Abstract
Estrogen receptor α (ERα) is a ligand-regulated transcription factor with a broad range of physiological functions and one of the most important classifiers in breast cancer. MicroRNAs (miRNAs) are small noncoding RNAs that have emerged as important regulators of gene expression in a plethora of physiological and pathological processes. Upon binding the 3′ untranslated region (UTR) of target mRNAs, miRNAs typically reduce their stability and/or translation. The ERα mRNA has a long 3′ UTR of about 4.3 kb which has been reported to reduce mRNA stability and which bears evolutionarily conserved miRNA target sites, suggesting that it might be regulated by miRNAs. We have performed a comprehensive and systematic assessment of the regulatory role of all miRNAs that are predicted to target the 3′ UTR of the ERα mRNA. We found that miR-22 represses ERα expression most strongly and by directly targeting the ERα mRNA 3′ UTR. Of the three predicted miR-22 target sites in the 3′ UTR, the evolutionarily conserved one is the primary target. miR-22 overexpression leads to a reduction of ERα levels, at least in part by inducing mRNA degradation, and compromises estrogen signaling, as exemplified by its inhibitory impact on the ERα-dependent proliferation of breast cancer cells.
The steroid hormone 17β-estradiol (E2) regulates a number of developmental and physiological processes, such as growth and differentiation, in a range of tissues, including the male and female reproductive tracts, breast epithelium, and a plethora of other organs (9, 23). These physiological effects are mediated, at least in part, by two nuclear receptors, estrogen receptor α (ERα) and ERβ. Upon binding E2, they are activated as transcription factors and regulate target genes by binding directly to specific regulatory sequences or indirectly to other DNA-bound transcription factors (23). In addition, rapid nongenomic effects of E2 can be elicited by the same ERs as membrane-associated receptors (22). ERα is the more widely expressed of the two ER isoforms, and its levels are regulated by multiple mechanisms in a development- and tissue-specific manner (23). ERα levels are regulated at all levels of gene expression, beginning with transcriptional control by several transcription factors (48) and continuing with regulation at the posttranscriptional (26, 27) and posttranslational levels (2), including the control of protein turnover (44).
Breast cancer is the most frequent form of cancer in women, and ERα is still the most important classifier of breast tumors. ERα-positive tumors, which represent nearly 70% of all breast tumors, respond to E2 for growth and survival, and consequently, ERα-positive tumors can be inhibited with antiestrogens such as tamoxifen. Why and how most tumors eventually become resistant to antiestrogen therapy and why some tumors do not express ERα from the outset remain hotly debated questions. Multiple mechanisms have been invoked to explain the loss of ERα from initially ERα-positive breast tumors. These include epigenetic modifications such as the hypermethylation of CpG islands in the 5′-regulatory regions of the ERα gene (19) and/or mutations in the open reading frame of the ERα gene (24).
MicroRNAs (miRNAs) are a new class of small noncoding RNAs, 20 to 25 nucleotides in length, that have been shown to regulate gene expression in many physiological and developmental pathways in a multitude of different organisms (5). According to the September 2008 release of miRBase (http://microrna.sanger.ac.uk), the number of human miRNAs is 695 (20), but several studies indicate that the number of human miRNA genes could be well over 1,000 (7, 8). Typically, miRNAs regulate gene expression by lowering protein levels by repressing target mRNA translation and/or by inducing the degradation of the mRNA (17). In metazoans, miRNAs target mRNAs by imperfect complementary base pairing (6, 17, 46), for which the most important requirement is a continuous and perfect base pairing of miRNA nucleotides 2 to 8, known as the seed region, to the 3′ untranslated region (UTR) of the target mRNA (21, 38). A few exceptions have been reported in which the miRNA associates with the coding region and/or the 5′ UTR of the target mRNA (47). There are different computational approaches to predict the targets of miRNAs, and nearly all of these programs predict that a large number of human genes, possibly one-third of all genes, are targets of miRNAs. These predicted target genes represent a broad diversity of molecular functions and biological processes, including development, differentiation, apoptosis, and proliferation (5, 6, 21, 38). Furthermore, a large number of studies have documented the nearly ubiquitous deregulation of miRNA expression in cancer cells (12, 28, 29, 58). These changes in miRNA expression are highly informative for the classification and prognosis of cancer (12). In addition, altered expression of specific miRNAs has been shown to contribute to tumorigenesis and miRNAs have been shown to function as both tumor suppressors and oncogenes (12).
Despite a growing number of studies documenting the miRNA profiles of breast tumors (3, 12, 28, 29, 51, 57), there have been few attempts, and no comprehensive one, to find a mechanistic link between the deregulation of miRNA expression and estrogen signaling through ERα. The ERα mRNA has a long 3′ UTR of about 4.3 kb which has been shown to be involved in regulating mRNA levels of ERα. For example, E2 downregulates ERα levels in human MCF7 breast cancer cells by destabilizing the mRNA through sequences present in the 3′ UTR (49) whereas E2 upregulates ERα levels by stabilizing the ERα mRNA in sheep endometrium via the 3′ UTR (27). Another study supported a role for the 3′ UTR in destabilizing the ERα mRNA. With reporter gene assays, it was found that a 1-kb fragment of the 3′ UTR retained the destabilizing activity (32). However, the mechanism through which this fragment or the full-length 3′ UTR of the ERα mRNA regulates expression has remained elusive. The large size of the ERα 3′ UTR makes it likely that it is a target of miRNAs, and indeed, there are recent reports suggesting that miR-206, miR-221, miR-222, and miR-18a repress ERα expression by targeting the ERα 3′ UTR (1, 40, 59). Here, we report a comprehensive and systematic survey of the miRNAs predicted to target the 3′ UTR of the ERα mRNA by using two different screening approaches. We found that miR-22 is the miRNA that represses ERα expression most significantly and directly through the 3′ UTR and that this leads to a reduction in estrogen signaling.
MATERIALS AND METHODS
Cell culture.
HEK 293T and HepG2 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) without phenol red and supplemented with 10% fetal calf serum (FCS). MCF7-SH cells, a variant of MCF7 cells which are estrogen independent but ERα dependent (31), were maintained in DMEM without phenol red and supplemented with 10% charcoal-treated FCS.
Plasmids.
miRNAs were expressed as short hairpin RNAs (shRNAs) with the vector pRetroSuper (10). Control shRNAs against green fluorescent protein, luciferase, and ERα were expressed in a similar fashion. They contain, respectively, the following sequences that are complementary to the targets: 5′-CGGCAAGCTGACCCTGAAGTTC-3′ (15), 5′-CGTACGCGGAATACTTCGA-3′ (35), and 5′-TCAAGGACATAACGACTAT-3′ (41). To generate the miRNA/shRNA constructs, two complementary oligonucleotides (see Table S1A in the supplemental material) with ends compatible with BglII and XhoI restrictions sites were annealed, phosphorylated, and ligated into pRetroSuper. To construct the luciferase reporter plasmids, the full-length ERα 3′ UTR and various subfragments thereof were amplified from plasmid pfGH-UTR (32) with appropriate primers (see Table S1B in the supplemental material), generating SpeI restriction sites at both ends, and inserted into plasmid pGL3-basic (Promega) at the XbaI site downstream of the luciferase coding region. To introduce the two point mutations in the seed region of the full-length 3′ UTR and of subfragment UTR2 of the ERα gene, the QuikChange site-directed mutagenesis method was used (for the oligonucleotides used, see Table S1B in the supplemental material).
Reporter assays.
For the luciferase reporter assays, 293T, HepG2, and MCF7-SH cells were seeded into 24-well plates. 293T cells were transfected with 2 ng of cytomegalovirus (CMV)-Renilla (Promega), 10 ng of pGL3.3′UTR constructs, and 500 ng of miRNA/shRNA constructs by the calcium phosphate coprecipitation method. HepG2 cells were transfected with 2 ng of CMV-Renilla, 10 ng of pGL3.3′UTR constructs, and 20 nM of anti-miR-22 and control oligonucleotides (AM10203 and AM17010, respectively; Ambion) by using Fugene HD (Roche) in accordance with the manufacturer's instructions. 293T and HepG2 cells were harvested at 40 h posttransfection. MCF7-SH cells were transfected with 2 ng of CMV-Renilla, 200 ng of the ER reporter XETL (11), and 200 ng of miRNA/shRNA constructs by using Fugene HD. At 12 to 16 h posttransfection, the cells were induced with 100 nM 17-β-estradiol for 24 h before harvesting for luciferase assays. For all cells, luciferase activity was measured with the Dual-Luciferase reporter system (Promega). Firefly luciferase activities were standardized to the Renilla luciferase activities as an internal standard of transfection efficiency. All experiments were carried out with triplicate samples, and the data shown are averages of those triplicate samples.
Production of retroviruses and cells stably expressing miRNA/shRNAs.
Retroviruses expressing miRNAs/shRNAs were made as described previously (45). Briefly, 293T were cotransfected with 20 μg pSuperRetro miRNA/shRNA constructs, 5 μg VSV-G envelope plasmid pMD2.G, and 10 μg packaging plasmid pCMV.Gag-Pol (vectors were generously provided by Patrick Salmon, Geneva, Switzerland) in 10-cm plates by the calcium phosphate coprecipitation method. At 16 h posttransfection, the transfected cells were washed with Tris-buffered saline (TBS) and further incubated in regular growth medium. At 24 h later, the medium was collected and used to infect cells. To generate cells stably expressing miRNA/shRNAs, MCF7-SH cells were transduced twice overnight with appropriate retroviral supernatants, washed three times with TBS, and then subjected to selection with 3 μg/ml puromycin for at least 24 h to eliminate the noninfected cells. To minimize clone-to-clone variations, pools of very large numbers of resistant clones were collected.
Immunoblotting.
Cells were collected in ice-cold TBS and resuspended in 20 mM Tris-HCl (pH 8)-100 mM NaCl-10% glycerol-0.1% NP-40-1 mM monovanadate-1 mM dithiothreitol-1× protease inhibitors. DNA was sheared by seven or eight passages through a 25-gauge needle. The lysates were cleared by centrifugation at 10,000 rpm for 10 min, and protein concentrations were determined by the Bradford assay (Bio-Rad). Extracts (20 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, immunoblotted with primary antibodies, incubated with secondary antibodies, and revealed by chemiluminescence. For ERα, monoclonal antibody ER17 was used at a 1:5,000 dilution, and for glyceraldehyde-3-phosphate dehydrogenase (GAPDH), antibody ab8245 (Abcam) was used at a 1:10,000 dilution. Immunoblots were imaged and quantified with the GeneGnome (Syngene). The intensity of ERα bands was standardized to the intensity of the GAPDH bands.
Proliferation assays.
Twenty thousand MCF7-SH cells stably expressing miRNA/shRNAs were seeded into 12-well plates. On the following day (day 0), the cells were washed with warm TBS and further incubated with DMEM supplemented with 1% charcoal-treated FCS. The medium was changed every third day. The cells were counted on day 6, and the cell counts were normalized to MCF7-SH cells expressing a control shRNA. The experiments were carried out in triplicate with one set of pools of stable transformants.
Quantitative PCR (Q-PCR).
Endogenous ERα mRNA and exogenously expressed luciferase mRNA were quantitated by real-time PCR after reverse transcription (for the primers used, see Table S1C in the supplemental material). To assess ERα mRNA levels in MCF7-SH cells stably expressing miR-22, an shRNA directed against the ERα mRNA or a control shRNA, triplicate samples were processed, and since the mRNAs of the housekeeping genes for EEF1A1 and TFRC were found to vary the least across all of the samples, all measurements were normalized to the geometric mean of these two control genes. To quantitate exogenously expressed luciferase mRNA levels, 293T cells were cotransfected in six-well plates in triplicate with 60 ng of the luciferase reporter pGL3.3′UTR (full-length ERα 3′ UTR) and 3 μg of the miRNA/shRNA expression constructs per well by the calcium phosphate coprecipitation method. RNA was extracted at 24 h posttransfection and treated with DNase before Q-PCR analysis. Luciferase mRNA levels were normalized to the mRNA of the puromycin resistance gene carried by the cotransfected miRNA/shRNA constructs.
RESULTS
miRNAs targeting the 3′ UTR of the ERα mRNA.
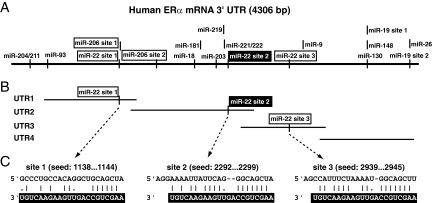
To identify the putative miRNAs targeting the 3′ UTR of the ERα mRNA, we used the three most widely used computational programs, TargetScan (37), PicTar (34), and miRANDA (30). Only the miRNAs found by all of the programs were considered for further analysis. One criterion to filter the miRNAs identified in silico is the evolutionary conservation of the predicted miRNA target(s) in the 3′ UTR (21, 38). Based on the bioinformatic prediction and conservation in mammals, we found that the 3′ UTR of the ERα mRNA was predicted to be targeted by 13 miRNAs, as depicted in Fig. 1A. While this work was in progress, another study reported that miR-206 targets the 3′ UTR of the ERα mRNA (1). Although we had initially excluded it from our list of miRNAs because its target sites are not conserved, we decided to investigate it in parallel in some of the key experiments.
FIG. 1.
Schematic representation of the ERα 3′ UTR and the conserved miRNA target sites. (A) Scheme showing all of the predicted and conserved miRNA target sites of the full-length 3′ UTR. For miR-22 and miR-206, all of the predicted sites are shown. The conserved miR-22 target site is highlighted with white letters on a black background. (B) Scheme of 3′ UTR subfragments that were tested experimentally. miR-22 target sites are indicated. (C) Base pairing of the miR-22 target sites with miR-22.
miR-22 targets the 3′ UTR of the ERα mRNA.
In order to find out which miRNAs target/repress ERα expression, two different screens based on luciferase reporter assays were performed. First, we performed an assay that addresses the ability of a miRNA to inhibit target mRNA expression specifically and most likely by direct interaction. To this end, we cloned the entire ERα 3′ UTR downstream of the open reading frame of the firefly luciferase and cotransfected this reporter with a panel of miRNA expression constructs. We exploited the same type of luciferase reporters to map the 3′ UTR subfragments that are targeted by a miRNA that scored positive with the full-length 3′ UTR. Second, we assessed the ability of miRNAs to downregulate endogenous ERα expression and thereby to affect indirectly the activity of an ERα target gene. We cotransfected the ER luciferase reporter with the miRNA expression constructs into MCF7-SH cells, which express endogenous ERα, and measured reporter activity after a 24-h induction with E2. In both screens, we focused primarily on the miRNAs that had been predicted in silico to target the 3′ UTR of ERα at conserved target sites; however, we also tested a few additional miRNAs whose target sites are not conserved. For example, according to the TargetScan prediction, miR-129-5p, miR-30-3p, and miR-17-5p have three or more putative target sites in the 3′ UTR of ERα, but none of these sites are conserved. We did not observe any downregulation of the luciferase reporter activity with these miRNAs (data not shown), emphasizing further the importance of the conservation of target sites. While the reduction in the luciferase reporter activity by overexpression of those miRNAs which we considered to be positive may appear relatively modest, these findings are in agreement with recent reports indicating that a miRNA may target many genes but with relatively modest impact on protein output (4, 50).
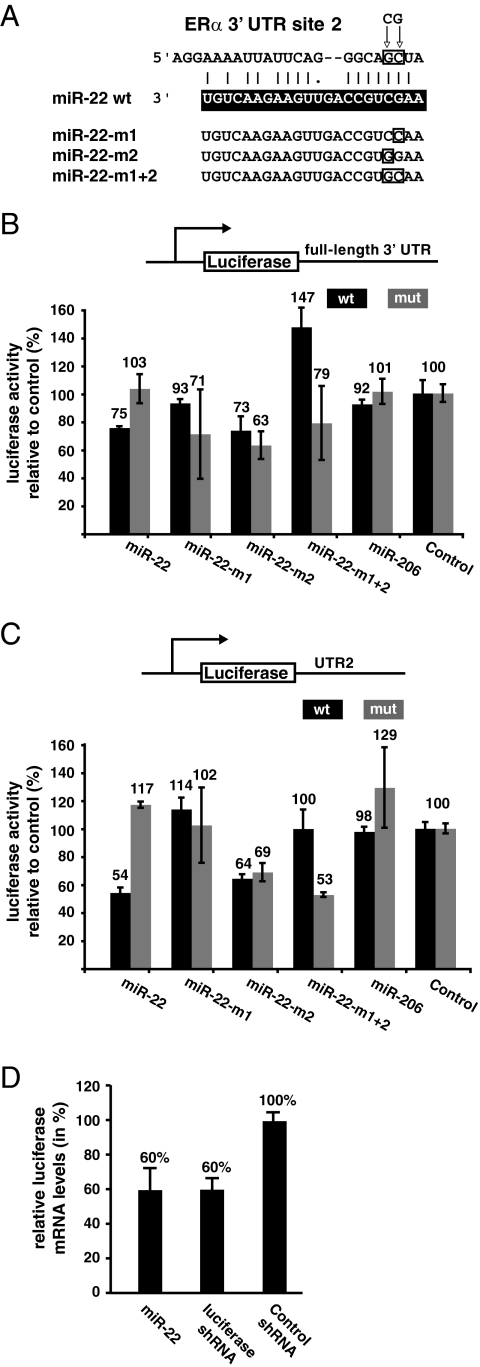
With both screens, we found that miR-22 downregulates the reporter activities most strongly (see Fig. S1 in the supplemental material). The 3′ UTR of the ERα mRNA contains three putative target sites for miR-22 (sites 1, 2, and 3), one conserved and two nonconserved ones (Fig. 1). In order to find out which one of them is functional, the 3′ UTR of the ERα mRNA was divided into four different subfragments of nearly equal lengths, designated UTR1, UTR2, UTR3, and UTR4 (Fig. 1B), which were cloned into the luciferase reporter. Note that the intrinsic activities of these reporters vary widely (see Fig. S2A in the supplemental material) and cannot be directly compared, considering the large scale differences of the 3′ UTRs of the chimeric mRNAs produced from them. UTR1, UTR2, and UTR3 each have one miR-22 target site. Of these, only UTR2 has a conserved target site for miR-22, whereas UTR1 and UTR3 have nonconserved target sites. Upon cotransfection of these reporter constructs with the expression constructs for wild-type miR-22, we found that miR-22 targets the 3′ UTR of the ERα mRNA primarily through UTR2, the second subfragment of the 3′ UTR (see Fig. S2 in the supplemental material; Fig. 2). The seed region of the miRNA, which typically base pairs perfectly with the target mRNA and involves 5′-most nucleotides 2 to 8 of the miRNA, is the most important determinant of miRNA targeting (38). Based on the results obtained with the 3′ UTR subfragments, we focused on putative target site 2 contained in UTR2. We generated three constructs to express different seed mutant forms of miR-22 and generated target site 2 mutant forms in the luciferase reporters with the full-length 3′ UTR or with only UTR2. The site 2 mutants should no longer be targeted by wild-type miR-22 but may still be inhibited by the complementary miR-22 mutant (Fig. 2). We found that miR-22 seed mutant form m1 was severely defective in inhibiting the expression of the luciferase reporter genes. Mutant form m2 remained largely competent for repression. Importantly, miR-22-mediated repression of the ERα 3′ UTR was abolished when the mRNA target site (site 2) was mutated in the context of either the full-length 3′ UTR (Fig. 2B) or subfragment UTR2 (Fig. 2C). Repression of the mutant targets was restored with the miR-22 mutant forms that contained complementary changes (Fig. 2). This time, the nucleotide changed in m2 seemed to be more important. It is worth noting that the pattern of repression by wild-type miR-22 and the rescue of repression by the mutant miR-22 variants are very similar for the full-length 3′ UTR and UTR2. Taken together, these results emphasize the specificity of the targeting of the ERα 3′ UTR by miR-22. Interestingly, we did not observe any effects of miR-206 in the context of either the full-length 3′ UTR of the ERα mRNA or subfragments UTR1 and UTR2 in any of our assays (see Fig. S1 in the supplemental material; Fig. 2B and C).
FIG. 2.
miR-22 targets the 3′ UTR of the ERα mRNA. (A) Detailed view of the conserved target site of miR-22. The target site mutations are boxed and indicated by arrows above the sequence. Details of the mutant miR-22 variants are given below the wild-type sequence, which is in white letters on a black background. (B) miR-22 represses the luciferase activity of the wild-type full-length 3′ UTR reporter (wt) but not that of the corresponding reporter with a target site mutation (mut). The graph shows the results obtained with 293T cells cotransfected with the indicated reporter plasmids and miRNA constructs. (C) Subfragment 2 (UTR2) of the 3′ UTR of ERα contains the conserved target site of miR-22 and mediates miR-22 repression of the luciferase reporter. The graph shows the same type of experiment as in panel B. Data points are averages of triplicate samples, which were standardized to the values obtained with the control shRNA construct, which were set to 100%. Error bars represent standard deviations. (D) Q-PCR analysis of luciferase mRNA levels of the full-length 3′ UTR reporter gene in 293T cells cotransfected with the indicated miRNA/shRNA constructs. Shown are averages of triplicate samples with standard deviations.
To address the mechanism of repression, we quantitated the amount of luciferase mRNA of the 3′ UTR reporter constructs cotransfected with or without miR-22 by real-time PCR (Fig. 2D). The extent of reduction was comparable to that seen at the level of luciferase enzyme activity and to that obtained with an shRNA targeted against the luciferase mRNA. This result suggests that miR-22 represses through the ERα 3′ UTR primarily by inducing the degradation of the target mRNA.
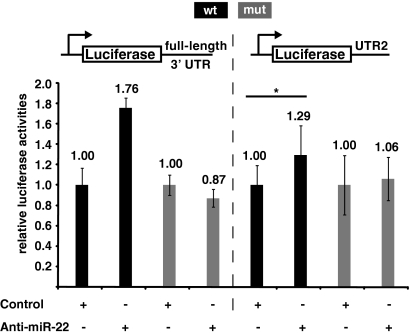
Inhibition of miR-22 derepresses the 3′ UTR of the ERα mRNA.
Our experiments up to this point involved the overexpression of wild-type and mutant miRNAs. Next, we investigated the effect of inhibiting endogenous miR-22 on ERα 3′ UTR function by using the same luciferase reporter system. Since 293T and MCF7 cells, as well as MCF7 derivative MCF7-SH cells, do not express miR-22, we chose human HepG2 hepatoma cells, which do express miR-22 (36, 39; data not shown). We cotransfected HepG2 cells with an anti-miR-22 oligonucleotide and the 3′ UTR luciferase reporters. As expected, the anti-miR-22 oligonucleotide derepressed the expression of the luciferase reporters containing the wild-type full-length 3′ UTR or the UTR2 subfragment, while there were no significant changes with the reporters containing the full-length 3′ UTR or UTR2 with mutations in the miR-22 target sequence (site 2) (Fig. 3).
FIG. 3.
Inhibition of miR-22 derepresses the ERα 3′ UTR. The anti-miR-22 oligonucleotide or a control oligonucleotide was cotransfected with the indicated reporters into HepG2 hepatoma cells. The hashed line through the middle of the graph separates the data for the two reporters, and the data for each pair of samples (a given reporter without or with the anti-miR-22 oligonucleotide) were standardized to the control sample, which were arbitrarily set to 1.0. In the case of the UTR2 reporter, the luciferase activities obtained in the transfections with the anti-miR-22 and control oligonucleotides are significantly different (P < 0.05), as symbolized by the asterisk. Note that there is no significant difference between the luciferase activities represented by the two rightmost bars. wt, wild type; mut, mutant.
miR-22 represses estrogen signaling.
As mentioned above, our second approach consisted of determining whether the downregulation of endogenous ERα levels by miR-22 has any effect on estrogen signaling mediated by ERα. MCF7-SH cells were cotransfected with an ER-reporter construct and constructs expressing wild-type miR-22 or mutant miR-22 variants. Wild-type miR-22 significantly downregulated the expression of the luciferase reporter, whereas the repression was diminished by the mutant miR-22 variants (Fig. 4). Even though the reporter gene activity is not fully restored in the presence of the mutant miR-22 variants, this pattern of repression was similar to the pattern of repression of the full-length 3′ UTR and UTR2 luciferase reporters observed in 293T cells.
FIG. 4.
miR-22 represses estrogen signaling by ERα. Constructs expressing wild-type and mutant versions of miR-22 were cotransfected with the ERα reporter into MCF7-SH cells to assay endogenous ERα activity. The results obtained with an shRNA control were arbitrarily set to 100%.
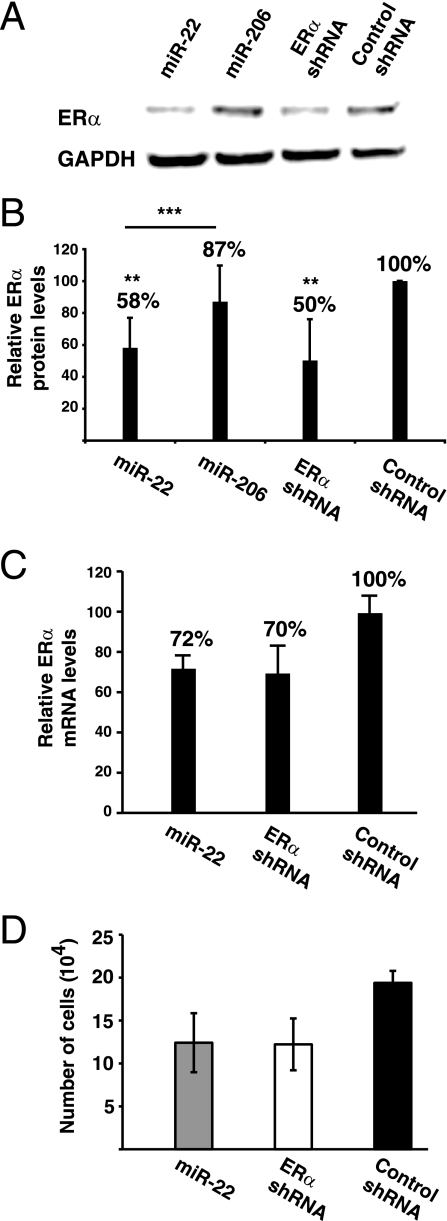
miR-22 downregulates endogenous ERα protein levels and inhibits the proliferation of MCF7-SH cells.
A clear prediction from our experiments is that endogenous ERα protein levels should be regulated by miR-22. In turn, this should affect not only the activity of a transfected reporter gene but also a physiological ERα-dependent function. In order to investigate the effect of miR-22 overexpression on endogenous ERα levels, MCF7-SH cells were transduced with retroviruses expressing miR-22, miR-206, an ERα-specific shRNA, and a control shRNA. As the immunoblot assay in Fig. 5 shows, miR-22 overexpression downregulates ERα protein levels to nearly half of those of cells expressing the control shRNA. This is comparable to the downregulation obtained with the specific shRNA that targeted ERα. Again, no significant changes in ERα levels were observed following miR-206 overexpression (Fig. 5). Moreover, the other miRNAs which repressed the luciferase reporter activities less than miR-22 (see Fig. S1 in the supplemental material), notably, miR-19a and miR-181, did not affect ERα levels (data not shown). As previously observed for the exogenously expressed luciferase reporter mRNA, endogenous ERα mRNA levels were significantly reduced in MCF7-SH cells stably expressing either miR-22 or a specific shRNA (Fig. 5C). Clearly, miR-22-induced degradation of the ERα mRNA accounts for much of the repression, but we cannot exclude the possibility that translational repression contributes to the overall repression of ERα expression.
FIG. 5.
Overexpression of miR-22 reduces ERα protein levels and inhibits the proliferation of ERα-dependent MCF7-SH cells. (A) Representative immunoblot analysis of ERα for MCF7-SH transformants stably expressing the indicated miRNA/shRNA. The GAPDH immunoblot is shown as a loading control. (B) Quantitation of ERα protein levels. The data shown represent five independent experiments (that is, five independent pools of stable transformants each). Averages, standardized to GAPDH and the control shRNA samples (set to 100%), are shown with error bars indicating standard deviations. Double asterisks indicate values that are significantly different from those of the control shRNA samples (P < 0.01), while triple asterisks indicate a significant difference between the values obtained with the miR-22- and miR-206-expressing cell lines (P < 0.001). (C) Quantitation of ERα mRNA levels in corresponding stable transformants. Shown are averages of triplicate samples with standard deviations. (D) Proliferation assay of MCF7-SH cells stably expressing miR-22, ERα shRNA, and the control shRNA. Shown are averages of triplicate samples with standard deviations.
Since MCF7-SH cells are dependent on ERα for proliferation (31), we determined whether reducing the protein levels of ERα by miR-22 affects their proliferation. We did proliferation assays of MCF7-SH cells infected with retroviruses expressing miR-22, the ERα-specific shRNA, and a control shRNA and found that expression of miR-22 impairs the proliferation of MCF7-SH cells to an extent similar to that of the shRNA directed at the ERα mRNA (Fig. 5D). Thus, the effects of miR-22 expression on proliferation correlate with the reduced levels of ERα in miR-22-expressing MCF7-SH cells (Fig. 5A and B).
DISCUSSION
We found that miR-22 represses ERα expression by directly targeting the 3′ UTR of the ERα mRNA. To identify miR-22 as a regulator of ERα, we have used two complementary approaches based on luciferase reporters to systematically screen all of the miRNAs that might target the 3′ UTR of the ERα mRNA. The first approach explored the ability of a miRNA to inhibit mRNA expression by directly targeting the 3′ UTR of the reporter mRNA. In the second approach, we measured the activity of an ER-reporter construct, which reflects the ability of the miRNAs to suppress estrogen signaling by reducing ERα levels. Both approaches identified miR-22 as the strongest repressor (see Fig. S1 in the supplemental material). We confirmed the specificity of the miR-22-mediated repression by mutational analyses and observed that inhibition of miR-22 action derepresses the luciferase reporter with a wild-type ERα 3′ UTR but not reporters with mutated target sites. Furthermore, we observed that miR-22 overexpression reduces endogenous ERα protein (and mRNA) levels in MCF7-SH cells, resulting in impaired ERα-dependent proliferation (Fig. 5).
miR-22 has three putative target sites in the ERα 3′ UTR, only one of which is conserved among mammals. Although seed pairing involving nucleotides 2 to 8 of the 5′ end of the miRNA is the most important determinant to predict miRNA targets, this information is not sufficient to predict the miRNA targets with high confidence. Therefore, nearly all of the bioinformatic resources used to predict miRNA targets extensively use the conservation of targets among all mammalian species as a criterion. Perhaps not surprisingly, we found that miR-22 targets the ERα 3′ UTR through the single conserved site in subfragment UTR2 (Fig. 1, 2, and 4). An earlier study, performed before miRNAs were recognized as important regulators, reported that nearly the same 1-kb region as UTR2 retains the ability of the full-length 3′ UTR of ERα to destabilize a reporter mRNA (32). However, at the time, the factors mediating this inhibitory effect could not be identified. Our data provide an explanation of the role of UTR2 in conferring mRNA instability by the 3′ UTR of ERα and argue that inhibition is mediated, at least in part, by miR-22.
Recently, several miRNA profiling studies have been performed with breast tumors and/or breast cancer cell lines to understand the role of deregulation of miRNA expression in the development and regulation of breast cancer (3, 12, 28, 29, 51). Some reports have sought to correlate miRNAs levels with ERα status. miR-206, miR-221, and miR-222 appeared to be upregulated in ERα-negative breast tumors and cell lines (1, 33, 59) and repress ERα expression (1, 59). In contrast to these publications, we were not able to observe the repression of any of the 3′ UTR reporters by miR-206, nor did miR-206 overexpression significantly downregulate the protein levels of endogenous ERα in MCF7-SH cells. Furthermore, Adams et al. (1) reported that the inhibition of miR-206 upregulates ERα protein levels in a nearly linear and dose-dependent manner in MCF7 cells, which is somewhat puzzling since MCF7 cells, like the MCF7-SH cells derived from them (data not shown), do not even express miR-206 (36). A very recently published analysis of the relationship between miRNA expression profiles and ERα levels in hepatocellular carcinoma cells also failed to find a correlation with miR-206 levels (40). However, the same study did highlight miR-18a, which had also scored positive in our survey with the luciferase reporter gene (see Fig. S1A in the supplemental material) as a regulator of ERα levels both in hepatocellular carcinoma cells and in MCF7 cells. It will be interesting to determine the combinatorial effects of miR-18a and miR-22, whose target sites are in the same region of the ERα 3′ UTR (Fig. 1A).
Two very recent reports have suggested that the two closely related miRNAs miR-221 and miR-222 might be involved in conferring tamoxifen resistance on breast cancer cells (43, 59). miR-221/222, which is upregulated in tamoxifen-resistant MCF7 cells compared to tamoxifen-sensitive ones, confers tamoxifen resistance by targeting the cell cycle inhibitor p27/Kip1 (43). miR-221/222 levels are also elevated in ERα-negative breast cancer cell lines (59). Furthermore, the same report indicated that miR-221/222 might confer tamoxifen resistance by directly targeting the ERα mRNA (59). In contrast, we have observed that the miRNAs miR-219 and miR-221/222 upregulate both the ERα 3′ UTR reporter and the standard ERα-reporter (see Fig. S1 in the supplemental material). Several recent studies have shown that miRNAs can upregulate gene expression by targeting the promoter (47) or the 3′ UTR of the target gene (55, 56). Therefore, it is conceivable that miRNA-219 and/or miR-221/222 upregulate ERα expression by targeting the ERα 3′ UTR. Our ongoing investigations are aimed at clarifying the mechanism of these effects and its relevance to the aforementioned correlations between miR-221/222 expression profiles and estrogen signaling. We are at a loss to explain why we did not see repression of ERα expression by miR-206 and miR-221/222. Although we have used slightly different assays and cell lines, it is difficult to explain these apparent discrepancies. An attractive speculation is that subtle cell-intrinsic or -extrinsic parameters not only affect the overall miRNA complement but also the regulatory impact of individual miRNAs.
There is little information about other genes targeted by miR-22, and only very recently have some miRNA profiling studies shed light on the probable functions of miR-22. Whether miR-22 regulates the expression of other nuclear receptors remains to be seen, but it is unlikely that it affects the expression of the other ER isoform, ERβ, since the comparatively short ERβ 3′ UTR does not contain a recognizable miR-22 target site. miR-22 is part of a set of miRNAs which are significantly more highly expressed in ErbB2-negative breast tumors than in ErbB2-positive ones, in ERα-positive tumors than in ERα-negative ones, and in tumors positive for progesterone receptor than in progesterone receptor-negative ones (42). Yet other studies involving miRNA expression profiling with similar samples did not notice anything with regard to miR-22 (51). Another miRNA profiling study has reported that curcumin, a naturally occurring flavonoid and proapoptotic compound derived from the rhizome of Curcuma longa, upregulates miR-22 expression in human pancreatic cells (54). The same authors predicted the ERα mRNA as a target and demonstrated that the transfection of miR-22 mimetics into BxBC-3 pancreatic cancer cells repressed ERα expression. However, the molecular mechanism of this effect was not further characterized and indirect effects could not be ruled out.
There is additional circumstantial evidence for a yin-yang relationship between miR-22 and ERα that fits all of these and our own observations. Of six breast cancer cell lines whose miRNA profiles were established as part of a larger effort to characterize the miRNA profiles of the NCI-60 panel of human tumor-derived cell lines, the only two ERα-positive ones had the lowest levels of miR-22 (18). miR-22 was found to be overexpressed in mouse mammary progenitor cells (25), which may not express ERα (52), and the tissues, such as uterus and ovary, which in the mouse express the highest levels of ERα mRNA (9) have the lowest levels of miR-22 expression (36, 39) (see Fig. S3 in the supplemental material). All of this fits well with the hypothesis that genes which are the target of a particular miRNA have evolved to avoid being expressed at the same time and in the same place as the miRNA (16, 53). The oncogenic transcription factor c-Myc directly represses miR-22 expression in both human and mouse lymphomas (14). Since the c-myc gene is induced by ERα, ERα, c-Myc, and miR-22 might be part of a regulatory feedback loop. The fact that there is an ERα binding site 25 kb upstream of the miR-22 transcription start site (13) suggests additional regulatory interactions, but it will require further studies to establish whether miR-22 is an estrogen-responsive gene.
In summary, we have shown that miR-22 targets ERα and represses estrogen signaling. Additional miRNAs may contribute individually (see Fig. S1 in the supplemental material) or in combination to the fine-tuning of ERα levels and allow continuous adaptation to a variety of signals. The precise combination of miRNAs that intervene to set ERα levels is likely to depend on the particular cell type and conditions.
Supplementary Material
Acknowledgments
We are grateful to F. Gannon and D. Trono for gifts of reagents. We thank Peter Dudek, Julian Pakay, Asvin Lakkaraju, and Jurgi Camblong for critical reading of the manuscript. We are indebted to the team of the genomics platform of the NCCR Frontiers-in-Genetics at the University of Geneva for help with miRNA expression profiling and Q-PCR.
Funding was provided by the Fondation Medic, the Swiss National Science Foundation, and the Canton de Genève.
Footnotes
Published ahead of print on 4 May 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, B. D., H. Furneaux, and B. A. White. 2007. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-α (ERα) and represses ERα messenger RNA and protein expression in breast cancer cell lines. Mol. Endocrinol. 211132-1147. [DOI] [PubMed] [Google Scholar]
- 2.Ali, S., and R. C. Coombes. 2002. Endocrine-responsive breast cancer and strategies for combating resistance. Nat. Rev. Cancer 2101-112. [DOI] [PubMed] [Google Scholar]
- 3.Ambs, S., R. L. Prueitt, M. Yi, R. S. Hudson, T. M. Howe, F. Petrocca, T. A. Wallace, C. G. Liu, S. Volinia, G. A. Calin, H. G. Yfantis, R. M. Stephens, and C. M. Croce. 2008. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 686162-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baek, D., J. Villen, C. Shin, F. D. Camargo, S. P. Gygi, and D. P. Bartel. 2008. The impact of microRNAs on protein output. Nature 45564-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. [DOI] [PubMed] [Google Scholar]
- 6.Bartel, D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136215-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentwich, I., A. Avniel, Y. Karov, R. Aharonov, S. Gilad, O. Barad, A. Barzilai, P. Einat, U. Einav, E. Meiri, E. Sharon, Y. Spector, and Z. Bentwich. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37766-770. [DOI] [PubMed] [Google Scholar]
- 8.Berezikov, E., V. Guryev, J. van de Belt, E. Wienholds, R. H. Plasterk, and E. Cuppen. 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 12021-24. [DOI] [PubMed] [Google Scholar]
- 9.Bookout, A. L., Y. Jeong, M. Downes, R. T. Yu, R. M. Evans, and D. J. Mangelsdorf. 2006. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296550-553. [DOI] [PubMed] [Google Scholar]
- 11.Bunone, G., P.-A. Briand, R. J. Miksicek, and D. Picard. 1996. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 152174-2183. [PMC free article] [PubMed] [Google Scholar]
- 12.Calin, G. A., and C. M. Croce. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6857-866. [DOI] [PubMed] [Google Scholar]
- 13.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 381289-1297. [DOI] [PubMed] [Google Scholar]
- 14.Chang, T. C., D. Yu, Y. S. Lee, E. A. Wentzel, D. E. Arking, K. M. West, C. V. Dang, A. Thomas-Tikhonenko, and J. T. Mendell. 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 4043-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donzé, O., and D. Picard. 2002. RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res. 30e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farh, K. K., A. Grimson, C. Jan, B. P. Lewis, W. K. Johnston, L. P. Lim, C. B. Burge, and D. P. Bartel. 2005. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 3101817-1821. [DOI] [PubMed] [Google Scholar]
- 17.Filipowicz, W., S. N. Bhattacharyya, and N. Sonenberg. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9102-114. [DOI] [PubMed] [Google Scholar]
- 18.Gaur, A., D. A. Jewell, Y. Liang, D. Ridzon, J. H. Moore, C. Chen, V. R. Ambros, and M. A. Israel. 2007. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 672456-2468. [DOI] [PubMed] [Google Scholar]
- 19.Giacinti, L., P. P. Claudio, M. Lopez, and A. Giordano. 2006. Epigenetic information and estrogen receptor alpha expression in breast cancer. Oncologist 111-8. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths-Jones, S., H. K. Saini, S. van Dongen, and A. J. Enright. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36D154-D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimson, A., K. K. Farh, W. K. Johnston, P. Garrett-Engele, L. P. Lim, and D. P. Bartel. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 2791-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammes, S. R., and E. R. Levin. 2007. Extranuclear steroid receptors: nature and actions. Endocr. Rev. 28726-741. [DOI] [PubMed] [Google Scholar]
- 23.Heldring, N., A. Pike, S. Andersson, J. Matthews, G. Cheng, J. Hartman, M. Tujague, A. Strom, E. Treuter, M. Warner, and J.-Å. Gustafsson. 2007. Estrogen receptors: how do they signal and what are their targets. Physiol. Rev. 87905-931. [DOI] [PubMed] [Google Scholar]
- 24.Herynk, M. H., and S. A. Fuqua. 2007. Estrogen receptors in resistance to hormone therapy. Adv. Exp. Med. Biol. 608130-143. [DOI] [PubMed] [Google Scholar]
- 25.Ibarra, I., Y. Erlich, S. K. Muthuswamy, R. Sachidanandam, and G. J. Hannon. 2007. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 213238-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ing, N. H. 2005. Steroid hormones regulate gene expression posttranscriptionally by altering the stabilities of messenger RNAs. Biol. Reprod. 721290-1296. [DOI] [PubMed] [Google Scholar]
- 27.Ing, N. H., D. A. Massuto, and L. A. Jaeger. 2008. Estradiol up-regulates AUF1p45 binding to stabilizing regions within the 3′-untranslated region of estrogen receptor α mRNA. J. Biol. Chem. 2831764-1772. [DOI] [PubMed] [Google Scholar]
- 28.Iorio, M. V., M. Ferracin, C. G. Liu, A. Veronese, R. Spizzo, S. Sabbioni, E. Magri, M. Pedriali, M. Fabbri, M. Campiglio, S. Menard, J. P. Palazzo, A. Rosenberg, P. Musiani, S. Volinia, I. Nenci, G. A. Calin, P. Querzoli, M. Negrini, and C. M. Croce. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 657065-7070. [DOI] [PubMed] [Google Scholar]
- 29.Iorio, M. V., R. Visone, G. Di Leva, V. Donati, F. Petrocca, P. Casalini, C. Taccioli, S. Volinia, C. G. Liu, H. Alder, G. A. Calin, S. Menard, and C. M. Croce. 2007. MicroRNA signatures in human ovarian cancer. Cancer Res. 678699-8707. [DOI] [PubMed] [Google Scholar]
- 30.John, B., A. J. Enright, A. Aravin, T. Tuschl, C. Sander, and D. S. Marks. 2004. Human MicroRNA targets. PLoS Biol. 2e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalkhoven, E., E. Beraldi, M. L. Panno, J. P. De Winter, J. H. Thijssen, and B. Van Der Burg. 1996. Growth inhibition by anti-estrogens and progestins in TGF-β-resistant and -sensitive breast-tumor cells. Int. J. Cancer 65682-687. [DOI] [PubMed] [Google Scholar]
- 32.Kenealy, M. R., G. Flouriot, V. Sonntag-Buck, T. Dandekar, H. Brand, and F. Gannon. 2000. The 3′-untranslated region of the human estrogen receptor α gene mediates rapid messenger ribonucleic acid turnover. Endocrinology 1412805-2813. [DOI] [PubMed] [Google Scholar]
- 33.Kondo, N., T. Toyama, H. Sugiura, Y. Fujii, and H. Yamashita. 2008. miR-206 expression is down-regulated in estrogen receptor α-positive human breast cancer. Cancer Res. 685004-5008. [DOI] [PubMed] [Google Scholar]
- 34.Krek, A., D. Grun, M. N. Poy, R. Wolf, L. Rosenberg, E. J. Epstein, P. MacMenamin, I. da Piedade, K. C. Gunsalus, M. Stoffel, and N. Rajewsky. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37495-500. [DOI] [PubMed] [Google Scholar]
- 35.Lakkaraju, A. K., P. P. Luyet, P. Parone, T. Falguieres, and K. Strub. 2007. Inefficient targeting to the endoplasmic reticulum by the signal recognition particle elicits selective defects in post-ER membrane trafficking. Exp. Cell Res. 313834-847. [DOI] [PubMed] [Google Scholar]
- 36.Landgraf, P., et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 1291401-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 12015-20. [DOI] [PubMed] [Google Scholar]
- 38.Lewis, B. P., I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel, and C. B. Burge. 2003. Prediction of mammalian microRNA targets. Cell 115787-798. [DOI] [PubMed] [Google Scholar]
- 39.Liang, Y., D. Ridzon, L. Wong, and C. Chen. 2007. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, W.-H., S.-H. Yeh, C.-C. Lu, S.-L. Yu, H.-Y. Chen, C.-Y. Lin, D.-S. Chen, and P.-J. Chen. 2009. microRNA-18a prevents estrogen receptor-α expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology 136683-693. [DOI] [PubMed] [Google Scholar]
- 41.Matthews, J., B. Wihlen, J. Thomsen, and J.-Å. Gustafsson. 2005. Aryl hydrocarbon receptor-mediated transcription: ligand-dependent recruitment of estrogen receptor α to 2,3,7,8-tetrachlorodibenzo-p-dioxin-responsive promoters. Mol. Cell. Biol. 255317-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattie, M. D., C. C. Benz, J. Bowers, K. Sensinger, L. Wong, G. K. Scott, V. Fedele, D. Ginzinger, R. Getts, and C. Haqq. 2006. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, T. E., K. Ghoshal, B. Ramaswamy, S. Roy, J. Datta, C. L. Shapiro, S. Jacob, and S. Majumder. 2008. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 28329897-29903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nawaz, Z., D. M. Lonard, A. P. Dennis, C. L. Smith, and B. W. O'Malley. 1999. Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl. Acad. Sci. USA 961858-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parone, P. A., D. I. James, S. Da Cruz, Y. Mattenberger, O. Donze, F. Barja, and J. C. Martinou. 2006. Inhibiting the mitochondrial fission machinery does not prevent Bax/Bak-dependent apoptosis. Mol. Cell. Biol. 267397-7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pillai, R. S., S. N. Bhattacharyya, and W. Filipowicz. 2007. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 17118-126. [DOI] [PubMed] [Google Scholar]
- 47.Place, R. F., L. C. Li, D. Pookot, E. J. Noonan, and R. Dahiya. 2008. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 1051608-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid, G., S. Denger, M. Kos, and F. Gannon. 2002. Human estrogen receptor-α: regulation by synthesis, modification and degradation. Cell. Mol. Life Sci. 59821-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saceda, M., M. E. Lippman, P. Chambon, R. L. Lindsey, M. Ponglikitmongkol, M. Puente, and M. B. Martin. 1988. Regulation of the estrogen receptor in MCF-7 cells by estradiol. Mol. Endocrinol. 21157-1162. [DOI] [PubMed] [Google Scholar]
- 50.Selbach, M., B. Schwanhausser, N. Thierfelder, Z. Fang, R. Khanin, and N. Rajewsky. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 45558-63. [DOI] [PubMed] [Google Scholar]
- 51.Sempere, L. F., M. Christensen, A. Silahtaroglu, M. Bak, C. V. Heath, G. Schwartz, W. Wells, S. Kauppinen, and C. N. Cole. 2007. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 6711612-11620. [DOI] [PubMed] [Google Scholar]
- 52.Sleeman, K. E., H. Kendrick, D. Robertson, C. M. Isacke, A. Ashworth, and M. J. Smalley. 2007. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J. Cell Biol. 17619-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stark, A., J. Brennecke, N. Bushati, R. B. Russell, and S. M. Cohen. 2005. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 1231133-1146. [DOI] [PubMed] [Google Scholar]
- 54.Sun, M., Z. Estrov, Y. Ji, K. R. Coombes, D. H. Harris, and R. Kurzrock. 2008. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol. Cancer Ther. 7464-473. [DOI] [PubMed] [Google Scholar]
- 55.Vasudevan, S., and J. A. Steitz. 2007. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 1281105-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasudevan, S., Y. Tong, and J. A. Steitz. 2007. Switching from repression to activation: microRNAs can up-regulate translation. Science 3181931-1934. [DOI] [PubMed] [Google Scholar]
- 57.Verghese, E. T., A. M. Hanby, V. Speirs, and T. A. Hughes. 2008. Small is beautiful: microRNAs and breast cancer—where are we now? J. Pathol. 215214-221. [DOI] [PubMed] [Google Scholar]
- 58.Yanaihara, N., N. Caplen, E. Bowman, M. Seike, K. Kumamoto, M. Yi, R. M. Stephens, A. Okamoto, J. Yokota, T. Tanaka, G. A. Calin, C. G. Liu, C. M. Croce, and C. C. Harris. 2006. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9189-198. [DOI] [PubMed] [Google Scholar]
- 59.Zhao, J. J., J. Lin, H. Yang, W. Kong, L. He, X. Ma, D. Coppola, and J. Q. Cheng. 2008. MicroRNA-221/222 negatively regulates estrogen receptor α and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem. 28331079-31086. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.