Abstract
The positive regulatory machinery in the microRNA (miRNA) processing pathway is relatively well characterized, but negative regulation of the pathway is largely unknown. Here we show that a complex of nuclear factor 90 (NF90) and NF45 proteins functions as a negative regulator in miRNA biogenesis. Primary miRNA (pri-miRNA) processing into precursor miRNA (pre-miRNA) was inhibited by overexpression of the NF90 and NF45 proteins, and considerable amounts of pri-miRNAs accumulated in cells coexpressing NF90 and NF45. Treatment of cells overexpressing NF90 and NF45 with an RNA polymerase II inhibitor, α-amanitin, did not reduce the amounts of pri-miRNAs, suggesting that the accumulation of pri-miRNAs is not due to transcriptional activation. In addition, the NF90 and NF45 complex was not found to interact with the Microprocessor complex, which is a processing factor of pri-miRNAs, but was found to bind endogenous pri-miRNAs. NF90-NF45 exhibited higher binding activity for pri-let-7a than pri-miR-21. Of note, depletion of NF90 caused a reduction of pri-let-7a and an increase of mature let-7a miRNA, which has a potent antiproliferative activity, and caused growth suppression of transformed cells. These findings suggest that the association of the NF90-NF45 complex with pri-miRNAs impairs access of the Microprocessor complex to the pri-miRNAs, resulting in a reduction of mature miRNA production.
MicroRNAs (miRNAs) constitute a class of noncoding small RNAs that function as repressors for eukaryotic gene regulation by binding to the 3′ untranslated regions of target mRNAs (2). This binding causes mRNA cleavage or translational inhibition of the mRNA, depending upon the degree of complementarity. The lengths of miRNAs are 21 to 23 nucleotides (nt), and over 500 miRNAs have been discovered in mammals. miRNAs regulate the expression of a large number of genes (38) that are involved in cell proliferation, apoptosis, hematopoietic differentiation, viral infection, and tumorigenesis (4, 5, 7, 22, 26, 32, 39, 45).
In mammals, miRNA genes are transcribed by RNA polymerase II as primary miRNAs (pri-miRNAs) (36). These pri-miRNAs are processed into precursor miRNAs (pre-miRNAs) by the Microprocessor complex (8, 13, 20, 31, 33). Another complex comprised of exportin-5 and RanGTP transports the pre-miRNAs from the nucleus to the cytoplasm (3, 40, 58). In the cytoplasm, Dicer, a cytoplasmic RNase III enzyme, cleaves the pre-miRNAs to approximate 22-nt mature miRNA duplexes with 2-nt 3′ overhangs (14, 24, 28). One strand of the duplex is incorporated into the RNA-induced silencing complex (12, 19, 29, 41, 51). The single strand of RNA guides the RNA-induced silencing complex to the target mRNA with sequence complementarity, which leads either to mRNA cleavage or to translational repression (12, 24, 41, 44).
The Microprocessor complex, which cleaves pri-miRNA to pre-miRNA during miRNA biogenesis, is comprised of a nuclear RNase III enzyme, Drosha, and its cofactor, DGCR8 (8, 13, 20). In addition to the Microprocessor complex, excessively expressed Drosha forms other larger complexes composed of a multitude of different RNA binding proteins (13). A family of the larger Drosha complex is the double-stranded RNA binding motif (dsRBM) proteins, including DGCR8, nuclear factor 90 (NF90), and NF45 (13). DGCR8 is an essential factor for the Drosha-mediated pri-miRNA processing reaction (13, 20). However, the functions of NF90 and NF45 in pri-miRNA processing remain unclear.
The NF90 family proteins consist of several different but closely related proteins generated through alternate splicing. Members of this family include NF90a/b and its longer form, NF110a/b. NF90b and NF110b have an NVKQ insert between their two dsRBMs, whereas NF90a and NF110a lack this insert. These proteins are identical at the N-terminal and central regions but diverge at their C-terminal regions (46). The central region includes a functional nuclear localization signal and two dsRBMs. In contrast, NF45 possesses a zinc-finger nucleic acid binding domain (DZF) and a glutamic acid-rich region. The NF90 family and NF45 proteins form a heterodimer and are predominantly localized in the nucleus.
NF90 and NF45 were first isolated as nuclear factors that bind to a cis element of the interleukin-2 promoter, known as an antigen receptor response element in the activated Jurkat T-cell line (27). Independently, we previously identified NF90 as a binding factor that recognizes a unique palindromic sequence in the DNase I-hypersensitive site of the HLA-DRα gene in monocytic leukemia THP-1 cells (47). The NF90 family proteins are also known to bind to a minihelix RNA derived from adenovirus VA RNA (17). Interestingly, the secondary structures of both the palindromic sequence within the HLA-DRα gene and the minihelix RNA are predicted to be small double-stranded structures, similar to the structures of miRNA precursors (pri- and pre-miRNA).
As mentioned above, the NF90 and NF45 proteins associate with overexpressed Drosha, and NF90 favorably binds to the small double-stranded structures contained in the miRNA precursors. Therefore, NF90 and NF45 are highly likely to be involved in miRNA biogenesis, especially during processing of pri-miRNAs into pre-miRNAs. To investigate this possibility, we have examined the functions of the NF90 and NF45 proteins in the pri-miRNA processing step. Using an in vitro pri-miRNA processing assay, we found that forced expression of NF90 and NF45 inhibited the cleavage of pri-miRNAs into pre-miRNAs. Furthermore, pri-miRNAs accumulated in cells that coexpressed NF90 and NF45. In vivo binding analyses demonstrated that NF90 did not interact with the endogenous Drosha-DGCR8 complex, but the NF90-NF45 complex was bound to endogenous pri-miRNAs. Furthermore, this protein complex exhibited a higher affinity for pri-let-7a than pri-miR-21 in vitro. Interestingly, knockdown of NF90 caused a decrease in pri-let-7a and an increase in mature let-7a. Therefore, we propose that the association of the NF90-NF45 complex with pri-miRNAs may impair access of the Microprocessor complex to pri-miRNAs, resulting in the inhibition of the mature miRNA production.
MATERIALS AND METHODS
Sequences of all the oligonucleotides and PCR protocols are listed in Table S1 of the supplemental material.
Cells, plasmids, and antibodies.
Human embryonic kidney 293T cells and 293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. For expression of Flag-tagged NF90a, NF90b, NF110a, or NF110b proteins, cDNA fragments corresponding to the full length of these proteins were obtained by PCR from pcDNA3.1-NF90a,- NF90b, -NF110a, and -NF110b plasmids (provided by M. B. Mathews, University of Medicine and Dentistry of New Jersey, Newark). The cDNA fragments were subcloned into the EcoRI sites of the pcDNA3-Flag vector. To construct the Flag-tagged NF45 protein, a full-length NF45 cDNA was PCR amplified from HeLa cell cDNA and inserted into the EcoRI and XhoI sites of the pcDNA3-Flag vector. To construct an expression plasmid of Drosha, a full-length Drosha cDNA was obtained by PCR from HeLa cell cDNA and subcloned into BamHI and EcoRV sites of the pcDNA3.1 vector. Anti-Drosha, anti-DGCR8, anti-Ku70, and antitubulin antibodies were obtained from Upstate Biotechnology, ProteinTech Group, Inc., Santa Cruz Biotechnology, and CalBiochem, respectively. Anti-NF90 antibody was produced by immunizing New Zealand White rabbits with glutathione S-transferase-NF90 (residues 419 to 604) as previously described (53).
Transfections.
293T cells were transfected using Lipofectamine Plus reagent according to the manufacturer's instructions (Invitrogen).
In vitro pri-miRNA processing assay.
An in vitro pri-miRNA processing assay was performed as described by Lee et al. (35) with some modifications. Briefly, pri-miR-30a was amplified from 293T cDNA by PCR. The PCR product was subcloned into the pGEM-T-easy vector (Promega). The plasmid was linearized with SpeI and used for in vitro transcription to make a 32P-radiolabeled RNA probe. Whole-cell extracts (WCEs) were prepared from 293T cells by sonication in lysis buffer (20 mM HEPES-KOH, pH 8.0, 100 mM KCl, 0.2 mM EDTA, 5% glycerol, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride [PMSF]), followed by centrifugation. Each 30-μl reaction mixture contained the radiolabeled RNA probe, 100 μg of WCE, 1 U RNase Out (Invitrogen), and 6.4 mM MgCl2 in reaction buffer (20 mM HEPES-KOH, pH 8.0, 100 mM KCl, 0.2 mM EDTA, 5% glycerol). After incubation of the mixture at 37°C for 90 min, the resulting RNA was extracted and electrophoresed on a 12% denaturing polyacrylamide gel. The processed miRNAs were visualized by autoradiography. Intensities of the produced pre-miR-30a were measured using a BAS 2000 imaging system (Fuji Film).
Western blot analysis.
Western blot analysis was performed as previously described (48).
RNA interference.
For the RNA interference experiment, 293 cells were transfected with small interfering RNAs (siRNAs) using Lipofectamine RNAiMAX according to the manufacturer's instructions (Invitrogen). siRNA duplexes targeting the NF90 family proteins were obtained from Invitrogen. As a negative control, Negative Universal Control Med (Invitrogen) was used.
qRT-PCR analysis.
Total RNA was prepared by using TRIzol according to the manufacturer's instructions (Invitrogen). RNA was treated with DNase I (Ambion) for 20 min at 37°C, and DNase I was inactivated using DNase I Inactive reagent (Ambion). Two micrograms of DNase I-treated RNA was used to synthesize cDNA using a cDNA synthesis kit (Invitrogen). For quantitative reverse transcription-PCR (qRT-PCR), the PCR sample was comprised of diluted (1/160) cDNA, QuantiTect SYBR green PCR master mix (Qiagen), and 1 mM each of forward and reverse primers in a total volume of 12.5 μl. PCRs were performed on a DNA Engine OPTICON 2 (MJ Research) using a cycling program. SYBR green PCR master mix (Applied Biosystems) was used when PCRs were performed on an ABI 7000 sequence detection system (Applied Biosystems). All runs included the β-actin gene as an internal control. Samples were normalized to β-actin RNA, giving arbitrary values representing a ratio of experimental to control results. Results are expressed as relative mRNA levels. qRT-PCR for detection of miRNA was carried out using a TaqMan MicroRNA assay kit (Applied Biosystems) according to the manufacturer's protocol. RNU6B small nuclear RNA was used as an internal control in the qRT-PCR to normalize RNA input.
Immunocytochemical staining.
Immunocytochemical staining was performed as described by Muraki et al. (43). The primary and secondary antibodies used were anti-NF90 antibody (10 μg/ml) and fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (IgG; 1:1,000; ICN), respectively. 4′,6-diamidino-2-phenylindole (DAPI) was used for nuclear staining. Stained cells were mounted with ProLong Gold antifade reagent (Invitrogen). Images were taken using a fluorescence microscope (Axiophoto; Carl Zeiss).
Immunoprecipitation.
WCEs were prepared by lysing cells in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 0.1 mM PMSF, and protein inhibitor cocktail (Roche) and immunoprecipitated with anti-Drosha antibody or normal rabbit IgG. Immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene difluoride membrane. The membrane was probed with anti-Drosha, anti-DGCR8, or anti-NF90 antibodies. Following incubation, the membrane was washed and incubated with peroxidase-conjugated anti-rabbit IgG and subsequently developed by chemiluminescence using Western Lightning (Perkin-Elmer).
Blue native PAGE.
Cells were resuspended with 1× sample buffer (Invitrogen) including 1% n-dodecyl-β-d-maltoside and incubated for 15 min at 0°C. After centrifugation (15,000 × g) at 4°C for 20 min, the supernatants were collected as WCEs. The WCEs were mixed with 0.25% G-250 sample additive (Invitrogen) and subjected to electrophoresis on a native PAGE 4 to 16% bis-Tris gel (Invitrogen) and analyzed by Western blotting.
Detection of the interaction between the NF90-NF45 complex and pri-miRNA in vivo.
WCEs from cells were immunoprecipitated with anti-FLAG affinity gel (Sigma), and the coprecipitated RNA was purified by proteinase K digestion, extracted with phenol-chloroform, and ethanol precipitated. The precipitated RNA was annealed with 250 ng/μl random hexamers and reverse transcribed with the Reverse SuperScript III first-strand synthesis system (Invitrogen). cDNA was amplified by PCR using the specific primers for pri-miRNA. The primer sequences are described in Table S1 of the supplemental material.
Preparation of recombinant proteins.
The full-length (FL) cDNAs of NF90 and NF45 were synthesized by PCR using pcDNA3.1-NF90b and pcDNA3.1-NF45 as templates, respectively. The DGCR8 FL cDNA was obtained by PCR using HeLa cDNA as a template. The PCR products were subcloned into pIVEX 2.4d (Roche). The dsRNA-binding domains (dsRBDs) of DGCR8 (residues 484 to 773) were generated by PCR using pIVEX 2.4d-DGCR8 FL as a template. The PCR product was subcloned into pIVEX 2.4d. The pIVEX 2.4d-NF90, the pIVEX 2.4d-DGCR8 FL, and the pIVEX 2.4d-DGCR8 dsRBDs were transformed into Escherichia coli Origami(DE3) pLacI (Novagen) and the pIVEX 2.4d-NF45 was transformed into E. coli BL21(DE3, rep4). After induction with isopropyl-β-d-thiogalactopyranoside, clones were resuspended in 50 mM KPB, pH 7.0, and 300 mM NaCl, followed by sonication on ice. After centrifugation, the supernatants were loaded on a TALON metal affinity column (Clontech). Recombinant proteins were eluted with the same buffer including 200 mM imidazole. The eluted proteins were dialyzed against 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM dithiothreitol, 10% glycerol, and 0.5 mM PMSF and were stored at −80°C until use.
EMSAs.
pri-let-7a-1, pre-let-7a-1, and pri-miR-21 were amplified from 293T cDNA by PCR with specific primers. The PCR products were subcloned into the pGEM-T-easy vector (Promega). The plasmids were linearized with SpeI and used for in vitro transcription to make 32P-radiolabeled RNA probes. An electrophoretic mobility shift assay (EMSA) was performed in a 20-μl reaction mixture containing 50 mM Tris-HCl, pH 7.6, 100 mM NaCl, 5% glycerol, 2 mM MgCl2, 0.2% bovine serum albumin, 20 units of RNase Out (Invitrogen), 30 μg yeast Saccharomyces cerevisiae tRNA, and ∼25,000 cpm labeled probe. The mixture was incubated at 0°C for 15 min and then electrophoresed on a 4% polyacrylamide gel containing 10% glycerol in 0.5× Tris-borate-EDTA. For supershift assays, recombinant proteins were preincubated with antibodies at 0°C for 20 min followed by the addition of the labeled probe. Images were captured and intensities of specific bands were measured using a BAS-2500 imaging system (Fuji Film).
Cell viability assay.
Three days posttransfection, siRNA-transfected 293 cells were seeded at 1,000 cells/well in 96-well plates. Viability of the cells following 0, 1, 3, and 4 days of culture was assayed by staining with 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) reagent (Promega) and measuring the optical density at 490 nm.
RESULTS
Overexpression of NF90 and NF45 inhibits the processing step to produce pre-miRNA during miRNA biogenesis.
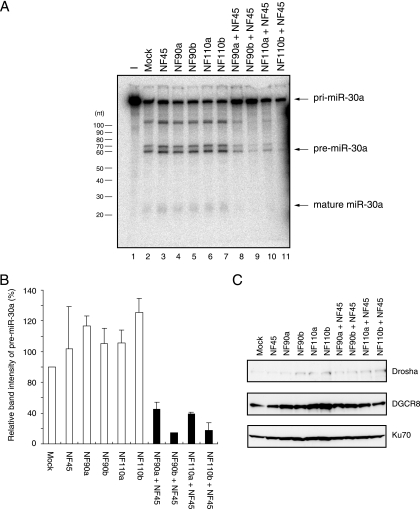
To examine the effect of the NF90 and NF45 proteins on the Drosha-mediated processing step in miRNA biogenesis, we performed an in vitro pri-miRNA processing assay. WCEs were prepared from 293T cells transfected with expression plasmids for the NF90 family (NF90a, NF90b, NF110a, or NF110b) and/or NF45. After radiolabeled pri-miR-30a was incubated with WCEs and RNAs were extracted and loaded onto a denaturing gel (Fig. 1A). The levels of pre-miR-30a were measured with a densitometer (Fig. 1B). This analysis showed that simultaneous expression of the NF90 and NF45 proteins markedly inhibited the cleavage of pri-miR-30a to pre-miR-30a (Fig. 1A, compare lanes 2 to 7 and 8 to 11, and B). In contrast, neither NF90 nor NF45 alone affected the processing step relative to the mock-transfected control (Fig. 1A, compare lane 2 and lanes 3 to 7, and B). Immunoblot analysis showed that expression of Drosha and DGCR8, which play a critical role in cleavage of pri-miRNA to pre-miRNA, was not altered by the overexpression of the NF90 and/or NF45 proteins (Fig. 1C). These results suggest that the NF90-NF45 complex may function as a negative regulator in the processing step to produce pre-miRNA during miRNA biogenesis.
FIG. 1.
Inhibition of pri-miRNA processing by coexpression of NF90 and NF45. (A) In vitro processing of pri-miR30a. Radiolabeled pri-miR-30a was incubated with WCEs from 293T cells transfected with the indicated expression plasmids. RNA was isolated and separated on a 12% polyacrylamide urea gel. Processing of pri-miR-30a was visualized by autoradiography. (B) Band intensities corresponding to the pre-miR-30a in panel A were measured with a densitometer. Data represent the average of two independent experiments. Error bars indicate standard deviation. (C) Immunoblot analyses of Drosha and DGCR8 in WCEs used for the in vitro pri-miR-30a processing assay. WCEs from 293T cells transfected with the indicated expression plasmids were probed with anti-Drosha or anti-DGCR8 antibodies. Anti-Ku70 antibody was used as loading control.
Pri-miRNAs accumulate in mammalian cells that overexpress NF90 and NF45.
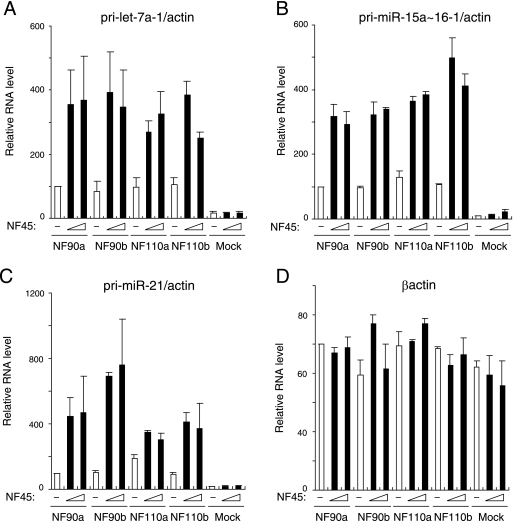
To assess the ability of the NF90-NF45 complex to function as a negative regulator in the pri-miRNA processing step, we measured the levels of pri-miRNAs in cells that overexpressed NF90 and/or NF45. 293T cells were transfected with expression plasmids for mock transfection or NF45 and/or NF90. RNAs were extracted and analyzed by qRT-PCR with specific primers for three different, randomly chosen pri-miRNAs. β-Actin was used as an internal control. Cells transfected with NF45 did not show induction of pri-miRNAs, and the forced expression of NF90 resulted in a slight increase in the pri-miRNAs (Fig. 2). Importantly, the amounts of all pri-miRNAs were strikingly increased by overexpression of both the NF90 and NF45 proteins (Fig. 2), whereas β-actin expression was not affected by these proteins (Fig. 2, compare A to C with D). These results indicate that pri-miRNAs specifically accumulated upon overexpression of NF90 and NF45. We also measured the levels of mature miRNAs, which were processed from the pri-miRNAs analyzed in the experiment shown in Fig. 2, in NF90-NF45-overexpressing cells. However, we could not observe any significant change in miRNA production by the overexpression of NF90, either with or without NF45 (data not shown). Several previous studies reported that mature miRNAs are highly stable in cells (33, 40). Therefore, a decrease of mature miRNAs at a steady-state level may be difficult to detect.
FIG. 2.
Pri-miRNAs accumulate upon coexpression of NF90 and NF45. RNAs isolated from 293T cells transfected with the indicated expression plasmids were analyzed for expression of pri-miRNAs by qRT-PCR with specific primers for (A) pri-let-7a-1, (B) pri-miR-15a∼16-1, (C) pri-miR-21, or (D) β-actin. β-Actin was used as an internal control and for normalization of the data. These results were reproducible in three independent transfection experiments. Data represent the averages of duplicate PCRs and the range.
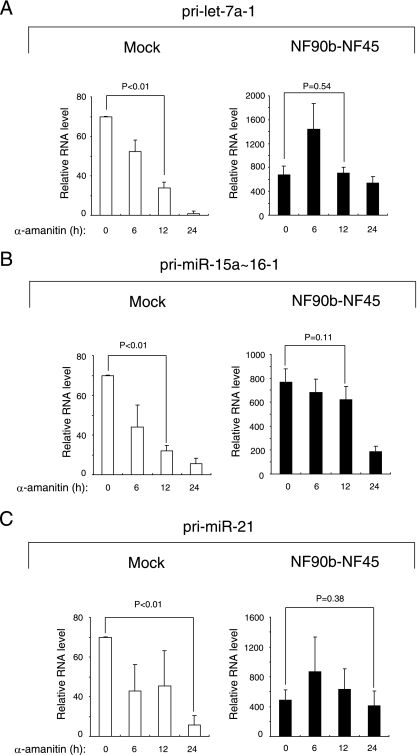
We next tested whether the increase in pri-miRNAs by NF90-NF45 overexpression is due to transcriptional activation. For this purpose, we used a universal inhibitor of RNA polymerase II, α-amanitin. Cells were mock transfected or transfected with both NF90b and NF45 plasmids and subsequently treated with α-amanitin for 0, 6, 12, and 24 h. RNAs were prepared from the cells and analyzed by RT-qPCR with specific primers for selected pri-miRNAs. These analyses indicated that all the pri-miRNAs examined were significantly decreased in mock-transfected cells treated with α-amanitin for 12 h (Fig. 3A and B) or 24 h (Fig. 3C), indicating that the steady-state levels of pri-miRNAs are maintained by transcriptional activation. In contrast, the α-amanitin treatment of cells overexpressing NF90b and NF45 did not significantly reduce the levels of pri-miRNAs (Fig. 3), suggesting that transcriptional activation is not the sole factor for the elevation of pri-miRNAs by NF90-NF45 overexpression. These findings together with results of the in vitro pri-miRNAs processing assay (Fig. 1) suggest that the inhibition of the Drosha-mediated pri-miRNAs processing step by the NF90-NF45 complex caused the accumulation of pri-miRNAs.
FIG. 3.
Accumulation of pri-miRNAs by coexpression of NF90 and NF45 is not caused by transcriptional activation. 293T cells transfected with the indicated expression plasmids were treated with α-amanitin for 6, 12, or 24 h. The expression of pri-miRNAs was analyzed by qRT-PCR with the specific primers for (A) pri-let-7a-1, (B) pri-miR-15a∼16-1, and (C) pri-miR-21. β-Actin was used as an internal control and for normalization of the data. These results were reproducible in two independent transfection experiments. Data represent the averages of triplicate PCRs with ranges. P values were calculated by using the two-tailed Student's t test.
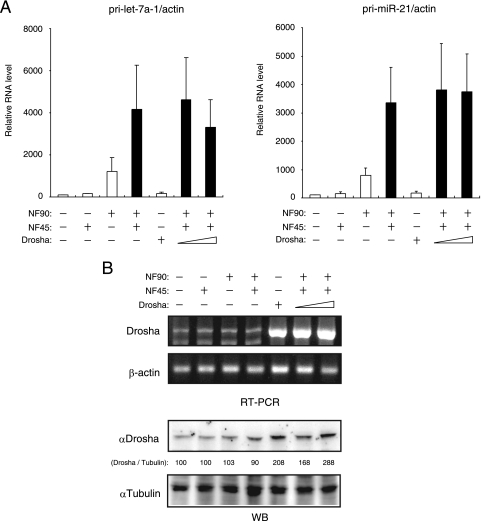
A previous study indicated that overexpressed Drosha associates with NF90 and NF45 (13). This finding prompted us to examine whether the accumulation of pri-miRNAs by overexpressed NF90-NF45 is influenced by the forced expression of Drosha. However, no significant change in the accumulated level of pri-miRNAs was observed in cells transfected with NF90-NF45 and Drosha compared with NF90-NF45-overexpressing cells (Fig. 4A). We verified that Drosha was expectedly overexpressed upon transfection at the RNA and protein levels (Fig. 4B). These results suggest that overexpressed NF90-NF45 may disturb the accessibility of Drosha to pri-miRNAs.
FIG. 4.
Forced expression of Drosha does not affect the accumulation of pri-miRNAs by NF90-NF45. (A) RNAs isolated from 293T cells transfected with the indicated expression plasmids were analyzed for expression of pri-miRNAs by qRT-PCR with specific primers for pri-let-7a-1 and pri-miR-21. β-Actin was used as an internal control and for normalization of the data. These results were reproducible in three independent transfection experiments. Data represent the averages of triplicate PCRs with ranges. (B) RNA and WCEs were prepared from 293T cells transfected with the indicated expression plasmids and analyzed by RT-PCR and immunoblot analysis, respectively. Intensities of specific bands in the immunoblotting assay were measured with a densitometer and are presented as relative band intensities.
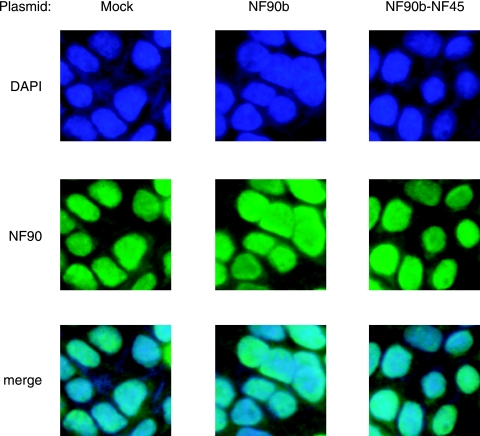
NF90 is retained in the nucleus during overexpression of the NF90-NF45 complex.
To examine the subcellular localization of the NF90 family proteins in cells that overexpressed NF90 and NF45, we performed immunocytochemical staining. 293T cells were mock transfected or transfected with plasmids for NF90b or both NF90b and NF45 and stained with DAPI for nuclear staining (Fig. 5, upper panels) and anti-NF90 antibody (Fig. 5, middle panels). The lower panels show overlays of these two signals (Fig. 5, merge). As previously reported, endogenous NF90 proteins were predominantly localized in the nucleus (Fig. 5, first column). Importantly, enforced expression of NF90b or both NF90b and NF45 did not affect the nuclear localization of the NF90 family proteins (Fig. 5, second and third columns). This indicates that the NF90 family proteins are retained in the nucleus, even if NF90 and NF45 are excessively expressed.
FIG. 5.
Localization of endogenous and overexpressed NF90 family proteins. 293T cells transfected with the indicated expression plasmids were stained with DAPI (upper panels) and anti-NF90 antibody (middle panels). Lower panels show overlays of these two signals.
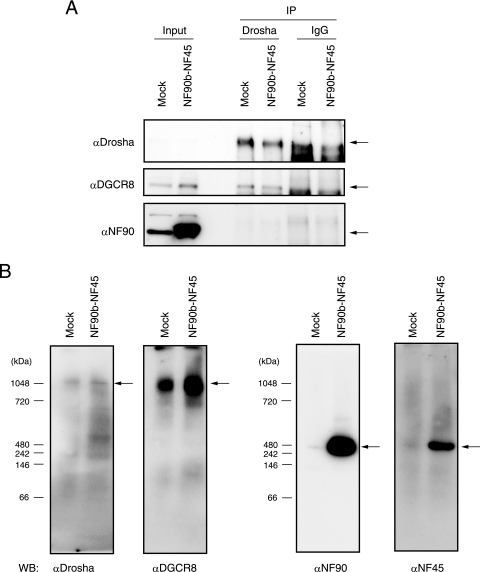
NF90 does not interact with the endogenous Drosha-DGCR8 complex, but the NF90-NF45 complex binds endogenous pri-miRNAs.
To test whether the NF90 proteins associate with endogenous Drosha in vivo, WCEs were prepared from 293T cells that were mock transfected or transfected with plasmids for both NF90b and NF45 and subjected to immunoprecipitation with anti-Drosha antibody or normal rabbit IgG. The immunoprecipitates were analyzed by Western blotting with anti-Drosha, anti-DGCR8, or anti-NF90 antibodies. As expected, Drosha interacted with DGCR8 in mock-transfected cells and cells transfected with both NF90b and NF45 (Fig. 6A, middle panels). However, NF90 did not associate with the endogenous Drosha-DGCR8 complex in vivo (Fig. 6A, lower panel). To confirm these results, we carried out a blue native electrophoresis analysis, which is a powerful tool for analyzing native protein complexes. WCEs prepared from 293T cells that were mock transfected or transfected with both NF90b and NF45 were subjected to native PAGE and immunoblotting to determine whether the Drosha-DGCR8 complex and the NF90-NF45 complex are distinct. As shown in Fig. 6B, the mobility of the Drosha-containing complex was completely consistent with that of DGCR8, and the NF90- and NF45-containing complexes exhibited the same mobility (Fig. 6B). These results show that Drosha forms a complex with DGCR8, while NF90 associates with NF45. However, the mobility of NF90-NF45 was clearly different from that of Drosha-DGCR8 (Fig. 6B). These observations, together with previously reported data (13), suggest that the NF90-NF45 complex is not included in the endogenous Drosha-DGCR8 complex, although forced expression of Drosha associates with NF90 and NF45 (13).
FIG. 6.
NF90 and NF45 do not interact with endogenous Drosha-DGCR8. (A) WCEs from 293T cells transfected with the indicated expression plasmids were immunoprecipitated with anti-Drosha antibody or normal rabbit IgG. The immunoprecipitates were probed with anti-Drosha, anti-DGCR8, or anti-NF90 antibodies. Arrows indicate the specific target proteins. (B) WCEs from 293T cells transfected with the indicated expression plasmids were subjected to blue native page analysis, followed by immunoblot analysis using anti-Drosha, anti-DGCR8, anti-NF90, or anti-NF45 antibodies. Arrows indicate the complexes containing the specific target proteins.
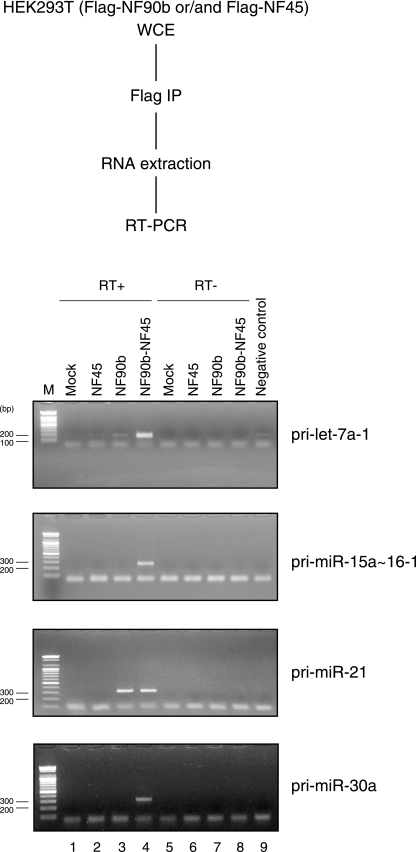
We next examined if the NF90-NF45 complex associates with endogenous pri-miRNAs in vivo. WCEs from 293T cells that were mock transfected or transfected with plasmids for Flag-tagged NF90b and/or Flag-tagged NF45 were immunoprecipitated with anti-Flag antibody. RNAs were recovered from the immunoprecipitates and analyzed by RT-PCR with the specific primers for pri-miRNAs. All the pri-miRNAs examined were undetectable in no-template and no-reverse transcriptase samples (Fig. 7, lanes 5 to 9). Of note, the NF90b-NF45 complex associated with all pri-miRNAs (Fig. 7, lane 4), whereas the NF45 or NF90b proteins alone were not able to bind to the majority of pri-miRNAs (Fig. 7, lanes 2 and 3). Interestingly, the number of PCR cycles needed to detect pri-let-7a-1 was much lower than that for other pri-miRNAs tested (35, versus 45 for other pri-miRNAs), although the efficiency of PCR amplification for pri-let-7a-1 was not higher than that for other pri-miRNAs. These results suggest that the higher amount of pri-let-7a-1 was associated with NF90-NF45 compared to other pri-miRNAs. In addition, we measured the absolute amounts of pri-miRNAs in cells transfected with NF90 and NF45 by qRT-PCR using plasmids harboring cDNA for pri-miRNAs as standards. The results indicated that the absolute amount of pri-let-7a-1 was about 3- to 30-fold higher than that of other pri-miRNAs (see Table S2 in the supplemental material), implying that overexpressed NF90-NF45 has the ability to associate with endogenous pri-miRNAs dependent on the abundance of each pri-miRNA.
FIG. 7.
The NF90-NF45 complex binds to endogenous pri-miRNAs in vivo. The experimental procedure is indicated in the upper part. WCEs from 293T cells transfected with indicated FLAG-tagged proteins were immunoprecipitated with anti-FLAG antibody. RNAs were isolated from the immunoprecipitates and analyzed by RT-PCR with specific primers for pri-let-7a-1, pri-miR-15a∼16-1, pri-miR-21, or pri-miR-30a. The numbers of PCR cycles for detection of pri-let-7a-1 and pri-miR-15a∼16-1, pri-miR-21, and pri-miR-30a were 35 and 45 cycles, respectively.
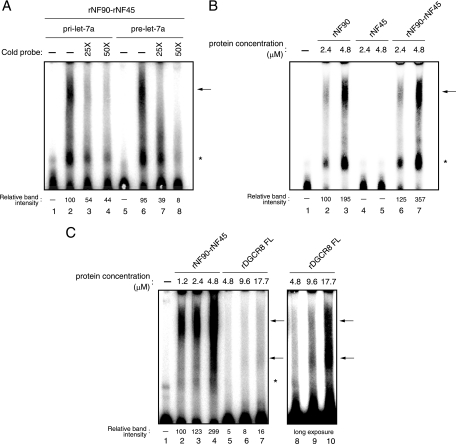
The NF90-NF45 complex binds to pri- and pre-let-7a-1, and the affinity of NF90-NF45 complex for pri-let-7a-1 is higher than that of DGCR8 in vitro.
To confirm the data from the in vivo RNA-protein binding analysis (Fig. 7), we performed an EMSA using recombinant proteins. We generated recombinant NF90 (rNF90), rNF45, FL DGCR8, and dsRBDs of DGCR8 in E. coli (see Fig. S1A in the supplemental material) and used these proteins for an EMSA with pri-miRNAs or pre-miRNA probe. Immunoblot analysis with anti-DGCR8 antibody confirmed the existence of DGCR8 in our purified rDGCR8 FL sample (see Fig. S1A in the supplemental material). The EMSA probed with pri-let-7a-1 clearly showed that rNF90-rNF45 bound to these RNAs in vitro (Fig. 8; see also Fig. S1B in the supplemental material). However, we found two major bands in these analyses (Fig. 8; see Fig. S1B [arrows and asterisks] in the supplemental material). To determine which band is the complex between rNF90 or rNF90-rNF45 and RNAs, we carried out a supershift assay using anti-NF90 antibody. Addition of NF90 antibody resulted in the formation of supershifted complex with the upper band (see Fig. S1B, lanes 3 and 4, in the supplemental material), indicating that the upper band is the complex of rNF90-rNF45 with RNAs. Under this experimental condition, EMSA probed with pri-let-7a-1 and pri-miR-21 showed that the rNF90-rNF45 complex exhibited 4.3-fold-higher binding activity for pri-let-7a-1 than pri-miR-21 (see Fig. S1D, arrow, compare lanes 4 and 9, in the supplemental material). We next examined if the NF90-NF45 complex discriminates between pri-miRNAs and pre-miRNAs. An EMSA with pri- or pre-let-7a-1 indicated that the binding activity of the rNF90-rNF45 complex to pre-let-7a-1 was comparable to that of this complex to pri-let-7a-1 (Fig. 8A, compare lanes 2 and 6). Recent studies also showed that NF90, either with or without NF45, binds to pre-let-7 in vitro (23, 57). Therefore, these results suggest that the NF90-NF45 complex selectively binds the stem-loop structure, which is a common secondary structure of pri- and pre-miRNAs. Furthermore, we tested whether the NF90-NF45 complex exhibits higher binding to pri-miRNA than with NF90 alone as shown in Fig. 7. Figure 8B showed that the binding activity of rNF90-rNF45 to pri-let7a-1 was 1.8-fold higher than that of rNF90 alone (compare lanes 3 and 7). NF45 alone did not exhibit the binding to pri-let-7a-1 (Fig. 8B, lanes 4 and 5). To confirm this result, we performed rNF45 titrations in the presence of rNF90. As expected, the binding activity of rNF90 was increased according to the elevation of rNF45 concentration, but not that of a control protein (GST) (see Fig. S1C in the supplemental material, compare lanes 2 to 4 and 5 to 7).
FIG. 8.
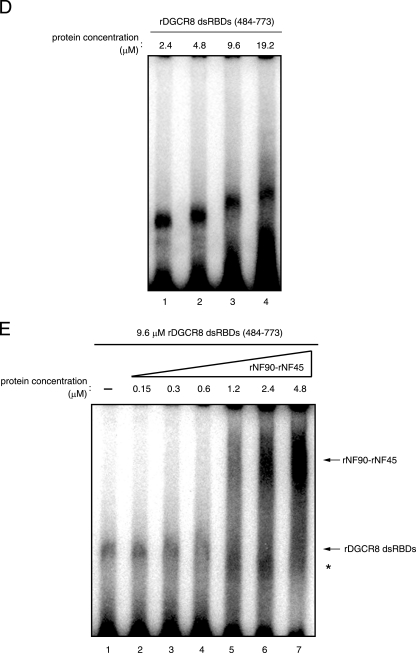
NF90 binds to pri- and pre-miRNAs in vitro. (A) EMSA performed with pri- and pre-let-7a probes and 2.4 μM each rNF90 and rNF45, in the absence (-) or presence of a 25- or 50-fold excess of unlabeled competitor RNA as indicated. (B) EMSA performed with the pri-let-7a probe and rNF90 alone, rNF45 alone, or both rNF90 and rNF45 proteins at the indicated protein concentrations. (C) EMSA performed with the pri-let-7a probe and rNF90-rNF45 and rDGCR8 FL at the indicated protein concentrations. Intensities of bands corresponding to the specific RNA-protein complexes indicated by upper arrows were measured with a densitometer and are presented as relative band intensities. Lanes 8 to 10 indicate the same picture after a long exposure. (D) EMSA performed with the pri-let-7a probe and rDGCR8 dsRBDs at the indicated protein concentrations. (E) EMSA performed using rDGCR8 dsRBDs and rNF90-rNF45 at the indicated protein concentrations. The specific RNA-protein complexes are indicated by arrows. Asterisks indicate nonspecific bands.
Since the NF90-NF45 complex impairs pri-miRNA processing and binds to pri-miRNA while these proteins do not associate with endogenous Drosha-DGCR8, we hypothesized that the binding of NF90-NF45 to pri-miRNAs reduces the accessibility of Drosha-DGCR8 to pri-miRNAs, resulting in the inhibition of pri-miRNA processing. It is known that DGCR8 recognizes the substrate pri-miRNAs, whereas Drosha is responsible for cleavage in Drosha-DGCR8-mediated pri-miRNA processing (21). Therefore, we compared the difference in the binding activity to pri-let-7a-1 between NF90-NF45 and DGCR8. rDGCR8 FL showed two bands on the EMSA when probed with pri-let-7a-1 (Fig. 8C, lanes 9 and 10). Thus, we performed a supershift assay using anti-DGCR8 antibody, which recognizes the DGCR8 C-terminal region containing dsRBDs. In this analysis, both bands disappeared with the addition of anti-DCGR8 antibody (see Fig. S1B in the supplemental material, compare lanes 5 and 6 with 7 and 8), demonstrating that the two bands were the complex of rDGCR8 FL with pri-miRNA. Under this experimental condition, we compared the binding activities of rNF90-rNF45 and rDGCR8 FL to pri-miRNA. As shown in Fig. 8C, rNF90-rNF45 clearly exhibited a greater binding activity to pri-let-7a-1 than rDGCR8 FL (compare lanes 2 to 4 and 5 to 7). To confirm this result, we performed an EMSA using rDGCR8-dsRBDs. Titration of rDGCR8-dsRBDs indicated the formation of a complex between rDGCR8-dsRBDs and pri-let-7a-1 (Fig. 8D). As shown in Fig. 8E, the complex between rDGCR8-dsRBDs and pri-miRNA was replaced by the NF90-NF45-pri-miRNA complex at lower concentrations of rNF90-rNF45 (1.2 to 4.8 μM) compared with that of rDGCR8-dsRBDs (9.6 μM). These results suggest that NF90-NF45 might preferentially bind to pri-let-7a-1 even in the presence of Drosha-DGCR8 under the condition of NF90-NF45 overexpression.
Knockdown of NF90 leads to a reduction of pri-let-7a-1 and an increase of mature let-7a-1 and causes growth suppression of transformed cells.
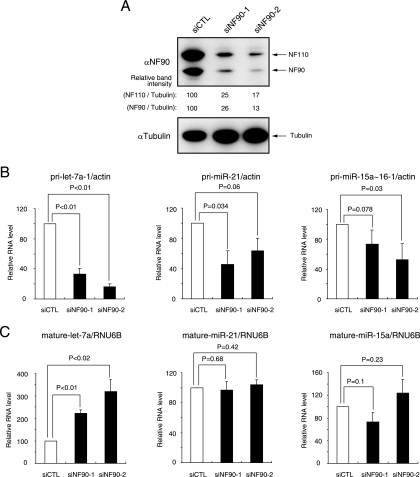
To confirm the function of the NF90-NF45 complex as a negative regulator in miRNA biogenesis, we measured the levels of pri-miRNAs and mature miRNAs which were produced from tested pri-miRNAs in NF90 knockdown cells. Transfection of siRNAs targeting the NF90 family proteins resulted in ∼75% inhibition of the expression of endogenous NF90 family proteins (Fig. 9A). As expected, we observed a significant decrease in pri-let-7a-1 in the NF90 knockdown cells (Fig. 9B). In accordance with the decrease in pri-let-7a-1, mature let-7a was increased in cells that were depleted of NF90 (Fig. 9C). However, knockdown of NF90 did not significantly affect the levels of other pri-miRNAs and mature miRNAs (Fig. 9B and C). These results, together with protein-RNA binding assays (see Fig. S1D in the supplemental material), suggest that processing of pri-miRNAs having high affinity for the NF90-NF45 complex into mature miRNAs may be specifically prevented by these proteins.
FIG. 9.
Knockdown of NF90 decreases pri-miRNA and increases mature miRNA. (A) WCEs were prepared from 293 cells transfected with the indicated siRNAs and analyzed by immunoblotting with anti-NF90 and anti-α-tubulin antibodies. Intensities of specific bands were measured with a densitometer and are presented as relative band intensities. (B) RNAs were isolated from 293 cells transfected with the indicated siRNAs and analyzed for the amount of pri-miRNAs by qRT-PCR with specific primers for pri-let-7a-1, pri-miR-21 and pri-miR-15a∼16-1. β-Actin was used as an internal control and for normalization of the data. siCTL, negative control siRNA; siNF90-1 and -2, siRNA targeting NF90. These results were reproducible in two independent transfection experiments. Data represent the averages of triplicate PCRs with ranges. P values were calculated by a two-tailed Student's t test. (C) RNAs were isolated from 293 cells transfected with the indicated siRNAs and analyzed for the amount of mature let-7a, miR-21 and miR-15a miRNAs using the TaqMan microRNA assay. RNU6B was used as an internal control and for normalization of the data. These results were reproducible in two independent transfection experiments. Data represent the averages of triplicate PCRs with ranges. P values were calculated by the two-tailed Student's t test.
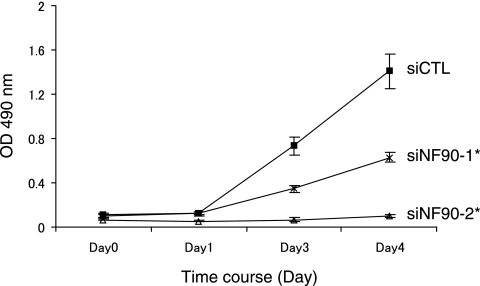
Several reports have indicated that let-7a has a potent antiproliferative activity in cultured cell lines (1, 25). This finding prompted us to test if NF90-NF45 participates in the control of cell proliferation. Knockdown of NF90 resulted in a significant reduction in growth rates of 293 cells (Fig. 10). Since the level of mature let-7a was elevated in NF90 knockdown cells, as shown in Fig. 9C, these observations suggest that NF90-NF45 might control cell proliferation via the regulation of mature let-7a production.
FIG. 10.
Knockdown of NF90 causes growth retardation in cells. Three days posttransfection, siRNA-transfected 293 cells were plated at 1,000 cells/well in 96-well plates. After the indicated days of culture, an MTS assay was performed according to the manufacturer's instructions, and absorbance was measured at 490 nm. Data represent the averages of two independent experiments. Error bars represent standard deviations. *, P < 5 × 10−5 relative to a control, by a two-tailed Student's t test.
DISCUSSION
The miRNA processing machinery has been relatively well characterized. The basic miRNA factors include Drosha, DGCR8, Exportin-5, Dicer, and Argonaute (Ago). In addition, p68 and p72 DEAD-box RNA helicase subunits, DDX5 and DDX17, human immunodeficiency virus transactivating response RNA binding protein, and PKR-activating protein have been reported as miRNA processing factors (6, 9, 18, 34). Like NF90 and NF45, DDX5 and DDX17 associate with overexpressed Drosha (13), but DDX5 and DDX17 facilitate the Drosha-DGCR8-mediated pri-miRNA processing reaction (9), while NF90 and NF45 inhibit the pri-miRNA processing (this study). Transactivating response RNA binding protein and PKR-activating protein function as bridging molecules between Dicer and Ago, but they do not show any effect on the Dicer processing reaction (6, 34). Notably, knockdown of these proteins reduces mature miRNA production (6, 9, 34), indicating that these miRNA processing factors are positive regulators for miRNA biogenesis. In contrast, negative regulatory events in the miRNA processing pathways are largely unknown.
In this study, we have shown that the forced expression of NF90 and NF45 impairs the Drosha-mediated pri-miRNA processing step in vitro (Fig. 1). Furthermore, pri-miRNAs accumulated in cells that overexpressed NF90 and NF45, and the accumulation was not due to transcriptional activation (Fig. 2 and 3). Interestingly, the enforced expression of Drosha, which plays a crucial role in cleavage of pri-miRNA to pre-miRNA, had no apparent effect on the accumulation (Fig. 4). In addition, the NF90-NF45 complex did not interact with the endogenous Drosha-DGCR8 complex (Fig. 6). On the other hand, in vivo and in vitro RNA-protein binding assays demonstrated that the NF90-NF45 complex bound to pri-miRNAs (Fig. 7 and 8; see also Fig. S1 in the supplemental material). Particularly, pri-let-7a-1 was found to show higher binding with NF90-NF45 than pri-miR-21 (see Fig. S1D in the supplemental material). Notably, knockdown of NF90 caused the decrease of pri-let-7a-1 and the increment in mature let-7a (Fig. 9). Therefore, these findings suggest that the binding of NF90-NF45 to pri-miRNAs may reduce the accessibility of the Microprocessor complex to pri-miRNAs, resulting in the reduction of miRNA production.
The NF90 and NF45 mRNAs are ubiquitously expressed in human and murine tissues (50, 60). However, protein expression levels of NF90 and NF45 are restricted. In both humans and mice, the NF90 family proteins are minimally expressed in lung, spleen, skeletal muscle, kidney, and liver, whereas high expression is seen in brain, testis, ovary, and thymus (50, 52) (S. Sakamoto, unpublished data). NF45 is also minimally expressed in heart, lung, spleen, skeletal muscle, and liver, but it is abundantly expressed in brain, testis, kidney, and thymus (60; Sakamoto, unpublished). The highest amounts of both proteins are seen in immortalized cell lines derived from uterus, kidney, leukocyte, fibroblast, stomach, and liver (50, 60; Sakamoto, unpublished). These observations suggest that the expression of the NF90 and NF45 proteins in undifferentiated cells may be much higher than that in differentiated cells. Indeed, the thymus and testis, in which the highest levels of the NF90 and NF45 proteins are seen, are comprised largely of cells undergoing differentiation, while lung, spleen, skeletal muscle, kidney, and liver are comprised largely of terminally differentiated cells. Furthermore, our previous results showed that higher levels of NF90 expression were detected in undifferentiated leukemia cells compared with phorbol ester-induced differentiated cells (47).
Numerous studies have demonstrated that miRNAs are associated with tumorigenesis. It is noteworthy that expression of a high copy number of miRNAs is reduced in tumor tissues and in transformed cell lines compared with normal tissues (10, 39). Interestingly, primary miRNA transcripts are not reduced in tumors, suggesting that the reduction of miRNAs in cancers is due to the altered regulation of the pri-miRNA processing (56). In fact, knockdown of Drosha, DGCR8, or Dicer, which are crucial factors for pri- and pre-miRNA processing, enhances tumor formation (30). However, expression of these factors is not dependent upon cytodifferentiation (39, 56). These observations suggest that the reduction of miRNAs in tumors might be due either to posttranslational modifications of the miRNA processing machinery or to the involvement of another miRNA-processing regulatory factor(s).
let-7 miRNA, which is a founding member of the miRNA family, is composed of 12 variants, let-7a-1 to let-7i and miR-98. Among them, mature let-7a, -7d, -7f-1, -7f-2, -7i, and miR-98 are detectable only in highly differentiated human and mouse embryonic cells, although the primary transcripts of those miRNAs are expressed in both undifferentiated and differentiated cells (54, 56). Several studies also reported that the level of let-7 family is decreased in tumor tissues compared with adjacent normal tissues (1, 26, 55, 59) and has the ability to inhibit proliferation of transformed cell lines (1, 25). Furthermore, it is known that a decrease in the level of let-7 family causes an increase in the expressions of RAS, c-MYC, and HMGA2, resulting in tumorigenesis (1, 26, 37, 42, 49). It has been thought that the alteration of let-7 family level according to cytodifferentiation is due to posttranscriptional regulation (54, 56). Indeed, recent studies reported that lin28 functions as a negative regulator for production of mature let-7 family by blockade of Drosha processing or Dicer processing (23, 57).
The present study has shown that depletion of NF90 results in a reduction of pri-let-7a-1 accompanied by an increase in mature let-7a (Fig. 9) and growth retardation in transformed cells (Fig. 10). A recent study also indicated that growth suppression is observed in NF90 or NF45 knockdown HeLa cells (15). Analyses of NF90 and NF45 expression profiles suggest that these proteins could be more highly expressed in undifferentiated than differentiated cells (47, 50, 52, 60). In addition, we have found that NF90-NF45 expression exhibits significant elevation in human tumor tissues bearing hepatocellular carcinoma compared with adjacent normal tissues (unpublished data). The elevation of NF90 has been also found in lung cancer tissues (16). Moreover, mature let-7a is downregulated in lung cancer and hepatocellular carcinoma compared with adjacent normal tissues or liver cirrhosis (11, 26, 55; Sakamoto, unpublished). It is therefore possible that the repression of let-7 is likely due to the high expression of the NF90 and NF45 proteins in tumor cells, together with lin28, resulting in the accelerated growth of the cells. Elucidation of the roles for these proteins in the downregulation of let-7, followed by the control of cell growth in tumors, will be of interest but will require extensive work in the future.
Supplementary Material
Acknowledgments
We thank M. B. Mathews for providing pcDNA3.1-NF90a, NF90b, NF110a, and NF110b plasmids, S. Kai for technical assistance, M. Takahashi for critical reading of the manuscript, and H. Kimura for helpful discussion.
This work was supported by a Grant-in-Aid for Young Scientists (B) from The Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 27 April 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akao, Y., Y. Nakagawa, and T. Naoe. 2006. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 29903-906. [DOI] [PubMed] [Google Scholar]
- 2.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. [DOI] [PubMed] [Google Scholar]
- 3.Bohnsack, M. T., K. Czaplinski, and D. Gorlich. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennecke, J., D. R. Hipfner, A. Stark, R. B. Russell, and S. M. Cohen. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 11325-36. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. Z., L. Li, H. F. Lodish, and D. P. Bartel. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 30383-86. [DOI] [PubMed] [Google Scholar]
- 6.Chendrimada, T. P., R. I. Gregory, E. Kumaraswamy, J. Norman, N. Cooch, K. Nishikura, and R. Shiekhattar. 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436740-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimmino, A., G. A. Calin, M. Fabbri, M. V. Iorio, M. Ferracin, M. Shimizu, S. E. Wojcik, R. I. Aqeilan, S. Zupo, M. Dono, L. Rassenti, H. Alder, S. Volinia, C. G. Liu, T. J. Kipps, M. Negrini, and C. M. Croce. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 10213944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denli, A. M., B. B. Tops, R. H. Plasterk, R. F. Ketting, and G. J. Hannon. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432231-235. [DOI] [PubMed] [Google Scholar]
- 9.Fukuda, T., K. Yamagata, S. Fujiyama, T. Matsumoto, I. Koshida, K. Yoshimura, M. Mihara, M. Naitou, H. Endoh, T. Nakamura, C. Akimoto, Y. Yamamoto, T. Katagiri, C. Foulds, S. Takezawa, H. Kitagawa, K. Takeyama, B. W. O'Malley, and S. Kato. 2007. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 9604-611. [DOI] [PubMed] [Google Scholar]
- 10.Gaur, A., D. A. Jewell, Y. Liang, D. Ridzon, J. H. Moore, C. Chen, V. R. Ambros, and M. A. Israel. 2007. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 672456-2468. [DOI] [PubMed] [Google Scholar]
- 11.Gramantieri, L., M. Ferracin, F. Fornari, A. Veronese, S. Sabbioni, C. G. Liu, G. A. Calin, C. Giovannini, E. Ferrazzi, G. L. Grazi, C. M. Croce, L. Bolondi, and M. Negrini. 2007. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 676092-6099. [DOI] [PubMed] [Google Scholar]
- 12.Gregory, R. I., T. P. Chendrimada, N. Cooch, and R. Shiekhattar. 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123631-640. [DOI] [PubMed] [Google Scholar]
- 13.Gregory, R. I., K. P. Yan, G. Amuthan, T. Chendrimada, B. Doratotaj, N. Cooch, and R. Shiekhattar. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432235-240. [DOI] [PubMed] [Google Scholar]
- 14.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 10623-34. [DOI] [PubMed] [Google Scholar]
- 15.Guan, D., N. Altan-Bonnet, A. M. Parrott, C. J. Arrigo, Q. Li, M. Khaleduzzaman, H. Li, C. G. Lee, T. Pe'ery, and M. B. Mathews. 2008. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol. Cell. Biol. 284629-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, N. L., Y. W. Wan, K. Tosun, H. Lin, Z. Msiska, D. C. Flynn, S. C. Remick, V. Vallyathan, A. Dowlati, X. Shi, V. Castranova, D. G. Beer, and Y. Qian. 2008. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin. Cancer Res. 148213-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gwizdek, C., B. Ossareh-Nazari, A. M. Brownawell, S. Evers, I. G. Macara, and C. Dargemont. 2004. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J. Biol. Chem. 279884-891. [DOI] [PubMed] [Google Scholar]
- 18.Haase, A. D., L. Jaskiewicz, H. Zhang, S. Laine, R. Sack, A. Gatignol, and W. Filipowicz. 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6961-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates transcriptional gene silencing in Drosophila cells. Nature 404293-296. [DOI] [PubMed] [Google Scholar]
- 20.Han, J., Y. Lee, K. H. Yeom, Y. K. Kim, H. Jin, and V. N. Kim. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 183016-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, J., Y. Lee, K. H. Yeom, J. W. Nam, I. Heo, J. K. Rhee, S. Y. Sohn, Y. Cho, B. T. Zhang, and V. N. Kim. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125887-901. [DOI] [PubMed] [Google Scholar]
- 22.He, L., J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon, and S. M. Hammond. 2005. A microRNA polycistron as a potential human oncogene. Nature 435828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heo, I., C. Joo, J. Cho, M. Ha, J. Han, and V. N. Kim. 2008. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 32276-284. [DOI] [PubMed] [Google Scholar]
- 24.Hutvagner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293834-838. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, C. D., A. Esquela-Kerscher, G. Stefani, M. Byrom, K. Kelnar, D. Ovcharenko, M. Wilson, X. Wang, J. Shelton, J. Shingara, L. Chin, D. Brown, and F. J. Slack. 2007. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 677713-7722. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, S. M., H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K. L. Reinert, D. Brown, and F. J. Slack. 2005. RAS is regulated by the let-7 microRNA family. Cell 120635-647. [DOI] [PubMed] [Google Scholar]
- 27.Kao, P. N., L. Chen, G. Brock, J. Ng, J. Kenny, A. J. Smith, and B. Corthesy. 1994. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J. Biol. Chem. 26920691-20699. [PubMed] [Google Scholar]
- 28.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 152654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khvorova, A., A. Reynolds, and S. D. Jayasena. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115209-216. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, M. S., J. Lu, K. L. Mercer, T. R. Golub, and T. Jacks. 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39673-677. [DOI] [PubMed] [Google Scholar]
- 31.Landthaler, M., A. Yalcin, and T. Tuschl. 2004. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 142162-2167. [DOI] [PubMed] [Google Scholar]
- 32.Lecellier, C. H., P. Dunoyer, K. Arar, J. Lehmann-Che, S. Eyquem, C. Himber, A. Saib, and O. Voinnet. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308557-560. [DOI] [PubMed] [Google Scholar]
- 33.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425415-419. [DOI] [PubMed] [Google Scholar]
- 34.Lee, Y., I. Hur, S. Y. Park, Y. K. Kim, M. R. Suh, and V. N. Kim. 2006. The role of PACT in the RNA silencing pathway. EMBO J. 25522-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, Y., K. Jeon, J. T. Lee, S. Kim, and V. N. Kim. 2002. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 214663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, Y., M. Kim, J. Han, K. H. Yeom, S. Lee, S. H. Baek, and V. N. Kim. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 234051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, Y. S., and A. Dutta. 2007. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 211025-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis, B. P., I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel, and C. B. Burge. 2003. Prediction of mammalian microRNA targets. Cell 115787-798. [DOI] [PubMed] [Google Scholar]
- 39.Lu, J., G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz, and T. R. Golub. 2005. MicroRNA expression profiles classify human cancers. Nature 435834-838. [DOI] [PubMed] [Google Scholar]
- 40.Lund, E., S. Guttinger, A. Calado, J. E. Dahlberg, and U. Kutay. 2004. Nuclear export of microRNA precursors. Science 30395-98. [DOI] [PubMed] [Google Scholar]
- 41.Martinez, J., A. Patkaniowska, H. Urlaub, R. Luhrmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110563-574. [DOI] [PubMed] [Google Scholar]
- 42.Mayr, C., M. T. Hemann, and D. P. Bartel. 2007. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 3151576-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muraki, M., B. Ohkawara, T. Hosoya, H. Onogi, J. Koizumi, T. Koizumi, K. Sumi, J. Yomoda, M. V. Murray, H. Kimura, K. Furuichi, H. Shibuya, A. R. Krainer, M. Suzuki, and M. Hagiwara. 2004. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J. Biol. Chem. 27924246-24254. [DOI] [PubMed] [Google Scholar]
- 44.Olsen, P. H., and V. Ambros. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216671-680. [DOI] [PubMed] [Google Scholar]
- 45.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304734-736. [DOI] [PubMed] [Google Scholar]
- 46.Reichman, T. W., A. M. Parrott, I. Fierro-Monti, D. J. Caron, P. N. Kao, C. G. Lee, H. Li, and M. B. Mathews. 2003. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J. Mol. Biol. 33285-98. [DOI] [PubMed] [Google Scholar]
- 47.Sakamoto, S., K. Morisawa, K. Ota, J. Nie, and T. Taniguchi. 1999. A binding protein to the DNase I hypersensitive site II in HLA-DR alpha gene was identified as NF90. Biochemistry 383355-3361. [DOI] [PubMed] [Google Scholar]
- 48.Sakamoto, S., and T. Taniguchi. 2001. Identification of a phorbol ester-responsive element in the interferon-gamma receptor 1 chain gene. J. Biol. Chem. 27637237-37241. [DOI] [PubMed] [Google Scholar]
- 49.Sampson, V. B., N. H. Rong, J. Han, Q. Yang, V. Aris, P. Soteropoulos, N. J. Petrelli, S. P. Dunn, and L. J. Krueger. 2007. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 679762-9770. [DOI] [PubMed] [Google Scholar]
- 50.Saunders, L. R., D. J. Perkins, S. Balachandran, R. Michaels, R. Ford, A. Mayeda, and G. N. Barber. 2001. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 27632300-32312. [DOI] [PubMed] [Google Scholar]
- 51.Schwarz, D. S., G. Hutvagner, T. Du, Z. Xu, N. Aronin, and P. D. Zamore. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115199-208. [DOI] [PubMed] [Google Scholar]
- 52.Shi, L., G. Zhao, D. Qiu, W. R. Godfrey, H. Vogel, T. A. Rando, H. Hu, and P. N. Kao. 2005. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J. Biol. Chem. 28018981-18989. [DOI] [PubMed] [Google Scholar]
- 53.Song, D., S. Sakamoto, and T. Taniguchi. 2002. Inhibition of poly(ADP-ribose) polymerase activity by Bcl-2 in association with the ribosomal protein S3a. Biochemistry 41929-934. [DOI] [PubMed] [Google Scholar]
- 54.Suh, M. R., Y. Lee, J. Y. Kim, S. K. Kim, S. H. Moon, J. Y. Lee, K. Y. Cha, H. M. Chung, H. S. Yoon, S. Y. Moon, V. N. Kim, and K. S. Kim. 2004. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270488-498. [DOI] [PubMed] [Google Scholar]
- 55.Takamizawa, J., H. Konishi, K. Yanagisawa, S. Tomida, H. Osada, H. Endoh, T. Harano, Y. Yatabe, M. Nagino, Y. Nimura, T. Mitsudomi, and T. Takahashi. 2004. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 643753-3756. [DOI] [PubMed] [Google Scholar]
- 56.Thomson, J. M., M. Newman, J. S. Parker, E. M. Morin-Kensicki, T. Wright, and S. M. Hammond. 2006. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 202202-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viswanathan, S. R., G. Q. Daley, and R. I. Gregory. 2008. Selective blockade of microRNA processing by Lin28. Science 32097-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yi, R., Y. Qin, I. G. Macara, and B. R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 173011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu, F., H. Yao, P. Zhu, X. Zhang, Q. Pan, C. Gong, Y. Huang, X. Hu, F. Su, J. Lieberman, and E. Song. 2007. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 1311109-1123. [DOI] [PubMed] [Google Scholar]
- 60.Zhao, G., L. Shi, D. Qiu, H. Hu, and P. N. Kao. 2005. NF45/ILF2 tissue expression, promoter analysis, and interleukin-2 transactivating function. Exp. Cell Res. 305312-323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.