Abstract
Boron is a micronutrient in plants and animals, but its specific roles in cellular processes are not known. To understand boron transport and functions, we screened a yeast genomic DNA library for genes that confer resistance to the element in Saccharomyces cerevisiae. Thirty boron-resistant transformants were isolated, and they all contained the ATR1 (YML116w) gene. Atr1 is a multidrug resistance transport protein belonging to the major facilitator superfamily. C-terminal green fluorescent protein-tagged Atr1 localized to the cell membrane and vacuole, and ATR1 gene expression was upregulated by boron and several stress conditions. We found that atr1Δ mutants were highly sensitive to boron treatment, whereas cells overexpressing ATR1 were boron resistant. In addition, atr1Δ cells accumulated boron, whereas ATR1-overexpressing cells had low intracellular levels of the element. Furthermore, atr1Δ cells showed stronger boron-dependent phenotypes than mutants deficient in genes previously reported to be implicated in boron metabolism. ATR1 is widely distributed in bacteria, archaea, and lower eukaryotes. Our data suggest that Atr1 functions as a boron efflux pump and is required for boron tolerance.
Boron has been proposed as an important micronutrient in plants and animals. Studies have shown the presence of several genes associated with boron transport and tolerance in plants (18, 25, 27); however, boron transport mechanisms in other organisms, including animals, remain unclear. In plants, boron functions as a cross-linker for rhammogalacturanon II in the cell membrane (9, 14, 21) and also as a structural component in cytoskeleton assembly (1). Arabidopsis thaliana BOR1 was the first gene shown to play a role in boron tolerance (28). Homologs of BOR1 were found in many organisms, including yeasts, plants, and mammals (22, 25, 29). A high level of boron leads to degradation of its own exporter, BOR1, in A. thaliana (27), and A. thaliana BOR1 cannot be used to produce genetically modified plants that grow in soil with high boron levels. However, transgenic plants expressing BOR4, one of six paralogs of BOR1, showed high tolerance to toxic levels of boron (18). Multicopy expression of BOT1, a BOR1 ortholog, provided boron tolerance to barley (25).
The yeast Saccharomyces cerevisiae has been used as a model organism for characterization of plant boron tolerance genes (19, 20, 25, 26, 29). While 10 mM boric acid is lethal to Arabidopsis (18), yeast can grow in the presence of 80 mM boron and is considered a boron-tolerant organism (19, 20). Yeast Bor1 was characterized in detail (10). This protein is localized to the plasma membrane and functions as a boric acid exporter (26). The bor1Δ yeast strain overaccumulates boron (20, 28), and cells that overexpress BOR1 have less intracellular boron and show resistance to boron treatment (20). In addition to Bor1, two other proteins, Dur3 and Fps1, have been implicated in boron tolerance in yeast, but their functions are not clear (20). Dur3 is a plasma membrane transporter that plays a role in urea and polyamine transport (5, 31), and Fps1 is a member of the major intrinsic protein family and plays a role in glycerol, acetic acid, arsenite, and antimonite transport (16, 30, 33). Overexpression of FPS1 and DUR3 showed controversial effects on cellular boron levels. While FPS1 expression lowered the protoplasmic boron concentration, DUR3 expression led to a small increase in boron (20).
The objective of this study was to identify proteins that are primarily responsible for boron transport in yeast. ATR1 was identified as a boron tolerance gene by screening a yeast DNA expression library. Yeast Atr1 is a member of the DHA2 family of drug-H+ antiporters with 14 predicted membrane-spanning segments (7). It was first characterized in a genetic screen as a high-copy-number suppressor of the 3-amino-1,2,4-triazole sensitivity of gcn4Δ mutants (11). It also conferred resistance to the DNA-damaging agent 4-nitroquinoline-N-oxide in a separate genetic screen (17). In this study, we demonstrated that high-copy-number expression of ATR1 conferred extreme resistance to boron and reduced intracellular levels of the element, whereas cells lacking the ATR1 gene were hypersensitive to boron and increased its intracellular levels. We analyzed changes in the global gene expression profile in response to boron and found that ATR1 is the most induced transporter gene. The Atr1-green fluorescent protein (GFP) fusion protein localized to the plasma membrane and vacuole. Taken together, our data show that Atr1 functions as a major boron efflux pump and provides tolerance of the element by pumping boron out of cells.
MATERIALS AND METHODS
Yeast strains.
The wild-type strain BY4741 (MATa his3 leu2 met15 ura3) and its isogenic deletion mutants were obtained from the yeast deletion library (Invitrogen). The W303-1a (MATa ade2 ura3 leu2 trp1 his3 can1) and Y252 (MATa his3 leu2 met15 ura3-52 lys2-801 amber ade2,101ochre trp1 leu2) strains were previously described by Wallis et al. (32) and Spector et al. (23), respectively. A single-step gene replacement procedure was applied to delete the ATR1 gene in W3031-a cells. A disruption cassette containing the URA3 marker gene was prepared using Pfx polymerase (Invitrogen) with the p426GAL vector as the template. Both primers contained homologous regions for recombination with the ATR1 gene. The primers used were 5′-GTGTACGATTGTAAAAAGAGAGGCAATTAGAGAATCTCAAACAGGTAATAACTTTTTTTTGATTCCGGTTTCTTTGA-3′ and 5′-CGCATGACAGCGGTGTATTTCTATCTATTTACCTTAATAACCGCTTTCCGCTAATTTGTGAGTTTAGTATACATGC-3′. The purified PCR fragment was transformed into the W303-1a strain using a standard lithium acetate method. Transformants were confirmed for deletion of ATR1 by PCR using the primers ATR1-F (5′-TGGGCAATCAGTCATTAGTTGTGC) and ATR1-B (5′-TGCAATCGTATTCTGCAGCA). YPD medium (2% glucose, 2% peptone, 1% yeast extract, and 2% agar for solid medium) or yeast nitrogen base (YNB) medium (Bio101, Vista, CA) supplemented with the required amino acids or bases were used for cell growth.
Yeast genomic DNA library screening.
The yeast genomic DNA library in YEp13 was obtained from ATCC (no. 37323). Library plasmids were purified from Escherichia coli with a MaxiPrep kit (Invitrogen). Wild-type yeast (BY4741) was transformed with purified library DNA using the standard LiAc method, and cells were incubated on YNB medium containing 100 mM boric acid. Plates were incubated at 30°C for 5 days, and 30 colonies that grew in the presence of 100 mM boric acid were picked and regrown with higher concentrations of boron (125 to 200 mM) to confirm that the transformants were boron tolerant. Plasmids were isolated from selected clones and transferred to E. coli for amplification purposes. The isolated plasmids were then sequenced with vector-specific primers.
DNA microarray analysis.
Cells were grown to an optical density at 600 nm (OD600) of 0.2 to 0.3 in 200 ml of YPD medium, with or without treatment for 1 hour with 20 mM boric acid; harvested by centrifugation; and kept at −80°C. Total RNA was isolated using the Ambion RiboPure yeast kit according to the manufacturer's instructions. The Agilent two-color low RNA input linear amplification kit was used to generate fluorescently labeled cRNA for two-color microarray hybridizations. Fluorescently labeled cRNA molecules were purified from the reaction mixture using the Qiagen RNeasy minikit. cRNA samples (825 ng each) were combined with Agilent Hi-RPM hybridization buffer. Microarray hybridizations were performed using Agilent SureHyb hybridization chambers. The hybridization chambers were loaded onto a rotisserie in an Agilent hybridization oven and were incubated at 65°C for 17 h with a rotational speed of 10 rpm. Following incubation, the microarray slides were washed for 1 min each in Gene Expression wash buffer 1 (6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 0.005% N-lauroylsarcosine at room temperature) and Gene Expression wash buffer 2 (0.06× SSPE, 0.005% N-lauroylsarcosine at 31°C) for 1 min each. The microarray slides were briefly dipped in a solution of acetonitrile, dried, and scanned in an Agilent Technologies G2505B microarray scanner at 5-μm resolution.
RNA isolation and real-time PCR analyses.
Total RNA was isolated using the RNeasy minikit (Qiagen). Traces of genomic DNA were removed by DNase treatment (DNase RQ1; Promega). cDNA synthesis was performed using the First Strand cDNA synthesis kit (Fermentas) according to the manufacturer's instructions. The cDNA was used as a template to amplify a 186-bp fragment for the ATR1 gene and a 103-bp fragment for the internal control ACT1 gene. Real-time PCR assays were performed with the IQ5 real-time PCR system (Bio-Rad). The primers used for amplification of ATR1 were ATR1F (5′-ACGCGTATAGCATAGCCGCTTTCA-3′) and ATR1B (5′-TGTAAGCCTGGTTCCAACCCGATA-3′), and those used for amplification of the ACT1 gene were ACT1F (5′-ACGTTCCAGCCTTCTACGTTTCCA-3′) and ACT1B (5′-ACGTGAGTAACACCATCACCGGAA-3′). The conditions for PCR amplification were as follows: 30 cycles at 94°C for 30 s, 62°C for 30 s, and 72°C for 30 s.
Overexpression of the ATR1 gene.
The wild-type allele of the ATR1 gene was amplified by Pfu polymerase (Fermentas) from yeast genomic DNA using the primers ATR1F (5′-TGGATCCGAGGCAATTAGAGAATCTCAAACAGG-3′) and ATR1B (5′-ACTCGAGCTTTCCGCTAAGCCACAGTGCAATCG-3′) containing BamHI and XhoI restriction sites, respectively. The amplified fragment was first cloned into the SmaI site of pBluescript II KS (Stratagene) and then moved into the BamHI/XhoI sites of the yeast high-copy-number expression vector p426GPD to generate a p426-ATR1 expression construct. Yeast cells were transformed with the empty vector p426 or with p426-ATR1 and selected for uracil prototrophy.
Determination of intracellular boron concentrations.
Exponentially growing cells were treated with 50 mM boric acid for 1 hour and harvested by centrifugation. The cells were washed with distilled water three times and either kept at −80°C or disrupted in 500 μl lysis buffer (20 mM Tris, pH 7.6, 100 mM NaCl, 1 mM EDTA, pH 8, 2% Triton X-100, 1% sodium dodecyl sulfate, 10 mg/ml protease inhibitor cocktail [Sigma], 500 μl glass beads) using a vortex. The supernatants were transferred to new tubes, and the protein concentrations were determined by the Bradford protein detection assay (2). The boron concentrations of diluted cell extracts were determined using the boron cell test kit (Merck, Germany) as described in the manufacturer's instructions and normalized by the amount of total protein in the cell extracts. The measurement sensitivity of the kit was 8 × 10−7 M (0.05 mg boron/liter) as stated by the manufacturer.
Protein localization.
A previously described procedure (13) was followed for tagging the chromosomal ATR1 gene with GFP at the C terminus. Primers 116S3 (5′-AGGCCGCAGGGCAAGAGCTGCTGCAGAATACGATTGCACTGTG GCTCGTACGCTGCAGGTCGAC-3′) and 116S2 (5′-CATGACAGCGGTGTATTTCTATCTATTTACCTTAATAACCGCTTTCCGATCGATGAATTCG AGCTCG-3′) were used to amplify a cassette including GFP and the Kluyveromyces lactis TRP1 marker from plasmid pMY26 (13). The resulting PCR fragment was used to transform W3031-a (MATa ade2 ura3 leu2 trp1 his3) (32) wild-type cells, and transformants were selected on YNB-uracil dropout medium. Exponentially growing cells were analyzed with a confocal fluorescence microscope.
Western blot analysis.
Wild-type cells (W3031-a) and the isogenic ATR1-GFP strains were grown in YPD medium to logarithmic phase, and equal numbers of cells were harvested by centrifugation for each strain. Cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotting. The GFP signal was detected by primary and secondary antibodies from Sigma and SuperSignal West Pico chemiluminescent substrate from Pierce.
Comparative genomic analyses of ATR1, BOR1, DUR3, and FPS1 in prokaryotes and eukaryotes.
Sequenced genomes of 45 archaea and 547 bacteria and 53 annotated genomes of eukaryotes (37 completely sequenced and 16 unfinished) from NCBI (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) were analyzed for the occurrence of boron transporter genes. S. cerevisiae Atr1 protein (gi 798884) was used as the query in TBLASTN and BLASTP searches with default parameters. Orthologous proteins were defined by a bidirectional BLAST test. Hits were considered orthologous if S. cerevisiae Atr1 was among the three best hits in bidirectional BLAST tests with an E value of 1e−5.
Multiple-sequence alignments.
Multiple-sequence alignments were performed using CLUSTALW and T-Coffee with the default parameters. Phylogenetic trees were prepared using the Interactive Tree Of Life service (http://itol.embl.de/), which is based on the highly resolved tree of life project (3).
RESULTS
Identification of ATR1 as a boron resistance gene.
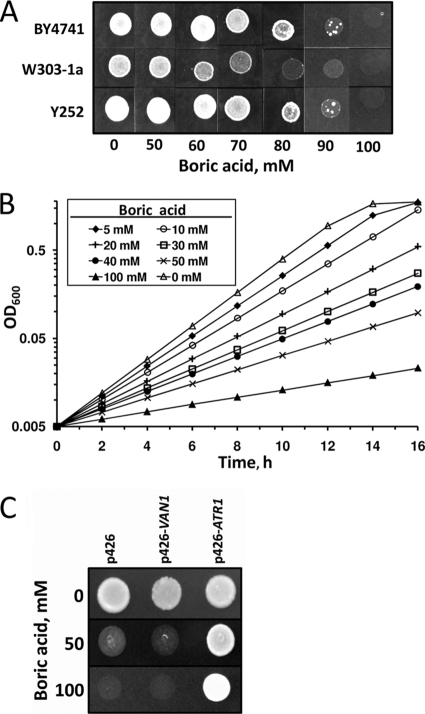
We first tested several wild-type strains for boron tolerance and found that cells did not grow well at concentrations of boric acid above 80 mM (Fig. 1A and B). We used this observation to screen a high-copy-number yeast genomic DNA library for genes that confer boron resistance on BY4741 wild-type cells. Transformed cells were exposed to 100 mM boric acid for 5 days. Thirty colonies grew in the presence of 100 mM boric acid. Each colony was regrown on plates with increasing amounts of boric acid. All identified colonies grew in the presence of 200 mM boric acid. We recovered plasmids from five randomly chosen colonies and sequenced the inserts using a pair of vector-based primers. All of the inserts had sequences derived from S. cerevisiae chromosome XIII, spanning a region that contained intact VAN1 (YML115c) and ATR1 (YML116w) genes. PCR analyses of the other library constructs showed that they also contained the same region of chromosome XIII. Van1 plays a role in mannan synthesis (24) and vanadium tolerance (12). Atr1 is a drug-H+ antiporter of the major facilitator superfamily with unknown function (7, 11).
FIG. 1.
Boric acid tolerance of yeast cells. (A) Three different wild-type strains were grown to logarithmic phase in rich (YPD) medium and diluted to an OD600 of 0.2, and 5 μl of each cell suspension was spotted on plates containing the indicated amounts of boric acid. The plates were incubated at 30°C for 5 days and photographed. (B) An overnight culture of wild-type yeast (BY4741) was diluted to an OD600 of 0.05 and incubated with the indicated amounts of boric acid in YPD medium. Cell growth was monitored by OD600 measurements. (C) Wild-type cells (BY4741) were transformed with an empty vector (p426) or with p426-VAN1 or p426-ATR1 plasmid, and the transformants were spotted on YNB plates containing 0, 50, or 100 mM boric acid.
To determine whether VAN1 or ATR1 conferred boron resistance on cells, the two genes were separately cloned into the yeast high-copy-number expression vector p426GPD and expressed in BY4741 wild-type cells. Overexpression of the ATR1 gene provided strong boron resistance (Fig. 1C), whereas overexpression of VAN1 did not. We concluded that the ATR1 gene was responsible for boron tolerance.
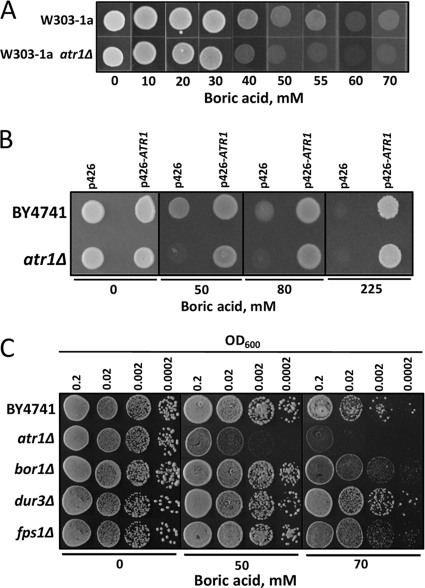
Next, we tested whether deletion of ATR1 leads to boron sensitivity. As shown in Fig. 2A and B, atr1Δ mutants in both the W3031-a and BY4741 backgrounds did not grow in the presence of boric acid in excess of 50 mM, whereas wild-type cells tolerated larger amounts of boric acid. Overexpression of ATR1 in the atr1Δ mutant enabled cell growth in the presence of boron. In fact, ATR1 expression provided very strong boron resistance to both atr1Δ and wild-type cells, as they could grow in the presence of 225 mM boric acid (Fig. 2B).
FIG. 2.
The ATR1 gene provides boron resistance. (A) Wild-type strain W3031-a and its isogenic atr1Δ mutant were grown to logarithmic phase in rich (YPD) medium, and the cultures were diluted to an OD600 of 0.2. Five microliters of each cell suspension was spotted on plates containing the indicated amounts of boric acid. The plates were incubated at 30°C for 5 days and photographed. (B) ATR1-overexpressing wild-type and atr1Δ cells were grown in the presence of the indicated concentrations of boron. Cells transformed with an empty vector were used as a control. (C) Comparisons of the boron tolerances of atr1Δ, bor1Δ, dur3Δ, and fps1Δ mutants. Overnight cultures were diluted to the indicated densities, and 5 μl of each cell suspension was spotted. The cells were grown for 3 days, and the plates were photographed.
To determine the overall contribution of ATR1 to boron toxicity resistance, we compared the boron tolerance of atr1Δ mutants to those of bor1Δ, dur3Δ, and fps1Δ mutants. These mutants are defective for the yeast genes that were previously reported to be involved in boron resistance. The boron tolerances of the four mutants were compared by growing cells on boron-containing medium (Fig. 2C). The atr1Δ mutant was the most sensitive to boron treatment. Its growth was inhibited by 50 mM boric acid, whereas the growth of the other mutants was unaffected. Thus, deletion of the ATR1 gene created stronger boron sensitivity than deletion of any other known boron resistance gene.
ATR1 paralogs in the S. cerevisiae genome.
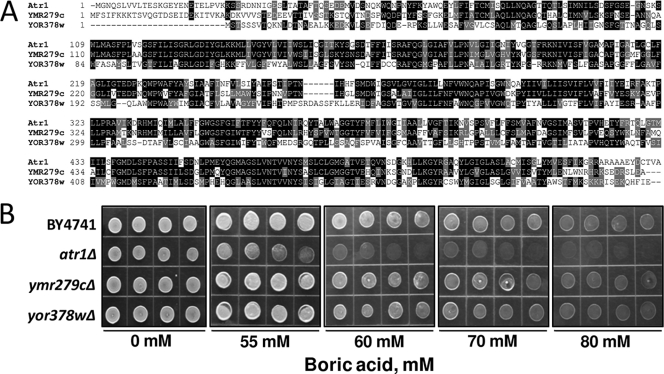
Two ATR1 paralogs were identified in the S. cerevisiae genome by sequence analyses (Fig. 3A). Atr1 and its paralogs share a pfam07690 domain and show more than 70% identity in their sequences. Strains with YMR279c and YOR378w deletions were tested for boron tolerance, but no difference from wild-type cells was found, suggesting that, among the three paralogs, only ATR1 provides boron resistance (Fig. 3B).
FIG. 3.
Sequence alignment and roles of ATR1 paralogs in boron tolerance. (A) Multiple-sequence alignment of yeast ATR1 paralogs. Conserved residues are highlighted with BOXSHADE 3.21. (B) Comparisons of the boron tolerances of the atr1Δ mutant and paralogs, YMR279c and YOR378w mutants. Overnight cultures were diluted, and 5 μl of each cell suspension was spotted. The cells were grown for 3 days, and the plates were photographed.
Atr1 localization.
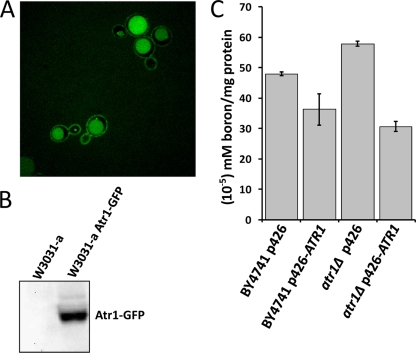
Atr1 is a predicted membrane protein containing 14 transmembrane segments (7, 8). We tagged the native ATR1 gene with a GFP tag and examined the localization of the fusion protein. Confocal microscopy analysis showed that the Atr1-GFP fusion protein localized to the plasma membrane and vacuole (Fig. 4A). To confirm that the vacuolar localization of the fusion protein was not the result of degradation of the intact protein, we performed Western blot analysis using an antibody against GFP. As seen in Fig. 4B, there was only one band for the Atr1-GFP fusion protein, suggesting that the protein was intact.
FIG. 4.
Atr1 localization and boron efflux function. (A) Atr1 localizes to the cell membrane and vacuole. The ATR1 gene was tagged with GFP at the C terminus, and cells expressing the Atr1-GFP fusion protein were photographed with a confocal fluorescence microscope. (B) Western blot analysis of the Atr1-GFP fusion protein with anti-GFP antibody showing a single band. (C) Intracellular boron levels in wild-type and atr1Δ cells that were exposed to 50 mM boric acid for 1 h. The cells were transformed with either empty vector or an ATR1 overexpression plasmid (p426-ATR1). Boron levels were normalized to protein levels. The error bars represent three separate measurements.
Atr1 is a boron efflux transporter.
To examine the mechanism of boron resistance by Atr1, we determined the intracellular boron levels in wild-type and atr1Δ cells. Exponentially growing cells were treated with 50 mM boric acid for 1 hour, and then their intracellular boron levels were determined. As shown in Fig. 4C, atr1Δ cells had 21% more boron than wild-type cells. Overexpression of ATR1 lowered the intracellular boron concentration by 25% in wild-type cells and by 47% in atr1Δ cells. Thus, the absence of the ATR1 gene caused accumulation of boron inside the cells, whereas its overexpression led to the exclusion of boron from the cells.
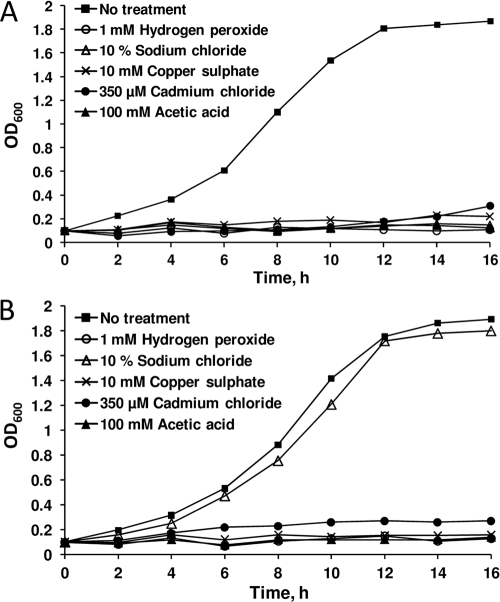
To check the possibility that Atr1 might be a general stress tolerance protein with multiple substrates, we tested wild-type cells that overexpressed ATR1 for different stress conditions, including oxidative stress, salt stress, heavy metal stress, and acetic acid stress (Fig. 5). Among these tested conditions, ATR1 overexpression provided resistance only to salt stress.
FIG. 5.
Role of ATR1 in yeast stress tolerance. Wild-type cells were transformed with p426 (A) and p426-ATR1 (B), and the transformants were grown in the presence of the indicated toxic substances. Growth rates were determined by OD600 measurements.
Transcriptional response to boron treatment.
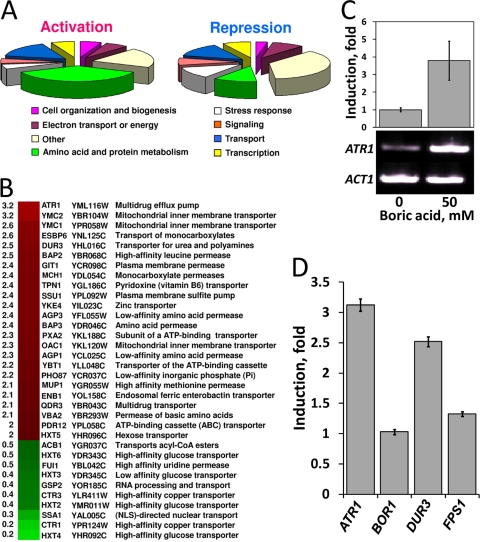
We analyzed the transcriptional response of wild-type yeast cells to 20 mM boric acid using DNA microarrays. Interestingly, groups of genes involved in amino acid biosynthesis and membrane transport were differentially expressed (Fig. 6A). Among transporter genes whose expression was induced by boric acid, ATR1 had the greatest increase (over threefold induction) (Fig. 6B). Expression of the ATR1 paralogs YMR279C and YOR378W was induced by 1.4- and 1.6-fold, respectively. Upregulation of the ATR1 gene in response to boron treatment was also observed by real-time PCR. ATR1 transcripts showed fourfold-higher expression in cells exposed to 50 mM boric acid for 1 hour than in control cells (Fig. 6C). Expression levels of the BOR1 and FPS1 genes did not change in response to boron treatment, but DUR3 was upregulated 2.5-fold (Fig. 6D). Thus, our expression analyses showed that the ATR1 gene is more strongly upregulated in the presence of boron than the three previously known boron transporters.
FIG. 6.
Genomic expression analyses of boron-treated cells. Logarithmically growing cells were split into two groups, one of which was treated with 20 mM boric acid for 1 hour while the other was not. The gene expression patterns of the two groups were then compared. (A) Functional categories of boron-responsive genes with known functions. Genes activated or repressed at least twofold are shown. (B) Transporter genes that were upregulated or downregulated at least threefold in response to boron treatment. (C) Real-time PCR transcriptional analysis of ATR1 in response to boron treatment. Wild-type cells were treated or not with 50 mM boric acid, and ATR1 mRNA levels were detected (top). PCR products were run on a 1% Tris-acetate-EDTA-agarose gel (bottom). Actin (ACT1) was used as the internal control. (D) Transcriptional regulation of ATR1, BOR1, DUR3, and FPS1 genes by boron treatment. The data are derived from microarray experiments. The error bars represent four replicates.
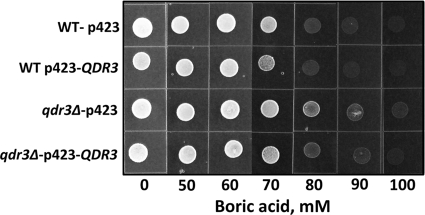
In addition to ATR1, transcription of the QDR3 (YBR043c) gene, whose product is a member of the DHA1 family of drug-H+ antiporters with 12 predicted membrane-spanning segments, was upregulated twofold in response to boron in the microarray analysis (Fig. 6B). To test whether the QDR3 gene plays a role in boron tolerance similar to that of ATR1, we cloned and overexpressed it in wild-type (BY4741) and qdr3Δ mutant cells. As seen in Fig. 7, neither overexpression nor deletion of the QDR3 gene changed the boron tolerance level of the cells.
FIG. 7.
Deletion or overexpression of QDR3 has no effect on boron tolerance. Wild-type (WT) strain BY4741 and its isogenic qdr3Δ mutant were transformed with an empty vector and p423-QDR3 plasmid. The transformants were grown to logarithmic phase in YNB-His medium, and the cultures were diluted to an OD600 of 0.2. Five microliters of each cell suspension was spotted on plates containing the indicated amounts of boric acid. The plates were incubated at 30°C for 5 days and photographed.
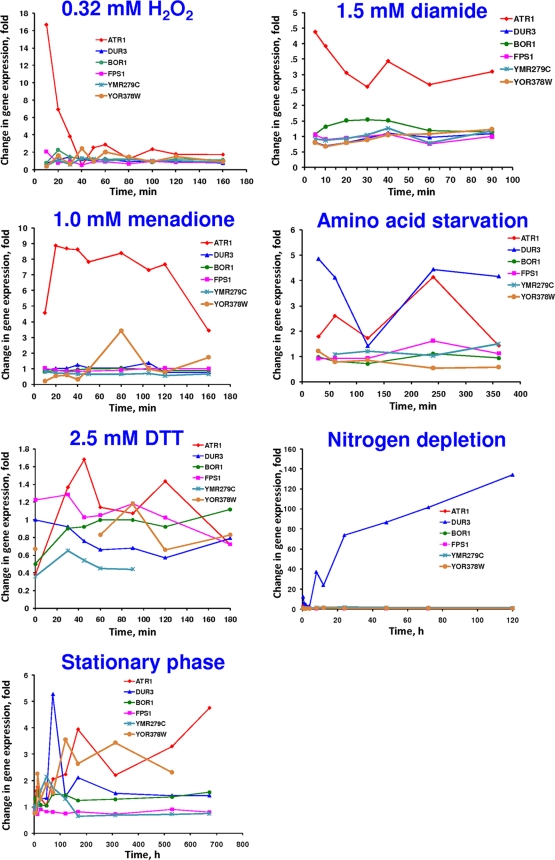
To better understand the regulation of ATR1, BOR1, DUR3, FPS1, and the two ATR1 paralogs, YMR279c and YOR378w, we analyzed their expression changes in response to various stresses (Fig. 8) and participation in the yeast environmental stress response (6). There was significant upregulation of ATR1 expression in response to hydrogen peroxide, diamide, and menadione, whereas the expression of the other boron transporters was not changed.
FIG. 8.
Time course of changes in expression of the boron transporter gene and ATR1 paralogs in response to the indicated chemical stressors, 0.32 mM H2O2, 1.5 mM diamide, 1 mM menadione, and 2.5 mM dithiothreitol (DTT), and to the indicated stress conditions, amino acid starvation, nitrogen depletion, and stationary phase.
Widespread occurrence of ATR1 homologs in fungi, bacteria, and archaea.
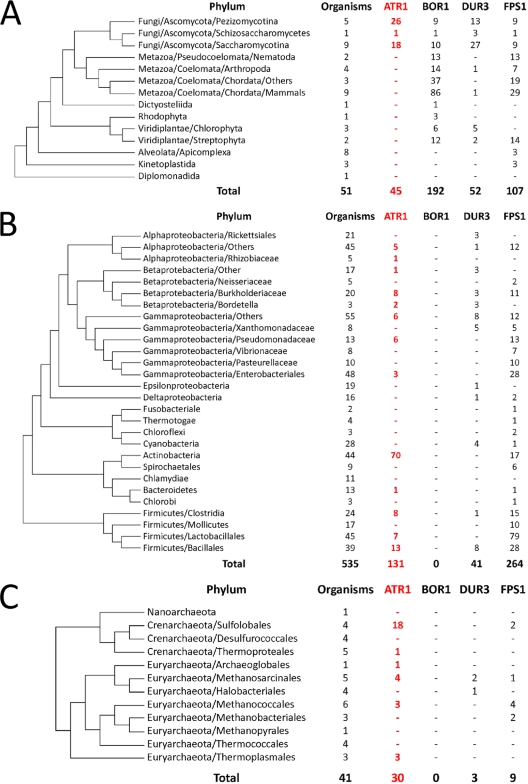
ATR1 homologs were detected in fungi, bacteria, and archaea (Fig. 9; see Fig. S1 in the supplemental material), but not in animals, vascular plants, and algae. Vascular plants and Dictyostelium discoideum contained distantly related ATR1 homologs with unknown functions. Actinobacteria had the highest content of ATR1 orthologs among bacteria and Sulfolobales among archaea. ATR1 distribution in eukaryotes was limited to fungi. We also analyzed the occurrence of other boron transporters in the three domains of life. FPS1 and DUR3 homologs showed scattered occurrence, whereas BOR1 was mainly found in eukaryotes.
FIG. 9.
Occurrence of ATR1 in the three domains of life. Distribution of ATR1 in Eukaryota (A), Bacteria (B), and Archaea (C). The phylogenetic trees were derived from a highly resolved tree of life (3).
DISCUSSION
We identified ATR1 as a major boron resistance gene in yeast by screening a genomic DNA library for genes that support the growth of yeast cells in the presence of high concentrations of boric acid. Two ATR1 paralogs, YMR279c and YOR378w, could not be linked to boron detoxification, as the corresponding mutants were not boron sensitive. Overexpression of ATR1 provided boron resistance to wild-type cells, and its deletion reduced boron tolerance levels. In boron-containing media, the intracellular boron concentration of atr1Δ mutants was higher than that of wild-type cells, whereas overexpression of the ATR1 gene reduced intracellular boron levels. We conclude that Atr1 functions as a boron efflux transporter and helps cells to keep intracellular boron below toxic levels.
Several genes have been shown to have roles in boron resistance in yeast. Among these, the functions of DUR3 and FPS1 in boron export are not clear (20). DUR3 and FPS1 deletion mutants did not show elevated levels of intracellular boron; however, deletion of BOR1 increased boron levels in yeast cells (20). Overexpression of the BOR1 and FPS1 genes resulted in lower cytoplasmic boron levels, while overexpression of DUR3 resulted in slightly increased amounts of boron (20). Among all boron transporters, Atr1 showed the widest phyletic distribution (Fig. 9; see Fig. S1 in the supplemental material). The diverse occurrence of these boron transporters (characterized by unrelated protein sequences) suggests the possibility of independent evolution of their functions in boron detoxification.
Our microarray data showed that expression of the BOR1 and FPS1 genes did not change in response to boron treatment, while DUR3 was upregulated 2.5-fold (Fig. 6D). Jennings et al. also showed that BOR1 transcription is not induced by exposure of cells to boron (10). We compared the growth rates of atr1Δ, bor1Δ, fps1Δ, and dur3Δ mutants in the presence of boron and found that only deletion of the ATR1 gene led to boron hypersensitivity (Fig. 2C), suggesting that ATR1 has the most critical function in eliminating boron. The atr1Δ mutant was hypersensitive to boron treatment, and cells overexpressing ATR1 survived in the presence of 225 mM boric acid, which is the largest amount of boron shown to be tolerated by a eukaryotic system. ATR1 expression was also significantly upregulated by boron exposure (Fig. 6). Thus, ATR1 is a physiologically relevant boron tolerance gene in yeast. Larger amounts of intracellular boron in atr1Δ mutants and smaller amounts in ATR1-overexpressing cells suggest that Atr1 controls boron traffic in one direction, toward the extracellular space.
Transcriptional control of ATR1 has not been studied in detail, but it is known that Gcn4 and Yap1 transcription factors play roles in ATR1 induction (4). Apart from the other boron tolerance genes, ATR1 expression was upregulated in response to hydrogen peroxide, diamide, and menadione treatments (Fig. 8), which could be explained by a Yap1-dependent mechanism. Yap1 is a transcription factor that regulates expression of oxidative-stress response genes (15). The expression patterns of the ATR1 and DUR3 genes in response to amino acid starvation were similar (Fig. 8).
ATR1 could also be viewed as a stress tolerance gene in lower eukaryotes and prokaryotes; however, except for boron, its substrate spectrum is not clear. The role of Atr1 in 3-amino-1,2,4-triazole and 4-nitroquinoline-N-oxide resistance was shown previously. To investigate the possibility that the ATR1 product may be a general stress tolerance protein with multiple substrates besides boron, we subjected wild-type cells that overexpressed ATR1 to different stress conditions (Fig. 5). ATR1 overexpression provided resistance to salt stress but not to oxidative, heavy metal, or acetic acid stress. Thus, although ATR1 may not be a general stress tolerance gene, it may have a role in the detoxification of more than one substrate.
In summary, four properties of ATR1 provided evidence for its role in boron resistance: (i) deletion of ATR1 led to boron sensitivity, (ii) overexpression of ATR1 increased boron tolerance, (iii) ATR1 expression was regulated by boron, and (iv) the concentration of intracellular boron was controlled by ATR1. Compared to other proteins previously shown to play roles in boron metabolism in yeast, Atr1 emerges as the most effective.
Supplementary Material
Acknowledgments
We thank Karim Labip (Paterson Institute for Cancer Research, United Kingdom) for providing the pMY12 vector, Serdar Özçelik (Izmir Institute of Technology, Izmir, Turkey) for his help with confocal fluorescence microscopy, and the biotechnology core facility of the Izmir Institute of Technology for help with instruments. We also thank Anne Frary for critical review of the manuscript and Irem Uluisik, Banu Demir, and Elise Hacioglu for their help with some of the experiments.
This work was supported by Turkish Scientific and Technical Research Council (TUBITAK) Grant no. 104T213 and a Turkish Academy of Science GEBIP grant to A.K. and in part by NIH grants to V.N.G. Alaattin Kaya was supported by the Graduate Student Support Program of TUBITAK-BIDEP.
Footnotes
Published ahead of print on 4 May 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bassil, E., H. Hu, and P. H. Brown. 2004. Use of phenylboronic acids to investigate boron function in plants. Possible role of boron in transvacuolar cytoplasmic strands and cell-to-wall adhesion. Plant Physiol. 1363383-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 3.Ciccarelli, F. D., T. Doerks, C. von Mering, C. J. Creevey, B. Snel, and P. Bork. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 3111283-1287. [DOI] [PubMed] [Google Scholar]
- 4.Coleman, S. T., E. Tseng, and W. S. Moye-Rowley. 1997. Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J. Biol. Chem. 27223224-23230. [DOI] [PubMed] [Google Scholar]
- 5.ElBerry, H. M., M. L. Majumdar, T. S. Cunningham, R. A. Sumrada, and T. G. Cooper. 1993. Regulation of the urea active transporter gene (DUR3) in Saccharomyces cerevisiae. J. Bacteriol. 1754688-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 114241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gbelska, Y., J. J. Krijger, and K. D. Breunig. 2006. Evolution of gene families: the multidrug resistance transporter genes in five related yeast species. FEMS Yeast Res. 6345-355. [DOI] [PubMed] [Google Scholar]
- 8.Goffeau, A., J. Park, I. T. Paulsen, J. L. Jonniaux, T. Dinh, P. Mordant, and M. H. Saier, Jr. 1997. Multidrug-resistant transport proteins in yeast: complete inventory and phylogenetic characterization of yeast open reading frames with the major facilitator superfamily. Yeast 1343-54. [DOI] [PubMed] [Google Scholar]
- 9.Iwai, H., A. Hokura, M. Oishi, H. Chida, T. Ishii, S. Sakai, and S. Satoh. 2006. The gene responsible for borate cross-linking of pectin rhamnogalacturonan-II is required for plant reproductive tissue development and fertilization. Proc. Natl. Acad. Sci. USA 10316592-16597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jennings, M. L., T. R. Howren, J. Cui, M. Winters, and R. Hannigan. 2007. Transport and regulatory characteristics of the yeast bicarbonate transporter homolog Bor1p. Am. J. Physiol. Cell Physiol. 293C468-C476. [DOI] [PubMed] [Google Scholar]
- 11.Kanazawa, S., M. Driscoll, and K. Struhl. 1988. ATR1, a Saccharomyces cerevisiae gene encoding a transmembrane protein required for aminotriazole resistance. Mol. Cell. Biol. 8664-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanik-Ennulat, C., and N. Neff. 1990. Vanadate-resistant mutants of Saccharomyces cerevisiae show alterations in protein phosphorylation and growth control. Mol. Cell. Biol. 10898-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15963-972. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi, M., T. Matoh, and J. Azuma. 1996. Two chains of rhamnogalacturonan II are cross-linked by borate-diol ester bonds in higher plant cell walls. Plant Physiol. 1101017-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 27416040-16046. [DOI] [PubMed] [Google Scholar]
- 16.Luyten, K., J. Albertyn, W. F. Skibbe, B. A. Prior, J. Ramos, J. M. Thevelein, and S. Hohmann. 1995. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 141360-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mack, M., P. Gompel-Klein, E. Haase, J. Hietkamp, A. Ruhland, and M. Brendel. 1988. Genetic characterization of hyperresistance to formaldehyde and 4-nitroquinoline-N-oxide in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 211260-265. [DOI] [PubMed] [Google Scholar]
- 18.Miwa, K., J. Takano, H. Omori, M. Seki, K. Shinozaki, and T. Fujiwara. 2007. Plants tolerant of high boron levels. Science 3181417. [DOI] [PubMed] [Google Scholar]
- 19.Nozawa, A., K. Miwa, M. Kobayashi, and T. Fujiwara. 2006. Isolation of Arabidopsis thaliana cDNAs that confer yeast boric acid tolerance. Biosci. Biotechnol. Biochem. 701724-1730. [DOI] [PubMed] [Google Scholar]
- 20.Nozawa, A., J. Takano, M. Kobayashi, N. von Wiren, and T. Fujiwara. 2006. Roles of BOR1, DUR3, and FPS1 in boron transport and tolerance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 262216-222. [DOI] [PubMed] [Google Scholar]
- 21.O'Neill, M. A., T. Ishii, P. Albersheim, and A. G. Darvill. 2004. Rhamnogalacturonan II: structure and function of a borate cross-linked cell wall pectic polysaccharide. Annu. Rev. Plant Biol. 55109-139. [DOI] [PubMed] [Google Scholar]
- 22.Park, M., Q. Li, N. Shcheynikov, W. Zeng, and S. Muallem. 2004. NaBC1 is a ubiquitous electrogenic Na+-coupled borate transporter essential for cellular boron homeostasis and cell growth and proliferation. Mol. Cell 16331-341. [DOI] [PubMed] [Google Scholar]
- 23.Spector, D., J. Labarre, and M. B. Toledano. 2001. A genetic investigation of the essential role of glutathione: mutations in the proline biosynthesis pathway are the only suppressors of glutathione auxotrophy in yeast. J. Biol. Chem. 2767011-7016. [DOI] [PubMed] [Google Scholar]
- 24.Stolz, J., and S. Munro. 2002. The components of the Saccharomyces cerevisiae mannosyltransferase complex M-Pol I have distinct functions in mannan synthesis. J. Biol. Chem. 27744801-44808. [DOI] [PubMed] [Google Scholar]
- 25.Sutton, T., U. Baumann, J. Hayes, N. C. Collins, B. J. Shi, T. Schnurbusch, A. Hay, G. Mayo, M. Pallotta, M. Tester, and P. Langridge. 2007. Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 3181446-1449. [DOI] [PubMed] [Google Scholar]
- 26.Takano, J., M. Kobayashi, Y. Noda, and T. Fujiwara. 2007. Saccharomyces cerevisiae Bor1p is a boron exporter and a key determinant of boron tolerance. FEMS Microbiol. Lett. 267230-235. [DOI] [PubMed] [Google Scholar]
- 27.Takano, J., K. Miwa, L. Yuan, N. von Wiren, and T. Fujiwara. 2005. Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc. Natl. Acad. Sci. USA 10212276-12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takano, J., K. Noguchi, M. Yasumori, M. Kobayashi, Z. Gajdos, K. Miwa, H. Hayashi, T. Yoneyama, and T. Fujiwara. 2002. Arabidopsis boron transporter for xylem loading. Nature 420337-340. [DOI] [PubMed] [Google Scholar]
- 29.Takano, J., M. Wada, U. Ludewig, G. Schaaf, N. von Wiren, and T. Fujiwara. 2006. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 181498-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamas, M. J., K. Luyten, F. C. Sutherland, A. Hernandez, J. Albertyn, H. Valadi, H. Li, B. A. Prior, S. G. Kilian, J. Ramos, L. Gustafsson, J. M. Thevelein, and S. Hohmann. 1999. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 311087-1104. [DOI] [PubMed] [Google Scholar]
- 31.Uemura, T., K. Kashiwagi, and K. Igarashi. 2007. Polyamine uptake by DUR3 and SAM3 in Saccharomyces cerevisiae. J. Biol. Chem. 2827733-7741. [DOI] [PubMed] [Google Scholar]
- 32.Wallis, J. W., G. Chrebet, G. Brodsky, M. Rolfe, and R. Rothstein. 1989. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58409-419. [DOI] [PubMed] [Google Scholar]
- 33.Wysocki, R., C. C. Chery, D. Wawrzycka, M. Van Hulle, R. Cornelis, J. M. Thevelein, and M. J. Tamas. 2001. The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Mol. Microbiol. 401391-1401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.