Abstract
p21Cip1 is an inhibitor of cell cycle progression that promotes G1-phase arrest by direct binding to cyclin-dependent kinase and proliferating cell nuclear antigen. Here we demonstrate that mitogenic stimuli, such as epidermal growth factor treatment and oncogenic Ras transformation, induce p21Cip1 downregulation at the posttranslational level. This downregulation requires the sustained activation of extracellular signal-regulated kinase 2 (ERK2), which directly interacts with and phosphorylates p21Cip1, promoting p21Cip1 nucleocytoplasmic translocation and ubiquitin-dependent degradation, thereby resulting in cell cycle progression. ERK1 is not likely involved in this process. Phosphopeptide analysis of in vitro ERK2-phosphorylated p21Cip1 revealed two phosphorylation sites, Thr57 and Ser130. Double mutation of these sites abolished ERK2-mediated p21Cip1 translocation and degradation, thereby impairing ERK2-dependent cell cycle progression at the G1/S transition. These results indicate that ERK2 activation transduces mitogenic signals, at least in part, by downregulating the cell cycle inhibitory protein p21Cip1.
The cyclin-dependent kinase (CDK) inhibitor p21Cip1 is important in the control of cell proliferation, differentiation, senescence, and apoptosis. p21Cip1 was initially identified as a component of a quaternary complex containing CDK, cyclin, and proliferating cell nuclear antigen (PCNA) that regulates cell cycle progression and DNA replication. Overexpression of p21Cip1 results in cell cycle arrest (11), and p21Cip1 expression is induced at the transcriptional level by activation of p53 (10). Although the inhibitory role of p21Cip1 is well established, a positive role for p21Cip1 as an assembly factor for cyclin D1-CDK4/6 complexes has also been shown (8, 18). In addition to transcriptional regulation, p21Cip1 function can be regulated at the posttranslational level. AKT, protein kinase C zeta, CDK2, and glycogen synthase kinase 3β (GSK-3β) phosphorylate p21Cip1 at Thr145, Ser146, Ser130, and Thr57/Ser114, respectively, resulting in inhibition, translocation, or destabilization of p21Cip1 (19, 27, 28, 31, 40, 41). Paradoxically, phosphorylation of Ser130 (by JNK1 or p38α) or Ser146 (by AKT) has also been reported to enhance p21Cip1 stability (16, 20). p21Cip1 is a highly unstable protein (7, 21) that has been shown to accumulate following proteasome inhibition (3, 29). Multiple mechanisms appear to be involved in the proteasomal degradation of p21Cip. Some of these mechanisms are ubiquitination dependent, and others are ubiquitination independent (33), including mechanisms mediated by an Skp2-containing SCF (Skp1, Cullin, and F-box protein) complex (2, 5) and by N-terminal ubiquitination (4) and a mechanism mediated by direct p21Cip1 interaction with the C8 subunit of the 20S proteasome (34). We previously demonstrated that nucleocytoplasmic translocation of p21Cip1, mediated by two nuclear export sequences (NES), is required for p21Cip1 degradation (13).
The Ras-dependent extracellular signal-regulated kinase 1/2 (ERK1/2) pathway plays a central role in controlling cell proliferation (22). Various mechanisms have been proposed to explain this action of the ERK1/2 pathway. For example, the ERK pathway has been shown to induce cyclin D1 transcription (1, 38) and to enhance the stability of the c-Myc protein (32), which play a central role in cell cycle progression and cell growth. A recent study has revealed that ERK associates with and phosphorylates GSK-3β, resulting in inactivation of GSK-3β and upregulation of β-catenin, which in turn stimulates c-Myc and cyclin D1 transcription (9). ERK also directly interacts with and phosphorylates FOXO3a, downregulating it by enhancing its degradation, thereby promoting cell proliferation (39). While these observations have provided tantalizing mechanisms, a complete picture of ERK1/2 regulation of cell proliferation has yet to emerge (22).
Recently we observed that p21Cip1 protein levels were decreased in hepatocytes from H-RasV12-transgenic mice, which contain high levels of constitutively activated ERK. Here we focus on p21Cip1 downregulation as an alternative mechanism of Ras-ERK signaling-mediated cell proliferation. We demonstrate that ERK2 phosphorylates p21Cip1 on both Thr57 and Ser130 and show that this phosphorylation leads to cytoplasmic translocation, ubiquitination, and proteasome-dependent degradation of p21Cip1, thereby resulting in cell cycle progression.
MATERIALS AND METHODS
Reagents.
DNase-free RNase A and protein A-agarose were purchased from Sigma. MG-132 was purchased from EMD Biosciences. Nickel affinity agarose from Qiagen and 4′,6′-diamidino-2-phenylindole (DAPI) from Roche were used. Cycloheximide, U0126, LY294002, and epidermal growth factor (EGF) were purchased from Calbiochem. Blasticidin, zeocin, and tetracycline were purchased from Invitrogen. The antibodies against green fluorescent protein (GFP) (FL), H-Ras (C20), lamin A/C (N18), p21Cip1 (C19 and F5), ubiquitin (P4D1), MEK1/2 (12B), and β-actin (I19) were all obtained from Santa Cruz Biotechnology. Antibodies against hemagglutinin (HA) (H6908 [Sigma] and 12CA5 [Roche]), Myc (Invitrogen), FLAG (M2; Sigma), tubulin (DM1A; Calbiochem), ERK2 (Qiagen), tubulin (Calbiochem), and phospho-ERK1/2 and phospho-MEK1/2 (Cell Signaling Technology) were used.
Cell culture.
HCT116 p21Cip1−/− (human colon cancer), HeLa (human cervical cancer), HEK293/HEK293T (human embryonic kidney), NIH 3T3 (mouse fibroblast), and H-Ras-transformed NIH 3T3 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (Invitrogen), 20 mM HEPES, and antibiotics (Invitrogen) at 37°C in a humidified atmosphere containing 5% CO2. HCT116 p21Cip1−/−, HeLa, HEK293, and HEK293T cells were transfected with various plasmids using calcium phosphate or Lipofectamine/Plus reagents (Invitrogen).
Construction of plasmids.
C-terminally HA-epitope-tagged human p21Cip1 cDNA was generated by PCR and subcloned into the BamHI and XhoI sites of pcDNA3 (Invitrogen). Site-directed mutagenesis was carried out according to the manufacturer's protocol (Stratagene). The Thr57 and Ser130 residues in p21Cip1 were replaced with alanine (p21Cip1 T57A S130A). Two phenylalanines in the FXF docking motif (51FDF) and two arginines in the KIM docking motif were together converted to alanine (p21Cip1 F51A F53A R93A R94A). Human ERK1 and mouse ERK2 cDNAs were cloned into the pFLAG-CMV2 vector (Sigma). Mouse ERK2 was also inserted into the pCMV (Clontech) vector with an N-terminal HA tag. Human ubiquitin cDNA was inserted into the pCMV (Clontech) vector with an N-terminal His6 tag. For bacterial expression, human p21Cip1 cDNA was cloned into the pET-28a vector (Novagen). Short hairpin RNAs (shRNAs) targeting human ERK2 mRNA were designed according to the manufacturer's protocol (Invitrogen). To construct an shRNA expression vector, oligonucleotides (5′-CACCGGACCTCATGGAAACAGATCTCGAAAGATCTGTTTCCATGAGGTCC-3′ and 5′-AAAAGGACCTCATGGAAACAGATCTTTCGAGATCTGTTTCCATGAGGTCC-3′) were annealed and ligated into the pENTR/H1/TO vector.
Immunoprecipitation and immunoblotting.
Cells washed in phosphate-buffered saline (PBS) were lysed with lysis buffer [20 mM HEPES, 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM NaF, 0.1 mM Na3VO4, 0.1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride, 2 μg/ml leupeptin, and 5 μg/ml aprotinin] and centrifuged to remove insoluble debris. Cell lysates were incubated with anti-HA or anti-FLAG M2 agarose (Sigma) for 4 h at 4°C. The immobilized proteins were collected by centrifugation, washed three times with lysis buffer, and solubilized by boiling for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (10 mM Tris-HCl [pH 6.8], 1 mM EDTA, 10% glycerol, 0.0004% bromophenol blue, 4% SDS, 20 mM dithiothreitol). After electrophoresis, proteins were transferred onto a nitrocellulose membrane (Schleicher & Schuell), blocked with 5% skim milk, washed briefly, and incubated with specific antibodies. Blots were washed three times with TTBS buffer (20 mM Tris [pH 7.4], 150 mM NaCl, and 0.05% Tween 20) and incubated with horseradish peroxidase-conjugated anti-mouse (Pierce), anti-rabbit (Pierce), or anti-goat imunoglobulin G (Sigma) antibody and then developed using a chemiluminescence detection system (Pierce).
Protein purification.
Recombinant His6-tagged p21Cip1 was expressed in Escherichia coli and purified by an inclusion body refolding method (14). Briefly, the inclusion body fraction was solubilized in 8 M urea and dialyzed against a buffer containing 20 mM Tris [pH 8.0], 150 mM NaCl, 0.1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride, 2 μg/ml leupeptin, and 5 μg/ml aprotinin. The dialysate containing refolded proteins was passed over a nickel column to increase homogeneity. The purity of the resulting His6-p21Cip1 preparation as assessed by SDS-PAGE was >90%.
In vitro kinase assay.
For in vitro kinase assays, recombinant active ERK2 (kindly provided by E. J. Woo) and His6-tagged p21Cip1 purified from E. coli were used. His6-p21Cip1 immobilized on nickel agarose was incubated with ERK2 in a kinase reaction mixture containing 25 mM Tris-HCl, 10 mM MgCl2, 0.1 mM EGTA, 2 mM dithiothreitol, 0.1 mM Na3VO4, 50 μM ATP, and 5 μCi of [γ-32P]ATP for 30 min at 30°C. The reaction was terminated by the addition of SDS sample buffer, and proteins were separated by SDS-PAGE and visualized by autoradiography. Immunoprecipitates from mammalian cell lysates were also used in kinase assays. Accordingly, HEK293T cells were transfected with either HA-tagged wild-type p21Cip1, T57A S130A mutant p21Cip1, or the HA-ERK2 expression vector, and 36 h after transfection each population of cells was lysed and separately immunoprecipitated with an anti-HA antibody. HA-p21Cip1 immunoprecipitates and HA-ERK2 immunoprecipitates or recombinant active ERK2 were incubated in kinase reaction buffer containing [γ-32P]ATP. Phosphorylated proteins were resolved by SDS-PAGE and analyzed by autoradiography.
Phosphoamino acid analysis.
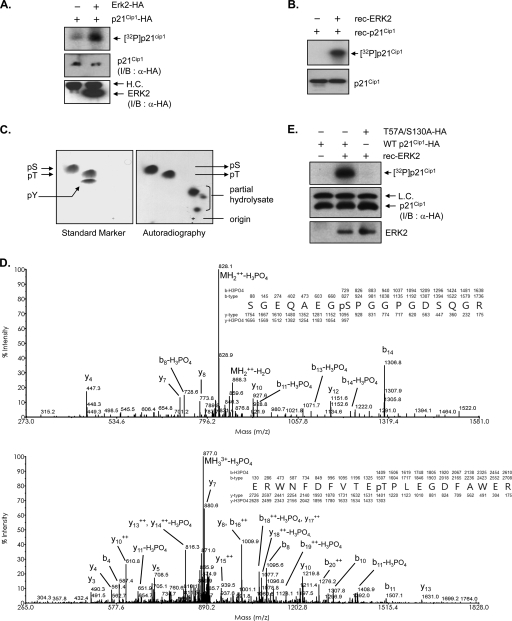
Recombinant His6-p21Cip1 protein purified from E. coli was incubated with the active fraction of purified recombinant ERK2 in the presence of [γ-32P]ATP. 32P-labeled proteins were separated by SDS-PAGE on 15% acrylamide gels and blotted onto a polyvinylidene difluoride membrane. Radiolabeled protein was excised from the membrane, washed several times with water, and hydrolyzed for 1 h at 110°C in 6 M HCl. The acid was evaporated, and the sample was dissolved in 10 μl of water and mixed with 0.1 μg cold phosphoserine, phosphothreonine, and phosphotyrosine standards. Phosphoamino acids were separated on cellulose-thin-layer-chromatography plates (Merck) using the HTLE 7000 (CBS Scientific) apparatus. The first-dimension separation was at 1.5 kV for 20 min in TLE (pH 1.9) buffer containing 2.2% formic acid and 7.8% acetic acid; the second-dimension separation was at 1.3 kV for 13 min in TLE (pH 3.5) buffer containing 5% acetic acid and 0.5% pyridine. Phosphoamino acids were located by ninhydrin staining, and the position of 32P-amino acids was determined by autoradiography.
MS/MS.
The electrophoretically separated p21Cip1 protein band was excised and stain stripped in 50% acetonitrile-25 mM ammonium bicarbonate, dehydrated with 100% acetonitrile, and dried in a vacuum evaporator. Gel pieces were rehydrated in 25 mM ammonium bicarbonate and treated with 12 ng/μl trypsin overnight at 37°C. Peptides were extracted with 50% acetonitrile-25 mM ammonium bicarbonate and then with 50% acetonitrile containing 0.3% trifluoroacetic acid. The extracts were combined, dried, and reconstituted in 5% acetonitrile containing 0.1% trifluoroacetic acid. Protein digests were analyzed using an LTQ ion trap mass spectrometer (ThermoFinnigan) coupled with a nano-electrospray ionization source and an Agilent 1100 Series Capillary LC inlet system (Agilent Technologies). Peptides were separated on a capillary column (75-μm inside diameter by 150 mm) prepared by packing a fused-silica capillary with 200-Å Magic C18AQ resin (Michrom BioResources Inc.). The chromatographic system was configured to generate a flow rate of approximately 200 nl/min using a precolumn split. Peptides were eluted over 60 min using a 5 to 40% linear gradient of acetonitrile containing a constant 0.1% formic acid. The eluted peptides were analyzed in data-dependent MS experiments employing dynamic exclusion. Each analytical event consisted of four consecutive scans: the first, full MS (m/z 300 to 2,000) scan was followed by three tandem mass spectrometry (MS/MS) scans on the three most intense peptide ions from the full MS spectrum. The spray voltage was set at 1.4 kV, and the normalized collision energy for MS/MS peptide decomposition was set at 30%. The resultant spectra were analyzed using SEQUEST and then manually inspected using the Bioworks 3.1 software program (ThermoFinnigan).
Immunofluorescence.
HeLa cells were plated onto glass coverslips at a density of 7 × 104/ml in six-well dishes. The cells were transfected with 0.5 μg of wild-type p21Cip1 or T57A S130A mutant p21Cip1 together with a FLAG-ERK2 expression construct or control vector. At 42 h after transfection, cells were treated with MG132 for 2 h, fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and incubated with rabbit polyclonal anti-HA and mouse monoclonal anti-FLAG antibodies for 1 h. Cells were then incubated with fluorescein isothiocyanate-conjugated antirabbit secondary antibody (Sigma) or tetramethylrhodamine isothiocyanate-conjugated antimouse secondary antibody (Sigma) plus DAPI. Coverslips were mounted in 10% glycerol and examined under a fluorescence microscope (Axioskop; Zeiss).
Flow cytometry analysis.
Cell cycle progression was assayed by DNA content using propidium iodide and flow cytometry. HEK293 and HCT116 p21−/− cells were cotransfected with wild-type p21Cip1 or T57A S130A mutant p21Cip1 and an ERK2 expression plasmid or control vector using Lipofectamine/Plus reagents. A vector encoding membrane-bound GFP was cotransfected to identify transfected cells. Approximately 1 × 106 cells were trypsinized and washed twice with ice-cold PBS and then fixed overnight at −20°C in 70% ethanol. Immediately before flow cytometry, the cells were resuspended in PBS containing propidium iodide (50 μg/ml) and DNase-free RNase (10 μg/ml). Flow cytometry was performed using a FACScalibur (BD biosciences) system with CELLquest software. The percentages of cells in different phases of the cell cycle within the GFP-positive population were determined using the software program ModFit.
RESULTS
ERK2 activation downregulates p21Cip1 at the posttranslational level.
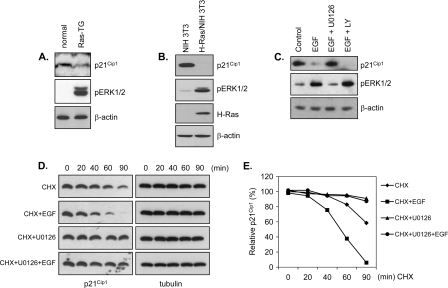
In hepatocytes from H-RasV12-transgenic mice expressing a constitutively active form of Ras (36), p21Cip1 levels were decreased and inversely correlated with ERK1/2 phosphorylation (Fig. 1A). Consistent with this observation, we found that p21Cip1 levels were decreased in H-Ras-transformed NIH 3T3 cells in association with elevated levels of phosphorylated ERK1/2 (Fig. 1B). We next tested whether activating the ERK pathway with EGF would produce similar effects on p21Cip1 levels. HeLa cells were incubated with or without the MEK1/2 inhibitor, U0126, and then treated with EGF. EGF stimulation decreased p21Cip1 levels, an effect that was abrogated by pretreatment with U0126 (Fig. 1C, upper panel), which inhibits EGF-induced ERK activation (Fig. 1C, middle panel), but not by pretreatment with LY294002, which inhibits EGF-induced phosphatidylinositol-3 kinase activation. These data indicate that EGF-induced p21Cip1 downregulation requires the ERK pathway. To determine whether the ERK pathway-mediated downregulation of p21Cip1 was due to effects on p21Cip1 protein stability, we examined the half-life of the p21Cip1 protein in cells treated with U0126. Following inhibition of protein synthesis with cycloheximide, p21Cip1 levels were measured in the presence and absence of EGF, U0126, and EGF plus U0126 (Fig. 1D and E). Blocking de novo p21Cip1 protein synthesis with cycloheximide resulted in a rapid decrease in the p21Cip1 protein (half-life, ∼90 min), a decrease that was accelerated by EGF treatment (half-life, ∼50 min) but dramatically attenuated in the presence of the MEK inhibitor, U0126. These data indicate that the ERK pathway regulates p21Cip1 by reducing its protein stability.
FIG. 1.
Downregulation of p21Cip1 by ERK activation. (A) p21Cip1 levels are decreased in Ras transgenic mice. Hepatic tissue from normal and H-RasV12-transgenic mice was lysed and immunoblotted with anti-p21Cip1 and anti-phospho-ERK antibodies and normalized with anti-β-actin antibody. (B) Ras overexpression reduces p21Cip1 level. H-Ras-transformed and mock-transformed NIH 3T3 cells were lysed and immunoblotted with anti-p21Cip1, anti-phospho-ERK, and anti-H-Ras antibodies and normalized with anti-β-actin antibody. (C) ERK inhibition blocks EGF stimulation-induced p21Cip1 downregulation. HeLa cells were pretreated with U0126 (20 μM, 1 h) or LY294002 (5 μM, 1 h) and then incubated with or without EGF. Cell lysates were prepared 40 min after stimulation, analyzed by immunoblotting with anti-p21Cip1 and anti-phospho-ERK antibodies, and normalized with anti-β-actin antibody. (D and E) ERK inhibition prolongs p21Cip1 half-life. HeLa cells were pretreated with U0126 (10 μM) or dimethyl sulfoxide (vehicle) for 1 h and were incubated with cycloheximide (CHX) (10 μg/ml) in the absence or presence of EGF (50 ng/ml). At the indicated times, cell lysates were prepared, analyzed by immunoblotting with anti-p21Cip1 antibody, and normalized with anti-β-actin antibody. A representative Western blot (D) was quantified by densitometry (E). All signals are normalized to the signal at time zero (100%).
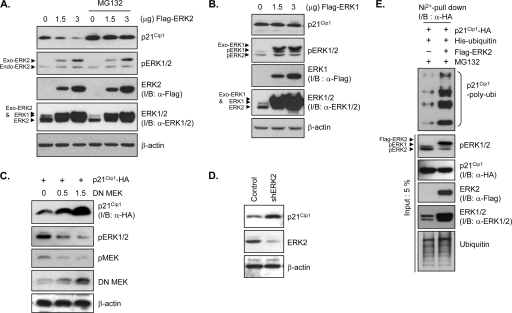
To further dissect this signaling pathway, we measured p21Cip1 levels in cells transiently overexpressing ERK1, ERK2, dominant-negative (DN) MEK1, or ERK2 shRNA. Overexpression of ERK1 or ERK2 significantly increased the levels of phosphorylated ERK in a dose-dependent manner (Fig. 2A and B), indicating ERK activation. ERK2 overexpression induced a pronounced and dose-dependent decrease in p21Cip1 protein levels (Fig. 2A). In contrast, ERK1 overexpression had little effect on p21Cip1 levels (Fig. 2B). Furthermore, overexpression of DN MEK1, which inhibits downstream ERK activity, led to a dose-dependent increase in the level of the p21Cip1 protein (Fig. 2C). This link was confirmed by RNA interference-mediated silencing of ERK2. Transfection of ERK2 shRNA constructs, which significantly decreased ERK2 levels (Fig. 2D, middle panel), resulted in an increase in the level of endogenous p21Cip1 (Fig. 2D, upper panel). Collectively, these results indicate that p21Cip1 destabilization is specifically dependent on ERK2 activity.
FIG. 2.
ERK2 induces proteasome-dependent p21Cip1 degradation. (A and B) ERK2 expression decreases the p21Cip1 level, an effect that is blocked by the proteasome inhibitor, MG-132. HeLa cells were transfected with a plasmid encoding FLAG-ERK2 (A) or FLAG-ERK1 (B). After 36 h, cells were incubated with or without MG132 for 4 h prior to preparing cell extracts. Extracts were analyzed by immunoblotting (I/B) with anti-p21Cip1, anti-phospho-ERK1/2 (pERK1/2), anti-FLAG (α-FLAG), anti-ERK1/2 (α-ERK1/2), and anti-β-actin antibodies. (C) DN MEK increases p21Cip1 levels. HEK293T cells were cotransfected with HA-tagged p21Cip1-encoding and increasing amounts of DN-MEK-encoding plasmids. After 40 h, cells were harvested and lysed. Extracts were analyzed by immunoblotting with anti-HA (α-HA), anti-phospho-ERK1/2, anti-phospho-MEK1/2 (pMEK), anti-MEK1/2, and anti-β-actin antibodies. (D) shRNA-mediated ERK2 knockdown increases p21Cip1 levels. HeLa cells were transfected with an ERK2 shRNA (shERK2) expression plasmid or control vector. After 48 h, cells were harvested and lysed. Extracts were analyzed by immunoblotting with anti-p21Cip1, anti-ERK2, and anti-β-actin antibodies. (E) ERK2 expression induces p21Cip1 ubiquitination. HEK293 cells were cotransfected with HA-tagged p21Cip1- and His6-tagged ubiquitin-encoding plasmids together with a FLAG-ERK2 expression plasmid or control vector. After 36 h, cells were incubated with MG132 for 4 h prior to preparation of cell extracts. His6-ubiquitin-conjugated HA-p21Cip1 proteins were purified by nickel affinity chromatography and analyzed by immunoblotting with an anti-HA antibody. Total cell extracts were analyzed by immunoblotting with anti-phospho-ERK1/2, anti-HA, anti-FLAG, anti-ERK1/2, and antiubiquitin antibodies.
ERK2 activation induces proteasome-dependent p21Cip1 degradation.
p21Cip1 protein levels are known to be regulated by the proteasome pathway (23). In HeLa cells, treatment with the proteasome inhibitor MG-132 prevented the decrease in p21Cip1 protein levels induced by ERK2 overexpression (Fig. 2A), indicating that ERK2 activation triggers proteasome-dependent degradation of p21Cip1. To determine whether ubiquitination is involved in ERK2 activation-induced p21Cip1 degradation, we coexpressed His6-tagged ubiquitin and HA-tagged p21Cip1 in HEK293 cells with and without FLAG-ERK2 coexpression. We then purified ubiquitin-conjugated p21Cip1 from MG-132-pretreated cells by nickel affinity chromatography and probed for p21Cip1 by immunoblotting with an anti-HA antibody. High-molecular-weight bands corresponding to polyubiquitinated proteins appeared in the ERK2-overexpressing cells (Fig. 2E), suggesting that ERK2-mediated p21Cip1 degradation is ubiquitination dependent.
ERK2 physically interacts with p21Cip1.
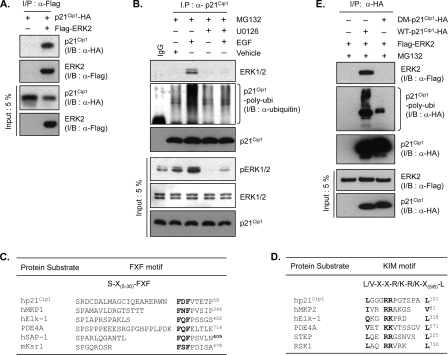
To explore the mechanism of ERK2-mediated p21Cip1 downregulation, the interaction between ERK2 and p21Cip1 was characterized. p21Cip1 physically interacts with several protein kinases that are capable of phosphorylating the protein (26, 37, 40). To detect an association between ERK2 and p21Cip1, we cotransfected HEK293T cells with HA-p21Cip1 and FLAG-ERK2 expression plasmids and probed immunoprecipitates by immunoblotting. p21Cip1 was consistently detected in immunoblots of FLAG-ERK2 immunoprecipitates (Fig. 3A); similar results were obtained by probing HA-p21Cip1 immunoprecipitates with anti-FLAG antibodies to detect associated ERK2 (Fig. 3E). In the absence of stimulation, the association of endogenous ERK2 with endogenous p21Cip1 was barely detectable (Fig. 3B, lane 2), but it was prominent in cells treated with EGF (Fig. 3B, lane 3). This endogenous association was blocked by U0126 (Fig. 3B, lane 5), suggesting that only the active form of ERK2 is capable of associating with p21Cip1. Inhibition of proteasome-mediated degradation by MG132 enabled us to detect not only this endogenous ERK2-p21Cip1 association in EGF-treated cells but also the increase of endogenously polyubiquitinated p21Cip1 complex (Fig. 3B, second panel). U0126 also blocked p21Cip1 ubiquitination, suggesting that p21Cip1 ubiquitination is dependent on ERK activation.
FIG. 3.
p21Cip1 interacts with ERK2. (A) HEK293T cells were cotransfected with HA-p21Cip1 and FLAG-ERK2 or control vector. After 36 h, cells were lysed and ERK2 was immunoprecipitated (I/P) with anti-FLAG (α-FLAG) antibody. p21Cip1 and ERK2 were detected by immunoblotting with anti-HA (α-HA) and anti-FLAG antibodies, respectively. (B) HeLa cells were pretreated with MG132 alone and along with U0126 (10 μM) for 2 h and were treated with EGF (50 ng/ml) or left untreated for 30 min. Cells were lysed, and p21Cip1 was immunoprecipitated (I/P) with an anti-p21Cip1 (α-p21Cip1) antibody. ERK1/2, polyubiquitinated p21Cip1, and native p21Cip1 were detected by immunoblotting with anti-ERK1/2, antiubiquitin (α-ubiquitin), and anti-p21Cip1 antibodies, respectively. Total cell extracts were analyzed by immunoblotting with anti-phospho-ERK1/2, anti-ERK1/2, and anti-p21Cip1 antibodies. (C) Aligned amino acid sequences of the ERK-docking FXF motifs of human p21Cip1 and other known ERK substrates: human mitogen-activated protein kinase phosphatase 1 (hMKP1), Elk-1, phosphodiesterase 4A (PDE4A), serum response factor accessory protein 1 (hSAP-1), and mouse kinase suppressor of Ras 1 (mKsr1). Conserved residues are highlighted. Numbers on the right indicate the positions of the final residues shown in each case. (D) Aligned amino acid sequences of the ERK-docking KIM motifs of human p21Cip1 and other known ERK substrates: hMKP2, Elk-1, PDE4A, striatally enriched tyrosine phosphatase (STEP), and p90 ribosomal S6 kinase 1 (RSK1). (E) HEK293T cells were cotransfected with either HA-tagged wild-type (WT) p21Cip1 (1 μg) or docking mutant (DM) p21Cip1 (0.4 μg) and FLAG-ERK2 or control vector. Different amounts of p21Cip1 constructs were used to adjust the levels of protein expression equally. After 36 h, cells were incubated with MG132 for 4 h prior to preparing cell extracts. p21Cip1 was immunoprecipitated with an anti-HA antibody. p21Cip1 and ERK2 were detected by immunoblotting with anti-HA and anti-FLAG antibodies, respectively.
Proteins that physically interact with ERK share conserved FXF (or DEF, docking site for ERK) and/or KIM (kinase interacting motif) docking motifs (15, 25). As the name suggest, FXF itself is the conserved amino acid sequence; KIM shares an L/V-X2-R/K-R/K-X6-L consensus sequence. Sequence alignment identified putative FXF (51FDF) and KIM (89LX3RRX6L) docking motifs within the p21Cip1 molecule (Fig. 3C and D). To determine whether these putative docking sites were required for the interaction of p21Cip1 with ERK2, we generated p21Cip1 variants in which the two phenylalanines in 51FDF (F51A and F53A) and two arginines in 89LX3RRX6L were changed to alanines (R93A/R94A), separately and together, and tested the effects of these mutations on ERK2 interaction in coimmunoprecipitation experiments in the presence of MG132. While variants containing mutations of each docking site alone showed partial defects in ERK2 interaction (data not shown), the double mutation totally abrogated ERK2 interaction (Fig. 3E, top panel) and further impaired ERK2-mediated ubiquitination (Fig. 3E, second panel; also see below in Fig. 6A) and subsequent degradation (see below in Fig. 6C). We thus conclude that p21Cip1 interacts directly with ERK2, presumably through the concerted action of the FXF and KIM docking sites.
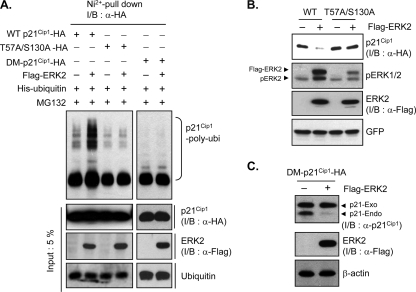
FIG. 6.
ERK2 phosphorylation of p21Cip1 at Thr57 and Ser130 induces p21Cip1 ubiquitination and subsequent degradation. (A) HEK293 cells were cotransfected with HA-tagged wild-type (WT) p21Cip1, T57A S130A mutant p21Cip1, or docking mutant (DM) p21Cip1 together with His6-ubiquitin and FLAG-ERK2 or a control vector. After 36 h, cells were incubated with MG132 for 4 h prior to preparation of cell extracts. His6-ubiquitin conjugates were purified by nickel affinity chromatography and analyzed by immunoblotting (I/B) with an anti-HA (α-HA) antibody. Total cell extracts were analyzed by immunoblotting with anti-HA, anti-FLAG (α-FLAG), and antiubiquitin antibodies. (B) HEK293 cells were cotransfected with either HA-tagged wild-type (WT) p21Cip1 or T57A S130A mutant p21Cip1 and FLAG-ERK2 or control vector. Cells were cotransfected with an enhanced-GFP vector for normalization. After 40 h, cells were harvested and lysed, and extracts were analyzed by immunoblotting with anti-HA, anti-phospho-ERK1/2, anti-FLAG, and anti-GFP antibodies. (C) HeLa cells were transfected with HA-tagged docking mutant p21Cip1 together with FLAG-ERK2 or a control vector. After 48 h, cells were harvested, and cell extracts were analyzed by immunoblotting with anti-p21Cip1 (α-p21Cip1), anti-FLAG, and anti-β-actin antibodies.
ERK2 phosphorylates p21Cip1 at Thr57 and Ser130.
Because a number of ERK2 substrates contain the docking motif, we performed in vitro kinase assays to determine whether p21Cip1 is a direct substrate of ERK2. HA-p21Cip1 and HA-ERK2 were immunoprecipitated from lysates of HEK293T cells expressing either HA-p21Cip1 or HA-ERK2 using an anti-HA antibody and incubated together in the presence of [γ-32P]ATP. HA-p21Cip1 was clearly phosphorylated under these conditions (Fig. 4A). These results were confirmed by in vitro kinase assays using purified recombinant His6-p21Cip1 and active recombinant ERK2 (Fig. 4B). Collectively, these results strongly suggest that p21Cip1 is indeed a substrate for ERK2.
FIG. 4.
ERK2 phosphorylates Thr57 and Ser130 residues in p21Cip1 in vitro. (A) HEK293T cells transfected with either HA-p21Cip1 or HA-ERK2 were lysed and separately immunoprecipitated with anti-HA antibody. p21Cip1 and ERK2 immunoprecipitates were incubated in kinase reaction buffer containing [γ-32P]ATP. Phosphorylated proteins were resolved by SDS-PAGE and analyzed by autoradiography. The lower two panels show immunoblots (I/B) for the p21Cip1 and ERK2 proteins used in the kinase reactions, detected with an anti-HA (α-HA) antibody. (B) Recombinant ERK2 and p21Cip1 proteins were purified from E. coli and incubated in kinase reaction buffer containing [γ-32P]ATP. Phosphorylated proteins were resolved by SDS-PAGE and analyzed by autoradiography. The lower panel shows an immunoblot with anti-p21Cip1 antibody as a loading control. (C) The samples in panel B resolved by SDS-PAGE were transferred onto a polyvinylidene difluoride membrane. Radioactive protein was cut from the membrane, acid hydrolyzed, mixed with cold phosphoserine, phosphothreonine, and phosphotyrosine standards, and separated on cellulose-thin-layer chromatography plates (Merck) in two dimensions using an HTLE 7000 (CBS Scientific) apparatus. The positions of standard phosphoamino acids were determined by ninhydrin staining (left panel), and 32P-amino acids were located by autoradiography (right panel). (D) Identification of the ERK-mediated phosphorylation sites in p21Cip1. Recombinant p21Cip1 protein was phosphorylated in vitro with ERK2. After the sample was resolved on SDS-PAGE, the p21Cip1 protein band was excised and subjected to proteolysis with trypsin, and the resulting peptides were analyzed by LC-MS/MS. Two different phosphopeptides were detected. The presence of y- and b-type fragment ions in MS/MS spectra enabled identification of the tryptic peptides, SGEQAEGpSPGGPGDSQGR and ERWNFDFVTEpTPLEGDFAWER; phosphorylated Thr57 and Ser130 are indicated. (E) HEK293T cells transfected with either HA-tagged wild-type or T57A S130A p21Cip1 were lysed and immunoprecipitated with anti-HA antibody. p21Cip1 immunoprecipitates were incubated with recombinant active ERK2 (rec-ERK2) in kinase reaction buffer containing [γ-32P]ATP. Phosphorylated proteins were resolved by SDS-PAGE and analyzed by autoradiography. The lower two panels show immunoblots for the p21Cip1 and ERK2 proteins used in the kinase reactions, detected with anti-HA and anti-ERK2 antibodies, respectively.
Phosphoamino acid analysis of in vitro-phosphorylated His6-p21Cip1 revealed that serine and threonine residues were phosphorylated whereas tyrosine residues were not (Fig. 4C). To identify the ERK2-mediated phosphorylation sites, in vitro-phosphorylated His6-p21Cip1 was purified by SDS-PAGE and digested with trypsin, and the resulting peptides were analyzed by tandem mass spectrometry with a liquid chromatography system (LC-MS/MS) (see Table S1 and Fig. S1 in the supplemental material). Two different phosphopeptides, SGEQAEGpSPGGPGDSQGR (Fig. 4D, upper panel) and ERWNFDFVTEpTPLEGDFAWER (Fig. 4D, lower panel), were reproducibly identified. The MS/MS spectra of the peptides indicated that the fragment ion from the neutral loss was notably abundant and clearly showed a decrease of 49.0 m/z resulting from the loss of phosphoric acid from the doubly charged precursor (Fig. 4D, upper panel) and a decrease of 33.7 m/z from the triply charged precursor (Fig. 4D, lower panel). The presence of the intense peaks y10 (927.6 m/z) and y4 (447.3 m/z) along with the neutral loss fragments of y12 (y12 − H3PO4 at 1,053.8 m/z) and b8 (b8 − H3PO4 at 728.6 m/z) in the MS/MS spectrum of SGEQAEGpSPGGPGDSQGR verified that Ser130, and not any of the other serines in the peptide, is the specific phosphorylation site. Likewise, generation of y10 (1,219.8 m/z), b10 (1,326.0 m/z), y11 − H3PO4 (1,303.0 m/z), and b11 − H3PO4 (1,408.9 m/z) from the precursor ERWNFDFVTEpTPLEGDFAWER verified that Thr57 and not Thr55 is the specific phosphorylation site. We therefore conclude that ERK2 phosphorylates Thr57 and Ser130 of p21Cip1 in vitro. This result is consistent with the phosphoamino acid analysis data, which demonstrated dual Thr and Ser phosphorylation (Fig. 4C). p21Cip1 T57A S130A, a mutant in which both phosphorylation residues were substituted for alanines to mimic a nonphosphorylated status, was resistant to phosphorylation by ERK2 in the kinase assay in vitro compared with wild-type p21Cip1 (Fig. 4E). Taken together, these data support that Thr57 and Ser130 of p21Cip1 are specific phosphorylation sites for ERK2.
Phosphorylation-dependent nucleocytoplasmic translocation and degradation.
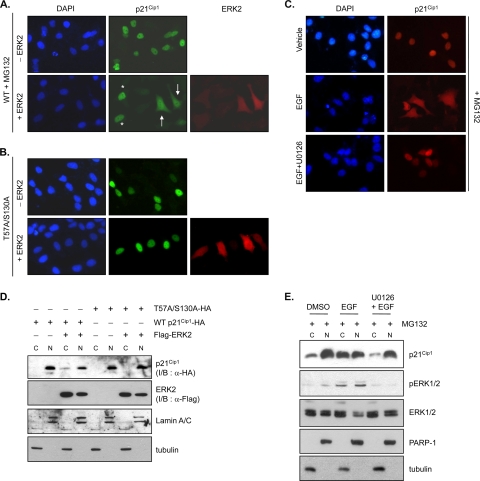
We previously showed that nucleocytoplasmic translocation is required for ubiquitination and subsequent degradation of p21Cip1 (13). To determine whether ERK2-mediated p21Cip1 degradation is dependent on p21Cip1 translocation, we used immunoassaying (Fig. 5A and B) and fractionation experiments (Fig. 5D) in the presence of MG132 to analyze p21Cip1 subcellular localization. When expressed alone in HeLa cells, ectopic p21Cip1 was predominantly present in the nucleus (Fig. 5A, upper panels) but exhibited both cytoplasmic and nuclear localization when ERK2 was coexpressed (Fig. 5A, lower panel). About 80% of ERK2-coexpressing cells showed cytoplasmic localization of p21Cip1. In contrast, double mutation of ERK2 phosphorylation sites (T57A S130A) in p21Cip1 caused the retention of p21Cip1 in the nucleus, even in ERK2-coexpressing cells (Fig. 5B, lower panels). Similar results were obtained using cell fractionation and immunoblot experiments, which showed that ERK2 coexpression could induce cytoplasmic localization of wild-type p21Cip1 (Fig. 5D, lane 3) but had no effect on T57A S130A p21Cip1 localization (Fig. 5D, lane 7). These results suggest that phosphorylation at Thr57 and Ser130 of p21Cip1 by ERK2 are critical in determining its cellular localization. To investigate whether endogenous p21Cip1 can be regulated in a similar way, we fractionated cells after activating the ERK pathway with EGF treatment. p21Cip1 was also located in both the cytoplasm and the nucleus after EGF treatment (Fig. 5C, middle panel). Cotreatment with U0126, which inhibits EGF-induced ERK activation, abolished cytoplasmic localization (Fig. 5C, lower panel). Cell fractionation experiments confirmed the effects of EGF and U0126 on endogenous p21Cip1 localization (Fig. 5E). p21Cip1 levels were reduced in the nucleus but increased in the cytoplasm following EGF treatment, suggesting that p21Cip1 was translocated from the nucleus to the cytoplasm following ERK2-mediated phosphorylation.
FIG. 5.
ERK2 alters the cellular localization of p21Cip1. (A and B) HeLa cells were cotransfected with HA-tagged wild-type p21Cip1 (WT) or T57A S130A mutant p21Cip1 together with FLAG-ERK2 or the control vector. At 42 h after transfection, cells were treated with MG132 for 2 h prior to fixation. The cellular localization of p21Cip1 was detected using an anti-HA antibody. After washing extensively in PBS, samples were further incubated with an anti-FLAG antibody to detect ERK2. Nuclei were visualized by DAPI staining. Cells were observed by fluorescence microscopy. A representative microscopic field in the lower middle panel in A shows that nuclear and cytoplasmic localization of p21Cip1 is indicated by two cells (arrows) coexpressing FLAG-ERK2 (red signals in ERK2 staining; right panel) and HA-p21Cip1 and that nuclear localization of p21Cip1 is indicated by two cells (starred) expressing HA-p21Cip1 only (no signal in ERK2 staining; right panel). (C) HeLa cells were transfected with HA-tagged wild-type p21Cip1. At 42 h after transfection, cells were treated with MG132 and U0126 (10 μM) or DMSO (vehicle) for 2 h prior to treatment with EGF (50 ng/ml) for 30 min. After fixation, the cellular localization of p21Cip1 was detected using an anti-HA antibody. Nuclei were visualized by DAPI staining. Cells were observed by fluorescence microscopy. (D) Cells in A and B were fractionated, and cytoplasmic (C) and nuclear (N) proteins were resolved by SDS-PAGE and analyzed by immunoblotting with anti-HA (α-HA), anti-FLAG (α-FLAG), antitubulin, and anti-lamin A/C antibodies. Lamin A/C and tubulin are nuclear and cytoplasmic markers, respectively. (E) HeLa cells were treated with MG132 and U0126 (10 μM) or with DMSO (vehicle) for 2 h prior to treatment with EGF (50 ng/ml) for 30 min. Cytoplasmic (C) and nuclear (N) proteins were resolved by SDS-PAGE and analyzed by immunoblotting with anti-p21Cip1, anti-pERK1/2, anti-ERK1/2, antitubulin, and anti-PARP-1 antibodies. PARP-1 and tubulin are nuclear and cytoplasmic markers, respectively.
We next tested whether phosphorylation had an effect on p21Cip1 ubiquitination and subsequent degradation. The T57A S130A double mutation abolished ERK2-dependent p21Cip1 ubiquitination (Fig. 6A) and degradation (Fig. 6B). Given that enzyme-substrate association is a prerequisite for ERK-mediated protein phosphorylation (15, 25), we tested whether ERK association affected p21Cip1 ubiquitination and degradation. Mutation of the ERK docking sites of p21Cip1 (F51A, F53A, R93A, and R94A) also abolished p21Cip1 ubiquitination and degradation (Fig. 6A and C). Collectively, these results suggest that interaction with and phosphorylation by ERK2 is necessary for p21Cip1 nucleocytoplasmic translocation, which may initiate the p21Cip1 degradation pathway.
ERK2 promotes cell cycle progression through p21Cip1 degradation.
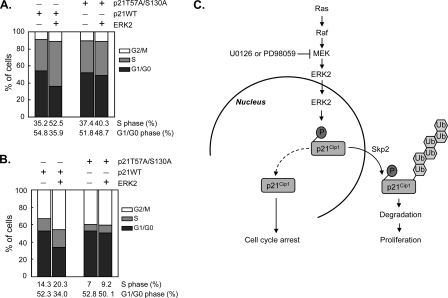
To determine the physiological relevance of ERK2-mediated p21Cip1 downregulation in vivo, we investigated the effects of ERK2 on p21Cip1-mediated cell cycle regulation. Whereas ectopic expression of p21Cip1 alone in HEK293 cells efficiently inhibited entry into S phase and promoted G1 arrest, ERK2 coexpression rescued cells from G1 arrest, inducing S phase entry (Fig. 7A). Overexpression of the phosphorylation-deficient p21Cip1 T57A S130A mutant also induced G1 arrest, but in this case ERK2 coexpression had no effect (Fig. 7A), confirming that phosphorylation of p21Cip1 at Thr57 and Ser130 is required for ERK2-induced S-phase entry. Although HEK293 cells have a very low level of endogenous p21Cip1 compared with normal human cells, it is possible that even these low levels of p21Cip1 could have affected the outcome. To completely exclude interference from endogenous p21Cip1, we used HCT116 p21Cip1-null cells. Consistent with the results obtained in HEK293 cells, ERK2 rescued the G1 arrest induced by ectopic expression of wild-type p21Cip1 in HCT116 p21Cip1-null cells but did not rescue that induced by p21Cip1 T57A S130A (Fig. 7B). These results suggest that ERK2 induces S-phase progression by phosphorylating p21Cip1 and promoting its subsequent degradation.
FIG. 7.
ERK2 promotes cell cycle progression via degradation of p21Cip1. (A) HEK293 cells were transfected with HA-tagged wild-type (WT) p21Cip1 or T57A S130A mutant p21Cip1 together with FLAG-ERK2 or a control vector. A vector encoding membrane-bound GFP was cotransfected, and cells were incubated for 32 h. During the last 16 h, cells were starved in serum-free medium and then returned to normal medium for an additional 8 h before harvesting. The cell cycle profile of GFP-positive cells was determined by fluorescence-activated cell sorting analysis. (B) HCT116 p21−/− cells were transfected with HA-tagged wild-type (WT) p21Cip1 or T57A S130A mutant p21Cip1 together with FLAG-ERK2 or a control vector and a membrane-bound GFP vector. Cells were treated and analyzed as for panel A. The levels of HA-p21Cip1 and FLAG-ERK2 were analyzed by immunoblotting (see Fig. S2 in the supplemental material). (C) Schematic representation of the proposed pathway, showing that ERK2 interacts with and phosphorylates p21Cip1 at Thr57 and Ser130; phosphorylation of these residues enhances p21Cip1 ubiquitination and subsequent degradation through the proteasome pathway, leading to cell proliferation.
DISCUSSION
Here we propose that the oncoprotein ERK2 acts through a novel signaling pathway to downregulate the tumor suppressor p21Cip1 and thereby regulate cell cycle progression (Fig. 7C). We have shown that ERK2 interacts with and phosphorylates p21Cip1, promoting p21Cip1 ubiquitination. We identified two ERK2 phosphorylation sites, Thr57 and Ser130, in p21Cip1 and showed that phosphorylation of these residues increases p21Cip1 cytoplasmic distribution and proteasome-dependent degradation. Moreover, the phosphorylation-deficient T57A S130A p21Cip1 mutant, which is resistant to ERK2-mediated downregulation, retains the ability to potently inhibit the G1/S transition, providing direct evidence that phosphorylation by ERK2 regulates p21Cip1 function. Our data thus support an essential role for ERK2-mediated p21Cip1 regulation in mitogenic signaling.
ERK has been shown to promote cell growth and tumorigenesis (17). Sustained ERK activation arising from Ras mutation, which is present in 30% of human cancers, is known to result in posttranslational inactivation of GSK-3β (9) and FOXO3a (39), leading to resistance to apoptosis and enhancement of cell proliferation. We propose here that downregulation of the p21Cip1 protein is also involved in the Ras-MEK-ERK signaling pathway in cancer cells. Thus, orchestrated inhibition of GSK-3β, FOXO3a, and p21Cip1 by activated ERK may be responsible for the constitutive survival signaling cascades that underlie tumorigenesis. An added level of complexity is provided by several reports suggesting that ERK induces an increase in p21Cip1 mRNA expression (6). It is possible that ERK exerts opposing transcriptional and posttranslational influences on p21Cip1 levels such that the net effect of ERK activation might depend on the cellular context, determined by the accompanying signaling pathways triggered in parallel by ERK activation. Under our experimental conditions, however, ERK activation did not affect the p21Cip1 mRNA level (see Fig. S3 in the supplemental material). Without the potential confounding influence of transcriptional changes, we were thus able to focus on changes in p21Cip1 protein levels to elucidate the mechanism by which the ERK cascade drives cell cycle progression upon mitogenic stimulation.
Our finding that overexpression of ERK2 but not ERK1 reduces p21Cip1 protein levels (Fig. 2A and B) is consistent with recent reports that ascribe subtype-specific functions to the two major ERK isoforms. The prevailing view maintains that ERK1 and ERK2 are regulated similarly and contribute to intracellular signaling by phosphorylating a largely common subset of substrates. This seems reasonable given the extensive amino acid identity between the two isoforms. However, it has been reported that ERK2-deficient mice show embryonic lethality (12), whereas ERK1-deficient mice are viable, fertile, and normal in size (24). ERK2 targeting was shown to impair mitogenic signaling in trophectoderm cells (30) and to abolish Ras-dependent cell proliferation (35). In contrast, ERK1 silencing either had no effect on embryonic fibroblast proliferation (24), or increased the proliferation rate of fibroblasts (35). Thus, the ERK1 and ERK2 isoforms may have unique roles at least in certain cellular settings (22). Our demonstration that ERK2 but not ERK1 modulates cell cycle progression via p21Cip1 phosphorylation lends support to the idea that these two isoforms have distinct, as well as overlapping, roles.
Several kinases phosphorylate specific target sites on p21Cip1 and modulate its fate by governing p21Cip1 interaction potential, stability, and subcellular localization. GSK-3β and CDK2 are known to phosphorylate Thr57 (27) and Ser130 (41), respectively, resulting in p21Cip1 destabilization. Consistent with this observation, we found that simultaneous phosphorylation of Thr57 and Ser130 by ERK2 destabilizes p21Cip1. In contrast, phosphorylation of Thr57 by JNK1 and p38α kinase (16) or of Ser146 by Akt (20) was shown to stabilize p21Cip1. Thus, phosphorylation is not strictly a p21Cip1 destabilizing mechanism but rather may positively or negatively regulate stability depending on the kinases that act and the cell type. We have shown in this study that ERK2 is among the protein kinases that are capable of phosphorylating and destabilizing p21Cip1. However, although overexpression of ERK2 was correlated with increased phosphorylation of p21Cip1 at Thr57 and Ser130, we cannot exclude the possibility that the degradation of p21Cip1 requires additional ERK2-stimulated signaling events that act in concert with ERK2-mediated p21Cip1 phosphorylation.
Previously we found that p21Cip1 contains the NES 68VRGLGLPKLYL and 102LQGTAEEDHVDLSLSCTL, which are required for nuclear export and subsequent degradation (13). On the basis of the location of the phosphorylation sites, Thr57 and Ser130, which are in close proximity to the corresponding NES, we propose a model of phosphorylation-triggered nucleocytoplasmic translocation of p21Cip1. Although the three-dimensional structure of the entire p21Cip1 protein is not yet known, one possibility is that ERK2-mediated phosphorylation causes conformational changes that expose buried NES structures, making NES accessible to the export factor, CRM1/exportin-1, leading to phosphorylation-dependent nucleocytoplasmic translocation. The molecular mechanism by which ERK2 phosphorylation activates NES is an open question that will require further research.
Here we have identified a novel p21Cip1 phosphorylation/degradation pathway playing a role in the ERK-mediated cell proliferation signal, in addition to the known GSK-3β and FOXO3a inactivation pathways (9, 39). Our study provides new insight into the mechanism of tumor proliferation via p21Cip1 modulation and further supports the idea that ERK2 kinase activity may be a promising target for developing therapeutic agents that inhibit tumor progression or increase the sensitivity of cancer cells to chemotherapy.
Supplementary Material
Acknowledgments
This work was supported by grants from the KRCF and KOSEF (M10503010002, M10601000164, and M10642040003; to K.-S.K.) and a grant from the Functional Proteomics Research Center of the 21st Century Frontier Research Program (to C.L.).
We thank Bert Vogelstein for HCT116 p21 null cells, Michele Pagano and Rati Fotedar for the p21Cip1 constructs, Dae-Yeul Yu for hepatocytes from H-RasV12-transgenic mice, Eui-Jeon Woo for the purified active ERK2 protein, Young-Joo Jang for two-dimensional phosphoamino acid analysis, and Un-Beom Kang for LC-MS/MS analysis. We thank Hong-Duk Youn for critical reading of the manuscript.
Footnotes
Published ahead of print on 13 April 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 27023589-23597. [DOI] [PubMed] [Google Scholar]
- 2.Bendjennat, M., J. Boulaire, T. Jascur, H. Brickner, V. Barbier, A. Sarasin, A. Fotedar, and R. Fotedar. 2003. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell 114599-610. [DOI] [PubMed] [Google Scholar]
- 3.Blagosklonny, M. V., G. S. Wu, S. Omura, and W. S. el-Deiry. 1996. Proteasome-dependent regulation of p21WAF1/CIP1 expression. Biochem. Biophys. Res. Commun. 227564-569. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, J., V. Amador, F. Bartolini, G. DeMartino, and M. Pagano. 2003. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 11571-82. [DOI] [PubMed] [Google Scholar]
- 5.Bornstein, G., J. Bloom, D. Sitry-Shevah, K. Nakayama, M. Pagano, and A. Hershko. 2003. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 27825752-25757. [DOI] [PubMed] [Google Scholar]
- 6.Bottazzi, M. E., X. Zhu, R. M. Bohmer, and R. K. Assoian. 1999. Regulation of p21(cip1) expression by growth factors and the extracellular matrix reveals a role for transient ERK activity in G1 phase. J. Cell Biol. 1461255-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cayrol, C., and B. Ducommun. 1998. Interaction with cyclin-dependent kinases and PCNA modulates proteasome-dependent degradation of p21. Oncogene 172437-2444. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, M., P. Olivier, J. A. Diehl, M. Fero, M. F. Roussel, J. M. Roberts, and C. J. Sherr. 1999. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 181571-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding, Q., W. Xia, J. C. Liu, J. Y. Yang, D. F. Lee, J. Xia, G. Bartholomeusz, Y. Li, Y. Pan, Z. Li, R. C. Bargou, J. Qin, C. C. Lai, F. J. Tsai, C. H. Tsai, and M. C. Hung. 2005. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol. Cell 19159-170. [DOI] [PubMed] [Google Scholar]
- 10.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75817-825. [DOI] [PubMed] [Google Scholar]
- 11.Harper, J. W., S. J. Elledge, K. Keyomarsi, B. Dynlacht, L. H. Tsai, P. Zhang, S. Dobrowolski, C. Bai, L. Connell-Crowley, E. Swindell, et al. 1995. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell 6387-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatano, N., Y. Mori, M. Oh-hora, A. Kosugi, T. Fujikawa, N. Nakai, H. Niwa, J. Miyazaki, T. Hamaoka, and M. Ogata. 2003. Essential role for ERK2 mitogen-activated protein kinase in placental development. Genes Cells 8847-856. [DOI] [PubMed] [Google Scholar]
- 13.Hwang, C. Y., I. Y. Kim, and K. S. Kwon. 2007. Cytoplasmic localization and ubiquitination of p21(Cip1) by reactive oxygen species. Biochem. Biophys. Res. Commun. 358219-225. [DOI] [PubMed] [Google Scholar]
- 14.Hwang, C. Y., Y. S. Ryu, M. S. Chung, K. D. Kim, S. S. Park, S. K. Chae, H. Z. Chae, and K. S. Kwon. 2004. Thioredoxin modulates activator protein 1 (AP-1) activity and p27Kip1 degradation through direct interaction with Jab1. Oncogene 238868-8875. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs, D., D. Glossip, H. Xing, A. J. Muslin, and K. Kornfeld. 1999. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13163-175. [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, G. Y., S. E. Mercer, D. Z. Ewton, Z. Yan, K. Jin, and E. Friedman. 2002. The stress-activated protein kinases p38 alpha and JNK1 stabilize p21(Cip1) by phosphorylation. J. Biol. Chem. 27729792-29802. [DOI] [PubMed] [Google Scholar]
- 17.Kolch, W. 2005. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6827-837. [DOI] [PubMed] [Google Scholar]
- 18.LaBaer, J., M. D. Garrett, L. F. Stevenson, J. M. Slingerland, C. Sandhu, H. S. Chou, A. Fattaey, and E. Harlow. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11847-862. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. Y., S. J. Yu, Y. G. Park, J. Kim, and J. Sohn. 2007. Glycogen synthase kinase 3β phosphorylates p21WAF1/CIP1 for proteasomal degradation after UV irradiation. Mol. Cell. Biol. 273187-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y., D. Dowbenko, and L. A. Lasky. 2002. AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J. Biol. Chem. 27711352-11361. [DOI] [PubMed] [Google Scholar]
- 21.Maki, C. G., and P. M. Howley. 1997. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol. Cell. Biol. 17355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meloche, S., and J. Pouyssegur. 2007. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 263227-3239. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama, K. I., and K. Nakayama. 2006. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6369-381. [DOI] [PubMed] [Google Scholar]
- 24.Pages, G., S. Guerin, D. Grall, F. Bonino, A. Smith, F. Anjuere, P. Auberger, and J. Pouyssegur. 1999. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science 2861374-1377. [DOI] [PubMed] [Google Scholar]
- 25.Pulido, R., A. Zuniga, and A. Ullrich. 1998. PTP-SL and STEP protein tyrosine phosphatases regulate the activation of the extracellular signal-regulated kinases ERK1 and ERK2 by association through a kinase interaction motif. EMBO J. 177337-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romero-Oliva, F., and J. E. Allende. 2001. Protein p21(WAF1/CIP1) is phosphorylated by protein kinase CK2 in vitro and interacts with the amino terminal end of the CK2 beta subunit. J. Cell Biochem. 81445-452. [DOI] [PubMed] [Google Scholar]
- 27.Rossig, L., C. Badorff, Y. Holzmann, A. M. Zeiher, and S. Dimmeler. 2002. Glycogen synthase kinase-3 couples AKT-dependent signaling to the regulation of p21Cip1 degradation. J. Biol. Chem. 2779684-9689. [DOI] [PubMed] [Google Scholar]
- 28.Rossig, L., A. S. Jadidi, C. Urbich, C. Badorff, A. M. Zeiher, and S. Dimmeler. 2001. Akt-dependent phosphorylation of p21(Cip1) regulates PCNA binding and proliferation of endothelial cells. Mol. Cell. Biol. 215644-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rousseau, D., D. Cannella, J. Boulaire, P. Fitzgerald, A. Fotedar, and R. Fotedar. 1999. Growth inhibition by CDK-cyclin and PCNA binding domains of p21 occurs by distinct mechanisms and is regulated by ubiquitin-proteasome pathway. Oncogene 184313-4325. [DOI] [PubMed] [Google Scholar]
- 30.Saba-El-Leil, M. K., F. D. Vella, B. Vernay, L. Voisin, L. Chen, N. Labrecque, S. L. Ang, and S. Meloche. 2003. An essential function of the mitogen-activated protein kinase Erk2 in mouse trophoblast development. EMBO Rep. 4964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott, M. T., A. Ingram, and K. L. Ball. 2002. PDK1-dependent activation of atypical PKC leads to degradation of the p21 tumour modifier protein. EMBO J. 216771-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sears, R., F. Nuckolls, E. Haura, Y. Taya, K. Tamai, and J. R. Nevins. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 142501-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheaff, R. J., J. D. Singer, J. Swanger, M. Smitherman, J. M. Roberts, and B. E. Clurman. 2000. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell 5403-410. [DOI] [PubMed] [Google Scholar]
- 34.Touitou, R., J. Richardson, S. Bose, M. Nakanishi, J. Rivett, and M. J. Allday. 2001. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 202367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vantaggiato, C., I. Formentini, A. Bondanza, C. Bonini, L. Naldini, and R. Brambilla. 2006. ERK1 and ERK2 mitogen-activated protein kinases affect Ras-dependent cell signaling differentially. J. Biol. 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, A. G., H. B. Moon, M. R. Lee, C. Y. Hwang, K. S. Kwon, S. L. Yu, Y. S. Kim, M. Kim, J. M. Kim, S. K. Kim, T. H. Lee, E. Y. Moon, D. S. Lee, and D. Y. Yu. 2005. Gender-dependent hepatic alterations in H-ras12V transgenic mice. J. Hepatol. 43836-844. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Z., N. Bhattacharya, P. F. Mixter, W. Wei, J. Sedivy, and N. S. Magnuson. 2002. Phosphorylation of the cell cycle inhibitor p21Cip1/WAF1 by Pim-1 kinase. Biochim. Biophys. Acta 159345-55. [DOI] [PubMed] [Google Scholar]
- 38.Weber, J. D., D. M. Raben, P. J. Phillips, and J. J. Baldassare. 1997. Sustained activation of extracellular-signal-regulated kinase 1 (ERK1) is required for the continued expression of cyclin D1 in G1 phase. Biochem. J. 32661-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, J. Y., C. S. Zong, W. Xia, H. Yamaguchi, Q. Ding, X. Xie, J. Y. Lang, C. C. Lai, C. J. Chang, W. C. Huang, H. Huang, H. P. Kuo, D. F. Lee, L. Y. Li, H. C. Lien, X. Cheng, K. J. Chang, C. D. Hsiao, F. J. Tsai, C. H. Tsai, A. A. Sahin, W. J. Muller, G. B. Mills, D. Yu, G. N. Hortobagyi, and M. C. Hung. 2008. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol. 10138-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, B. P., Y. Liao, W. Xia, B. Spohn, M. H. Lee, and M. C. Hung. 2001. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol. 3245-252. [DOI] [PubMed] [Google Scholar]
- 41.Zhu, H., L. Nie, and C. G. Maki. 2005. Cdk2-dependent inhibition of p21 stability via a C-terminal cyclin-binding motif. J. Biol. Chem. 28029282-29288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.