by comparing the 80-kDa ICD level at 45 min and 4 h after PMA-induced cleavage. E) Y1035A ICD also demonstrated increased stability in response to lactacystin than Y1056A and Y1301A.
We also investigated whether this altered Nedd4 binding ability affected CYT-1 stability. For this purpose ErbB4 was cleaved by PMA, and protein levels were measured at different time points. As shown in Fig. 5D, Y1035A mutation did not change CYT-1 stability detectably from its parental CYT-1 isoform. In contrast, both Y1056A and Y1301A mutations partially decreased CYT-1 ICD degradation. Similarly there was greater accumulation of the Y1035A mutation in the face of proteasomal inhibition than either the Y1056A or Y1301A mutations (Fig. 5E). These results suggested that compared to the PY1 motif (PPIY1035), both the PY2 (PPAY1056) and PY3 (PPPY1301) motifs appear to be more important for Nedd4-CYT-1 binding and subsequent CYT-1 degradation. These results are also consistent with the lack of Nedd4 interaction with CYT-2, since this isoform does not contain the PY2 motif (Fig. 4A).
Nedd4 is an E3 ubiquitin ligase directly mediating ErbB4 CYT-1 ubiquitination
Nedd4 is known to possess E3 ubiquitin ligase activity (12). The interaction between Nedd4 and ErbB4 CYT-1 both in vitro and in vivo suggested that Nedd4 might target ErbB4 CYT-1 for ubiquitination. For direct evidence of ubiquitination of CYT-1 ICD by Nedd4, an in vitro ubiquitination assay was performed using recombinant GST-Nedd4, ErbB4 isoforms, E1, E2 (UbcH5b), and ubiquitin. At the completion of the reaction, ErbB4 was immunoprecipitated, and ubiquitination of ErbB4 was evaluated by immunoblotting with an anti-ubiquitin antibody. The CYT-1 ubiquitin level was dramatically increased in the presence of GST-Nedd4 (Fig. 6, upper panel, lane 6). Both E1 and E2 are required in this process, as minimal CYT-1 ubiquination was detectable in their absence (Fig. 6, top panel, lanes 2 and 3). The small amount of ubiquitination of CYT-1 in the absence of addition of exogenous E1 and E2 or GST-Nedd4 may be explained by the presence of residual endogenous E1, E2, and Nedd4 in the cell lysates used for ErbB4 protein in those reactions.
Figure 6.
In vitro ubiquitination assay. A) Nedd4 provided E3 ubiquitin ligase activity in vitro. In vitro ubiquitin assays were performed as described in Material and Methods, with individual components added as indicated. ErbB4 CYT-1 was used as the substrate. B) Comparison of ubiquitination levels of CYT-1, CYT-1 PY motif mutations and CYT-2 mediated by Nedd4. Lanes 7, 9, 11, 13, and 15 were ErbB4 protein lysates incubated without in vitro ubiquitination components and are used as controls. Lanes 8, 10, 12, 14, and 16 had full ubiquitin assay components, as indicated in lane 6 of A. ErbB4 ubiquitination was detected by IP with anti-ErbB4 antibody, followed by IB with anti-ubiquitin antibody and anti-ErbB4 antibody. Bottom panel of lanes 1-16 indicates Nedd4 level in each reaction.
Similar to the in vivo assay, GST-Nedd4 did not induce significant ubiquitination of the CTY-2 isoform or of CYT-1 PY mutations, especially Y1056A or Y1301A (Fig. 6, top panel, lanes 12, 14, 16).
Inhibition of both PI3 kinase activity and WW binding is required for ErbB4 JM-a/CYT-1 nuclear translocation
Our previous studies in MDCK II cells found that compared to CYT-2, CYT-1 does not translocate to the nucleus after PMA or HB-EGF stimulation (4). To explore the possible roles of the PI3 kinase and/or WW binding motif in the regulation of the nuclear translocation of the ErbB4 isoforms, we studied the subcellular localization of the different PY motif mutations in CYT-1 ICD and found that only Y1056A showed enhanced nuclear translocation after HB-EGF stimulation, which was comparable to the nuclear staining pattern of CYT-2 cells (Fig. 7A, C), while there was minimal nuclear translocation of Y1035A and Y1301A (data not shown). Unlike the other 2 PY motifs, the PY2 mutation with Y1056A disrupted the binding site for both WW domain-containing proteins and for PI3 kinase. We examined an additional PY2 mutation (PPtoAA of PPAY1056, Fig. 4A), which affects only WW domain binding and maintains the integrity of the binding motif of PI3K SH2 domain. The PPtoAA mutation failed to translocate to the nucleus, even though it did block Nedd4-CYT-1 binding (data not shown). When cells expressing PPtoAA mutation were pretreated with the specific PI3 kinase inhibitor, LY294002, HB-EGF administration significantly induced its nuclear translocation (Fig. 7B, D), while the same treatment did not induce either the parental CYT-1 or the other CYT-1 mutations to translocate to the nucleus (data not shown).These results indicated that the inhibition (or absence) of both the PI3 kinase and WW domain-binding motifs are required for ErbB4 nuclear translocation.
Figure 7.
CYT-1 PY2 motif mutation, Y1056A, promoted CYT-1 nuclear translocation following HB-EGF treatment. After HB-EGF treatment, cells were fixed and immunostained with anti-ErbB4 antibody (green). TO-PRO-3 was used for nuclear staining (red). A) Compared to parental CYT-1, Y1056A mutation promoted nuclear translocation, which was comparable to the pattern in CYT-2 cells. B) In the CYT-1 PPtoAA mutation, which blocks the WW binding domain but maintains the integrity of SH2 binding domain of PI-3 kinase, HB-EGF induced its nuclear translocation only in the presence of the PI-3 kinase inhibitor, LY294002 (LY). C, D) Cells with nuclear staining were quantitated from at least 100 ErbB4-expressing cells, and the percentage of cells with nuclear staining was calculated and compared to the control group. Data represent means ± se from 3 independent studies. *P < 0.05 vs. control group; **P < 0.05 vs. HB-EGF treatment alone.
DISCUSSION
It is now well accepted that the ErbB4 JM-a isoform undergoes RIP and releases a soluble ICD on PMA administration or ligand stimulation (20). This ICD harbors an active tyrosine kinase and a C terminus with several potential tyrosine phosphorylation sites as well as several protein-protein interaction domains, such as WW binding domains, an PI3K SH2 domain, and a postsynaptic density/disc large/zona occludens (PDZ) domain-binding motif (21, 22). All these features play a role in ICD function and subcellular localization. Unlike the CYT-1 ICD, which has an extra-WW binding domain and a binding site for PI3K SH2 domain, the CYT-2 ICD exhibits enhanced nuclear localization and transcriptional coactivatior activity (4, 23), while the exact function and destination of the CYT-1 ICD remain unclear.
In the current study, we demonstrated that after cleavage the CYT-1 ICD is rapidly degraded through both proteasomal and lysosomal pathways, as indicated by increased ErbB4 ICD levels following inhibition of either pathway. However, compared to the effect of proteasome inhibitor treatment, lysosome inhibitor treatment produced a less pronounced and later onset blocking effect on CYT-1 ICD degradation, suggesting that the primary pathway for degradation requires the proteosome, and lysosomal degradation may serve as a secondary or default pathway. In this regard, the CYT-1 ICD of ErbB4 may be similar to EGFR, as it has been shown that the proteasomal activity is required for ultimate lysosomal degradation of EGFR (24, 25). Our results showed that the majority of ErbB4 CYT-1 ICDs were polyubiquinated (Fig. 2A), which might be the reason why its degradation was primarily via the proteasome pathway. In general, proteins targeted to the proteasome carry multiubiquitin chains, whereas those targeted for lysosomal degradation are modified by 1–4 ubiquitin moieties (26).
Based on the presence of 2–3 WW domain-binding sites in the ErbB4 C terminus, several WW-containing domain proteins have been shown to interact with ErbB4 and possibly regulate its subcellular localization and functions. These interacting proteins include Yes associated protein (YAP), WW domain-containing oxidoreductase (WWOX), and more recently NIP4/Itch (8, 9, 27,28,29). In the present study we now report that Nedd4 also mediates ErbB4 processing in MDCK II cells.
Nedd4 is highly expressed in kidney, liver, muscle, brain, and heart. The identified substrates so far include ENaC, pTEN, IGF-1R, p63, VEGF-R2, Cbl-b, and EPS15, indicating that it plays an important role in degradation, endocytosis, and protein sorting of multiple intrinsic membrane proteins. Our results showed Nedd4 interacts with ErbB4 CYT-1 both in vitro and in vivo and promotes CYT-1 ubiquitination. The interaction of endogenous Nedd4 with CYT-1 in MDCK II cells also contributes to the degradation of CYT-1 ICD. Of note, in the kidney Nedd4 isoforms have been localized predominantly to the collecting ducts (30), which corresponds to the intrarenal localization of ErbB4 (4). As the predominant ErbB4 isoform in rat kidney is CYT-2 (4), the lack of Nedd4 binding to CYT-2 might play a role in increasing the half-life of this isoform and thereby allowing its nuclear translocation.
We also showed that PY motif mutations in CYT-1 ICD impaired Nedd4-CYT-1 binding with subsequent decreases in CYT-1 ubiquitination. Compared to mutation of the PY1 motif, PY2 and PY3 mutations produced more significant inhibition of Nedd4 binding. The PY1 mutation did not significantly alter CYT-1 stability, while both PY2 and PY3 mutations decreased CYT-1 ICD degradation. Similarly, there were significant increases in the levels of the CYT-1 ICD protein with the PY1 mutation following proteasome inhibition, but only minimal increases of CYT-1 ICD with either the PY2 or PY3 mutations. These results suggested that compared to the PY1 motif, PY2 and PY3 motifs are more crucial for Nedd4-ErbB4 binding and its subsequent degradation. We propose that the necessity for the PY2 motif is the reason Nedd4 fails to bind CYT-2.
We also investigated the subcellular localization of ErbB4 JM-a/CYT-1 mutations after cleavage in response to HB-EGF ligand binding to ErbB4. Interestingly, only Y1056A, which disrupts the binding sites for both WW domain-containing proteins and the PI3K SH2 domain, showed enhanced nuclear translocation after HB-EGF stimulation, whereas PPtoAA mutations at this site did not translocate to the nucleus unless there was concomitant PI-3 kinase inhibition. Unlike Y1056A (PPxY to PPxA) mutation, the PPtoAA (PPxY to AAxY) mutation abrogated only the WW domain binding without affecting the binding site for PI3K SH2 domain. Furthermore, the function of tyrosine on phosphorylation at the 1056 position is different from the tyrosines in the other PY motifs at positions 1035 and 1301, as only phophorylation of tyrosine 1056 serves as the binding site for the SH2 domain of PI3K, which is critical for certain biological functions of ErbB4, as none of the other tyrosine mutations affected the proliferation inhibition related to PI3K activation induced by ErbB4 (31). Traditionally, it has been thought that PI3K is a prosurvival protein and that PI3K signaling prevents apoptosis. However, a recent report suggests that the exact cellular response to PI3K activation may be isoform dependent and cell type specific, which can range from growth inhibition to apoptosis (32, 33). In any event, our results indicate that blockade of this WW binding domain by itself is not sufficient to induce the nuclear translocation of CYT-1, but rather the absence of both the WW domain-binding site and the PI3K SH2 domain-binding site in the CYT-2 ICD are required for nuclear translocation to occur.
While this manuscript was in preparation, two other groups also reported that CYT-1 and CYT-2 differ in endocytosis and degradation by a mechanism mediated by CYT-1 specific PPxY motif interacting with another WW domain-containing E3 ubiquitin ligase, NIP4/Itch, in HEK 293 cells (8, 9). Of note, in both of these studies the association was seen in cells transfected with NIP4/Itch. In contrast, in the present study we found that endogenous Nedd4 interacted with ErbB4 in MDCK II cells.
In conclusion, ErbB4 CYT-1 ICD is a substrate of the E3 ubiquitin ligase, Nedd4. Nedd4 mediates CYT-1 ICD ubiquitination and subsequent degradation in MDCK II cells. All 3 PY motifs in the CYT-1 ICD are important for the binding by Nedd4, while for the ErbB4 nuclear translocation, preventing binding to both the PY motif and the PI3K SH2 domain are required. We propose that after cleavage released soluble CYT-1 ICD is recognized and bound by Nedd4, which transfers ubiquitin to CYT-1 ICD, promoting its degradation and blocking its nuclear translocation. In contrast, the absence of the PY2 motif in the CYT-2 isoform impairs Nedd4 binding and blocks its degradation, making the CYT-2 ICD available for interaction with other WW domain-containing proteins, such as YAP (the effector of the Hippo pathway) (27), which may facilitate CYT-2 nuclear translocation.
Acknowledgments
This work was supported by funds from the Department of Veterans Affairs (R.H.) and National Institutes of Health grant DK51265 (R.H.).
References
- Olayioye M A, Neve R M, Lane H A, Hynes N E. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni C Y, Murphy M P, Golde T E, Carpenter G. γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- Maatta J A, Sundvall M, Junttila T T, Peri L, Laine V J, Isola J, Egeblad M, Elenius K. Proteolytic cleavage and phosphorylation of a tumor-associated ErbB4 isoform promote ligand-independent survival and cancer cell growth. Mol Biol Cell. 2006;17:67–79. doi: 10.1091/mbc.E05-05-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Zhang M Z, Singh A B, Zent R, Harris R C. ErbB4 isoforms selectively regulate growth factor induced Madin-Darby canine kidney cell tubulogenesis. Mol Biol Cell. 2007;18:4446–4456. doi: 10.1091/mbc.E07-03-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor M D, Yarden Y. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene. 2004;23:2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- Baulida J, Kraus M H, Alimandi M, Di Fiore P P, Carpenter G. All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. J Biol Chem. 1996;271:5251–5257. doi: 10.1074/jbc.271.9.5251. [DOI] [PubMed] [Google Scholar]
- Roepstorff K, Grovdal L, Grandal M, Lerdrup M, van Deurs B. Endocytic downregulation of ErbB receptors: mechanisms and relevance in cancer. Histochem Cell Biol. 2008;129:563–578. doi: 10.1007/s00418-008-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundvall M, Korhonen A, Paatero I, Gaudio E, Melino G, Croce C M, Aqeilan R I, Elenius K. Isoform-specific monoubiquitination, endocytosis, and degradation of alternatively spliced ErbB4 isoforms. Proc Natl Acad Sci U S A. 2008;105:4162–4167. doi: 10.1073/pnas.0708333105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omerovic J, Santangelo L, Puggioni E M, Marrocco J, Dall'Armi C, Palumbo C, Belleudi F, Di Marcotullio L, Frati L, Torrisi M R, Cesareni G, Gulino A, Alimandi M. The E3 ligase Aip4/Itch ubiquitinates and targets ErbB-4 for degradation. FASEB J. 2007;21:2849–2862. doi: 10.1096/fj.06-7925com. [DOI] [PubMed] [Google Scholar]
- Kaiser P, Huang L. Global approaches to understanding ubiquitination. Genome Biol. 2005;6:233.1–233.8. doi: 10.1186/gb-2005-6-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham R J, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23:1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- Chen H I, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M, Kitamura K, Adachi M, Miyoshi T, Wakida N, Ura N, Shikano Y, Shinshi Y, Sakamoto K, Hayashi M, Satoh N, Nishitani T, Tomita K, Shimamoto K. Liddle’s syndrome caused by a novel mutation in the proline-rich PY motif of the epithelial sodium channel beta-subunit. J Clin Endocrinol Metab. 2005;90:340–344. doi: 10.1210/jc.2004-1027. [DOI] [PubMed] [Google Scholar]
- Sudol M, Hunter T. NeW wrinkles for an old domain. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Jensen J P, Weissman A M. Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases. J Biol Chem. 1997;272:15085–15092. doi: 10.1074/jbc.272.24.15085. [DOI] [PubMed] [Google Scholar]
- Vecchi M, Baulida J, Carpenter G. Selective cleavage of the heregulin receptor ErbB-4 by protein kinase C activation. J Biol Chem. 1996;271:18989–18995. doi: 10.1074/jbc.271.31.18989. [DOI] [PubMed] [Google Scholar]
- Yagi R, Chen L F, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter G. ErbB-4: mechanism of action and biology. Exp Cell Res. 2003;284:66–77. doi: 10.1016/s0014-4827(02)00100-3. [DOI] [PubMed] [Google Scholar]
- Jones F E. HER4 Intracellular Domain (4ICD) Activity in the developing mammary gland and breast cancer. J Mammary Gland Biol Neoplasia. 2008;13:247–258. doi: 10.1007/s10911-008-9076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia R A, Vasudevan K, Buonanno A. The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Proc Natl Acad Sci U S A. 2000;97:3596–3601. doi: 10.1073/pnas.070042497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundvall M, Peri L, Maatta J A, Tvorogov D, Paatero I, Savisalo M, Silvennoinen O, Yarden Y, Elenius K. Differential nuclear localization and kinase activity of alternative ErbB4 intracellular domains. Oncogene. 2007;26:6905–6914. doi: 10.1038/sj.onc.1210501. [DOI] [PubMed] [Google Scholar]
- Longva K E, Pedersen N M, Haslekas C, Stang E, Madshus I H. Herceptin-induced inhibition of ErbB2 signaling involves reduced phosphorylation of Akt but not endocytic down-regulation of ErbB2. Int J Cancer. 2005;116:359–367. doi: 10.1002/ijc.21015. [DOI] [PubMed] [Google Scholar]
- Alwan H A, van Zoelen E J, van Leeuwen J E. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR de-ubiquitination. J Biol Chem. 2003;278:35781–35790. doi: 10.1074/jbc.M301326200. [DOI] [PubMed] [Google Scholar]
- Levy F, Muehlethaler K, Salvi S, Peitrequin A L, Lindholm C K, Cerottini J C, Rimoldi D. Ubiquitylation of a melanosomal protein by HECT-E3 ligases serves as sorting signal for lysosomal degradation. Mol Biol Cell. 2005;16:1777–1787. doi: 10.1091/mbc.E04-09-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Nagai M, Navin N E, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Omerovic J, Puggioni E M, Napoletano S, Visco V, Fraioli R, Frati L, Gulino A, Alimandi M. Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level. Exp Cell Res. 2004;294:469–479. doi: 10.1016/j.yexcr.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Aqeilan R I, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce C M. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;65:6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- Staub O, Yeger H, Plant P J, Kim H, Ernst S A, Rotin D. Immunolocalization of the ubiquitin-protein ligase Nedd4 in tissues expressing the epithelial Na+ channel (ENaC) Am J Physiol. 1997;272:C1871–C1880. doi: 10.1152/ajpcell.1997.272.6.C1871. [DOI] [PubMed] [Google Scholar]
- Gallo R M, Bryant I, Fry R, Williams E E, Riese D J., 2nd Phosphorylation of ErbB4 on Tyr1056 is critical for inhibition of colony formation by prostate tumor cell lines. Biochem Biophys Res Commun. 2006;349:372–382. doi: 10.1016/j.bbrc.2006.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Sobkiw C L, Logsdon N M, Watt J M, Signoretti S, O'Connell F, Shin E, Shim Y, Pao L, Neel B G, Depinho R A, Loda M, Cantley L C. Modulation of epithelial neoplasia and lymphoid hyperplasia in PTEN+/− mice by the p85 regulatory subunits of phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2005;102:10238–10243. doi: 10.1073/pnas.0504378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger A W, Yoo J Y. Phosphatidylinositol 3-kinase mediates heregulin-induced growth inhibition in human epithelial cells. Anticancer Res. 1997;17:2197–2200. [PubMed] [Google Scholar]