Abstract
Insulator elements play a role in gene regulation that is potentially linked to nuclear organization. Boundary element-associated factors (BEAFs) 32A and 32B associate with hundreds of sites on Drosophila polytene chromosomes. We hybridized DNA isolated by chromatin immunoprecipitation to genome tiling microarrays to construct a genome-wide map of BEAF binding locations. A distinct difference in the association of 32A and 32B with chromatin was noted. We identified 1,820 BEAF peaks and found that more than 85% were less than 300 bp from transcription start sites. Half are between head-to-head gene pairs. BEAF-associated genes are transcriptionally active as judged by the presence of RNA polymerase II, dimethylated histone H3 K4, and the alternative histone H3.3. Forty percent of these genes are also associated with the polymerase negative elongation factor NELF. Like NELF-associated genes, most BEAF-associated genes are highly expressed. Using quantitative reverse transcription-PCR, we found that the expression levels of most BEAF-associated genes decrease in embryos and cultured cells lacking BEAF. These results provide an unexpected link between BEAF and transcription, suggesting that BEAF plays a role in maintaining most associated promoter regions in an environment that facilitates high transcription levels.
Insulator elements participate in gene regulation by limiting potential interactions between promoters and regulatory elements. In transgene assays, an insulator can block enhancer-promoter interactions, but only when located between the enhancer and the promoter (21, 35). Similarly, insulators can block repression mediated by Polycomb group proteins (41). They can also protect bracketed transgenes from chromosomal position effects (36, 52). Because of these properties, insulators are thought to participate in genome organization and gene regulation by defining the boundaries of discrete regulatory domains (9, 38, 61, 63). The mode of action of insulators is unclear but might involve acting as promoter decoys (20) or the formation of chromatin loops through interactions between insulators and perhaps also other nuclear substructures that remain to be biochemically defined (7, 10, 17, 67).
The scs and scs′ elements from the 87A hsp70 heat shock locus were two of the first DNA sequences shown to have insulator activity (35, 36, 60). Two boundary element-associated factors (BEAFs), 32A and 32B, were identified based on their interaction with the scs′ insulator element (but not the scs element) (29, 69). 32A and 32B are derived from the same gene and differ only by about 80 amino acids located at their amino termini. These unique regions harbor different atypical C2H2 zinc finger DNA binding domains, termed BED fingers (3). BEAF binding sites are essential for scs′ insulator activity (16), as is functional BEAF protein (22, 53). BEAF immunolocalizes to hundreds of sites on polytene chromosomes (69). Several of these genomic binding sites have been shown to have insulator activity (16), indicating that BEAF-dependent insulators are common in Drosophila rather than being a unique property of scs′. While the mechanism by which BEAF functions is not known, evidence from polytene chromosome morphology and position effect variegation assays indicates that BEAF affects chromatin structure, dynamics, or both (22, 53). Consistent with the proposed role of insulators, results obtained with a dominant-negative form of BEAF in a screen based on eye development imply that BEAF plays an important role in gene regulation (54). Genetic interactions between BEAF and several transcription factors expressed in the anterior portion of Drosophila were uncovered, which was interpreted as indicating that proper gene regulation breaks down in the absence of BEAF function.
To gain insight into the role of BEAF in chromatin domain organization and gene regulation, we have constructed a genome-wide map of BEAF binding sites. DNA isolated by chromatin immunoprecipitation was hybridized to genome tiling microarrays. Differences in binding patterns indicate that 32B plays a dominant role over 32A in binding to chromosomes. Surprisingly, we find that more than 85% of the centers of BEAF peaks are located within 300 bp of annotated transcription start sites (TSSs). About half of the peaks are between head-to-head gene pairs. We present evidence that most BEAF-associated genes are transcriptionally active and highly expressed and that the transcription levels of most of these genes drop in the absence of BEAF. Our results link BEAF to transcription, suggesting that BEAF plays a role in maintaining most associated promoter regions in an environment permissive for transcription.
MATERIALS AND METHODS
Chromatin immunoprecipitation (ChIP) and microarray hybridization (ChIP-chip).
Cross-linked chromatin was prepared from 6- to 16-h-old y1 w67c23 embryos, and ChIPs were performed by standard methods (13, 28). Chromatin was sonicated into fragments with an average size of 500 bp, as determined by reversing the cross-links and running DNA on agarose gels. Samples of approximately 30 μg chromatin were used as ChIP input. ChIPs were conducted with three different affinity-purified rabbit polyclonal antibodies. One antibody recognized the unique amino terminus of 32A, one recognized the unique amino terminus of 32B, and the third recognized the portion of the protein common to both 32A and 32B (BEAF) (29). Independent chromatin preparations were used for the 32B ChIP and one BEAF ChIP, and a third chromatin preparation was used for both the 32A ChIP and the second BEAF ChIP. ChIP DNA was validated by quantitative reverse transcription PCR (q-RT-PCR; ABI PRISM 7000) with SYBR green and primers amplifying two sequences with BEAF binding sites (scs′ and BE76) and two sequences lacking BEAF binding sites (scs and the actin5C promoter) (28). The scs region has been reported to indirectly associate with BEAF at a low level (7). Enrichment was calculated by ChIP/input cycle threshold change ratios (see Fig. S1 in the supplemental material).
Prior to ChIP-chip analysis, ChIP samples and equivalent amounts of input genomic DNA were amplified with the GenomePlex complete whole-genome amplification kit (WGA2; Sigma). Amplified products were purified with a QIAquick PCR purification kit (Qiagen). q-RT-PCR confirmed that the amplification was not biased (see Fig. S1 in the supplemental material). The average sizes of the amplified DNA samples were 400 to 500 bp. The amplified DNA was sent to NimbleGen (Madison, WI), where it was labeled and hybridized to genome tiling microarrays with 50-bp oligonucleotides spaced every 100 bp. Three samples had the ChIP DNA labeled with Cy5 and the genomic DNA labeled with Cy3, while dye swapping was used for the second BEAF sample.
Peak identification and data analysis.
After hybridization, the ChIP-chip data were analyzed with NimbleScan and SignalMap software (NimbleGen) with the Drosophila Release 5.1 genome sequence. NimbleScan was used to define peaks, and SignalMap was used to visualize the hybridization and peak data with the annotated genome sequence. Peaks of ChIP-enriched sequences [log2(ChIP/genomic)] were identified when the false discovery rate (FDR) in a 500-bp window was below 5%. Peak height is the log2(ChIP/genomic) value of the fourth highest probe inside the peak. As discussed in Results, these peaks include possible indirect associations such as at scs. This led us to use the following more stringent peak height and FDR criteria for peak selection: peak height of ≥2.0 and FDR of <1.0 for the 32B ChIP and the first BEAF ChIP, peak height of ≥2.4 and FDR of <1.0 for the second BEAF ChIP (dye swapping), and peak height of ≥0.8 and FDR of <1.5 for the 32A ChIP. To be considered a BEAF binding region, peaks must occur at the same location in both BEAF samples and one or both of the 32A and 32B samples.
Electrophoretic mobility shift assays (EMSAs).
DNA fragments for EMSAs were PCR amplified from Drosophila genomic DNA and purified with a QIAquick PCR purification kit. The fragments were end labeled with [γ-32P]ATP by T4 polynucleotide kinase. End-labeled DNA was incubated with affinity-purified BEAF from embryonic nuclear extracts or bacterially expressed 32A or 32B protein and 1 μg dI-dC at room temperature for 10 min. The amount of protein used was enough to give roughly a 50% shift of the scs′ D fragment, which was included on all gels as a positive control. The scs′ D fragment contains a high-affinity binding site for Drosophila BEAF, 32A, and 32B (29). Protein was purified and gel electrophoresis was performed as previously described (69). Briefly, reaction mixtures were loaded onto 4% polyacrylamide gels and electrophoresis was done at room temperature in 0.25× Tris-borate-EDTA.
Comparison with published data.
For comparison with published data, genome coordinates were converted with the University of California Santa Cruz Genome Browser liftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver). Gene names were converted with the G:Profiler Gene ID Converter (http://biit.cs.ut.ee/gprofiler/gconvert.cgi). As necessary, Excel files were converted into a format compatible for use with the NimbleGen software.
q-RT-PCR.
For q-RT-PCR from embryos, total RNA was isolated from 4- to 8-h-old embryos with TRIzol reagent (Invitrogen). Embryos with the BEAFAB-KO null mutation (53) or wild type for BEAF were used. To eliminate maternal BEAF, null embryos were collected from an inter se cross of BEAFAB-KO flies. About 40% of the resulting embryos could hatch into larvae (53), and most embryos develop beyond the germ band retraction stage, which is beyond 8 h of development (S. Roy and C. M. Hart, unpublished data). Superscript III (Invitrogen) was used to synthesize cDNA of each of the experimental and reference genes, primed by gene-specific primers. The level of Trf mRNA, which encodes a general transcription factor, was used as an internal control for the RNA samples (26). There are no BEAF peaks near Trf, and our results indicate that lack of BEAF does not affect the level of Trf RNA. Three independent RNA preparations of each genotype were used. Triplicate q-RT-PCRs for each RNA preparation were done with an ABI PRISM 7000 machine and SYBR green.
The q-RT-PCR analysis of small interfering RNA (siRNA)-treated Drosophila Schneider SL2 cells was done as previously described (18). Briefly, 400 μl of 2 μM BEAF or control double-stranded RNA was added to 10 ml of exponentially growing cells. The BEAF double-stranded RNA was synthesized with full-length cDNAs of BEAF as templates. The sequence was checked for potential off-target effects by performing searches with dsCheck (http://dscheck.rnai.jp/). Treated cells were incubated for 2 h at 25°C, and then 20 ml Schneider's Drosophila medium (GIBCO) containing 10% fetal bovine serum (Sigma) was added. Total RNA was isolated after 5 days and used for q-RT-PCR. A BEAF mRNA knockdown of about 10-fold was detected. All of the primer sequences used for EMSA and q-RT-PCR are available upon request.
Microarray data accession number.
The microarray data are available at ArrayExpress (www.ebi.ac.uk/arrayexpress/) under accession number E-TABM-597.
RESULTS
Genome-wide high-resolution identification of BEAF binding regions.
BEAF binding sites were identified throughout the Drosophila genome by hybridization of ChIP DNA to genome tiling microarrays with 50-bp oligonucleotides spaced every 100 bp (ChIP-chip). We performed four ChIPs with three different affinity-purified antibodies and chromatin from embryos. One antibody was specific for 32A, one was specific for 32B, and the third recognized the portion of the protein present in both 32A and 32B (BEAF).
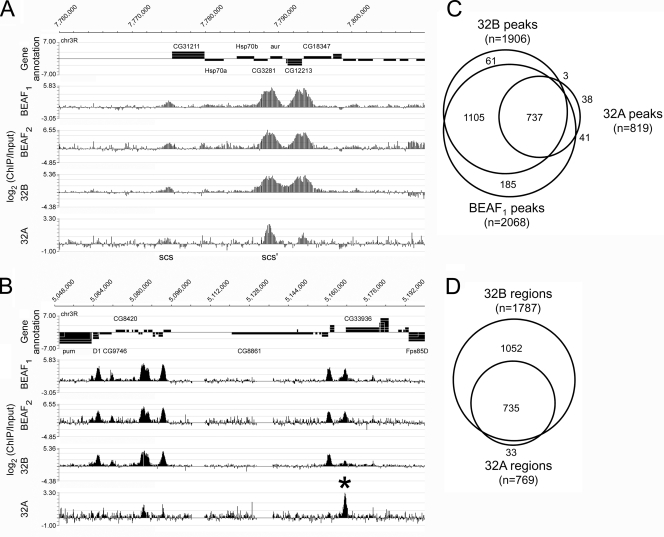
Inspection of the hybridization data for the 87A hsp70 region revealed a strong peak at scs′ and a smaller peak at scs in all four samples (Fig. 1A). With NimbleScan software to convert the hybridization data into peaks, the calculated FDR for scs was less than 5% for three of the samples. BEAF does not bind to scs in vitro. However, there is evidence that BEAF physically interacts with scs binding protein Zw5 and that this brings scs and scs′ into close proximity in vivo (7). To exclude peaks that might reflect such indirect, looping interactions between BEAF and other DNA binding proteins, we chose higher peak height cutoff values and lower FDR values than the scs peak data for each data set (see Materials and Methods for values). Depending on the data set, these criteria eliminated 30 to 50% of the peaks with an FDR of less than 5%.
FIG. 1.
ChIP-chip peaks are highly reproducible in the four samples and identify 1,820 BEAF binding regions. (A) Region of chromosome arm 3R including scs and scs′. The scs′ region (between head-to-head genes CG3281 and aur) is represented by a prominent peak in all four samples. The low but significant peak at scs (upstream of CG31211) presumably reflects an indirect interaction mediated by scs binding protein Zw5 (7). (B) A larger region from chromosome arm 3R showing several reproducible peaks, including one that has a major 32A peak but a weak 32B peak (asterisk, upstream of CG33936). In both panels A and B, select genes have been labeled for reference. (C) Representative Venn diagram showing the number of peaks, as defined in Materials and Methods, that overlap in the data for the 32A ChIP, the 32B ChIP, and the first BEAF ChIP. Peak start and end coordinates and peak height were determined with NimbleScan software, and these coordinates were used to identify overlapping peaks in the different data sets. See Table S1 in the supplemental material for overlap data for all four data sets. (D) Venn diagram showing the overlap in 32B peaks and 32A peaks, based on the definition that a peak is present in both BEAF ChIP-chip data sets in addition to isoform-specific data sets.
Comparison of the resulting set of peaks indicates that they largely coincide in the different ChIP samples (Fig. 1A to C; see Table S1 in the supplemental material). We define a BEAF binding region as a region that has a peak in both BEAF samples plus a peak in either or both of the 32A and 32B samples. By this definition, roughly 90% of the peaks in each data set correspond to a BEAF binding region. Less than 5% of the peaks are unique to one data set. This gives a minimum estimate of 1,820 BEAF binding regions in the Drosophila genome.
There are 1,052 32B-specific regions, 735 regions with both 32A and 32B peaks, and only 33 32A-specific regions (Fig. 1D). Figure 1B shows a rare region that includes a 32A-specific peak. The 32A peaks are generally lower than those in the other samples. Because more than 90% of the 32A peaks coincide with BEAF peaks, we believe the 32A data reflect a difference in the way 32A interacts with DNA compared to 32B, rather than low-quality data obtained with a poor antibody. It is likely that 32A is present at some regions we define as 32B specific but the 32A signals detected did not satisfy the peak selection criteria we used.
Validation of BEAF binding regions.
BEAF binding regions were validated in two ways, by PCR amplification of ChIP DNA and by EMSA. We designed 62 primer pairs for PCR amplification of 40 ChIP-chip peaks and 22 nonpeak regions. PCR was performed with genomic DNA and 32A, 32B, and BEAF ChIP DNA (Fig. 2A contains examples). Of the 40 peak region primers, 35 gave strong amplification with at least one source of ChIP DNA and another 4 gave weak amplification with at least two sources. Only one failed to amplify. In contrast, none of the 22 regions that did not correspond to peaks amplified. We conclude that the ChIP-chip data accurately represent sequences enriched by ChIP.
FIG. 2.
Validation of ChIP-chip results. (A) ChIPs were done with antibodies that recognize 32A or 32B, and PCRs were performed on ChIP DNA (Ch) and input genomic DNA (G). Primer set 2 amplifies a region that does not correspond to a peak; the other numbered primer sets correspond to peak regions. The regions amplified were upstream of or between the following genes: 1, hts and CalpA; 2, CG10862; 3, Fibp and Deaf1; 4, c(3)G and Acyp2; 5, janA and Sry-β; 6, CG11412; 7, RpS6 and bys; 8, Trc8. scs′ was included as a positive control; scs and act5C (the actin 5C promoter region) were included as negative controls. Similar results were obtained with ChIPs performed with an antibody that recognizes both forms of BEAF. (B) EMSA results with PCR-amplified sequences from the scs′ insulator, 32A plus 32B peaks (AB lanes), 32B-specific peaks (B lanes), 32A-specific peaks (A lanes), and the indicated protein. For details of the probes, see Table S2 in the supplemental material.
We next performed EMSAs on 61 peak regions and 4 nonpeak regions with 32A or 32B protein expressed in Escherichia coli or BEAF purified from embryonic nuclear extracts (Fig. 2B contains examples). As a positive control, we used the scs′ D fragment because it is bound by all three sources of protein with high affinity (29). Results are summarized in Table 1; for detailed information about each probe, see Table S2 in the supplemental material.
TABLE 1.
EMSA dataa
| Strong binding | Weak binding | n | 9-bp 32A | 32A model | 32B model | Mixed model | Does not fit model |
|---|---|---|---|---|---|---|---|
| 32A | 4 (1) | 3 (1) | 1 | 0 | 0 | 0 | |
| 32A | 4 | 2 | 1 | 0 | 0 | 1 | |
| 32A | NE | 3 | 1 | 2 | 0 | 0 | 0 |
| 32A, 32B, NE | 1 | 0 | 0 | 0 | 1 | 0 | |
| NE | 32A, 32B | 1 | 0 | 0 | 0 | 1 | 0 |
| 32A, 32B, NE | 3 | 0 | 0 | 1 | 0 | 2 | |
| 32B, NE | 10 | 0 | 0 | 9 | 0 | 1 | |
| 32B | NE | 4 | 0 | 0 | 2 | 0 | 2 |
| NE | 32B | 2 | 0 | 0 | 0 | 1 | 1 |
| 32B, NE | 10 | 0 | 0 | 2 | 3 | 5 | |
| NE | 5 | 0 | 0 | 2 | 3 | 0 | |
| No binding | 14 (3) | 0 | 1 (1) | 9 | 2 | 2 (2) | |
| Total | 61 (4) | 6 (1) | 5 (1) | 25 | 11 | 14 (2) |
9-bp 32A, one CGTGWCACG motif; 32A model, two or more CGTGA motifs, in either orientation, within 60 bp; 32B model, three or more CGATA motifs, in either orientation, within 100 bp; mixed model, at least two CGATA motifs and at least one CGTGA motif within 100 bp. Note that probes were counted in only one category of model even though they might fit more than one. Values in parentheses are for EMSA probes from nonpeak regions. NE, nuclear extract.
Confirming that BEAF binds to the peak regions we detected, 77% of the peak region probes showed some level of binding by at least one source of BEAF protein. However, these results were unexpectedly complex in terms of which protein sources bound and their variable binding affinities. Most probes were bound either by both 32B and nuclear extract BEAF or only by 32A. Only five were bound by all three sources of protein. In contrast, three of four probes from nonpeak regions were not bound by any of the BEAF sources. The fourth probe was tightly bound by 32A and was selected because it has a potential 9-bp 32A binding site (see below). As described in the next section, it is not clear why 23% of the probes were not bound and why the binding affinity was low for several others. Perhaps the binding affinity was below the level of detection of the assay. Under the conditions used, the scs′ D fragment gave strong shifts with all three protein sources, but the low-affinity site for 32B and nuclear extract BEAF present in the scs′ B fragment would have given weak shifts (69). Perhaps binding would have been detected if we had used different sequences from the peak regions. Another possibility is that BEAF binds better in vivo, perhaps due to phosphorylation (29, 48) or interactions with other, unknown, proteins.
Identification of potential BEAF binding sites in BEAF binding regions.
We inspected peak sequences for potential 32B binding sites based on the model in which 32B binds three copies of the motif CGATA clustered in 100 bp, with variable spacing and relative orientations between motifs (16). For 32A, we used the model in which it binds two copies of the motif CGTGA in a 60-bp region (C.M.H., unpublished data; 29) or a single copy of the 9-bp motif CGTGWCACG (see below). We counted a site as mixed if it had two 32B motifs and one 5-bp 32A motif in 100 bp. Each peak region was counted only once, even if it had motifs that fit more than one model. In addition, we counted the peak regions that had at least one 8-bp DNA replication-related element (DRE) motif (TATCGATA) that is recognized by the transcription factor DREF (30). DREF is a subunit of the TRF2 complex, a core-promoter recognition complex for a large number of TATA-less promoters (31, 33). The DRE motif contains a 32B motif (CGATA), and it has been shown that BEAF and DREF can compete for binding in vitro if a cluster of 32B motifs includes a DRE (28). As an indicator of how frequently this competition might occur, it was of interest to determine how many BEAF binding regions contain DREs.
EMSA probe sequences were inspected for potential binding sites based on the above binding site models (Table 1). Under the EMSA conditions we used, 10 of 11 peak region probes with potential 32A sites and 9 of 11 probes with potential mixed sites showed some level of binding by at least one protein source. In addition, the nonpeak probe with a 9-bp 32A motif was bound by 32A protein. However, 9 of 25 probes with potential 32B sites were not shifted and 12 of 14 probes that did not conform to our models of BEAF binding sites were shifted. There was no correlation between binding and the presence of a DRE. Five of the 13 sequences with a DRE gave weak or no shifts. While inspecting the probe sequences with potential 32B binding sites, we noted that 32B bound to most probes if they had what we term a “+ −” inverted repeat (CGATA-Nx-TATCG, where x ranged from 1 to 31 bp in the sequences we used) but not if they had a “− +” inverted repeat or only direct repeats. There was no obvious correlation between binding and integral numbers of helical turns between CGATA motifs.
We expanded this analysis to additional BEAF binding regions (Table 2). Potential 32A binding sites were present in 75% of the 33 32A-specific peaks. However, 57% of the 32B peaks on chromosome arm 2L (with and without colocalizing 32A peaks) were found to lack sites that fit our models. More peak regions had a DRE (39%) than a potential 32B binding site (33%). To pursue this analysis further, we inspected more than 100 of the highest 32B-specific and 32A plus 32B peaks. Compared to 2L, these regions had higher percentages of potential binding sites. The increase was particularly notable for potential 32B sites. This suggests that BEAF is more likely to bind tightly when these motifs are present.
TABLE 2.
Potential binding sites in peak regionsa
| Site type | All 32A peaks | % | 2L 32B & A+B | % | Top 32B peaks | % | Top A+B peaks | % |
|---|---|---|---|---|---|---|---|---|
| 32B site | 1 | 3 | 91 | 33 | 54 | 47 | 62 | 51 |
| 9-bp 32A | 14 | 42 | 3 | 1 | 0 | 0 | 1 | 1 |
| 32A site | 11 | 33 | 14 | 5 | 4 | 3 | 7 | 6 |
| Mixed | 1 | 3 | 12 | 4 | 8 | 7 | 13 | 11 |
| None | 6 | 18 | 157 | 57 | 49 | 43 | 39 | 32 |
| DRE | 4 | 12 | 108 | 39 | 34 | 30 | 56 | 46 |
| Total | 33 | 277 | 115 | 122 |
See the footnote to Table 1 for site type definitions. All 32A peaks, all 32A-specifc BEAF binding regions; 2L 32B and A+B, all BEAF binding regions on chromosome arm 2L with 32B peaks, with or without a 32A peak; top 32B peaks, 32B-specific regions selected based on having the highest peaks; top A+B peaks, binding regions with both 32A and 32B peaks selected based on having the highest peaks.
The ChIP-chip data were also compared to BEAF “dual-core” predictions (Table 3). By a bioinformatic approach, it was recently reported that predicted BEAF binding elements often occur in pairs termed dual cores (18). In this model, a BEAF binding element is defined as a cluster of three CGATA motifs within 200 bp. A dual core has two such binding elements separated by less than 800 bp of generally AT-rich DNA. A dual-core-like element has one BEAF element paired with a cluster of two CGATA motifs within 100 bp. Referring to these collectively as dual cores, 1,720 were identified in the genome. Of these, about 25% correspond to ChIP-chip peaks. If consideration is limited to dual cores located within 600 bp of a TSS, then nearly 70% correspond to ChIP-chip peaks. This suggests that there are many false positives in the dual-core predictions; the model is too simplistic. Also, BEAF binds to many regions lacking a dual core. Many BEAF binding sites are not predicted by the model. However, the dual-core model has merit since BEAF binds to the majority of the predicted dual-core regions close to TSSs.
TABLE 3.
Comparison of ChIP-chip data and dual-core predictionsa
| Location | No. of BEAF binding regions
|
Overlap | |
|---|---|---|---|
| ChIP-chip | Dual core | ||
| Genome | 1,820 | 1,720 | 434 |
| Within 600 bp of a TSS | 1,703 | 581 | 403 |
The number of BEAF binding regions based on ChIP-chip data or dual-core predictions is given for the genome or for regions centered within 600 bp of a TSS. Overlap is the number of regions in common to both data sets.
Combined with previous evidence that 32B and BEAF from nuclear extracts bind to CGATA motifs (16, 29, 69), we draw the following three conclusions. First, clusters of CGATA and CGTGA motifs can play a role in binding by the BEAF proteins, although they are neither necessary nor sufficient. However, binding of the BEAF proteins to sequences lacking clustered motifs is usually weak in vitro. Second, although 32B appears to prefer + − inverted repeats of CGATA, there is no consensus for spacing or relative orientations between motifs in binding sites. Third, because of this lack of consensus, binding affinities of the BEAF proteins cannot be accurately predicted by inspection of DNA sequences for clusters of CGATA and CGTGA motifs.
To identify additional features that might be present in BEAF binding regions, different groupings of sequences were analyzed with the Multiple Em for Motif Elicitation and Discriminating Motif Enumerator motif discovery programs (4, 59). We found two motifs. Using the 32A-specific peak sequences, we identified the 9-bp motif CGTGWCACG, which is related to the 5-bp 32A motif (CGTGA). 32A bound to all of the probes with this motif, even a sequence from a nonpeak region. However, there are 346 occurrences of this motif in the Drosophila genome and only a few are in BEAF binding regions. It is not clear why 32A apparently can bind to this motif yet does not do so at most sites in vivo. Using 32B-specific and 32A plus 32B regions, we found the 8-bp DRE motif. However, not all of the probes tested with a DRE were bound by 32B or nuclear extract BEAF, and other probes were only weakly shifted. Also, there are more than 3,000 DREs in the Drosophila genome and most are not in BEAF binding regions. As previously reported (28), it does not appear that BEAF directly recognizes the DRE but can use it as part of a binding site if other sequence requirements are met. These results reinforce the concept that there is great plasticity in the ability of BEAF complexes to bind various arrangements of short recognition motifs. They also suggest that there are recognition motifs yet to be identified, perhaps in combination with protein partners yet to be identified.
BEAF binding regions are located near TSSs of active genes.
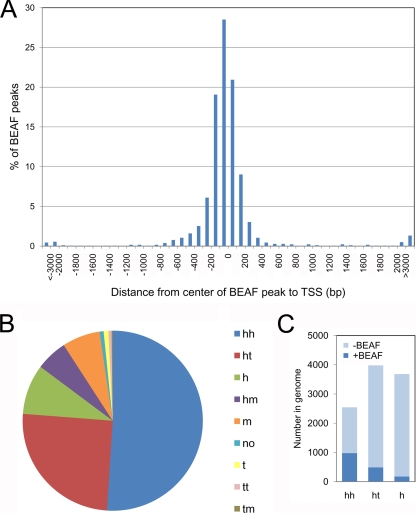
Because BEAF-dependent insulators are thought to play a role in gene regulation, it was of interest to determine where BEAF binding regions are localized with respect to genes. In addition, the comparison with dual cores indicated that most BEAF binding regions are near TSSs. We found a striking clustering, with the centers of more than 85% of the BEAF peaks being found within 300 bp of annotated TSSs (Fig. 3A). Less than 2% were found further than 3 kb from a TSS. About 50% of the BEAF binding regions were between head-to-head gene pairs separated by less than 3 kb, and another 25% were between head-to-tail gene pairs separated by less than 3 kb (Fig. 3B). We analyzed the Drosophila Release 5 annotations for gene pairs separated by less than 3 kb and found that more than one-third of the head-to-head gene pairs were associated with a BEAF peak (Fig. 3C). While this implies that there is one BEAF binding site for every five to six promoters organized this way, this is a minimum estimate because some regions could have two BEAF binding sites but one peak. For instance, scs′ has BEAF binding sites by both of its TSSs but has a single ChIP-chip peak. Also, more than half of the dual cores that correspond to BEAF peaks are between head-to-head gene pairs. Smaller percentages of promoters in head-to-tail gene pairs and promoters located more than 3 kb from a neighboring gene were associated with a BEAF binding region.
FIG. 3.
BEAF peaks are near TSSs, about half of which are organized in head-to-head gene pairs. (A) Positions of the centers of 1,820 BEAF peaks relative to the nearest annotated TSS. (B) Distribution of BEAF peaks relative to gene organization. With a 3-kb window, the two genes closest to the centers of the BEAF peaks were identified. It was then determined if a TSS (h, for head) or polyadenylation site (t, for tail) was nearest to the peak. If the center of the peak was within transcribed sequences more than 200 bp from either end, it was considered in the middle of the gene (m). These definitions also applied to overlapping or nested genes. This resulted in the following categories: hh, head-to-head gene pair with TSSs within 3 kb of each other; ht, head-to-tail gene pair with the TSS and polyadenylation site within 3 kb of each other; h, head, no adjacent gene within 3 kb; hm, head of one gene and middle of another (for instance, overlapping genes or alternative promoters); m, middle of gene, greater than 200 bp downstream of TSS, with no other gene within 3 kb; no, nearest gene at least 3 kb away; t, tail, no adjacent gene within 3 kb; tt, tail-to-tail pair of genes within 3 kb of each other; tm, tail of one gene and middle of another. (C) Number of genes within 3 kb of each other organized as head-to-head (hh) or head-to-tail (ht) gene pairs and genes whose TSS is more than 3 kb from the nearest neighboring gene (h). Light blue, number in genome lacking a BEAF peak; dark blue, number in genome with a BEAF peak.
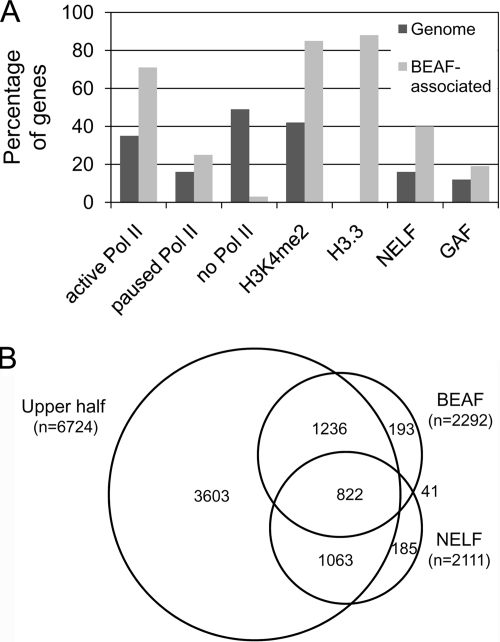
A comparison of our data with data from other studies indicates that most of the genes associated with BEAF are transcriptionally active. It has been reported that RNA polymerase II (Pol II) is associated with the bodies of 3,633 genes in 2- to 4-h-old Toll10b mutant embryos (“active genes”), with another 1,616 genes having a presumably paused Pol II at their 5′ ends. Another 4,997 “inactive genes” lack Pol II (68). Because many BEAF peaks are between head-to-head gene pairs, the centers of the 1,820 BEAF peaks are within −500 bp to +200 bp of the TSSs of 2,305 genes. We matched 1,343 of these genes to the Pol II data and found that 71% are active genes, 26% are paused, and 3% are inactive (Fig. 4A). An additional 861 genes matched 3,202 excluded Pol II-associated genes that belong in the active or paused category but could not unambiguously be assigned to either category. Similar Pol II data are available for S2 cells (47). In this case, 4,389 genes were classified as active, 1,014 genes had Pol II enriched at their promoters, and 7,700 genes were inactive. We matched 2,179 of our genes to these data and found that 73% are active, 15% have a paused Pol II, and 12% are inactive (see Table S3 in the supplemental material). Thus, for both data sets, BEAF is preferentially found at active and, to a lesser extent, paused genes. Two features associated with active genes are methylation of histone H3 on lysine 4, particularly at the 5′ end (55), and replacement of H3 with the alternative histone H3.3 in nucleosomes in transcribed sequences (2). Comparing our data to those for H3K4me2 on chromosome arm 2L (56) and H3.3 on chromosome arm 3R (44), we found that 85% of the genes associated with BEAF have H3K4me2 in their promoter regions and 88% have H3.3 in their promoter and/or transcribed regions (Fig. 4A). This is consistent with the correlation between BEAF-associated genes and the Pol II data.
FIG. 4.
BEAF is associated with active genes. (A) Percentage of genes in the genome (dark gray) and associated with BEAF (light gray) that are associated with active Pol II, paused Pol II, or no Pol II in Toll10b embryos (68); associated with the active chromatin marks H3K4me2 in Kc cells (56) and H3.3 in S2 cells (44); and associated with NELF or GAF in S2 cells (40). The data for H3K4me2 are only for chromosome arm 2L. The data for H3.3 are only for chromosome arm 3R, and the total number of genes associated with this alternative histone was not calculated. (B) Venn diagram showing the relationships among BEAF-associated genes, NELF-associated genes, and the upper half of the genes ranked by expression levels in Toll10b embryos.
It was recently reported that NELF and GAGA factor (GAF) are linked to many genes with a paused Pol II and that genes associated with NELF tend to be highly expressed (40). NELF represses transcription elongation (65) and has been implicated in promoter-proximal pausing by Pol II (64). GAF has been shown to play a role in pausing of Pol II on Drosophila hsp70 genes (58). There is a large overlap in the sets of genes associated with NELF and GAF (40), most of which are associated with active or paused Pol II. Because of the link with genes associated with Pol II, we compared these data to our data and the Pol II data for both Toll10b embryos and S2 cells (see Table S3 in the supplemental material). Considerable overlap of BEAF with NELF (39%) and GAF (18%) was found (Fig. 4A). There is a larger overlap with NELF, and nearly 65% of the genes associated with both BEAF and GAF are also associated with NELF, so we focused our attention on NELF. To examine relationships with gene expression, all genes were ranked by expression levels in Toll10b embryos. Consistent with reports that NELF-associated genes are highly expressed (23, 40), around 90% of the NELF-associated genes matched those in the upper half of these ranked genes. Around 90% of the BEAF-associated genes also matched the upper half of the ranked genes, as did 95% of the genes associated with both BEAF and NELF (Fig. 4B). Similar results were obtained when the comparison was done by using gene expression in S2 cells (see below).
BEAF does not colocalize with the insulator proteins Su(Hw) and CTCF.
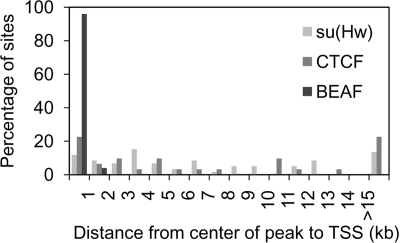
Two other insulator proteins in Drosophila that have been well studied are Suppressor of Hairy-wing [Su(Hw)] and CCCTC Binding Factor (CTCF) (21, 45). Su(Hw) has mainly been studied in the context of a 340-bp insulator sequence present in the gypsy retrotransposon (51), although non-gypsy binding sites have been shown to have insulator activity (39). Before the Drosophila homolog was discovered, CTCF was originally studied in vertebrates (37). Almost all of the characterized vertebrate insulators are associated with CTCF (6, 46), which has also been studied both as a transcriptional activator and repressor (8, 62) and as a participant in genomic imprinting (5, 27). Both Su(Hw) and CTCF have been localized by ChIP-chip to a 3-Mb region around the Adh gene, a 130-kb region around the achaete-scute complex, and the Bithorax complex (BX-C) (1, 32). We compared our data for BEAF to these data. Of 23 BEAF peaks, 18 CTCF peaks, and 60 Su(Hw) peaks in the Adh region, only 2 BEAF peaks localize within 1 kb of CTCF peaks. There are no BEAF peaks in the BX-C or achaete-scute region. We next compared the locations of Su(Hw), CTCF, and BEAF sites to TSSs in the Adh region and, in the case of CTCF, also in the BX-C region. As found for the whole genome, the centers of 96% of the 23 BEAF peaks were within 1 kb of a start site. In stark contrast, only 7 of 60 Su(Hw) binding sites and 7 of 31 CTCF binding sites were this close (Fig. 5). Thus, only BEAF is highly enriched near promoter regions.
FIG. 5.
BEAF does not colocalize with CTCF or Su(Hw). While the centers of most BEAF peaks (dark gray) are within 1 kb of the nearest TSS, most peaks for CTCF (medium gray) (32) and Su(Hw) (light gray) (1) are centered well over 1 kb from the nearest TSS.
The Su(Hw) protein has been reported to form “insulator bodies” that localize to the nuclear periphery (19) through interactions with other proteins that link it to the Drosophila B-type lamin (11). Although the functional significance of these aggregates is unclear (25), it has been proposed that insulator bodies play a role in insulator function by organizing chromatin into loop domains (10). Nearly 500 genes were mapped by DamID as being associated with lamin, and these genes were found to be transcriptionally silent and to replicate late in S phase (50). We found BEAF peaks near only 15 of the lamin-associated genes, which is consistent with BEAF being associated with active promoters and the nuclear lamina being associated with inactive genes. This also provides further evidence for the distinct localization of BEAF and Su(Hw).
BEAF binds upstream of highly expressed genes.
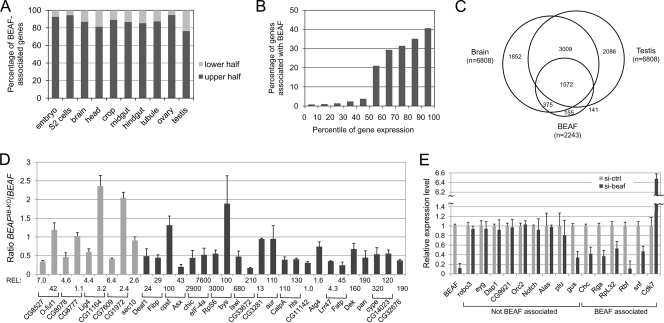
To pursue the link between BEAF and transcription further, we determined the relative transcription levels of BEAF-associated genes by comparing our data to published gene expression data for Toll10b embryos (23), S2 cells (47), and various adult tissues (14). We found that 75 to 95% of the BEAF-associated genes were among the upper half of the genes ranked by expression level (Fig. 6A). It is interesting that the lowest value, 76%, came from testes while the highest value, 95%, came from ovaries. Although flies homozygous for the BEAFAB-KO null mutation are difficult to keep alive, males are fertile while females are nearly sterile (53). To refine this analysis, the data from S2 cells were divided into bins of 10 percentile points based on gene expression levels. We then tabulated the percentage of genes in each bin that were associated with BEAF (Fig. 6B). About 40% of the genes above the 90th percentile in expression were associated with BEAF, which accounted for 24% of the genes associated with BEAF.
FIG. 6.
Most BEAF-associated genes are highly expressed, and lack of BEAF leads to reduced expression levels of many BEAF-associated genes. (A) More than 13,000 genes were ranked by expression levels for Toll10b embryos (68), S2 cells (47), and various adult tissues (14). We matched 2,204 genes with a BEAF peak centered within −500 bp to +200 bp with the Toll10b embryo data, 2,191 genes to the S2 cell data, and 2,243 genes to the adult tissue data. The percentages of these BEAF-associated genes in the upper half of the genes by expression level (dark gray) and the lower half by expression level (light gray) are shown. (B) The data for S2 cells were divided into 10 percentile bins based on expression levels, and the percentage of genes in each bin that is associated with BEAF was plotted. (C) Venn diagram showing the overlap between the upper half of the genes by expression level for brain and testis, together with BEAF-associated genes. (D) q-RT-PCR results for RNA isolated from 4- to 8-h-old embryos with wild-type BEAF or the BEAFAB-KO knockout allele. Ten genes not associated with BEAF (light gray bars plus Trf) and 23 BEAF-associated genes (dark gray bars) were assayed. Head-to-head gene pairs are indicated by brackets. Results with standard deviations are shown as the ratio of BEAFAB-KO samples to BEAF samples, with Trf used to normalize mRNA levels. There are no BEAF peaks near Trf, and our results indicate that lack of BEAF does not affect the level of Trf mRNA. DRE elements were located between the Deaf1/Fibp, chic/eIF-4a, RpS6/bys, Iswi/CG33672, CalpA/hts, and CG14023/CG32676 gene pairs. REL, relative expression levels of the genes in BEAF embryos, normalized to the lowest expression level. (E) q-RT-PCR results for RNA isolated from SL2 cells treated with control (light gray bars) or BEAF (dark gray bars) siRNA. In addition to BEAF, nine genes not associated with BEAF and six BEAF-associated genes were assayed, as indicated. Dsp1 and CG9921 form a head-to-head gene pair. Cdk7 and snf form a head-to-head gene pair with a DRE element in between. Results with standard deviations are shown, normalized to the control (ctrl) siRNA level for each gene.
It was of interest to determine if the same BEAF-associated genes are highly expressed in different tissues. As two very different tissues, we chose brain and testis for this comparison. When genes were ranked by expression level, we found that 67% of the genes in the upper half were the same in both brain and testis. About 70% of the genes we found to be BEAF associated were present in this set of genes (Fig. 6C). This indicates that many genes are expressed at high levels in diverse tissues, including the majority of BEAF-associated genes.
As another test of the relationship between BEAF and transcription, we performed q-RT-PCR on divergent gene pairs separated by BEAF peaks (Fig. 6D). We hypothesized that the transcription levels of two genes in a head-to-head pair should converge in the absence of BEAF if BEAF separates these genes into independent functional domains. Nine head-to-head gene pairs separated by BEAF peaks were tested, along with two single genes from two additional head-to-head gene pairs and two genes not organized in this fashion. As controls, four head-to-head gene pairs that lacked BEAF peaks were tested, along with one single gene (Trf) from an additional head-to-head gene pair and one gene lacking this organization. The levels of Trf mRNA were used to normalize the mRNA levels of the other genes because we found that Trf mRNA levels were not affected by a lack of BEAF. Genes to be tested were selected randomly, with the following two caveats. The CG3281 and aur genes were included because they are separated by scs′, and the selected genes lacking associated BEAF peaks were among the upper half of the genes ranked by expression level in S2 cells in order to roughly match expression levels with the BEAF-associated genes.
RNA was isolated from 4- to 8-h-old embryos that were homozygous for either wild-type BEAF or the BEAFAB-KO null mutation. The BEAFAB-KO embryos also lacked maternal BEAF. For six BEAF-associated gene pairs, the expression levels of both genes dropped by a factor of two to four in the absence of BEAF. Two other gene pairs showed a reduction in the expression of one gene, while the other gene showed no significant change or as much as a twofold increase. Only the gene pair bracketing scs′ showed no change in the expression level of either gene. All four of the single genes tested showed a two- to fourfold drop in expression level in the absence of BEAF. Overall, 19 of 23 BEAF-associated genes showed a decreased level of expression in the absence of BEAF, while 3 showed no change and only 1 showed an increase. There was a DRE between six gene pairs in this analysis, and the gene that showed an increase in expression was a member of one of these pairs. However, both genes showed decreased expression in the other five pairs. One might expect the opposite, that lack of BEAF would allow DREF to activate at least one gene in each of the six pairs. This effect was reported for siRNA-treated cultured cells and embryos producing a dominant-negative form of BEAF (18). But after longer siRNA treatments, these same genes showed decreased expression (O.C., unpublished data). Therefore, the prolonged growth that the BEAFAB-KO embryos underwent in the absence of BEAF could explain why expression levels dropped even for genes with a DRE. Repressive effects might dominate over time, perhaps via accumulation of high levels of H3 lysine 9 methylation (18).
Results for genes not associated with BEAF were more variable, as opposed to conforming to the prediction that their expression levels would not be affected by a lack of BEAF. For all four gene pairs, the expression of one gene dropped by a factor of two. For two gene pairs, expression of the other gene was unchanged, while expression increased twofold for the other two. The expression levels of Trf and the single gene were unchanged. Ignoring gene pairs, 4 of 10 genes showed decreased expression, 4 showed no change (including Trf), and 2 showed increased expression. This suggests that the absence of BEAF leads to a breakdown of gene regulation, causing indirect effects on gene expression levels for many genes that are not associated with BEAF.
Although the potential for indirect effects on gene expression levels in the absence of BEAF is a concern, the fairly consistent results obtained with BEAF-associated genes, as opposed to the variable results for genes not associated with BEAF, suggest that direct effects predominate at BEAF-associated genes. Based on this, our RT-PCR data do not support the hypothesis that BEAF separates adjacent genes into functionally independent domains. Rather, they suggest that BEAF helps maintain the expression levels of most of the genes it is associated with.
As an alternative method for examining the relationship between BEAF and gene expression, we used siRNA with SL2 cells (Fig. 6E). One reason for taking this approach was to test whether the variable findings on the expression levels of genes not associated with BEAF might be due to indirect effects in embryos. We reasoned that there would be less time for indirect effects to manifest themselves if gene expression were examined after a few cell generations rather than after prolonged growth and development without BEAF. Genes to be tested were randomly selected. The BEAF siRNA knocked down the level of BEAF mRNA about 10-fold. Of nine genes tested that are not associated with BEAF, eight showed no change in their expression levels. This includes both members of a head-to-head gene pair (Dsp1 and CG9921). One gene showed a threefold decrease. In contrast, five of six BEAF-associated genes showed a decrease similar to that observed in embryos. The sixth gene is part of a gene pair (Cdk7 and snf) that has a DRE and showed an increase in its expression level, as previously reported (18). These results are consistent with prior results (18) and support the view that the observed variable effects on the expression levels of genes not associated with BEAF could be due to indirect effects in BEAFAB-KO embryos. They also provide further support for the idea that BEAF helps maintain the expression levels of most of the genes it is associated with. An exception could be BEAF-associated genes regulated by DREF, where BEAF might antagonize activation by DREF. Even in this case, BEAF might help maintain the promoter region in an environment conducive to binding by DREF. Presumably, competition between BEAF and DREF would be regulated, perhaps in a cell cycle-related manner, considering the proposed role of DREF in the cell cycle and cell proliferation (43).
DISCUSSION
BEAF binds to more than 1,800 regions in the Drosophila genome, and 32B is dominant over 32A.
Using three different antibodies to perform ChIP-chip, we have localized BEAF to 1,820 regions in the Drosophila genome. This is in agreement with the broad distribution seen by immunostaining polytene chromosomes (29, 69). There was a clear difference between the association of 32A and 32B with chromosomes. 32B gave robust peaks, while 32A gave smaller peaks. By our peak selection criteria, only about 40% of the regions with 32B peaks also have 32A peaks. In contrast, more than 95% of the regions with 32A peaks also have 32B peaks. The dominant role of 32B in binding to chromosomes is consistent with results showing that flies producing only the 32B protein are viable but flies lacking both forms of BEAF are not (53). Also, we can rescue the BEAFAB-KO null mutation with a 32B transgene but not with a 32A transgene (C.M.H., unpublished), yet 32A is presumably performing an important function. Both 32A and 32B are highly conserved in all 12 sequenced Drosophila species, representing more than 40 million years of evolution (15).
In addition to the peaks we count as genuine BEAF binding regions, there are other, lower, peaks with an FDR of less than 5% that are present in three or four of our data sets. An example of this is found at scs. BEAF has been reported to indirectly associate with scs by interactions with Zw5, which directly binds scs (7). An intriguing possibility for future investigation is that these peaks represent interactions between BEAF and other chromatin-associated proteins such as Zw5. Although we did not tabulate the number of these peaks, there are certainly hundreds of them in our data. If they indeed represent the formation of chromatin loops by heterologous interactions, then investigating this phenomenon will provide valuable insight into nuclear organization.
We confirmed that BEAF binds to the identified regions by a combination of PCR and EMSA experiments. In agreement with previous footprinting, CGATA mutagenesis, and dual-core results (16, 18, 29, 69; C.M.H., unpublished), our results support the view that clusters of CGATA motifs (for 32B) and CGTGA motifs (for 32A) play a role in binding at some sites. However, they also indicate that this view is too simplistic. While 32B appears to have a preference for two of three CGATA motifs being arranged as + − inverted repeats, binding sites rarely look like the two in scs′ and no rules relating spacing and orientations of motifs to binding affinity in single sites emerged. Only about 25% of 1,720 dual cores correspond to the BEAF peaks described here. We used the FlyEnhancer program (42) with the more stringent definition for a single potential 32B binding element of three CGATA elements in a 100-bp window, with a DRE counted as a single CGATA, and found more than 2,800 clusters in the Drosophila genome. Most of these sites are not included in the set of BEAF peaks that met our selection criteria. The lack of strong binding of BEAF to these regions suggests that these motif clusters are not organized properly or are, for some reason, inaccessible to BEAF. In addition, examination of peak sequences indicates that these motifs are not necessary for binding by BEAF. Other, unknown, sequence features must play a role at many sites. Using the Multiple Em for Motif Elicitation and Discriminating Motif Enumerator motif discovery programs (4, 59) did not help identify consensus sequences. Presumably, this is because BEAF binds to short motifs with variable spacing and orientations between motifs, rather than a long, contiguous sequence. Refining models of BEAF binding sites so that they can be identified by inspection of DNA sequences will require performing additional experiments such as footprinting assays and perhaps identification of partner proteins.
BEAF is associated with promoter regions of active genes and helps maintain the expression levels of most of these genes.
The centers of BEAF peaks show a striking clustering near annotated TSSs. In addition, about half of the peaks are between head-to-head gene pairs so that the 1,820 peak centers are located within −500 bp to +200 bp of the TSSs of 2,305 genes. The scs′ insulator is one example of a head-to-head gene pair associated with a BEAF peak. It has two BEAF binding sites, one near each TSS. One is a high-affinity binding site (KD, ∼25 pM) that gives a prominent shift, and the other is a low-affinity binding site (KD, ∼600 pM) that gives a weak shift under the EMSA conditions we used (29). This could be a common theme. More than half of the 434 dual-core regions that correspond to BEAF peaks are between head-to-head gene pairs, suggesting that there are BEAF binding sites by both TSSs of these gene pairs. We cannot evaluate this possibility outside of the context of dual cores at this time because we cannot unambiguously identify BEAF binding sites based on DNA sequence.
A comparison with published data indicates that the majority of BEAF-associated genes are transcriptionally active or poised for activation and are highly expressed. One concern is that the Pol II, histone, and gene expression data came from a variety of tissues and cultured cells. However, the results consistently indicate that BEAF-associated genes are active. In fact, 70% of the BEAF-associated genes we identified are in the upper half of the genes ranked by expression levels in both brain and testis. This suggests that most BEAF-associated genes are expressed at high levels in a wide range of tissues, perhaps even ubiquitously. Based on this, BEAF is likely to constitutively bind to most sites, as has been reported for Su(Hw) (1). Therefore, the comparisons of these data to our BEAF peak data are likely to be relevant. This linkage of BEAF, TSSs, and high expression levels was not anticipated.
In support of the link between BEAF and transcription, our RT-PCR results with the BEAFAB-KO null mutation and siRNA in cultured cells indicate that BEAF is important for most BEAF-associated genes to maintain their expression levels. In the absence of BEAF, expression levels typically drop two- to fourfold. An exception to this appears to be activation of BEAF-associated genes regulated by DREF. However, our results with BEAFAB-KO embryos are not as clear on this point as previous results obtained with siRNA in cultured cells and the production of a dominant-negative form of BEAF in embryos (18). Perhaps prolonged growth without BEAF in the BEAFAB-KO embryos allowed repressive effects to dominate. This would be consistent with BEAF helping to keep promoter regions in an open configuration even at promoters where it competes with DREF.
The location of BEAF near TSSs of active genes is reminiscent of results recently reported for NELF, the negative regulator of elongation by Pol II (40). We found that 40% of BEAF-associated genes were also associated with NELF. Data indicate that NELF plays a role in pausing by Pol II and that this stimulates transcription levels by inhibiting promoter-proximal nucleosome assembly (23). Compared to the genome as a whole, NELF-associated genes are nearly threefold enriched for genes with paused Pol II as opposed to active Pol II in both Toll10b embryos and S2 cells. In contrast, BEAF-associated genes are about 1.1-fold enriched for genes with active Pol II as opposed to paused Pol II in both Toll10b embryos and S2 cells (see Table S3 in the supplemental material). This suggests that BEAF and NELF are functionally distinct. Perhaps their colocalization at a large number of genes provides complementary mechanisms of ensuring that the promoters of those genes are accessible to Pol II.
BEAF does not localize with Su(Hw) or CTCF.
Comparison of our data for BEAF with data for the Su(Hw) (1) and CTCF (32) insulator proteins indicates that BEAF does not colocalize with Su(Hw) and rarely colocalizes with CTCF. In fact, BEAF mainly localizes near TSSs while Su(Hw) and CTCF are usually found kilobases away from TSSs. In light of the possibility that the numerous minor BEAF peaks that we did not include in our analysis might represent indirect interactions of BEAF with DNA via interactions with other proteins, we checked minor peaks in the Adh region to see if they colocalized with Su(Hw) (60 peaks) or CTCF (18 peaks). Of 18 minor BEAF peaks in this region, 4 were within 1 kb of CTCF peaks. This indicates that BEAF and Su(Hw) do not physically interact. The results for CTCF are ambiguous, indicating that BEAF and CTCF might interact at a minor subset of CTCF binding sites. However, it is clear that if minor BEAF peaks represent interactions with other DNA binding proteins, neither Su(Hw) nor CTCF is a major target.
BEAF, insulator function, and transcription.
While it has been known for some time that transcripts emanate from scs′ (24), the relationship between this and BEAF binding is unknown. Our results indicate that BEAF normally binds near TSSs, suggesting that BEAF is performing the same function at scs′ as it is at the majority of its sites of association. In fact, like scs′, many BEAF binding regions are between closely spaced head-to-head gene pairs. Our data are not consistent with the model in which BEAF insulates these adjacent promoter regions from each other. Instead, our data suggest that BEAF helps to maintain most associated promoter regions in an environment that facilitates transcription. Insulator activity in transgene assays might be a consequence of this local open chromatin configuration. This is similar to the promoter decoy model of insulator function proposed by Geyer more than a decade ago (20). The accessible BEAF-associated promoter might trap upstream regulatory elements so that they do not affect downstream reporter transgenes. There are a variety of possible mechanisms that could be involved by which BEAF might affect promoter accessibility by positively or negatively influencing nucleosome modifications, structure, or positioning. Another interesting possibility is related to the report that nuclear matrix preparations retain 25% of BEAF (48). Both Su(Hw) and CTCF have also been reported to be retained in nuclear matrix preparations, leading to the proposal that they function in part by organizing chromatin into loop domains (10, 17, 66). It is possible that BEAF also organizes chromatin loop domains, but given its proximity to TSSs and relationship with gene expression, perhaps the nuclear matrix association is caused by BEAF-mediated targeting of promoters to transcription factories (12, 34, 57).
If BEAF is performing the same function at scs′ as at other sites, why did the expression levels of the genes in scs′ remain the same in the absence of BEAF? scs′ localizes to one end of an hsp70 domain (60), and recent results indicate that heat shock leads to a rapid, transcription-independent loss of nucleosomes over this domain that stops at scs′ (49). Depletion of BEAF by siRNA did not allow nucleosome loss to spread further, indicating that BEAF is not directly responsible for blocking the heat shock-induced nucleosome loss. As the authors suggest, perhaps the promoters in scs′ are responsible. The ability to rapidly lose nucleosomes suggests that the chromatin of the hsp70 domain is readily accessible. According to this reasoning, BEAF might then be redundant for keeping the promoters in scs′ in an open configuration. One way to test this would be to determine if transcripts initiate from scs′ in a transgenic context and, if so, if these transcript levels drop in the absence of BEAF.
The results presented here link BEAF to TSSs and highly expressed genes. They also provide the suggestion that interactions between BEAF and other chromatin-bound proteins could be widespread and contribute to nuclear organization. Future studies aimed at elucidating the relationships between BEAF and transcription and between BEAF and nuclear organization and the different roles of 32A and 32B will provide valuable insight into nuclear function.
Supplementary Material
Acknowledgments
This work was supported by NSF grant 0718898 to C.M.H. The laboratory of O.C. is supported by INSERM, by the CNRS, and by the UFR-SVT of Paul Sabatier University.
We thank NimbleGen for their expert technical support and the LSU CBS Genomics Facility for the use of their ABI Prism 7000.
Footnotes
Published ahead of print on 20 April 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adryan, B., G. Woerfel, I. Birch-Machin, S. Gao, M. Quick, L. Meadows, S. Russell, and R. White. 2007. Genomic mapping of Suppressor of Hairy-wing binding sites in Drosophila. Genome Biol. 8R167.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 91191-1200. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L. 2000. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem. Sci. 25421-423. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, T. L., and C. Elkan. 1995. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 321-29. [PubMed] [Google Scholar]
- 5.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405482-485. [DOI] [PubMed] [Google Scholar]
- 6.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98387-396. [DOI] [PubMed] [Google Scholar]
- 7.Blanton, J., M. Gaszner, and P. Schedl. 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17664-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burcin, M., R. Arnold, M. Lutz, B. Kaiser, D. Runge, F. Lottspeich, G. N. Filippova, V. V. Lobanenkov, and R. Renkawitz. 1997. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol. Cell. Biol. 171281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushey, A. M., E. R. Dorman, and V. G. Corces. 2008. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell 321-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrd, K., and V. G. Corces. 2003. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 162565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capelson, M., and V. G. Corces. 2005. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell 20105-116. [DOI] [PubMed] [Google Scholar]
- 12.Carter, D. R., C. Eskiw, and P. R. Cook. 2008. Transcription factories. Biochem. Soc. Trans. 36585-589. [DOI] [PubMed] [Google Scholar]
- 13.Cavalli, G., V. Orlando, and R. Paro. 1999. Mapping DNA target sites of chromatin-associated proteins by formaldehyde cross-linking in Drosophila embryos, p. 20-37. In W. A. Bickmore (ed.), Chromosome structural analysis: a practical approach. Oxford University Press, New York, NY.
- 14.Chintapalli, V. R., J. Wang, and J. A. Dow. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39715-720. [DOI] [PubMed] [Google Scholar]
- 15.Clark, A. G., M. B. Eisen, D. R. Smith, C. M. Bergman, B. Oliver, T. A. Markow, T. C. Kaufman, M. Kellis, W. Gelbart, V. N. Iyer, D. A. Pollard, T. B. Sackton, A. M. Larracuente, N. D. Singh, J. P. Abad, D. N. Abt, B. Adryan, M. Aguade, H. Akashi, W. W. Anderson, C. F. Aquadro, D. H. Ardell, R. Arguello, C. G. Artieri, D. A. Barbash, D. Barker, P. Barsanti, P. Batterham, S. Batzoglou, D. Begun, A. Bhutkar, E. Blanco, S. A. Bosak, R. K. Bradley, A. D. Brand, M. R. Brent, A. N. Brooks, R. H. Brown, R. K. Butlin, C. Caggese, B. R. Calvi, A. Bernardo de Carvalho, A. Caspi, S. Castrezana, S. E. Celniker, J. L. Chang, C. Chapple, S. Chatterji, A. Chinwalla, A. Civetta, S. W. Clifton, J. M. Comeron, J. C. Costello, J. A. Coyne, J. Daub, R. G. David, A. L. Delcher, K. Delehaunty, C. B. Do, H. Ebling, K. Edwards, T. Eickbush, J. D. Evans, A. Filipski, S. Findeiss, E. Freyhult, L. Fulton, R. Fulton, A. C. Garcia, A. Gardiner, D. A. Garfield, B. E. Garvin, G. Gibson, D. Gilbert, S. Gnerre, J. Godfrey, R. Good, V. Gotea, B. Gravely, A. J. Greenberg, S. Griffiths-Jones, S. Gross, R. Guigo, E. A. Gustafson, W. Haerty, M. W. Hahn, D. L. Halligan, A. L. Halpern, G. M. Halter, M. V. Han, A. Heger, L. Hillier, A. S. Hinrichs, I. Holmes, R. A. Hoskins, M. J. Hubisz, D. Hultmark, M. A. Huntley, D. B. Jaffe, S. Jagadeeshan, et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450203-218. [DOI] [PubMed] [Google Scholar]
- 16.Cuvier, O., C. M. Hart, and U. K. Laemmli. 1998. Identification of a class of chromatin boundary elements. Mol. Cell. Biol. 187478-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn, K. L., H. Zhao, and J. R. Davie. 2003. The insulator binding protein CTCF associates with the nuclear matrix. Exp. Cell Res. 288218-223. [DOI] [PubMed] [Google Scholar]
- 18.Emberly, E., R. Blattes, B. Schuettengruber, M. Hennion, N. Jiang, C. M. Hart, E. Kas, and O. Cuvier. 2008. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 62896-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerasimova, T. I., K. Byrd, and V. G. Corces. 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 61025-1035. [DOI] [PubMed] [Google Scholar]
- 20.Geyer, P. K. 1997. The role of insulator elements in defining domains of gene expression. Curr. Opin. Genet. Dev. 7242-248. [DOI] [PubMed] [Google Scholar]
- 21.Geyer, P. K., and V. G. Corces. 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 61865-1873. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert, M. K., Y. Y. Tan, and C. M. Hart. 2006. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics 1731365-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilchrist, D. A., S. Nechaev, C. Lee, S. K. Ghosh, J. B. Collins, L. Li, D. S. Gilmour, and K. Adelman. 2008. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 221921-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glover, D. M., M. H. Leibowitz, D. A. McLean, and H. Parry. 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 8195-105. [DOI] [PubMed] [Google Scholar]
- 25.Golovnin, A., L. Melnikova, I. Volkov, M. Kostuchenko, A. V. Galkin, and P. Georgiev. 2008. ‘Insulator bodies’ are aggregates of proteins but not of insulators. EMBO Rep. 9440-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen, S. K., S. Takada, R. H. Jacobson, J. T. Lis, and R. Tjian. 1997. Transcription properties of a cell type-specific TATA-binding protein, TRF. Cell 9171-83. [DOI] [PubMed] [Google Scholar]
- 27.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405486-489. [DOI] [PubMed] [Google Scholar]
- 28.Hart, C. M., O. Cuvier, and U. K. Laemmli. 1999. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma 108375-383. [DOI] [PubMed] [Google Scholar]
- 29.Hart, C. M., K. Zhao, and U. K. Laemmli. 1997. The scs′ boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol. 17999-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirose, F., M. Yamaguchi, H. Handa, Y. Inomata, and A. Matsukage. 1993. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J. Biol. Chem. 2682092-2099. [PubMed] [Google Scholar]
- 31.Hochheimer, A., S. Zhou, S. Zheng, M. C. Holmes, and R. Tjian. 2002. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 420439-445. [DOI] [PubMed] [Google Scholar]
- 32.Holohan, E. E., C. Kwong, B. Adryan, M. Bartkuhn, M. Herold, R. Renkawitz, S. Russell, and R. White. 2007. CTCF genomic binding sites in Drosophila and the organisation of the Bithorax complex. PLoS Genet. 3e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isogai, Y., S. Keles, M. Prestel, A. Hochheimer, and R. Tjian. 2007. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 212936-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson, D. A., A. B. Hassan, R. J. Errington, and P. R. Cook. 1993. Visualization of focal sites of transcription within human nuclei. EMBO J. 121059-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellum, R., and P. Schedl. 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 122424-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kellum, R., and P. Schedl. 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64941-950. [DOI] [PubMed] [Google Scholar]
- 37.Klenova, E. M., R. H. Nicolas, H. F. Paterson, A. F. Carne, C. M. Heath, G. H. Goodwin, P. E. Neiman, and V. V. Lobanenkov. 1993. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 137612-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuhn, E. J., and P. K. Geyer. 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15259-265. [DOI] [PubMed] [Google Scholar]
- 39.Kuhn-Parnell, E. J., C. Helou, D. J. Marion, B. L. Gilmore, T. J. Parnell, M. S. Wold, and P. K. Geyer. 2008. Investigation of the properties of non-gypsy Suppressor of Hairy-wing-binding sites. Genetics 1791263-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, C., X. Li, A. Hechmer, M. Eisen, M. D. Biggin, B. J. Venters, C. Jiang, J. Li, B. F. Pugh, and D. S. Gilmour. 2008. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 283290-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mallin, D. R., J. S. Myung, J. S. Patton, and P. K. Geyer. 1998. Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics 148331-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Markstein, M., and M. Levine. 2002. Decoding cis-regulatory DNAs in the Drosophila genome. Curr. Opin. Genet. Dev. 12601-606. [DOI] [PubMed] [Google Scholar]
- 43.Matsukage, A., F. Hirose, M. A. Yoo, and M. Yamaguchi. 2008. The DRE/DREF transcriptional regulatory system: a master key for cell proliferation. Biochim. Biophys. Acta 177981-89. [DOI] [PubMed] [Google Scholar]
- 44.Mito, Y., J. G. Henikoff, and S. Henikoff. 2007. Histone replacement marks the boundaries of cis-regulatory domains. Science 3151408-1411. [DOI] [PubMed] [Google Scholar]
- 45.Moon, H., G. Filippova, D. Loukinov, E. Pugacheva, Q. Chen, S. T. Smith, A. Munhall, B. Grewe, M. Bartkuhn, R. Arnold, L. J. Burke, R. Renkawitz-Pohl, R. Ohlsson, J. Zhou, R. Renkawitz, and V. Lobanenkov. 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 6165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukhopadhyay, R., W. Yu, J. Whitehead, J. Xu, M. Lezcano, S. Pack, C. Kanduri, M. Kanduri, V. Ginjala, A. Vostrov, W. Quitschke, I. Chernukhin, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 141594-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muse, G. W., D. A. Gilchrist, S. Nechaev, R. Shah, J. S. Parker, S. F. Grissom, J. Zeitlinger, and K. Adelman. 2007. RNA polymerase is poised for activation across the genome. Nat. Genet. 391507-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pathak, R. U., N. Rangaraj, S. Kallappagoudar, K. Mishra, and R. K. Mishra. 2007. Boundary element associated factor 32B connects chromatin domains to the nuclear matrix. Mol. Cell. Biol. 274796-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petesch, S. J., and J. T. Lis. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 13474-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pickersgill, H., B. Kalverda, E. de Wit, W. Talhout, M. Fornerod, and B. van Steensel. 2006. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 381005-1014. [DOI] [PubMed] [Google Scholar]
- 51.Roseman, R. R., E. A. Johnson, C. K. Rodesch, M. Bjerke, R. N. Nagoshi, and P. K. Geyer. 1995. A P element containing suppressor of hairy-wing binding regions has novel properties for mutagenesis in Drosophila melanogaster. Genetics 1411061-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roseman, R. R., V. Pirrotta, and P. K. Geyer. 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 12435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roy, S., M. K. Gilbert, and C. M. Hart. 2007. Characterization of BEAF mutations isolated by homologous recombination in Drosophila. Genetics 176801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roy, S., Y. Y. Tan, and C. M. Hart. 2007. A genetic screen supports a broad role for the Drosophila insulator proteins BEAF-32A and BEAF-32B in maintaining patterns of gene expression. Mol. Genet. Genomics 277273-286. [DOI] [PubMed] [Google Scholar]
- 55.Schneider, R., A. J. Bannister, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 673-77. [DOI] [PubMed] [Google Scholar]
- 56.Schübeler, D., D. M. MacAlpine, D. Scalzo, C. Wirbelauer, C. Kooperberg, F. van Leeuwen, D. E. Gottschling, L. P. O'Neill, B. M. Turner, J. Delrow, S. P. Bell, and M. Groudine. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 181263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sexton, T., D. Umlauf, S. Kurukuti, and P. Fraser. 2007. The role of transcription factories in large-scale structure and dynamics of interphase chromatin. Semin. Cell Dev. Biol. 18691-697. [DOI] [PubMed] [Google Scholar]
- 58.Shopland, L. S., K. Hirayoshi, M. Fernandes, and J. T. Lis. 1995. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev. 92756-2769. [DOI] [PubMed] [Google Scholar]
- 59.Smith, A. D., P. Sumazin, and M. Q. Zhang. 2005. Identifying tissue-selective transcription factor binding sites in vertebrate promoters. Proc. Natl. Acad. Sci. USA 1021560-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Udvardy, A., E. Maine, and P. Schedl. 1985. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 185341-358. [DOI] [PubMed] [Google Scholar]
- 61.Valenzuela, L., and R. T. Kamakaka. 2006. Chromatin insulators. Annu. Rev. Genet. 40107-138. [DOI] [PubMed] [Google Scholar]
- 62.Vostrov, A. A., and W. W. Quitschke. 1997. The zinc finger protein CTCF binds to the APBβ domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem. 27233353-33359. [DOI] [PubMed] [Google Scholar]
- 63.Wallace, J. A., and G. Felsenfeld. 2007. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17400-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu, C. H., Y. Yamaguchi, L. R. Benjamin, M. Horvat-Gordon, J. Washinsky, E. Enerly, J. Larsson, A. Lambertsson, H. Handa, and D. Gilmour. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 171402-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 9741-51. [DOI] [PubMed] [Google Scholar]
- 66.Yusufzai, T. M., and G. Felsenfeld. 2004. The 5′-HS4 chicken β-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. USA 1018620-8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yusufzai, T. M., H. Tagami, Y. Nakatani, and G. Felsenfeld. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13291-298. [DOI] [PubMed] [Google Scholar]
- 68.Zeitlinger, J., A. Stark, M. Kellis, J. W. Hong, S. Nechaev, K. Adelman, M. Levine, and R. A. Young. 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 391512-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao, K., C. M. Hart, and U. K. Laemmli. 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81879-889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.