Abstract
Over 2 billion people are estimated to be infected with virulent Mycobacterium tuberculosis, yet fewer than 10% progress to clinical tuberculosis within their lifetime. Twin studies and variations in the outcome of tuberculosis infection after exposure to similar environmental risks suggest genetic heterogeneity among individuals in their susceptibility to disease. In a mouse model of tuberculosis, we have established that resistance and susceptibility to virulent M. tuberculosis is a complex genetic trait. A new locus with a major effect on tuberculosis susceptibility, designated sst1 (susceptibility to tuberculosis 1), was mapped to a 9-centimorgan (cM) interval on mouse chromosome 1. It is located 10–19 cM distal to a previously identified gene, Nramp1, that controls the innate resistance of mice to the attenuated bacillus Calmette–Guérin vaccine strain. The phenotypic expression of the newly identified locus is distinct from that of Nramp1 in that sst1 controls progression of tuberculosis infection in a lung-specific manner. Mice segregating at the sst1 locus exhibit marked differences in the growth rates of virulent tubercle bacilli in the lungs. Lung lesions in congenic sst1-susceptible mice are characterized by extensive necrosis and unrestricted extracellular multiplication of virulent mycobacteria, whereas sst1-resistant mice develop interstitial granulomas and effectively control multiplication of the bacilli. The resistant allele of sst1, although powerful in controlling infection, is not sufficient to confer full protection against virulent M. tuberculosis, indicating that other genes located outside of the sst1 locus are likely also to be important for controlling tuberculosis infection.
Tuberculosis, historically the largest single cause of death of the human species, remains the seventh most important cause of global premature mortality and disability (1). Currently there are 8 million new cases and 2–3 million deaths annually from tuberculosis, and it is projected that a total of 225 million new cases and 79 million deaths will occur between 1998 and 2030 (2). These numbers are particularly striking because a vaccine and chemotherapeutic agents for tuberculosis have been available for more than a half of this century. Tuberculosis remains responsible for 26% of all preventable adult deaths on the planet.
Results of tuberculin testing indicate that infection with Mycobacterium tuberculosis imparts a 10% lifetime risk of developing clinical disease (3). This number is significantly higher when isolated ethnic groups with no previous history of tuberculosis encounter the pathogen. First-contact epidemics demonstrate the extreme susceptibility of the human species to M. tuberculosis (4). Clearly, environmental factors such as poor economic conditions, malnutrition, stress, and overcrowding play a role in determining the susceptibility to tuberculosis in human populations. Nevertheless, variations in the outcome of tuberculosis infection after exposure to similar risk factors (5, 6) and twin studies (7) support a significant role of the human host genetic background in predisposition to tuberculosis.
There is ample experimental evidence for the role of host genetic factors in the outcome of infection with virulent M. tuberculosis in several species of laboratory animals (8–13). The ability to control the establishment and progression of tuberculosis infection was shown to vary between, as well as within, species. Early work on crosses between resistant and susceptible animals proved that resistance to tuberculosis was a heritable trait with a complex, non-Mendelian inheritance (10, 14). By developing a mouse model, which significantly reduced the complexity of the disease-related phenotype, a gene on mouse chromosome 1 controlling early splenic multiplication of the avirulent vaccine strain derived from Mycobacterium bovis, bacillus Calmette–Guérin (BCG), was identified (15, 16). This gene (Nramp1) has been cloned (17) and shown to be identical to the one that controls experimental infection with Leishmania donovani and Salmonella typhimurium (18–20). Although a role for Nramp1 in controlling multiplication of BCG vaccine and several intracellular pathogens within murine macrophages is well documented both in vivo and in vitro, recent studies have failed to establish a role for Nramp1 in protection against virulent M. tuberculosis (21–23).
A significant step in understanding the genetic basis of susceptibility to mycobacterial diseases in humans derived from studies of patients with disseminated infections caused by poorly pathogenic mycobacterial species, such as Mycobacterium avium and BCG (24). Mutations in genes encoding IL-12, the IL-12 receptor, and the interferon-γ receptor were found to be responsible for selective susceptibility to mycobacterial species and highlight the essential role of interferon-γ-mediated immunity in the control of mycobacteria in man (25). These findings are consistent with results obtained earlier using IFN-γ, IFN-γR, and IL-12 gene-disrupted mice, indicating that an IFN-γ-dependent pathway is essential for protection against both BCG and M. tuberculosis, Erdman strain (MTB) infections (26–29). Nevertheless, genetic polymorphisms controlling susceptibility to infection caused by virulent M. tuberculosis in experimental systems have not yet been identified.
In this study, we demonstrate that susceptibility to tuberculosis infection in mice is a complex genetic trait and identify a locus on murine chromosome 1 that has a major effect on the development of the disease caused by virulent M. tuberculosis, especially in the lung.
Materials and Methods
Mice.
C3HeB/FeJ, C57BL/6J mice and their (C3HeB/FeJ × C57BL/6J) F1 hybrids were purchased from The Jackson Laboratory. The mice were housed and bred under specific pathogen-free conditions at the Albert Einstein College of Medicine Animal Institute (Bronx, NY). The mice routinely were tested and found to be free of common mouse pathogens and were used for experiments at 6 to 12 weeks of age. All manipulations with live animals were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Genotyping.
DNA was isolated from the tail tips by standard procedure (30). Genotyping for simple sequence length polymorphism markers polymorphic between C57BL and C3H (31) was performed by PCR with pairs of oligonucleotide primers (Mouse MapPairs) from Research Genetics (Huntsville, AL). PCRs were performed in 384-well plates (Robbins Scientific, Mountain View, CA) in a final volume of 11 μl using an Thermocycler PTC-100 (MJ Research, Watertown, MA) with a heated lid. The PCR mix contained 1× GeneAmp PCR Buffer II, 2.5 mM MgCl2, 200 μM of dNTP, 0.4 units of AmpliTaq Gold DNA polymerase (all obtained from Perkin–Elmer), 0.22 μM of forward and reverse primers, and 1× Rediload dye (Research Genetics). Five microliters of a 20-fold dilution of the stock mouse tail DNA in sterile deionized water was used per reaction. The Thermocycler was programmed as follows: 95°C for 10 min, followed by 35 cycles of 94°C for 20 sec, 50°C for 30 sec, 72°C for 40 sec, and a final step at 60°C for 10 min. Four microliters of reaction was loaded directly on 4% MetaPhor agarose gel (FMC) and run in 1× TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) at 10 V/cm with cooling and recirculation for 30–45 min. The bands were visualized with ethidium bromide staining.
Bacterial Strains and Infection.
Virulent M. tuberculosis (strain Erdman, TMC 107), M. bovis (strain Ravenel, TMC 401), and M. bovis BCG (vaccine strain Pasteur, TMC 1101) initially were obtained from the Trudeau Mycobacterial Culture Collection (TMC; Trudeau Institute, Saranac Lake, NY). To prepare frozen stocks for experiments, mycobacteria were isolated from the organs of infected mice, grown in Dubos oleic albumin complex-enriched Middlebrook 7H9 liquid medium (Difco) containing 0.05% Tween 80 in roller bottles, pelleted by centrifugation (800 × g for 10 min), and resuspended in PBS containing 0.05% Tween 80 (PBS-Tween 80). Glycerol was added to a final concentration of 10%, and the bacterial suspension was frozen and stored at −80°C. Viable bacterial counts were determined by plating serial 10-fold dilutions on Middlebrook 7H10 (Difco) agar plates. For infections, the stock solution was thawed, briefly sonicated (10 sec) to disaggregate clumps, and diluted to a desired concentration (50- to 200-fold) in PBS containing 0.05% Tween 80. Bacterial suspensions were injected through the lateral tail vein in a final volume of 100 μl. Bacterial counts in the organs of the infected mice were enumerated at various time points after infection by plating serial 10-fold dilutions of organ homogenates in PBS-Tween 80 on supplemented Middlebrook 7H10 agar plates. Colonies were counted after 3–4 weeks of incubation at 37°C. Five to seven mice per group were killed at each time point. In long-term experiments, mice that clearly were ill and moribund were killed and recorded as succumbing to infection.
Statistics.
The trait distribution of the time to death data are decidedly nonnormal and require nonparametrical statistical analyses for genetic linkage. Initially, the statistical analysis of the (C3H × B6) F2 intercross was performed by using the χ2 method with 2 degrees of freedom. The genotype distribution of the 25% most susceptible mice was compared with a theoretical expectation of Mendelian assortment. In a subsequent step, a χ2 method was used to compare the chromosome 1 genotypes of the most susceptible animals to all of the animals of the H × B F2 cross (a total of 367 mice were tested in four independent experiments). Next, the analyses of the (C3H × B6) F2 intercross data were performed with the Kruskal–Wallis nonparametric test. The K value (Table 1) gives the Kruskal–Wallis statistics. In addition, mapmaker/exp (version 3.0b) and mapmaker/qtl (version 1.9) software (32) were used to calculate map distances and logarithm of odds (lod) scores by using nonparametric statistics described by Kruglyak and Lander (33). The differences in survival times and bacterial colony-forming units (cfu) per organ between the sst1r/s and sst1s/s siblings in a progeny testing analysis were evaluated by the Student's t test.
Table 1.
Linkage of sst1 to chromosome 1
| Locus | Map position, cM | K | P |
|---|---|---|---|
| D1Mit156 | 34.7 | 119.759 | <0.0001 |
| D1Mit130 | 37.2 | 126.575 | <0.0001 |
| D1Mit46 | 40.2 | 154.258 | <0.0001 |
| D1Mit49 | 44.9 | 202.492 | <0.0001 |
| D1Mit187 | 52.5 | 135.45 | <0.0001 |
| D1Mit498 | 65.2 | 67.893 | <0.0001 |
The statistical analysis of the (C3H × B6) F2 intercross data was performed with the Kruskal–Wallis nonparametric test. A total of 368 (C3H × B6) F2 intercross progeny was analyzed in four independent experiments. The K value gives the Kruskal–Wallis statistics.
Results
Parental Strains and Their Hybrids.
Initially, susceptibility to tuberculosis infection of several standard inbred mouse strains was compared in animals infected i.v. with 106 cfu of virulent M. tuberculosis, Erdman strain (MTB). All tested inbred strains developed tuberculosis, and viable mycobacteria were detectable in their organs both by acid-fast staining of tissue sections and by plating of organ homogenates. Nevertheless, considerable differences between the strains were observed in the rates of bacterial growth as well as in strain survival times. Based on the median survival times, the inbred strains could be classified into highly susceptible (CBA/N, C3HeB/FeJ, and DBA/2), intermediate (BALB/c and 129/SvJ), and relatively resistant (C57BL/6J and C57BL/10J). Our results are consistent with the data of North and coworkers (11). All of the strains of MTB-susceptible mice used in our study (CBA, C3H, and DBA/2) carried the resistant allele of Nramp1, whereas the more resistant strains B6, B10, and BALB/c carried the susceptible allele of Nramp1(34). Dramatic pathology, including pneumonitis and necrosis, developed in the lung of MTB-susceptible C3H mice within 3–4 weeks after infection with MTB. In contrast, interstitial inflammation with very few live bacteria present was observed in the lung lesions of MTB-resistant B6 mice after 6 months of infection. Therefore, the two inbred strains C3HeB/FeJ (MTB-susceptible, Nramp1-resistant) and C57BL/6J (MTB-resistant, Nramp1-susceptible) were selected for further genetic analysis.
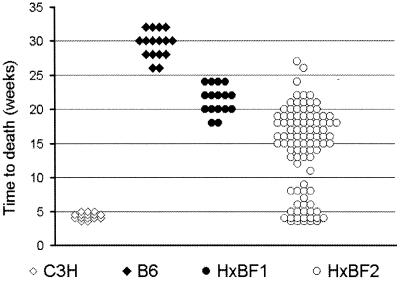
The survival of those strains as well as their F1 and F2 hybrids after infection with 106 cfu of virulent M. tuberculosis is shown in Fig. 1. All of the C3H mice succumbed to infection within the first 4–5 weeks, whereas the B6 mice survived for 6–8 months after the inoculation of the same dose, with some individuals surviving for >10 months. The survival of (C3H × B6) F1 hybrids was intermediate and significantly different from both parents. The F2 hybrids exhibited segregation of susceptibility. In four independent experiments, ≈25% of the F2 population survived for fewer than 10 weeks after infection. The survival of the remaining mice varied between 11 and 26 weeks with the majority of the F2 mice surviving between 14 and 20 weeks postinfection. No significant differences between the F2 hybrid males and females in tuberculosis susceptibility were observed.
Figure 1.
Survival of C3HeB/FeJ and C57BL/6J mice and their F1 and F2 hybrids after i.v. infection with 106 cfu of M. tuberculosis Erdman strain.
Mapping of a Tuberculosis Susceptibility Gene to Chromosome 1.
Analysis of (C3H × B6) F2 hybrids.
To identify genetic factors underlying susceptibility to tuberculosis, a total of 368 (C3H × B6) F2 mice were tested in four sequential experiments. At the initial step, we selected the 46 most susceptible F2 hybrids representing ≈25% of the 198 mice that had been tested in the first two experiments for whole genome scanning. A whole genome scanning was performed by using a total of 127 markers polymorphic between C3H and B6 (31) and spaced at 10- to 20-centimorgan (cM) intervals on 19 mouse autosomes. A χ2 analysis indicated clear linkage of susceptibility to D1Mit46 on chromosome 1. Next, all of the 367 F2 hybrid mice were genotyped for polymorphic markers on chromosome 1 at higher density, including the mice tested for their tuberculosis susceptibility in two additional experiments. The strongest evidence for linkage was observed with D1Mit49 (χ22DF = 170,65; P = 2.6 × 10−40). The results of a Kruskal–Wallis nonparametric test of the data are summarized in Table 1, which confirms linkage of susceptibility to tuberculosis to the region of mouse chromosome 1 around D1Mit49 (K value = 202.49; P < 0.0001). This analysis confines the responsible locus to a 33-cM interval between D1Mit156 and D1Mit498 (32–65 cM). In addition, nonparametric interval analysis using mapmaker/qtl software produced a lod score high of 10.4 in the interval between D1Mit46 and D1Mit49. This analysis also allowed us to define the 90% confidence limits (defined by a full 1-point lod drop) of the susceptibility locus to be in the 11-cM interval between the markers D1Mit46 and D1Mit187. We designated this locus as sst1 for susceptibility to tuberculosis. The B6-derived allele of the sst1 is dominant and confers resistance to infection with virulent M. tuberculosis. Curiously, the segment of mouse chromosome 1 established in our study contains the Nramp1 gene, which had been previously mapped to 39.2 cM on mouse chromosome 1. Therefore, the B6 mice carry the resistant allele of sst1 and the susceptible allele of Nramp1 on the same segment of mouse chromosome 1.
Fine mapping of the sst1.
A progeny testing strategy was used to refine the chromosomal location of sst1. This analysis infers the sst1 genotype of a recombinant parent from phenotypic analysis of its segregating progeny. The male mice produced either by F1 intercross or backcross onto the susceptible background (C3H) were genotyped to find chromosomal recombination within the region containing the sst1 locus. Recombinant males so identified subsequently were backcrossed to C3H females. A minimum of 20 progeny for each recombinant male were genotyped for chromosome 1 markers and were analyzed, after challenge, for cosegregation of B6-derived chromosome 1 loci retained after a recombination event with resistance to tuberculosis infection. In this way, the minimal interval for the sst1 locus was established between the chromosome 1 markers D1Mit334 and D1Mit45 (49.2–57.9 cM) distal to Nramp1 (Fig. 2).
Figure 2.
Segregation of sst1 alleles in the progeny of recombinant males. Males that carried recombinations within the sst1-containing segment of chromosome 1 were identified by genotyping with microsatellite markers using DNA obtained from tail biopsies. Recombinant males were crossed with C3H females, and their progeny was genotyped and tested for susceptibility to i.v. infection with M. tuberculosis. Each column of boxes represents a genotypic class. The number of recombinant males per each genotypic class is denoted by the digit under each column. Each box represents the genotype for loci corresponding to chromosome 1 map on the left. Solid boxes represent homozygotes for B6 alleles, striped boxes are heterozygotes, and open boxes are homozygotes for the C3H allele. The survival times of the progeny heterozygous or homozygous for each marker was compared by using Student's t test. Significant differences between those groups (P < 0.0001) indicated heterozygosity of the recombinant male for the sst1-resistant allele. The sst1 genotypes inferred by this method are at the top of the figure. The two horizontal lines drawn between D1Mit334 and D1Mit45 designate the location of the sst1-containing segment.
sst1-Dependent Control of the Mycobacterial Growth and Lung Inflammation.
In the progeny of the backcross 2 (N3) males, in which the effective size of the segregating B6 genome was reduced to an average of 6.25%, the penetrance of sst1 increased to 100%, possibly because of the reduction of the residual B6 genome. Therefore, the loci that might have significantly affected the phenotypic expression of sst1 in the F2 generation had been eliminated. In this setting, comparative studies of the sst1 heterozygous (relatively resistant) and homozygous for the C3H-derived allele (susceptible) mice offered an opportunity to investigate the phenotype conferred by the sst1 in the course of experimental infection with M. tuberculosis.
Control of bacterial growth.
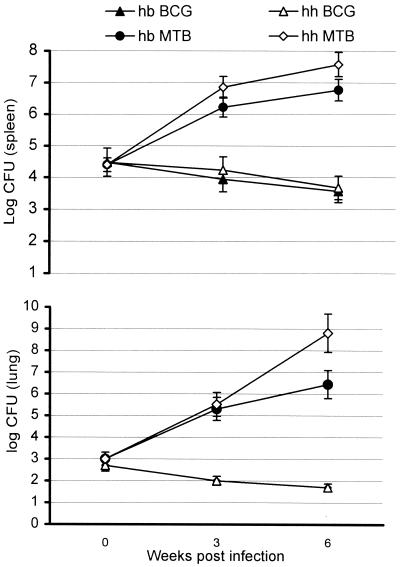
The backcross 3 (N4) population of mice homozygous for the resistant allele of Nramp1, but segregating at the sst1 locus, was infected with 2 × 104 cfu of either virulent M. tuberculosis or BCG vaccine strain (Fig. 3). The bacterial loads in the spleens of mice infected either with virulent or avirulent strain of mycobacteria were similar at 24 h after infection. Consistent with their Nramp1-resistant genotype, all mice, irrespective of the segregation at the sst1 locus, were able to control BCG infection. The numbers of viable BCG in their spleens and lungs never exceeded the primary load and progressively declined over the course of the experiment. In sharp contrast, virulent MTB grew progressively in the spleens and the lungs of both the sst1s/s and sst1r/s mice (Fig. 3). Initially, the growth rates of M. tuberculosis were similar in both groups, but the differences became obvious and significant between the third and the sixth weeks after infection. By that time, the growth rate of M. tuberculosis was diminished in the spleens and the lungs of sst1r/s mice. The increase in the cfu between the third and the sixth weeks of infection in that group was approximately 10-fold in both organs. In contrast, the bacterial burden in the lungs of the sst1-susceptible mice increased more than 1,000-fold during the same period, resulting in a >100-fold difference in the lung bacterial burden between the sst1-resistant and -susceptible mice at 6 weeks postinfection. Meanwhile, the difference between the two groups in spleen cfu, although statistically significant, was only 5-fold. Thus, the sst1 locus appears to be most important for the outcome of infection in the lung.
Figure 3.
Multiplication of M. tuberculosis Erdman and BCG in spleens (Upper) and lungs (Lower) of the backcross 3 (BC3H) mice segregating at the sst1 locus. The animals were infected with equal doses (2 × 104 cfu) of either virulent or vaccine strain of mycobacteria, and five to seven mice were killed per group at each time point.
Histopathology of the lung lesions.
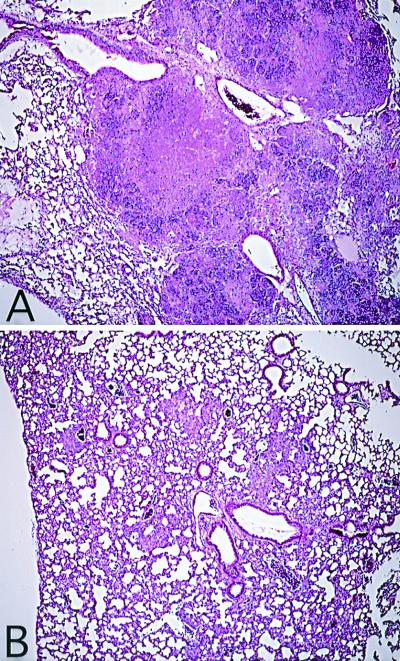
At each time point, the lesions in the lungs of the sst1-resistant siblings were confined exclusively to the lung interstitium and composed of mononuclear cells primarily of macrophage lineage (Fig.4B). The acid-fast bacilli were located intracellularly, and the infected cells were surrounded by significantly more numerous uninfected macrophages and lymphocytes. In contrast, the lesions in the lungs of the mice homozygous for the susceptible allele of the sst1 involved both interstitium and alveolar space (Fig.4A). In addition to macrophages and lymphocytes, the lesions contained polymorphonuclear leukocytes as well as macrophage-like cells with degenerate nuclei. At 6 weeks after infection the majority of the macrophage-like cells within the lesions contained acid-fast bacilli (data not shown). The numbers of the bacteria per cell varied between just a few and >20. The epithelial layer of the airways was disrupted, and masses containing cells and debris erupted into the lumen of alveoli and bronchi. Numerous mycobacteria were located extracellularly, densely populating the necrotic masses. At that time, although the sst1-susceptible mice exhibited symptoms of terminal illness, no weight loss or other visible symptoms of disease progression were observed yet among the heterozygous animals that carried the resistant allele of the sst1. The median survival times of the sst1s/s mice and sst1r/s mice in this experiment were 45 and 96 days, respectively (P < 0.001). These data provide clear evidence for the correlation between mouse survival, multiplication of M. tuberculosis in the lungs, and morphology of the lung lesions depending on the genetic polymorphism at the sst1 locus.
Multigenic Control of Resistance to Tuberculosis: Analysis of Backcross Populations.
Survival times of backcross 2 and backcross 3 animals carrying a single copy of the resistant allele of sst1 were significantly shorter than that of the B6 parental strain (10–16 weeks as compared with 25–32 weeks). The genetic basis of the above differences was addressed by comparing resistance to tuberculosis of the mice that carried one or two copies of the resistant allele of sst1 (Table 2). In the population of mice obtained after three backcrosses on B6 background (BC3B6), the content of the B6-derived genome was above 90%. No significant differences in tuberculosis resistance were detected between the BC3B6 subpopulations that carried either one or two copies of the sst1-resistant allele. The majority of the mice survived for 20–30 weeks after infection. In contrast, the progeny of intercross between heterozygous sst1r/s mice obtained after four backcrosses on C3H (BC4H) carrying less than 5% of the residual B6 genome and either one or two copies of the resistant allele of sst1 survived for a significantly shorter period. The difference between the heterozygous sst1r/s BC4H and BC3B6 mice was statistically significant (P < 10−4). Thus, resistance to tuberculosis was higher in the group of mice that carried a higher proportion of the B6 genome, whereas the sst1r/s and sst1r/r mice in both groups (either BC3B6 or BC4H IC) did not differ in their susceptibility, indicating that the number of the resistant alleles of the sst1 did not influence the survival significantly. These data suggest the presence of polymorphic loci other than the sst1 that also contribute to antituberculous resistance.
Table 2.
Median survival time of the backcross mice is dependent on genes located outside the sst1 locus
| Cross | sst1 genotype | Estimated % of B6 genome | Number of mice per group | MST, days | SD |
|---|---|---|---|---|---|
| (BC4H)IC | S/S | 3.12 | 14 | 28 | 2.5 |
| (BC4H)IC | R/S | 3.12 | 13 | 65 | 15.6 |
| (BC4H)IC | R/R | 3.12 | 12 | 69 | 16.8 |
| BC3B | R/S | 93.75 | 15 | 165 | 54.3 |
| BC3B | R/R | 93.75 | 18 | 172 | 56.5 |
The median survival times (MST) after intravenous infection with 2.5 × 105 cfu of M. tuberculosis Erdman were compared in five groups of the backcross mice differing in their genetic composition. The first three groups were the progeny of intercross between backcross 4 generation onto C3H background (BC4H IC) heterozygous at the sst1 locus. Each mouse in that population carried 3.12% of the B6 genome on average. Another two groups were obtained after three backcrosses onto the B6 background (BC3B), in which transfer of the C3H-derived susceptible allele of the sst1 was ensured by genotyping the breeder males in each backcross generation with the chromosome 1 markers flanking the sst1-containing interval. Because of the segregation at the sst1 locus in this backcross, half of the mice possessed two resistant alleles of the sst1 (R/R) and another half was heterozygous at that locus (R/S). Each mouse in that population was heterozygous at approximately 12.5% of their genomes on average, whereas the rest of their loci (87.5%) were homozygous for the B6-derived alleles. The difference between the groups with the same sst1 genotype but differing in proportion of the B6 genome was highly significant (P < 0.0001).
Discussion
The extent to which pathogenesis of tuberculosis is a function of the pathogen and to what extent it is a consequence of the immune response of the host remain fundamental questions in tuberculosis research. Although the mouse is not a natural host for M. tuberculosis, given the likelihood that all but the most remote human populations have been evolutionarily selected by tuberculosis infection, with a case fatality rate for untreated disease of 50%, the mouse may allow the delineation of genetic control of this infection at a level that may be difficult to study directly in humans. Our model is faithful to several key aspects of tuberculosis infection in its natural human host, specifically the development of extensive lung pathology in the susceptible C3H strain of mice, which correlated with the highest rate of bacterial multiplication, and the death of infected animals. In the human host, M. tuberculosis colonizes the lungs after hematogenous spread from the primary lesion formed at the initial site of bacterial infection, but it is usually the secondary lung lesions that progress, undergo disintegration, and lead to bacterial invasion of the airways that enables transmission to new hosts. Therefore, tropism of virulent M. tuberculosis for lungs and its ability to cause destructive inflammation in that organ are two essential, evolutionary strategic components of its pathogenesis. The genetic dissection of tuberculosis in our model allowed us to uncover a host-related mechanism specifically involved in regulation of the type of lung lesions caused by tuberculous infection, one that was determinative of the outcome of the disease.
The ability of virulent mycobacteria to produce remodeling of lung tissue and eventually extensive tissue damage in the absence of any known direct toxicity has long been considered as evidence for the important role of immunopathology in the pathogenesis of tuberculosis (35). Our studies of the lung lesions in the sst1-susceptible and -resistant backcross mice provide experimental support for that notion. Distinctive pathology starts to appear in the lungs of the susceptible mice after 2–3 weeks of infection, in parallel with the development of immune responses. Necrotic lesions that form within that period constitute an excellent environment for bacterial multiplication, which proceeds in the lungs of susceptible mice at almost the same rate as in logarithmic culture in vitro. In preliminary studies, this stage was characterized at the mRNA level by a burst in expression of proinflammatory chemokines (macrophage inflammatory protein-1α and -β, macrophage inflammatory protein-2, and monocyte chemoattractant protein-1) in the lungs, but not spleens, of the infected mice that was significantly higher in the sst1-susceptible siblings (data not shown).
Genetic mechanisms controlling infection with virulent M. tuberculosis are distinct from those that have been previously shown to play an important role in other diseases caused by intracellular pathogens. A resistant allele of Nramp1 controls intracellular multiplication of S. typhimurium, L. donovani (36), and several species of mycobacteria including BCG (9, 18), but is insufficient to control early multiplication of M. tuberculosis (21–23). Virulent MTB grew progressively in the spleens and livers of intravenously infected Nramp1-resistant mice within the first 2 weeks after infection (Fig. 3). Of interest, the susceptibility of mice to infection with M. bovis Ravenel, a virulent strain in the same species M. bovis from which attenuated BCG was derived, also is controlled by sst1, rather than the Nramp1 (data not shown). In contrast, the growth of attenuated BCG, which basically does not colonize the host's lung tissue well, is controlled by the Nramp1 but not the sst1 locus. The sst1 locus controls the progression of the lung disease caused by virulent mycobacteria, but has lesser impact on the growth of MTB in other organs, such as spleen and liver (Fig. 3). Therefore, we conclude that the sst1-dependent mechanism of resistance is largely organ (lung)-specific. It is currently unknown whether sst1 may control susceptibility to infection with other intracellular pathogens that target the lung.
How can one explain the findings that Nramp1 controls susceptibility to attenuated BCG, but not to virulent M. tuberculosis? The Nramp1 gene product is a 12-transmembrane domain molecule, highly homologous to Nramp2 recently found to be a plasma membrane divalent cation transporter, and it has been hypothesized that Nramp1, which is expressed on the phagosomal membrane, may transport iron required for bacterial growth (37). Because iron is limiting in vacuoles, a vacuolar iron transporter could influence survival of intracellular pathogens residing in the vacuolar compartment of infected cells, such as BCG. In the case of M. tuberculosis, however, there is evidence that the pathogen creates a channel in the vacuolar membrane that is of a molecular size that would allow nutrients and macromolecules to gain entry to the bacterium-containing vacuole and secreted antigens and putative toxic molecules to be released into the host cell cytoplasm (38, 39). Because M. tuberculosis gains access directly to the cytoplasmic compartment of infected cells, Nramp1 would be irrelevant to controlling the content of the MTB-containing phagosome and the growth of the virulent MTB within macrophage. Another possible explanation may be drawn from the observation that the Nramp1-encoded protein is localized to late endosomal and lysosomal compartment that fuses to the BCG-containing phagosome on its maturation (40). Therefore, the microenvironment of the BCG-containing vacuoles can be directly influenced by the Nramp1 function. In contrast, the MTB phagosome exhibits maturational arrest, which restricts its ability to fuse with late endosomes and lysosomes. The ability to alter phagosome maturation is characteristic only of the phagosomes containing live MTB, and it has been proposed that bacterial factors produced by virulent mycobacteria intracellularly may be responsible for that phenomenon (41, 42). Such factors may render the MTB-containing phagosomes spatially separated from the Nramp1-containing intracellular compartment, making the bacteria inaccessible to a putative effector function of Nramp1. However, it still remains to be shown that the Nramp1 protein is excluded from the MTB-containing phagosomes. In either case, although Nramp1 would be predicted to exert an effect on BCG replication, it would not on growth of M. tuberculosis.
A recent study in human populations has demonstrated association of the Nramp1 polymorphism with tuberculosis susceptibility. Nevertheless, the Nramp1 contribution accounted for less than 3% of the overall genetic component (43). Other studies failed to identify linkage of Nramp1 alleles with human tuberculosis (21). This finding is not entirely unexpected, because genetic control of tuberculosis is known to be a multigenic trait and, therefore, possibly dependent on epistatic interactions of functionally polymorphic loci. At least two genes (Nramp1 and sst1) were mapped less than 18 cM apart on mouse chromosome 1. Both of them are located within the region of syntenic homology with human chromosome 2 (44). Should a human homolog of the sst1 gene exist, it is likely to be located within the same interval. Accordingly, interpretation of the experimental data (obtained by studying congenic and knockout mice), as well as the data obtained in other species concerning the role of Nramp1 in susceptibility to tuberculosis, must now take into account the possible contribution of the closely linked sst1 locus. The present study demonstrates that the contribution of Nramp1 to tuberculosis resistance may be confounded by the presence of another closely linked locus (sst1) with a stronger effect on development of infection.
In addition to establishing a major effect of sst1 on the course of tuberculosis in mice, our experiments have provided evidence for multigenic control of tuberculosis resistance in a mouse model (45). As shown in Table 2, resistance to tuberculosis of mice that carry the resistant allele of sst1 is a quantitative trait and is not determined by the number of the sst1-resistant alleles. It depends rather on the presence of yet unknown loci outside the sst1-containing interval. Identification of those loci is in progress using recombinant congenic strains (46, 47) and a set of recently developed sst1-congenic inbred mouse strains (I.K., unpublished data).
It is to be expected that the analysis of gene interactions in the control of pathogenesis of tuberculosis can be a powerful means of understanding the interplay between host resistance and M. tuberculosis virulence factors. More generally, with a long history of evolutionary selection of human populations by infectious diseases, and with a multiplicity of genetic factors influencing the outcome of infections with different pathogens, perhaps each with small incremental effects, it may be difficult to identify statistically significant genetic associations and more difficult still to define gene interactions through studies carried out in humans. We believe that exploration of the genetics of infections in mice and characterization of the relevant genes and molecular pathways in such simpler systems have the potential to yield tools that permit more precise characterization of the genetic mechanisms controlling human infectious diseases, in some cases, than may be possible from direct studies in humans.
Figure 4.
Histopathology of lungs from M. tuberculosis-infected backcross 3 (BC3H) mice 6 weeks after infection with 2 × 104 cfu of M. tuberculosis (hematoxylin/eosin staining). (A) sst1s/s mice. (B) sst1r/s mice.
Acknowledgments
We thank Dr. Raju Kutcherlapati (Albert Einstein College of Medicine) for stimulating discussions and support, and Jeffrey Friedman (The Rockefeller University, New York) for helpful advice. We thank Dr. R. C. Jansen and Ms. A. de Jong from the Center for Plant Reproduction Research (Wageningen, The Netherlands) for the statistical analysis of the F2 data. We are grateful to Lola Okunola and Gerard Geelen for technical assistance. This work was supported by National Institutes of Health Grant HL59836 and by the Howard Hughes Medical Institute (B.R.B.).
Abbreviations
- cM
centimorgan
- BCG
bacillus Calmette–Guérin
- MTB
M. tuberculosis, Erdman strain
- cfu
colony-forming units
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150227197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150227197
References
- 1.Raviglione M C, Snider D E, Kochi A. J Am Med Assoc. 1995;273:220–226. [PubMed] [Google Scholar]
- 2.Murray C, Salomon J. Proc Natl Acad Sci USA. 1998;95:13881–13886. doi: 10.1073/pnas.95.23.13881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom B R, Murray C J. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 4.Sousa A O, Salem J I, Lee F K, Vercosa M C, Cruaud P, Bloom B R, Lagrange P H, David H L. Proc Natl Acad Sci USA. 1997;94:13227–13232. doi: 10.1073/pnas.94.24.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill A V. Annu Rev Immunol. 1998;16:593–617. doi: 10.1146/annurev.immunol.16.1.593. [DOI] [PubMed] [Google Scholar]
- 6.Stead W W. Ann Intern Med. 1992;116:937–941. doi: 10.7326/0003-4819-116-11-937. [DOI] [PubMed] [Google Scholar]
- 7.Kallmann F J, Reisner D. Am Rev Tuberc. 1943;47:549–574. [Google Scholar]
- 8.Allison, M. J., Zappasodi, P. & Lurie, M. B. (1962) 85, 553–569. [DOI] [PubMed]
- 9.Buschman E, Apt A S, Nickonenko B V, Moroz A M, Averbakh M H, Skamene E. Springer Semin Immunopathol. 1988;10:319–336. doi: 10.1007/BF02053844. [DOI] [PubMed] [Google Scholar]
- 10.Lynch C J, Pierce-Chase C H, Dubos R. J Exp Med. 1965;121:1051–1070. doi: 10.1084/jem.121.6.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medina E, North R J. Immunology. 1998;93:270–274. doi: 10.1046/j.1365-2567.1998.00419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikonenko B V, Apt A S, Mezhlumova M B, Avdienko V G, Yeremeev V V, Moroz A M. Clin Exp Immunol. 1996;104:37–43. doi: 10.1046/j.1365-2249.1996.d01-643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orrell J M, Brett S J, Ivanyi J, Coghill G, Grant A, Beck J S. J Pathol. 1992;166:77–82. doi: 10.1002/path.1711660112. [DOI] [PubMed] [Google Scholar]
- 14.Lurie M B, Zappasodi P, Dannenberg A M, Jr, Weiss G H. Am J Hum Genet. 1952;4:302–314. [PMC free article] [PubMed] [Google Scholar]
- 15.Gros P, Skamene E, Forget A. J Immunol. 1981;127:2417–2421. [PubMed] [Google Scholar]
- 16.Skamene E, Gros P, Forget A, Kongshavn P A L, St. Charles C, Taylor B A. Nature (London) 1982;297:506–509. doi: 10.1038/297506a0. [DOI] [PubMed] [Google Scholar]
- 17.Vidal S M, Malo D, Vogan K, Skamene E, Gros P. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 18.Vidal S, Tremblay M L, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. J Exp Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P. Infect Immun. 1996;64:2923–2929. doi: 10.1128/iai.64.8.2923-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barton C H, Whitehead S H, Blackwell J M. Mol Med. 1995;1:267–279. [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw M A, Collins A, Peacock C S, Miller E N, Black G F, Sibthorpe D, Lins-Lainson Z, Shaw J J, Ramos F, Silveira F, Blackwell J M. Tuber Lung Dis. 1997;78:35–45. doi: 10.1016/s0962-8479(97)90014-9. [DOI] [PubMed] [Google Scholar]
- 22.North R J, LaCourse R, Ryan L, Gros P. Infect Immun. 1999;67:5811–5814. doi: 10.1128/iai.67.11.5811-5814.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.North R J, Medina E. Trends Microbiol. 1998;6:441–443. doi: 10.1016/s0966-842x(98)01364-x. [DOI] [PubMed] [Google Scholar]
- 24.Altare F, Jouanguy E, Lamhamedi S, Doffinger R, Fischer A, Casanova J L. Curr Opin Immunol. 1998;10:413–417. doi: 10.1016/s0952-7915(98)80114-3. [DOI] [PubMed] [Google Scholar]
- 25.Jouanguy E, Lamhamedi-Cherradi S, Lammas D, Dorman S E, Fondaneche M C, Dupuis S, Doffinger R, Altare F, Girdlestone J, Emile J F, et al. Nat Genet. 1999;21:370–378. doi: 10.1038/7701. [DOI] [PubMed] [Google Scholar]
- 26.Cooper A M, Dalton D K, Stewart T A, Griffin J P, Russell D G, Orme I M. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper A M, Magram J, Ferrante J, Orme I M. J Exp Med. 1997;186:39–45. doi: 10.1084/jem.186.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flynn J L, Chan J, Triebold K J, Dalton D K, Stewart T A, Bloom B R. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalton D K, Pitts-Meek S, Keshav S, Figari I S, Bradley A, Stewart T A. Science. 1993;259:1739–1742. doi: 10.1126/science.8456300. [DOI] [PubMed] [Google Scholar]
- 30.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietrich W F, Miller J C, Steen R G, Merchant M, Damron D, Nahf R, Gross A, Joyce D C, Wessel M, Dredge R D, et al. Nat Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- 32.Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newburg L. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 33.Kruglyak L, Lander E S. Genetics. 1995;139:1421–1428. doi: 10.1093/genetics/139.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malo D, Vogan K, Vidal S, Hu J, Cellier M, Schurr E, Fuks A, Bumstead N, Morgan K, Gros P. Genomics. 1994;23:51–61. doi: 10.1006/geno.1994.1458. [DOI] [PubMed] [Google Scholar]
- 35.Rook G A, Stanford J L. Curr Top Microbiol Immunol. 1996;215:239–262. doi: 10.1007/978-3-642-80166-2_11. [DOI] [PubMed] [Google Scholar]
- 36.Glynn A A, Bradley D J, Blackwell J M, Plant J E. Lancet. 1982;2:151. doi: 10.1016/s0140-6736(82)91110-2. (lett.). [DOI] [PubMed] [Google Scholar]
- 37.Gruenheid S, Gros P. Curr Opin Microbiol. 2000;3:43–48. doi: 10.1016/s1369-5274(99)00049-1. [DOI] [PubMed] [Google Scholar]
- 38.Teitelbaum R, Cammer M, Maitland M L, Freitag N E, Condeelis J, Bloom B R. Proc Natl Acad Sci USA. 1999;96:15190–15195. doi: 10.1073/pnas.96.26.15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazzaccaro R J, Gedde M, Jensen E R, van Santen H M, Ploegh H L, Rock K L, Bloom B R. Proc Natl Acad Sci USA. 1996;93:11786–11791. doi: 10.1073/pnas.93.21.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Searle S, Bright N A, Roach T I, Atkinson P G, Barton C H, Meloen R H, Blackwell J M. J Cell Sci. 1998;111:2855–2866. doi: 10.1242/jcs.111.19.2855. [DOI] [PubMed] [Google Scholar]
- 41.Russell D G, Dant J, Sturgill-Koszycki S. J Immunol. 1996;156:4764–4773. [PubMed] [Google Scholar]
- 42.Clemens D L. Trends Microbiol. 1996;4:113–118. doi: 10.1016/0966-842X(96)81528-9. [DOI] [PubMed] [Google Scholar]
- 43.Bellamy R, Ruwende C, Corrah T, McAdam K P, Whittle H C, Hill A V. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 44.Schurr E, Skamene E, Morgan K, Chu M L, Gros P. Genomics. 1990;8:477–486. doi: 10.1016/0888-7543(90)90034-r. [DOI] [PubMed] [Google Scholar]
- 45.Kramnik I, Demant P, Bloom B B. Novartis Found Symp. 1998;217:120–131. doi: 10.1002/0470846526.ch9. [DOI] [PubMed] [Google Scholar]
- 46.Stassen A P, Groot P C, Eppig J T, Demant P. Mamm Genome. 1996;7:55–58. doi: 10.1007/s003359900013. [DOI] [PubMed] [Google Scholar]
- 47.Lipoldová M, Svobodová M, Krulová M, Havelková H, Badalová J, Nohýnková E, Holán V, Hart A, Volf P, Demant P. Genes Immun. 2000;1:200–206. doi: 10.1038/sj.gene.6363660. [DOI] [PubMed] [Google Scholar]