Abstract
The yeast AAA+ chaperone Hsp104 is essential for the development of thermotolerance and for the inheritance of prions. Recently, Hsp104, together with the actin cytoskeleton, has been implicated in the asymmetric distribution of carbonylated proteins. Here, we investigated the interplay between Hsp104 and actin by using a dominant-negative variant of Hsp104 (HAP/ClpP) that degrades substrate proteins instead of remodeling them. Coexpression of HAP/ClpP causes defects in morphology and the actin cytoskeleton. Taking a candidate approach, we identified Spa2, a member of the polarisome complex, as an Hsp104 substrate. Furthermore, we provided genetic evidence that links Spa2 and Hsp104 to Hof1, a member of the cytokinesis machinery. Spa2 and Hof1 knockout cells are affected in the asymmetric distribution of damaged proteins, suggesting that Hsp104, Spa2, and Hof1 are members of a network controlling the inheritance of carbonylated proteins.
The ensemble of molecular chaperones and proteases constitutes the cellular system that repairs and eliminates misfolded proteins. The activity of this system ensures not only the recovery of cells from protein-damaging stress conditions, but also the maintenance of protein homeostasis under normal growth conditions. The concomitant involvement of members of the Hsp70 and Hsp90 chaperone families in stress-related, regulatory, and housekeeping functions allows the integration of environmental stimuli into regulatory networks (4, 24, 39, 40). However, it has remained unclear whether other chaperones are also involved in regulatory processes.
One chaperone which so far has been connected only to stress-related protein quality functions is the oligomeric AAA+ chaperone Hsp104 of Saccharomyces cerevisiae. Hsp104 is essential for the development of thermotolerance by reactivating aggregated proteins after severe stress conditions and for prion propagation by severing prion fibrils (31). Yeast cells, when grown at 30°C, harbor approximately 5,000 copies of Hsp104 hexamers per cell, a number that is minor compared to other cytosolic chaperone machineries (e.g., Hsp70 and Hsp90) that are involved in general protein-folding events (10). The known cellular functions of Hsp104, however, cannot provide a rationale for the determined Hsp104 levels, since protein aggregation is hardly detectable in yeast cells at 30°C even in mutant cells lacking Hsp104 function. Furthermore, yeast prions occur de novo at a very low rate of 10−6 per cell. In consequence, both well-characterized Hsp104 activities are barely required at 30°C, suggesting that Hsp104 has additional, so far unknown housekeeping functions. On the other hand, an S. cerevisiae hsp104 knockout exhibits no obvious phenotype at 30°C (27), giving no clues to a potential involvement of Hsp104 in other cellular processes.
Recently, Hsp104 was demonstrated to influence the asymmetric distribution of oxidatively damaged (carbonylated) proteins (8). It remained unclear whether the role of Hsp104 in this process relies on its known activities in protein quality control or on an unknown involvement in other cellular processes. Here, we provide evidence that Hsp104 is part of a network that controls the inheritance of damaged proteins under physiological growth conditions.
MATERIALS AND METHODS
Media, strains, plasmids, antibodies, and reagents.
Yeast and bacterial media, as well as recombinant DNA methods, were as described previously (1, 20). Chromosomal green fluorescent protein (GFP) tagging was carried out using the versatile toolbox for PCR-based tagging of yeast genes (15), and promoter substitutions against the tetracycline-regulatable system were performed according to published protocols (5, 6). The genotypes of the strains and yeast plasmids used in this work are summarized in Tables 1 and 2, respectively. Polyclonal antisera were raised against purified Hsp104; antisera recognizing glucose-6-phosphate dehydrogenase (G6PDH) and monoclonal 9E10-anti-myc antibody were purchased from Sigma. Anti-Spa2 antibody was a gift from M. Snyder (Yale University). Anti-Mpk1 was purchased from Santa Cruz and anti-phospho-p42/p44 antibody from Cell Signaling Technology. Secondary antibodies for Western blotting were purchased from Rockland. Rhodamine-labeled phalloidin, anti-dinitrophenyl-KLH (A6430), and Alexa 546-labeled anti-rabbit antibody were obtained from Molecular Probes and α-factor from Zymoresearch. 2,4-Dinitrophenylhydrazine (DNPH) was purchased from Fluka.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pRS313 | pBluescript based; Ampr HIS3 CEN | 32 |
| pRS315 | pBluescript based; Ampr LEU2 CEN | 32 |
| pRS316 | pBluescript based; Ampr URA3 CEN | 32 |
| pRS303 | pBluescript based; Ampr His3 YIP | 32 |
| pRS425 | pBluescript based; Ampr LEU2 2μm | 7 |
| phs313 | pRS313; heat shock promoter (500 bp upstream of HSP104) | This study |
| pGAL425 | pRS425; Gal1,10 promoter | This study |
| phs313-Hsp104 | phs313; HSP104 | This study |
| phs313-HAP | phs313; HAP | This study |
| pGAL425-ClpP | pGAL425; ClpP | This study |
| pGAL425-ClpPS111A | pGAL425; ClpP(S111A) | This study |
| pmCUP425-ClpPtrap | pmCUP425; ClpPtrap (ClpPΔ1-13; S111A; SBP tagged) | 33 |
| pADHmyc316-Spa2 | pRS316; ADH1 promoter controlling myc6-Spa2 | This study |
TABLE 2.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| PT100 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 | W303/Lindquist laboratory |
| PT101 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 hsp104::kanMX4 | W303/Lindquist laboratory |
| PT102 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 hsp104::kanMX4 CDC3::GFP-hphNT1 | This study |
| PT104 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 hsp104::kanMX4 CDC12::GFP-hphNT1 | This study |
| PT105 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 CDC3::GFP-i | This study |
| PT106 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 CDC12::GFP-hphNT1 | This study |
| PT111 | matα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 | W303/Dobberstein laboratory |
| PT112 | mata/α can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 | W303/Dobberstein laboratory |
| PT132 | matα his3Δ leu2Δ ura3Δ met15Δ LYS2+can1::MFA1pr-HIS3 lyp1Δ | BY5563/Jentsch laboratory |
| PT133 | matα his3Δ leu2Δ ura3Δ met15Δ LYS2+can1::MFA1pr-HIS3 lyp1Δ hsp104::natMX4 | This study |
| PT150 | mata/α can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 hsp104::kanMX4−/−hof1::nat+/− | This study |
| PT152 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 hof1::natMX4 | This study |
| PT163 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 hsp104::kanMX4 Pea2-GFP::hphNT1 | This study |
| PT175 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 pea2::hphNT1 | This study |
| PT176 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 Pea2-GFP::hphNT1 | This study |
| PT185 | matα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 adh1p(tetR′-SSN6)::LEU2 tetO2::HOf1::kanMX4 hsp104::TRP1 | This study |
| PT186 | matacan1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 spa2::hphNT1 | This study |
RT-PCR.
mRNA was purified from S. cerevisiae using the YeaStar RNA preparation kit from Zymoresearch. Total mRNA (0.15 μg/μl) was used for the reverse transcription (RT) reaction using the Fermentas RevertAid RT-PCR kit. PCR was performed using Taq polymerase, and amplification cycles were adjusted to allow for logarithmic amplification of the template.
Microscopy.
Cells expressing GFP-tagged proteins were grown to the mid-logarithmic growth phase, recovered by centrifugation, washed in phosphate-buffered saline (PBS), and mounted in 50% glycerol in PBS. For actin staining, cells were grown to mid-logarithmic phase and fixed within the media with formaldehyde at a final concentration of 3.75% (vol/vol) for 10 min. Next, the cells were harvested and resuspended in PBS containing 3.75% (vol/vol) formaldehyde and incubated for one additional hour. Then, the cells were washed twice with PBS and subsequently incubated in PBS supplemented with 0.66 μM rhodamine-phalloidin for 1 hour. Finally, the cells were washed five times with PBS and mounted in 50% (vol/vol) glycerol in PBS. The staining of carbonylated proteins was carried out as described previously (3) with slight changes. Briefly, the DNPH solution was prepared by dissolving 20 mM DNPH in 10% (vol/vol) trichloroacetic acid, and the 2,4-dinitrophenol (DNP) moiety was detected using an anti-DNP antibody preabsorbed with 1% (wt/vol) acetone powder of an S. cerevisiae wild-type lysate. Microscopy was performed on a reverse-fluorescence microscope (Leica DMIRE 2, equipped with Openlab Software), and all images were processed with Adobe Photoshop.
Extract preparation and Western blotting.
For sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Western blotting, yeast cells were usually lysed using the NaOH/trichloroacetic acid method. Anti-Spa2 antibodies were used at a dilution of 1:300 after preabsorption with 1% (wt/vol) acetone powder of spa2Δ cells (13). Anti-G6PDH antibody was diluted 1:50,000. To detect phosphorylated Mpk1, cells were lysed according to the method of Martin et al. (21), and Western blots were probed with anti-Mpk1 (1:200) and anti-phospho-p42/p44 (1:1,000). Detection of Western blots was performed using the Licor system.
Co-IP using myc-tagged Spa2.
Cells were grown in synthetic complete medium supplemented with 2% glucose to late logarithmic growth phase and lysed by glass bead lysis in immunoprecipitation (IP) buffer (50 mM Tris/Cl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 5 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml pepstatin). The lysate was centrifuged for 5 min at 5,000 rpm, followed by 15 min at 13,000 rpm (each step at 4°C). The cleared lysate was adjusted to 250 μg protein in 500 μl IP buffer and incubated with 25 μl anti-myc agarose for 1 h at 4°C. The beads were subsequently washed with 5 ml IP buffer supplemented with 1% (vol/vol) Triton X-100, followed by a final washing step using 5 ml IP buffer. Bound proteins were eluted by boiling the beads in SDS sample buffer. Subsequently, the eluate was subjected to SDS-polyacrylamide gel electrophoresis and Western blotting.
Synthetic-lethal screen.
We used synthetic genetic array analysis as described previously (34, 35). Briefly, strain PTY133 (BY5563 hsp104Δ) was crossed to the complete knockout library of nonessential genes (11). After sporulation and selection for the respective double knockout, the latter was screened for viability. The screen was performed on a Beckman-Coulter Biomek FX.
RESULTS AND DISCUSSION
Coexpression of HAP and ClpP results in growth arrest and defects in the actin cytoskeleton.
As a first step toward understanding the role of Hsp104 under nonstress conditions, we used a genetically engineered variant of Hsp104, HAP, which is able to degrade Hsp104 substrates through association with the bacterial peptidase ClpP (33). The rationale for this approach was that the irreversible degradation of potential Hsp104 substrates by HAP/ClpP in yeast cells might result in detectable phenotypes. HAP/ClpP together represent a dominant-negative variant of Hsp104 and thereby might facilitate the identification of processes in which Hsp104 plays a role without having a strictly essential function (Fig. 1A). HAP and Hsp104 differ in only 3 amino acids at the C terminus of the protein (739GSK741 to 739IGF741), located in a region that is neither evolutionarily conserved nor involved in substrate selection. This largely excludes the possibility that HAP exhibits different substrate specificity than Hsp104 (19, 29). Moreover, HAP behaved identically to Hsp104 in the absence of ClpP with respect to protein disaggregation and prion propagation and did not induce growth phenotypes (33). Upon ClpP induction, we observed a severe growth defect of HAP-coexpressing cells, whereas cells coexpressing Hsp104 showed no defects (Fig. 1B). Notably, the plating efficiencies were the same for all plasmid combinations spotted, but the colony size was significantly decreased upon coexpression of HAP/ClpP, suggesting that HAP/ClpP coproduction is not lethal to yeast cells but leads to growth arrest. Similar results were obtained when we followed the plating efficiency after growth in liquid culture for 3 days (data not shown and Fig. 1C). These findings suggest that Hsp104 has a role in housekeeping functions and that the irreversible and processive degradation of previously unknown Hsp104 substrates by the proteolytic HAP/ClpP system exerts a dominant-negative effect on yeast. This interpretation was further substantiated by an experiment in which wild-type ClpP was replaced by the active-site mutant protein ClpP-S111A, which fails to degrade substrates (38). The coproduction of HAP and inactive ClpP-S111A did not result in growth arrest, indicating that the observed phenotype depends on the processive and irreversible degradation of Hsp104 substrates by HAP/ClpP (Fig. 1D).
FIG. 1.
Expression of HAP and ClpP induces specific growth arrest. (A) Rationale of the study. Hsp104 might be used to remodel protein complexes (left), but this activity is changed into degradation of the substrate in the case of the genetically modified variant of Hsp104 (HAP) carrying the recognition motif for the bacterial peptidase ClpP (indicated in red on the right). The latter case might lead to the development of phenotypes. (B) Growth of S. cerevisiae cells expressing the indicated plasmid-borne components. Serial dilutions of S. cerevisiae overnight cultures, adjusted to the same optical density, were spotted onto synthetic complete medium selecting for the transformed plasmids, supplemented with either 2% glucose or 2% galactose. The plates were scanned after 48 h of incubation at 30°C. Wild-type (WT) and hsp104Δ cells harbored empty plasmids carrying promoters only. (C) HAP/ClpP induces growth arrest. Overnight cultures of yeast cells were diluted to the same optical density at 600 nm of 0.1 in selective medium containing either glucose or galactose. After 72 h of growth, the cells were adjusted to the same optical density, fivefold serially diluted, and spotted onto yeast-peptone-dextrose agar. The plates were incubated for 48 h at 30°C prior to being scanned. The protein names represent plasmids encoding the respective proteins. (D) Processive protein degradation by HAP/ClpP is required for the observed phenotype, as a proteolytically inactive ClpP variant (ClpPS111A) abrogates the growth arrest. (E to G) Morphological inspection of S. cerevisiae wild-type and hsp104Δ mutant cells. The protein names indicate plasmid-borne genes expressed in hsp104Δ cells. WT and hsp104Δ cells harbored empty plasmids carrying promoters only. DAPI (4′,6′-diamidino-2-phenylindole) staining (E), actin visualization by rhodamine-labeled phalloidin (F), and localization of Pea2-GFP (G) in logarithmically growing cells incubated for 14 h in galactose-containing selective media are shown. Scale bar, 5 μm.
In order to identify the underlying cellular defects leading to the growth arrest of HAP/ClpP-expressing cells, we tested for possible morphological changes. Cells expressing HAP/ClpP were round and showed significantly increased size compared to control cells (Fig. 1E). This phenotype is typical for mutations affecting the actin cytoskeleton (14, 23). To further test whether HAP/ClpP expression affects this process, we monitored the actin cytoskeleton using rhodamine-labeled phalloidin. In contrast to wild-type and hsp104Δ cells, coexpression of HAP/ClpP severely affected the integrity of the actin cytoskeleton and lacked proper actin polarity (Fig. 1F). Actin polarity in budding yeast is mainly established by the rho family GTPase Cdc42 (2, 16). GTP-bound Cdc42 is responsible for the proper localization of the polarisome complex to the site of bud emergence. The polarisome complex recruits Bni1, one of two formins in yeast that have been shown to be responsible for actin cable polymerization (25, 26). To test whether the observed defects in cell polarity upon coexpression of HAP/ClpP are related to a potential mislocalization of the polarisome complex, we followed the consequences of HAP/ClpP production for this complex.
The polarisome complex consists of Bud6, Pea2, and Spa2. To investigate whether HAP/ClpP might influence the formation of this structure, we monitored the localization of GFP-tagged Pea2. Pea2-GFP localized properly to the bud tip in wild-type and hsp104Δ cells. However, in hsp104Δ cells coexpressing HAP and ClpP, no localized Pea2-GFP fluorescence was detectable (Fig. 1G), indicating that expression of HAP and ClpP interferes with the proper assembly of the polarisome complex. Taken together, coexpression of dominant-negative HAP/ClpP leads to defects in the actin cytoskeleton and mislocalization of the polarisome complex, suggesting a role of Hsp104 in processes leading to the establishment of a functional actin cytoskeleton.
Spa2 is a substrate of Hsp104.
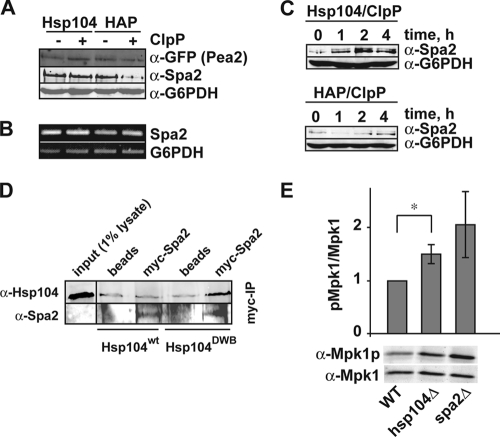
Pea2 localization is dependent on another polarisome component, Spa2 (30). Degradation of Spa2 by HAP/ClpP, therefore, might explain the improper positioning of Pea2. We monitored the degradation of Pea2 and Spa2 upon coexpression of HAP/ClpP. Coexpression had no significant influence on the levels of Pea2-GFP but led to reduced Spa2 levels (Fig. 2A). This effect can be directly attributed to the Spa2 protein, as Spa2 mRNA levels were not influenced (Fig. 2B). We also followed the cell cycle-dependent accumulation of Spa2 upon release from α-factor-induced arrest. In cells expressing Hsp104 and ClpP, Spa2 levels increased after entry into the cell cycle, peaking at around 2 h. In contrast, Spa2 levels were strongly reduced in cells expressing HAP/ClpP (Fig. 2C). Reduced Spa2 expression levels have been reported to be sufficient to cause defects in polarity formation (30), implying that defects of the actin cytoskeleton observed upon coexpression of HAP and ClpP might be caused, at least in part, by Spa2 degradation. However, at this point, we cannot rule out the possibility that HAP/ClpP has a more pleiotropic effect.
FIG. 2.
Hsp104 interacts with and remodels Spa2. (A and B) Steady-state levels of Pea2-GFP and Spa2 proteins (A) and Spa2 mRNA levels (B) of S. cerevisiae cells expressing the indicated components. A reprobe of the Western blot against G6PDH is shown as a loading control. (C) Cell cycle-dependent accumulation of Spa2 after release from an α-factor-mediated G1 arrest. Overnight cultures of yeast cells expressing either Hsp104/ClpP or HAP/ClpP were diluted to an optical density at 600 nm of 0.5 in selective medium containing 2% galactose and were grown for 3 hours. α-Factor was added to a final concentration of 5 μM, and the cells were incubated for an additional 3 hours. The cells were then washed three times in prewarmed selective medium, and samples were taken at the indicated times. Spa2 and G6PDH levels were determined by Western blotting. The protein names indicate plasmid-borne genes expressed in hsp104Δ cells. (D) myc-tagged Spa2 was used to coimmunoprecipitate Hsp104. To lock the binding of Hsp104 to Spa2, we used a variant of Hsp104 that stably interacts with substrates due to a mutation that allows ATP binding, but not hydrolysis (Hsp104-DWB). “Beads” indicates control cells expressing untagged Spa2 only. (E) Deletion of hsp104 induces the cell integrity pathway. The indicated yeast cells were grown to logarithmic phase, and total cell lysates were subjected to immunoblotting to determine the levels of phosphorylated (Mpk1p) and total Mpk1. The Mpk1p/Mpk1 ratio determined in wild-type (WT) cells was set as 1. Depicted are means and standard deviations from three independent experiments. The asterisk indicates a P value of <0.05 based on the unpaired two-tailed t test. α, anti.
The reduced levels of Spa2 in the presence of HAP/ClpP could formerly be explained by the degradation of a yet-unidentified factor rather than by a direct interaction of HAP (or Hsp104) with Spa2. To further substantiate our finding, we tested whether there is a direct interaction between Spa2 and Hsp104. The interactions between AAA+ proteins and substrates are transient in nature but can be stabilized by freezing the AAA+ protein in the ATP state (37). Therefore, we constructed an Hsp104 variant that was deficient in ATP hydrolysis (Hsp104 E285A/E687A, referred to as Hsp104-DWB) and hence led to stabilized Hsp104-substrate complexes (28, 37). We tested for an interaction between Hsp104-DWB and myc-tagged Spa2 by co-IP using anti-myc agarose (Fig. 2D). We observed nonspecific background binding of wild-type Hsp104 in cells expressing either wild-type or myc-tagged Spa2. In contrast, specific enrichment of Hsp104-DWB was obtained in the presence of myc-tagged Spa2, demonstrating a direct interaction of Hsp104 with Spa2. To further elucidate whether this interaction was due to chaperoning of Spa2 by Hsp104, we tested whether the loss of hsp104 might influence the so-called cell integrity pathway, in which Spa2 serves as a scaffold protein in a mitogen-activated protein (MAP) kinase signal transduction cascade (36). A deletion of spa2 leads to phosphorylation and thus activation of the MAP kinase Mpk1 as a compensatory mechanism (30). Thus, we expected higher levels of Mpk1 phosphorylation when Spa2 was, at least in part, not folded correctly in the absence of Hsp104. We did indeed observe increased phosphorylation of Mpk1 in hsp104Δ cells compared to that in wild-type cells (Fig. 2E), suggesting that fully functional Spa2 requires chaperoning of Hsp104.
Hsp104 and Spa2 are synthetically lethal with Hof1.
Our data indicate that the coexpression of HAP and ClpP induces defects in the actin cytoskeleton by degradation of at least Spa2. However, hsp104Δ cells exhibit only a weak polarity defect (as measured by upregulation of the cell integrity pathway) under physiological conditions. A potential explanation for the absence of phenotypes might be the existence of parallel pathways. To identify such potential parallel pathways, we took an unbiased approach and screened the Euroscarf deletion strain collection for synthetic growth defects with hsp104 (34, 35). This screen generated a candidate gene, HOF1, the homolog of cdc15 in S. cerevisiae. Tetrad dissections of combined homozygous hsp104 and heterozygous hof1 deletions confirmed the synthetic interaction between the two genes (Fig. 3A).
FIG. 3.
hsp104 and spa2 form synthetic-lethal pairs with hof1. (A) Tetrad analysis of a homozygous hsp104 (hsp104Δ−/−) and heterozygous hof1 deletion confirms synthetic lethality. The depicted strain was generated by deleting hof1 in a homozygous hsp104 deletion. After sporulation, the strain was dissected onto GNA plates using a micromanipulator.The plates were incubated at 23°C for 3 days prior to being scanned. (B) Similarly, a homozygous spa2 (spa2Δ−/−) and heterozygous hof1 deletion was generated, dissected, and treated as in panel A. (C) Serial dilutions of overnight cultures of the indicated yeast strains were spotted onto yeast-peptone-dextrose (YPD) plates containing 5 μg/ml doxycycline (Dox). HOF1 was placed under the control of the tetracycline/doxycycline-repressible promoter (hof1reg) in hsp104Δ and spa2Δ backgrounds, leading to reduced levels of Hof1 in the presence of doxycycline. Plasmid-encoded Hof1 and Hsp104 were under the control of their authentic promoter. The plates were incubated at 25°C and 34°C for 48 h prior to being scanned. WT, wild type. (D) Yeast strains as described in the legend to panel C were diluted in YPD with the additional presence of 5 μg/ml doxycycline to allow logarithmic growth at 25°C for 16 h. Then, the cells were shifted to 34°C for an additional 3 h, subsequently fixed, and stained with rhodamine-labeled phalloidin to visualize the actin cytoskeleton. Representative cells are shown. Scale bar, 5 μm.
Hof1 localizes to the septin ring during cell division and interacts with formins. Furthermore, cells knocked out for hof1 exhibit cytokinesis defects and have multiple nuclei (17, 18). Given the demonstrated interaction between Hsp104 and Spa2, this finding implies that both proteins are parts of a cellular pathway working parallel to Hof1. We followed this hypothesis by testing whether spa2 is also synthetically lethal with hof1. Indeed, by dissecting a homozygous spa2 and a heterozygous hof1 knockout, we could show a synthetically lethal interaction between the two genes (Fig. 3B). To further analyze the genetic interaction between hsp104, spa2, and hof1, we generated a strain in which hof1 is expressed under the control of a doxycycline-repressible promoter (hof1reg) (5) in an hsp104Δ or spa2Δ background. In the absence of doxycycline, cells of these strains grew comparably to those of the wild type, except for the hof1 deletion, which is thermosensitive (Fig. 3C). At 25°C and in the presence of doxycycline, hofreg spa2Δ cells showed slight growth defects, whereas hof1reg hsp104Δ cells grew comparably to those of the wild type. The lack of a strict nongrowth phenotype at 25°C is likely a consequence of the fact that Hof1 is expressed at very low levels in yeast cells (∼195 molecules per cell) (10) while the tetO2 promoter leads to higher copy numbers (5), leading to only a partial reduction of Hof1 levels compared to those of the wild type in the presence of doxycycline. Indeed, using RT-PCR, we could observe only a partial reduction of mRNA in this strain (data not shown). Importantly, the addition of doxycycline inhibited growth in the case of hof1reg spa2Δ and hof1reg hsp104Δ mutant cells at 34°C. The finding that spa2Δ had a greater impact on survival than hsp104Δ is in line with the idea of Hsp104 being a chaperone for Spa2 that is required for proper folding of Spa2.
One explanation for the synthetic interaction between hsp104, spa2, and hof1 might be defects in the organization of the actin cytoskeleton. We tested this prediction by growing hof1reg hsp104Δ spa2Δ, hof1reg hsp104Δ, and hof1reg spa2Δ cells in the presence of doxycycline at 25°C for 16 h, followed by shifting them to 34°C and subsequent staining of the actin cytoskeleton with rhodamine-labeled phalloidin. We observed a severe defect in the rearrangement of the actin cytoskeleton upon downregulation of Hof1 in spa2Δ and hsp104Δ cells (Fig. 3D), indicating that the three proteins function in parallel pathways that help to organize the actin cytoskeleton during cell division.
Hsp104-induced polarity is responsible for asymmetric segregation of oxidatively damaged proteins.
What might be the physiological implication of the involvement of Hsp104 in rearranging the actin cytoskeleton? Recently, Aguilaniu and coworkers identified a phenomenon by which yeast cells are able to segregate oxidatively damaged proteins in an asymmetric manner, leaving the daughter cell largely free of damaged proteins while the mother cell retains them (3). This asymmetry in damage distribution between mother and daughter cells is likely to have important implications for cellular aging and rejuvenation of progeny (8, 12). Intriguingly, a knockout of hsp104 lost the ability to asymmetrically distribute damaged proteins (8). Since the actin cytoskeleton also plays a major role in this process, the authors proposed a model in which Hsp104 might bind damaged proteins and attach them to actin filaments. Our findings provide the alternative explanation that Hsp104 can modulate the actin cytoskeleton either directly or indirectly through its interactions with components of the actin/septin machinery (Fig. 4A). Actin polarity is thought to be crucial for the inheritance of various organelles (22) and may be important for the asymmetric distribution of damaged proteins, as well. We tested this hypothesis by investigating whether the asymmetric segregation of damaged proteins is lost in yeast cells lacking components that exhibit either physical (Spa2) or genetic (Hof1) interactions with Hsp104. The distribution of oxidatively damaged proteins was visualized by staining of carbonylated proteins in logarithmically grown cells. Intriguingly, spa2 and hof1 cells failed to segregate carbonylated proteins in an asymmetric manner, comparable to the situation in cells deleted of hsp104 (Fig. 4B and C). Together, these findings unravel a network that ensures a functional actin cytoskeleton, which is a prerequisite for the asymmetric distribution of damaged proteins between mother and daughter cells. We suggest that the previously described role of Hsp104 in this process might not be simple attachment of damaged proteins to the actin cable but its role in the reported network. The data also suggest that even slight perturbations of the actin cytoskeleton in spa2 and hsp104 cells are already sufficient to abrogate the specific retention of damaged proteins in mother cells. In agreement with such a suggested impact of Spa2 and Hsp104 on the organizing of the actin cytoskeleton, we observed defects for spa2 and hsp104 cells in the dynamics of the actin cytoskeleton during the formation of mating projections (reference 9 and data not shown). More drastic consequences of missing Spa2 or Hsp104 functions for the actin cytoskeleton are probably impeded by parallel operating pathways (Hof1) or compensatory cascades (activated MAP kinase pathway), but can be unraveled by the dominant-negative variant HAP/ClpP.
FIG. 4.
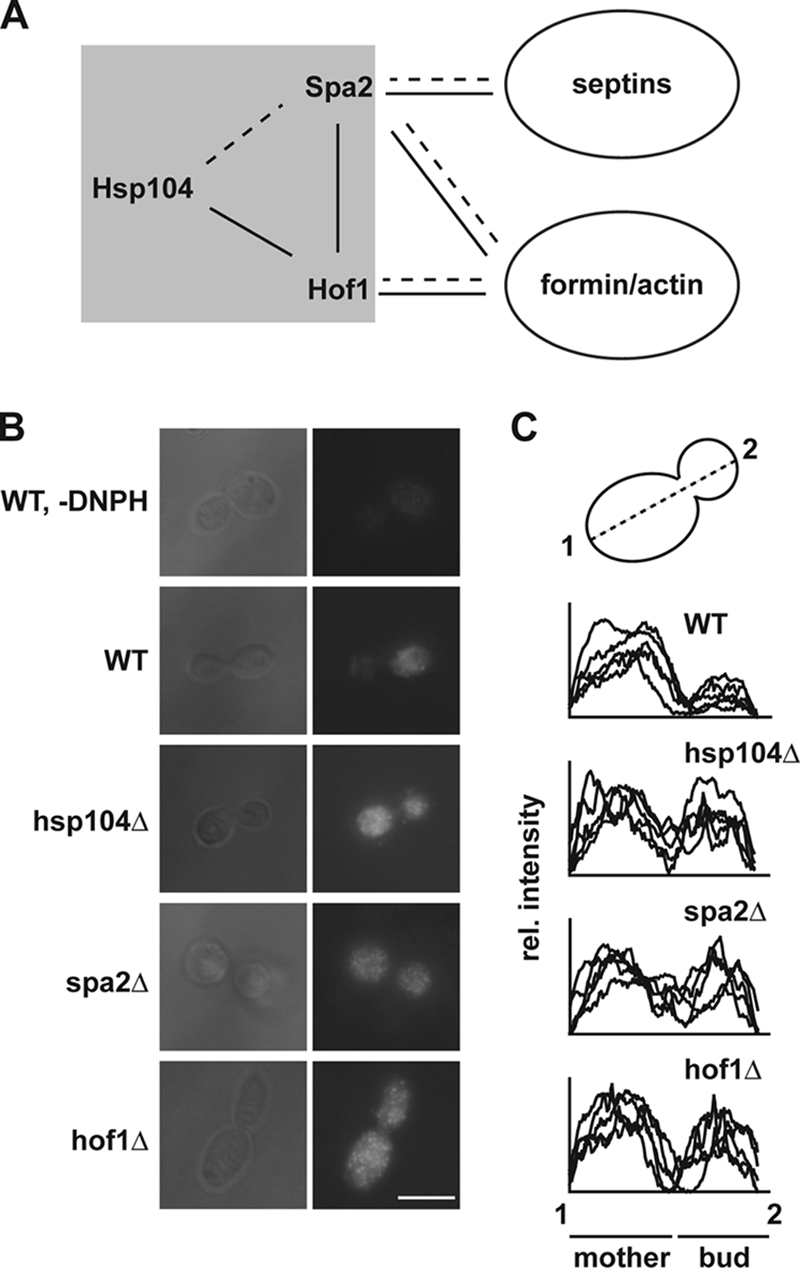
Asymmetric distribution of carbonylated proteins is dependent on polarity formation. (A) Genetic (solid lines) and physical (dashed lines) interactions reported in this study (shaded gray) and integration into the known interactome. (B) Carbonylated proteins were visualized by immunofluorescence following previously published protocols (4). Cells were grown to mid-logarithmic phase in yeast-peptone-dextrose prior to fixation. Carbonyl groups were derivatized using DNPH, which was subsequently detected by an antibody specific for the DNP moiety. A wild-type (WT) cell that was treated identically to the others but without DNPH is shown as a control. Representative cells are shown. Scale bar, 5 μm. (C) Quantification of five randomly picked cells of the fluorescence distribution through mother and daughter cells. The graph was produced using ImageJ. rel., relative.
Acknowledgments
We are grateful to S. Lindquist, B. Dobberstein, and E. Schiebel for strains and plasmids and to M. Snyder for Spa2 antibody. We thank Stefan Jentsch (Max Planck Institute of Biochemistry, Martinsried, Germany) for support and help with the synthetic-lethal screen.
M.S. is a Ph.D. student working with Stefan Jentsch. This work was supported by grants from the Fonds der Chemischen Industrie to B.B. and P.T. (Kekulé scholarship), a Heisenberg fellowship from the Deutsche Forschungsgemeinschaft to A.M., and the Network of Aging Research of the Land Baden-Württemberg.
Footnotes
Published ahead of print on 27 April 2009.
REFERENCES
- 1.Abelson, J. N., C. Guthrie, and G. R. Fink. 2003. Guide to yeast genetics and molecular biology. Academic Press Inc., New York, NY.
- 2.Adams, A. E., D. I. Johnson, R. M. Longnecker, B. F. Sloat, and J. R. Pringle. 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilaniu, H., L. Gustafsson, M. Rigoulet, and T. Nystrom. 2003. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science 2991751-1753. [DOI] [PubMed] [Google Scholar]
- 4.Arsene, F., T. Tomoyasu, and B. Bukau. 2000. The heat shock response of Escherichia coli. Int. J. Food Microbiol. 553-9. [DOI] [PubMed] [Google Scholar]
- 5.Belli, G., E. Gari, M. Aldea, and E. Herrero. 1998. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 141127-1138. [DOI] [PubMed] [Google Scholar]
- 6.Belli, G., E. Gari, L. Piedrafita, M. Aldea, and E. Herrero. 1998. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 26942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110119-122. [DOI] [PubMed] [Google Scholar]
- 8.Erjavec, N., L. Larsson, J. Grantham, and T. Nystrom. 2007. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 212410-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehrung, S., and M. Snyder. 1990. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J. Cell Biol. 1111451-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425737-741. [DOI] [PubMed] [Google Scholar]
- 11.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418387-391. [DOI] [PubMed] [Google Scholar]
- 12.Guarente, L., G. Ruvkun, and R. Amasino. 1998. Aging, life span, and senescence. Proc. Natl. Acad. Sci. USA 9511034-11036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 14.Hartwell, L. H. 1971. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69265-276. [DOI] [PubMed] [Google Scholar]
- 15.Janke, C., M. M. Magiera, N. Rathfelder, C. Taxis, S. Reber, H. Maekawa, A. Moreno-Borchart, G. Doenges, E. Schwob, E. Schiebel, and M. Knop. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21947-962. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, D. I., and J. R. Pringle. 1990. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol. 111143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamei, T., K. Tanaka, T. Hihara, M. Umikawa, H. Imamura, M. Kikyo, K. Ozaki, and Y. Takai. 1998. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J. Biol. Chem. 27328341-28345. [DOI] [PubMed] [Google Scholar]
- 18.Lippincott, J., and R. Li. 1998. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 1431947-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lum, R., M. Niggemann, and J. R. Glover. 2008. Peptide and protein binding in the axial channel of Hsp104. Insights into the mechanism of protein unfolding. J. Biol. Chem. 28330139-30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Martin, H., J. M. Rodriguez-Pachon, C. Ruiz, C. Nombela, and M. Molina. 2000. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2751511-1519. [DOI] [PubMed] [Google Scholar]
- 22.Moseley, J. B., and B. L. Goode. 2006. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 70605-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norden, C., D. Liakopoulos, and Y. Barral. 2004. Dissection of septin actin interactions using actin overexpression in Saccharomyces cerevisiae. Mol. Microbiol. 53469-483. [DOI] [PubMed] [Google Scholar]
- 24.Pratt, W. B. 1998. The hsp90-based chaperone system: involvement in signal transduction from a variety of hormone and growth factor receptors. Proc. Soc. Exp. Biol. Med. 217420-434. [DOI] [PubMed] [Google Scholar]
- 25.Pruyne, D., M. Evangelista, C. Yang, E. Bi, S. Zigmond, A. Bretscher, and C. Boone. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science 297612-615. [DOI] [PubMed] [Google Scholar]
- 26.Sagot, I., S. K. Klee, and D. Pellman. 2002. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 442-50. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez, Y., and S. L. Lindquist. 1990. HSP104 required for induced thermotolerance. Science 2481112-1115. [DOI] [PubMed] [Google Scholar]
- 28.Schaupp, A., M. Marcinowski, V. Grimminger, B. Bosl, and S. Walter. 2007. Processing of proteins by the molecular chaperone hsp104. J. Mol. Biol. 370674-686. [DOI] [PubMed] [Google Scholar]
- 29.Schlieker, C., J. Weibezahn, H. Patzelt, P. Tessarz, C. Strub, K. Zeth, A. Erbse, J. Schneider-Mergener, J. W. Chin, P. G. Schultz, B. Bukau, and A. Mogk. 2004. Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 11607-615. [DOI] [PubMed] [Google Scholar]
- 30.Sheu, Y. J., B. Santos, N. Fortin, C. Costigan, and M. Snyder. 1998. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 184053-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shorter, J., and S. Lindquist. 2005. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6435-450. [DOI] [PubMed] [Google Scholar]
- 32.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tessarz, P., A. Mogk, and B. Bukau. 2008. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol. Microbiol. 6887-97. [DOI] [PubMed] [Google Scholar]
- 34.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 2942364-2368. [DOI] [PubMed] [Google Scholar]
- 35.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303808-813. [DOI] [PubMed] [Google Scholar]
- 36.van Drogen, F., and M. Peter. 2002. Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr. Biol. 121698-1703. [DOI] [PubMed] [Google Scholar]
- 37.Weibezahn, J., C. Schlieker, B. Bukau, and A. Mogk. 2003. Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 27832608-32617. [DOI] [PubMed] [Google Scholar]
- 38.Weibezahn, J., P. Tessarz, C. Schlieker, R. Zahn, Z. Maglica, S. Lee, H. Zentgraf, E. U. Weber-Ban, D. A. Dougan, F. T. Tsai, A. Mogk, and B. Bukau. 2004. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119653-665. [DOI] [PubMed] [Google Scholar]
- 39.Whitesell, L., and S. L. Lindquist. 2005. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 5761-772. [DOI] [PubMed] [Google Scholar]
- 40.Young, J. C., J. M. Barral, and F. U. Hartl. 2003. More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 28541-547. [DOI] [PubMed] [Google Scholar]