Abstract
In mammalian circadian clockwork, the CLOCK-BMAL1 heterodimer activates E-box-dependent transcription, while its activity is suppressed by circadian binding with negative regulators, such as CRYs. Here, we found that the CLOCK protein is kept mostly in the phosphorylated form throughout the day and is partly hyperphosphorylated in the suppression phase of E-box-dependent transcription in the mouse liver and NIH 3T3 cells. Coexpression of CRY2 in NIH 3T3 cells inhibited the phosphorylation of CLOCK, whereas CIPC coexpression markedly stimulated phosphorylation, indicating that CLOCK phosphorylation is regulated by a combination of the negative regulators in the suppression phase. CLOCK-BMAL1 purified from the mouse liver was subjected to tandem mass spectrometry analysis, which identified Ser38, Ser42, and Ser427 as in vivo phosphorylation sites of CLOCK. Ser38Asp and Ser42Asp mutations of CLOCK additively and markedly weakened the transactivation activity of CLOCK-BMAL1, with downregulation of the nuclear amount of CLOCK and the DNA-binding activity. On the other hand, CLOCKΔ19, lacking the CIPC-binding domain, was far less phosphorylated and much more stabilized than wild-type CLOCK in vivo. Calyculin A treatment of cultured NIH 3T3 cells promoted CLOCK phosphorylation and facilitated its proteasomal degradation. Together, these results show that CLOCK phosphorylation contributes to the suppression of CLOCK-BMAL1-mediated transactivation through dual regulation: inhibition of CLOCK activity and promotion of its degradation.
Many physiological activities of living organisms show rhythmic changes, with a period of approximately 24 h, even under constant conditions without any external time cues. These daily variations, called circadian rhythms, are generated by the circadian clock, a cell-autonomous time-measuring system that has evolved in a wide variety of organisms, from cyanobacteria to higher plants and humans (9). In mammals, a master clock is located in the hypothalamic suprachiasmatic nucleus, while self-sustaining molecular clocks reside even in the peripheral tissues, such as the liver (18, 40, 45). In these central and peripheral clock cells, the clock genes form a transcription/translation-based negative-feedback loop to generate the molecular oscillation with 24-h periodicity. CLOCK and BMAL1 are basic helix-loop-helix (bHLH)-PAS transcription factors, and the CLOCK-BMAL1 heterodimer binds to CACGTG E-box (14) or E-box-like (25, 61) sequences for the transactivation of negative regulatory genes, such as Period (Per1 and Per2) and Cryptochrome (Cry1 and Cry2) genes. Newly translated PERs and CRYs enter the nuclei and bind to the CLOCK-BMAL1 heterodimer, leading to suppression of its transactivation (30).
In the mouse liver, the abundances and the phosphorylation states of both PER1 and PER2 show striking temporal changes (32). The total amount of each PER protein shows the lowest level at CT6 (CT is circadian time, with CT0 under the constant-dark [DD] condition corresponding to the lights-on time in the 12-h-light/12-h-dark [LD] cycle) and increases with time, reaching the maximal level at CT15 to CT18. On the other hand, the level of each Per transcript has its peak at around dusk (32). The time lag of the peaks between the Per transcripts and the PER protein levels is afforded mainly by PER phosphorylation, which is considered to play a central role in controlling subcellular localization and stability (1, 48, 49, 56, 57, 59, 60). The delayed timing of nuclear accumulation of PER1 and PER2 proteins relative to that of their transcripts is important in generating the molecular oscillation with a stable period of ∼24 h. The physiological importance of PER phosphorylation was strengthened by the identification of 21 phosphorylated residues of PER2, including Ser659 (55), which is mutated in patients suffering from familial advanced sleep phase syndrome (53).
In addition to PER proteins, many factors contribute to the negative regulation of E-box-dependent transactivation. These include CRY1 and CRY2 (30), DEC1 and DEC2 (20), NONO, WDR5 (5), and CIPC (63). The intriguing question is the mechanism by which these negative regulators suppress CLOCK-BMAL1-dependent transactivation in combination or independently. In the Neurospora crassa clock, a negative clock regulator, FRQ, recruits both CKI and CKII to WC1-WC2, a heterodimer of positive regulators, resulting in phosphorylation of the WC1-WC2 complex and inhibition of its DNA binding (16, 17, 21, 44). In Drosophila melanogaster, a negative clock regulator, PER, recruits DBT (CKI homolog) to positive clock regulators Drosophila CLOCK (dCLK) and CYC in a circadian manner, thereby causing multiple events with a possible causal relationship: circadian phosphorylation of dCLK, enhancement of its degradation, and inhibition of its DNA binding (22, 23, 37, 62). For mammals, on the other hand, little is known about the roles of the negative regulators in the control of E-box-dependent transactivation. Noticeably, the molecular activity of the CLOCK-BMAL1 heterodimer to bind to an E-box-containing DNA probe exhibits circadian changes (41), and hence it is thought that the circadian changes in the abundance of negative regulators cause functional variations of the CLOCK-BMAL1 complex. Some of the negative regulators may serve as adaptor proteins that recruit regulatory enzymes, such as protein kinase and phosphatase, to the CLOCK-BMAL1 complex on the E-box elements, and their catalyzing modifications may induce suppression of the transactivation.
In the present study, we produced anti-CLOCK monoclonal antibodies (MAbs) as molecular probes and found that immunoprecipitated CLOCK is hyperphosphorylated in the suppression phase. Two of the in vivo phosphorylation sites of CLOCK determined by an extensive proteomic analysis were in positions to regulate the transactivation activity of CLOCK-BMAL1. On the other hand, we found that a recently reported negative regulator, CIPC, stimulated BMAL1-dependent phosphorylation of CLOCK and that a mutant CLOCK protein lacking the CIPC-binding domain had less ability than the wild type to receive phosphorylation signals to be degraded. These data indicate that CLOCK phosphorylation governed by negative regulators dually contributes to the suppression of E-box-dependent transactivation through inhibition of CLOCK activity and promotion of its degradation.
MATERIALS AND METHODS
Animals.
Animals were treated in accordance with the guidelines of The University of Tokyo. Six-week-old male C57BL/BJ mice (see Fig. 2C) and female BALB/c mice (see other figures) were entrained for at least 2 weeks under the condition of the LD cycle, with food and water freely available. All mice were reared in a room kept at 23°C ± 1°C, with the light provided by fluorescent lamps (∼200 lx at the level of the cage). For experiments done under the DD condition, the animals entrained to the LD cycles for at least 2 weeks were placed under the DD condition. Zeitgeber time (ZT) is used for representing biological time in LD cycles, in which ZT0 and ZT12 correspond to the lights-on time and the lights-off time, respectively. CT is used for representing biological time under the DD condition, in which CT0 corresponds to the lights-on time in the LD cycle. Clock mutant mice from a BALB/c background were kindly supplied by Joseph S. Takahashi (Northwestern University, Evanston, IL).
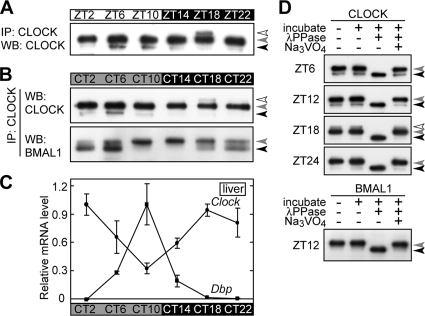
FIG. 2.
Phosphorylation rhythms of CLOCK and BMAL1 in the mouse liver. Daily variations (A) and circadian rhythms (B) of phosphorylation of CLOCK and BMAL1 in mouse liver nuclear extract. Nuclear extracts (100 μg of proteins) prepared at the indicated time points were immunoprecipitated with anti-CLOCK MAb. The immunoprecipitates were subjected to immunoblot analysis with CLSP3 anti-CLOCK MAb (A and B, top) and B1BH2 anti-BMAL1 MAb (B, bottom). WB, Western blotting. (C) Livers were isolated from mice (C57BL/BJ, male) at the indicated time points, and the relative mRNA levels of Dbp and Clock were determined by RT-PCR analysis. The signals obtained for each mRNA were normalized to those of Tbp mRNA, and the mean value at peak time was set to 1. Data are means with standard errors of the means from three experiments. (D) The immunoprecipitates were incubated at 30°C for 30 min with lambda protein phosphatase (λPPase) and 25 mM sodium vanadate as indicated. After the incubation, the samples were immunoblotted with B1BH2 anti-BMAL1 MAb (bottom) and CLSP3 anti-CLOCK MAb (top).
Preparation of MAbs.
Preparation of MAbs was performed as described previously (26), with minor modifications. The antigenic peptides were expressed from plasmids encoding mouse Clock or chicken Bmal1 as fusion proteins at the C terminus of glutathione S-transferase (GST) or of maltose-binding protein (MBP) in Escherichia coli, and the expressed proteins were purified by a glutathione-Sepharose column (Amersham Biosciences) or by an amylose-resin column (New England BioLabs), respectively. Female BALB/c mice were injected intraperitoneally with 50 to 100 μg of GST-NT (where NT is the N-terminal region Met1-Gly120 of CLOCK), MBP-SP (where SP is the Ser/Pro-rich region Ser377-Glu556 of CLOCK), or MBP-BH (where BH is the bHLH domain Ala73-Ala128 of BMAL1) in phosphate-buffered saline (PBS) emulsified with complete Freund's adjuvant (BD Biosciences). On day 14, each mouse was boosted intraperitoneally with the same antigen in PBS emulsified with incomplete Freund's adjuvant (BD Biosciences). Each mouse was bled on day 28, and the titer of the antiserum was tested by an enzyme-linked immunosorbent assay using MBP-NT, GST-SP, or GST-BH. The mice whose sera showed high immunoreactivities to the antigen were injected intraperitoneally with the same antigen in PBS. Three days after the final injection, the spleen cells were isolated from the mice and fused with myeloma cells (P3-X63-Ag8-U1) by use of polyethylene glycol. The hybridomas were selected with hypoxanthine-aminopterin-thymidine medium. The hybridomas were cloned by a limiting-dilution method and screened by enzyme-linked immunosorbent assay. The hybridomas thus cloned were injected intraperitoneally into pristane-primed BALB/c female mice, and the ascites were collected 10 to 14 days after inoculation. The antibodies were purified by precipitations with 50% saturated ammonium sulfate and were dialyzed against PBS. For coupling with an Affi-Gel Hz hydrazide gel (Bio-Rad), CLSP3 immunoglobulin G was purified by use of an Immuno Pure immobilized protein G column (Pierce) according to the manufacturer's protocol.
Preparation of mouse liver nuclear proteins.
Under dim red light (>640 nm), mice were sacrificed by cervical dislocation and their eyeballs were removed. Then, the livers were dissected under room light, and the liver nuclear proteins were prepared as described previously (7), with minor modifications. Briefly, the tissue (1 g [wet weight]) was washed with ice-cold PBS and homogenized at 4°C with 9 ml of ice-cold buffer A (10 mM HEPES-NaOH [pH 7.8], 10 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 4 μg/ml aprotinin, 4 μg/ml leupeptin, 50 mM NaF, and 1 mM Na3VO4). The homogenate was centrifuged twice (5 min each, 700 × g), and the unsedimented material was collected as a cytoplasmic fraction. The precipitate was resuspended in 2 ml of ice-cold buffer C (20 mM HEPES-NaOH [pH 7.8], 400 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 2% [vol/vol] glycerol, 1 mM DTT, 1 mM PMSF, 4 μg/ml aprotinin, 4 μg/ml leupeptin, 50 mM NaF, and 1 mM Na3VO4). After being gently mixed at 4°C for 30 min, the suspension was centrifuged twice (30 min each, 21,600 × g), and the final supernatant was used as a nuclear extract.
Antibodies and immunoblot analysis.
The anti-CLOCK polyclonal antibody used (see Fig. 1E) was purchased from Affinity BioReagents. The anti-CRY2 polyclonal antibody was kindly provided by Takeshi Todo from Osaka University. For immunoblot analysis, bound primary antibodies were detected by horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin G antibody (Kirkegaard & Perry Laboratories). The immunoreactivities were visualized by an enhanced chemiluminescence detection system (Western Lightning; Perkin Elmer Life Sciences).
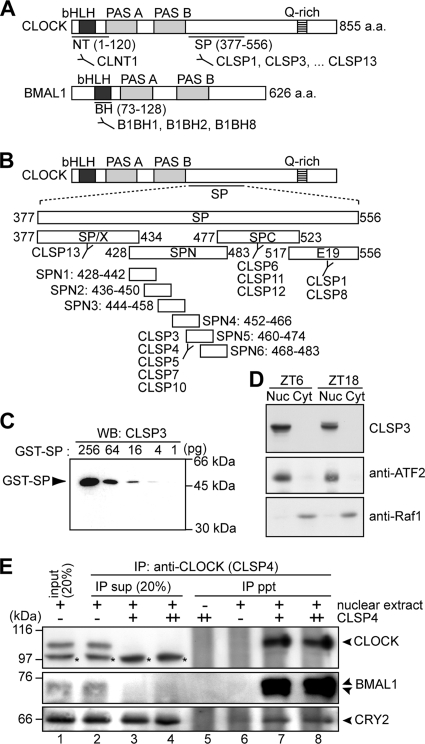
FIG. 1.
Characterization of anti-CLOCK and anti-BMAL1 MAbs. (A) Domain structures of CLOCK and BMAL1. Antigenic regions for anti-CLOCK and anti-BMAL1 MAbs are indicated. a.a., amino acids. (B) To localize epitopes of anti-SP antibodies, four partial peptides of SP, SP/X (Ser377-Ser434), SPN (Pro428-Glu483), SPC (Arg477-Met523), and E19 (Ser517-Glu556), were expressed as GST fusion proteins, which were recognized by CLSP13 (GST-SP/X); by CLSP3, -4, -5, -7, and -10 (GST-SPN); by CLSP6, -11, and -12 (GST-SPC); and by CLSP1 and -8 (GST-E19). For detailed determination of the epitopes, six shorter peptides of SPN, SPN1 (Pro428-His442), SPN2 (Ser436-Ser450), SPN3 (Ala444-Asp458), SPN4 (Pro452-His466), SPN5 (Pro460-Thr474), and SPN6 (Pro468-Glu483), were expressed as GST fusion proteins, among which GST-SPN5 was recognized by all of the SPN-immunoreactive antibodies (CLSP3, -4, -5, -7, and -10). (C) The immunizing peptide SP (Ser377-Glu556) was expressed as a GST fusion protein (GST-SP) and subjected to immunoblot analysis with CLSP3 anti-CLOCK MAb. The amounts of the fusion protein are indicated. WB, Western blotting. (D) The nuclear extract (Nuc) (40 μg of proteins) and the cytoplasmic fraction (Cyt) (40 μg of proteins) were prepared from the mouse liver at ZT6 and ZT18 and subjected to immunoblot analysis with CLSP3 anti-CLOCK MAb (top). Immunoblot analysis with anti-ATF2 antibody (middle) and anti-Raf1 antibody (bottom) supported the separation of nuclear and cytoplasmic proteins. (E) The nuclear extract (100 μg of proteins) prepared at ZT18 was immunoprecipitated with increasing amounts, 0.5 μg (+) and 1.0 μg (++), of CLSP4 anti-CLOCK MAb. The immunoprecipitates (ppt) and 20% of the remaining supernatants (sup) were subjected to immunoblot analysis with anti-CLOCK polyclonal antibody (Affinity BioReagents) (top), B1BH2 anti-BMAL1 MAb (middle), and anti-CRY2 polyclonal antibody (a kind gift of T. Todo) (bottom). The anti-CLOCK polyclonal antibody detected additional protein bands (shown by asterisks) that were invisible in the IP pellet in the presence of CLSP4 MAb, indicating that CLOCK was specifically immunoprecipitated by the antibody.
IP.
Immunoprecipitation (IP) was performed as described previously (46), with minor modifications. Mouse liver nuclear extract prepared in buffer C was diluted with 2 volumes of buffer D1 (20 mM HEPES-NaOH [pH 7.8], 5.5 mM NaCl, 1 mM EDTA, 6.5% [vol/vol] glycerol, 1.5% [vol/vol] Triton X-100, 1 mM DTT, 1 mM PMSF, 4 μg/ml aprotinin, 4 μg/ml leupeptin, 50 mM NaF, and 1 mM Na3VO4), incubated with protein G-Sepharose 4 Fast Flow (Amersham Biosciences) at 4°C for 30 min, and then centrifuged for 5 min at 2,400 × g. The clarified supernatant was mixed with anti-CLOCK antibody by gentle rotation at 4°C for 1 h. Protein G-Sepharose 4 Fast Flow was added to this mixture, and the resulting solution was mixed by gentle rotation at 4°C for 1 h. The beads were collected by centrifugation for 5 min at 2,400 × g as the immunoprecipitates.
Phosphatase treatment.
Endogenous CLOCK and BMAL1 in mouse liver nuclear extract were immunoprecipitated by anti-CLOCK MAb, and the beads were washed with phosphatase buffer P (50 mM Tris-HCl [pH 7.5], 0.1 mM EDTA, 5 mM DTT, 2 mM MnCl2, 0.01% Brij 35). The suspension was incubated with lambda protein phosphatase (Sigma) at 30°C for 30 min in the presence or absence of 25 mM vanadate as a phosphatase inhibitor and then subjected to immunoblot analysis.
Semiquantitative reverse transcription-PCR (RT-PCR) analysis.
Total RNA of the mouse liver or the cultured cells was prepared by using TRIzol reagent (Invitrogen) according to the manufacturer's directions. The total RNA was reverse transcribed with Superscript II (Invitrogen) by use of an anchored (dT)16 primer, and the reaction mixture was treated with RNase H (Takara, Japan). The reverse transcripts were subjected to PCR by use of AmpliTaq Gold and gene-specific primers as described previously (19, 27). The primers used were Tbp-Fw (TCATG GACCA GAACA ACAGC), Tbp-Rv (TGGTG TGGCA GGAGT GATAG), Clock-Fw (CACGA AAGTC ATCTC ACACC), Clock-Rv (TTACA GGTGT GAGTT GCTGG), Dbp-Fw (CCAAT CATGA AGAAG GCAAG G), and Dbp-Rv (AGGAT TGTGT TGATG GAGGC). The number of cycles was determined for each amplification reaction so as to maintain a linear correlation between the amplified products and the amounts of template DNA. The PCR products were subjected to polyacrylamide gel electrophoresis (PAGE), stained with SYBR green I (Cambrex), and detected with an FLA-2000 image analyzer (Fuji Film).
Cell culture and transfection.
NIH 3T3 cells (Riken Cell Bank) and human embryonic kidney 293T (HEK293T) cells were maintained at 37°C under conditions of 5% CO2 and 95% air in Dulbecco's modified Eagle's medium (Nissui) containing 1.8 mg/ml NaHCO3 and 4.5 mg/ml glucose supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum. For detection of endogenous CLOCK and BMAL1 (see Fig. 3A), the cells treated with 0.02 μM dexamethasone for 2 h were washed with PBS and were solubilized in ice-cold IP buffer (20 mM HEPES-NaOH [pH 7.8], 137 mM NaCl, 1 mM EDTA, 5% [vol/vol] glycerol, 1% [vol/vol] Triton X-100, 1 mM DTT, 2 mM PMSF, 4 μg/ml aprotinin, 4 μg/ml leupeptin, 50 mM NaF, and 1 mM Na3VO4). The cell extracts were then centrifuged for 10 min at 21,600 × g, and the supernatant was subjected to IP with CLNT1 anti-CLOCK MAb and protein G-Sepharose. For transient transfections, the cells were plated in the wells of 12-well plates (1 × 105 cells/well) 24 h before the experiments and were transiently transfected by use of Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's directions. Cells were washed with PBS 36 h after the transfections and solubilized in radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 2 mM EDTA, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM PMSF, 4 μg/ml aprotinin, 4 μg/ml leupeptin, 50 mM NaF, and 1 mM Na3VO4). The cell extracts were then centrifuged for 10 min at 21,600 × g, and the supernatant was used as a total cell extract.
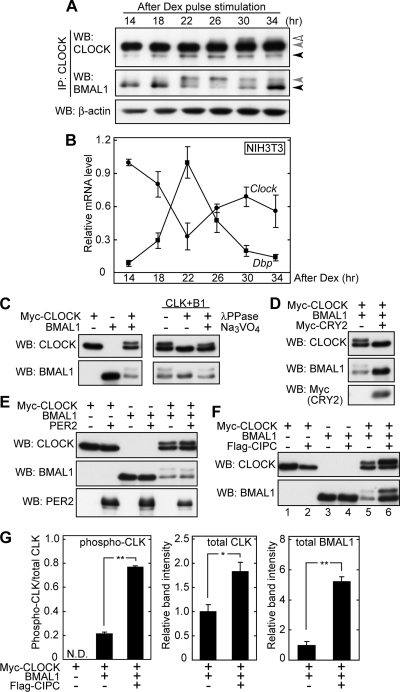
FIG. 3.
Negative regulators CRY2 and CIPC control phosphorylation states of CLOCK and BMAL1 in NIH 3T3 cells. (A) NIH 3T3 cells were cultured in a 10-cm dish, and dexamethasone (Dex) was added to the culture medium. After a 2-h incubation with dexamethasone, the medium was changed to normal culture medium, and this time point was defined as time zero. The cells were collected at the indicated time points, and total protein extracts were immunoprecipitated with CLNT1 anti-CLOCK MAb, followed by immunoblot analysis with CLSP3 anti-CLOCK MAb (top) and B1BH2 anti-BMAL1 MAb (middle). Total protein extracts were immunoblotted with anti-β-actin antibody as a sampling control (bottom). (B) NIH 3T3 cells cultured in a 35-mm dish were synchronized as described above. The cells were collected at the indicated time points, and the relative mRNA levels of Dbp and Clock were determined by RT-PCR analysis. The signals obtained for each mRNA were normalized to those of Tbp mRNA, and the mean value at the peak time was set to 1. Data are means with standard errors of the means (SEM) from three experiments. (C to F) NIH 3T3 cells were cultured in wells of 12-well plates and transiently transfected with various combinations of plasmids (each transfected plasmid is 200 ng) expressing the indicated proteins. The total amount of DNA was adjusted by adding pSG5 empty plasmid (for Myc-CLOCK and Flag-CIPC) and pcDNA3.1 empty plasmid (for BMAL1, Myc-CRY2, and PER2). The transfected cells were collected 36 h after the transfection, and 10% of the cell lysates were subjected to immunoblot analysis with CLSP3 anti-CLOCK MAb, B1BH2 anti-BMAL1 MAb, anti-Myc antibody, or anti-PER2 antibody (Alpha Di-agnostic International, Inc.). For phosphatase treatment, the cell lysates were incubated at 30°C for 30 min with lambda protein phosphatase (λPPase) and 25 mM sodium vanadate as indicated. (G) Quantitative analysis of CIPC effect on phosphorylation state and protein level of CLOCK and BMAL1. The upshifted CLOCK protein (phospho-CLK) band relative to the bands for total CLOCK protein was quantified by densitometry of the blots shown in panel F, lanes 1, 5, and 6 (left). Total protein amounts of CLOCK (middle) and BMAL1 (right) were quantified by densitometry of the blots shown in panel F, lanes 5 and 6. The mean values of the band intensities in lane 5 were set to 1. Data are means with SEM from four experiments. A single asterisk indicates a P value of 0.01, and a double asterisk indicates a P value of <0.01 (Student's t test). WB, Western blotting.
Plasmids.
The mammalian expression vector encoding the Myc epitope-tagged CLOCK proteins (Myc-CLOCK/pSG5) was a kind gift of Paolo Sassone-Corsi (8). BMAL1/pcDNA3.1 (a kind gift of Steve M. Reppert), PER2/pcDNA3 (a kind gift of Hitoshi Okamura), and Myc-CRY2/pCMV-Tag3B (15) were also used. All mutations were introduced into CLOCK and BMAL1 by using site-directed mutagenesis PCR.
Cloning of full-length mouse Cipc cDNA.
Full-length mouse Cipc cDNA was cloned from the total cDNA of NIH 3T3 cells by RT-PCR with gene-specific primers and verified by sequencing. The primers used were Cipc-Fw (GCGGC CGCAT GGAGA GGAAA ATCCC ATC) and Cipc-Rv (GCGGC CGCTA TACGT CTGGG TGATC AG). Mammalian expression vectors encoding Flag epitope-tagged CIPC protein were generated by inserting the Cipc cDNA into the pSG5 vector, with a slight modification to create NotI sites, a Kozak sequence, and a Flag epitope.
Identification of CLOCK phosphorylation sites by MS.
The nuclear extract was prepared from the mouse liver dissected at ZT18, followed by Toyopearl DEAE-650S column chromatography (Tosoh). Then, the CLOCK-containing fractions eluted with a linear gradient of NaCl were added to CLSP3 anti-CLOCK MAb-coupled Affi-Gel Hz hydrazide gel (Bio-Rad) in IP buffer, and the solution was mixed by gentle rotation at 4°C for 6 to 8 h. The CLSP3 was coupled with the gel according to the manufacturer's directions. To increase the purity, the bound proteins were eluted from the gel with elution buffer (20 mM HEPES-NaOH [pH 7.8], 0.5 mg/ml SPN5 peptide, 137 mM NaCl, 1 mM EDTA, 5% [vol/vol] glycerol, 0.1% [vol/vol] Triton X-100, 1 mM DTT, 2 mM PMSF, 4 μg/ml aprotinin, 4 μg/ml leupeptin) supplemented with SPN5, a pentadecapeptide corresponding to the epitope of CLSP3 (Pro460-Thr474 of CLOCK) (see Fig. 1B), after the column was washed with a scrambled sequence peptide of SPN5. The eluates were precipitated with trichloroacetic acid. The protein precipitate was treated with 50 mM DTT, followed by cysteine alkylation with monoacrylamide, and then directly subjected to SDS-PAGE. The protein bands at 100 to 110 kDa were excised from a strip of the gel, washed with water, and dried in vacuum. Then, 2 μg of modified trypsin (Promega) or 2 μg of endoproteinase Glu-C, sequencing grade (Roche), was added to the gel, which was incubated in 50 mM NH4HCO3 at 37°C overnight. The resultant gel was sonicated in 0.1% trifluoroacetic acid-50% CH3CN for 10 min, and the supernatant was collected. This step was repeated twice. The resultant solutions were desalted with InertSep RP-1 (GL Sciences Inc., Tokyo, Japan), concentrated in vacuum, and applied to nanoflow liquid chromatography-electrospray ionization-tandem mass spectrometry (MS-MS) (NanoFrontier LD; Hitachi High-Technologies Co., Tokyo). A part of the endoproteinase Glu-C digest was separated using a reversed-phase column (Cadenza CL C18, 1-mm inside diameter by 150-mm length; Imtakt Co. Ltd., Kyoto, Japan) in an Agilent 1100 high-performance liquid chromatography system (Agilent Technologies, Palo Alto, CA). A linear gradient using solvent A (0.1% trifluoroacetic acid in water) and solvent B (0.1% trifluoroacetic acid in acetonitrile) was used for the separation; the peptides were eluted by increasing solvent B from 2 to 50% over a period of 24 min at a flow rate of 70 μl/min. The effluents were monitored at 214 nm and were fractionated every 1 min, concentrated, and blotted onto a sample plate for matrix-assisted laser desorption ionization-MS-MS (MALDI-MS-MS) (4700 proteomics analyzer; Applied Biosystems, Framingham, MA). Thereafter, the matrix solution (5 mg/ml of α-CHCA [α-cyano-4-hydroxycinnamic acid]) was blotted manually onto each sample spot and then air dried. The MS-MS data were subjected to database searching with Mascot version 2.0 (Matrix Science, Manchester, United Kingdom). The resulting peptide candidates were judged significant hits (identified peptides) if the MS-MS ion scores were over 30.
EMSA.
Myc-CLOCK (wild type, Ser38Asp mutant, Ser42Asp mutant, or Ser38Asp/Ser42Asp mutant) and BMAL1 were synthesized in vitro by using a TNT quick-coupled transcription/translation system (Promega) according to the manufacturer's instructions. An electrophoretic mobility shift assay (EMSA) was performed by using a digoxigenin gel shift kit (Roche Diagnostics) according to the manufacturer's instructions. The short, double-stranded oligonucleotides were prepared by annealing a pair of synthetic oligonucleotides and subjected to digoxigenin-labeling procedures. The sequences of the two strands of the DNA probe were 5′-CGCG CAAGTCC ACGTGC AGGGAT-3′ and 5′-CGCG ATCCCT GCACGT GGACTTG-3′. This region corresponds to the Per1 promoter region (positions −142 to −155) and contains one E-box sequence. The DNA binding reaction mixture (20 μl) contained 20 mM HEPES-NaOH (pH 7.6), 30 mM KCl, 1 mM EDTA, 10 mM (NH4)2SO4, 1 mM DTT, 0.2% Tween 20, 1 μg poly[d(I-C)], 0.1 μg poly l-lysine, and proteins as indicated in Fig. 5F and G. For supershift experiments, the reaction mixtures were preincubated with antibody (1 μg) at 25°C for 15 min and then incubated with 100 fmol DNA probe at 25°C for 15 min.
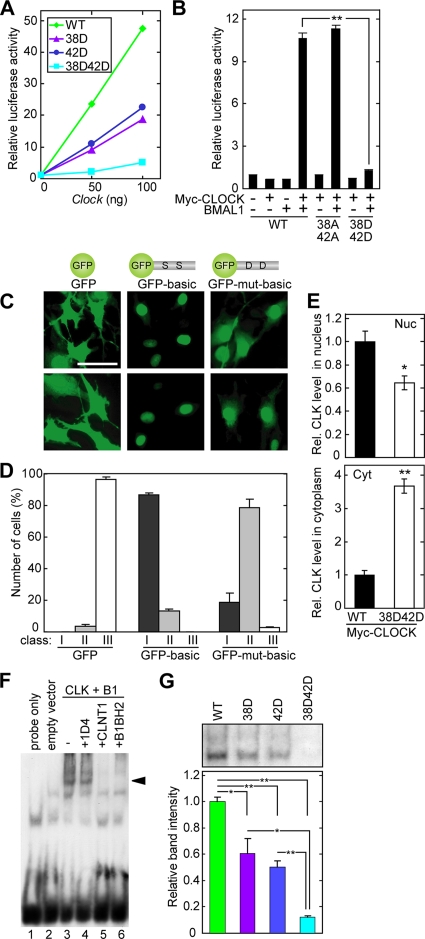
FIG. 5.
Ser38Asp and Ser42Asp mutations reduce transactivation ability. (A and B) HEK293T cells were cultured in wells of 12-well plates and transiently transfected with various combinations of plasmids expressing Myc-CLOCK (A, indicated amounts; B, 50 ng of plasmid) and BMAL1 (100 ng) as indicated, with 20 ng of firefly luciferase plasmid (mAVP E-box3-SV40-luc [38]) and 1 ng of Renilla luciferase plasmid (pRL-RSV) as an internal control. The total amount of DNA was adjusted by adding the empty expression plasmids. The transfected cells were collected 36 h after the transfection and subjected to a dual luciferase assay by luminometry (Promega) according to the manufacturer's protocol. The values were normalized to transfection efficiency with the help of the internal control for each cell culture. Data shown in panel B are means with standard errors of the means (SEM) from three experiments. (C) Representative fluorescence images demonstrating the subcellular localization of GFP-fused CLOCK hexadecapeptide Asp32-Arg47 (GFP-basic) or its Ser38Asp/Ser42Asp mutant (GFP-mut-basic) expressed in NIH 3T3 cells. Bar, 50 μm. (D) Quantitative analysis of the subcellular distribution of GFP fusion proteins. The subcellular-localization profile was categorized into three classes: class I, exclusively in the nucleus; class II, strongly in the nucleus and weakly in the cytoplasm; class III, similar intensities in the nucleus and the cytoplasm. The data were obtained from three independent experiments, and in each experiment more than 100 cells were counted. (E) Myc-CLOCK (wild type [WT] or Ser38Asp/Ser42Asp mutant) was expressed with BMAL1 in HEK293T cells. The nuclear extract (Nuc) and the cytoplasmic fraction (Cyt) were immunoblotted with CLSP3 anti-CLOCK MAb, and the band intensities were quantified. The mean value of the band intensities of WT CLOCK was set to 1 for each panel. Data are means with SEM from four experiments. A single asterisk indicates a P value of 0.02, and a double asterisk indicates a P value of <0.01 (Student's t test). (F and G) Myc-CLOCK (WT, Ser38Asp mutant, Ser42Asp mutant, or Ser38Asp/Ser42Asp mutant) and BMAL1 were synthesized in vitro, and their DNA-binding activities were examined by EMSA with a DNA probe containing the E-box sequence in the Per1 promoter. (F) A specific DNA-protein complex was detected (arrowhead) when WT CLOCK and BMAL1 (CLK+B1) were incubated with the DNA probe (lane 3). In supershift experiments, the DNA-protein complex disappeared when CLNT1 anti-CLOCK MAb or B1BH2 anti-BMAL1 MAb was added. 1D4 antirhodopsin antibody was used as an irrelevant MAb. (G) Quantification of the intensities of the bands representing the specific DNA-protein complex. Data are means with SEM from three experiments. A single asterisk indicates a P value of <0.05, and a double asterisk indicates a P value of <0.01 (Student's t test).
RESULTS
MAbs against CLOCK and BMAL1.
In order to trace circadian variations of the CLOCK-BMAL1 complex in vivo, a series of MAbs against CLOCK and BMAL1 were prepared as molecular probes. The NT and SP regions of CLOCK were selected as immunizing antigens (Fig. 1A). The NT region includes the bHLH domain, while the SP region is predicted by hydropathy analysis to be exposed to the molecular surface of the CLOCK protein. An anti-NT antibody, termed CLNT1, and 11 anti-SP antibodies, termed CLSP1, CLSP3 to -8, CLSP10 to -12, and CLSP13, were isolated. Among them, CLSP3 showed the highest sensitivity by immunoblot analysis, as it detected 4 pg (less than 0.1 fmol) of GST-SP, a fusion of the immunizing peptide with GST (Fig. 1C). Further analysis localized the epitopes of the antibodies CLNT1 and CLSP3 to Val26-Ser38 and Pro460-Thr474 of CLOCK, respectively (Fig. 1B and data not shown). In immunoblot analysis, CLNT1 and CLSP3 detected CLOCK protein expressed in HEK293T cells but showed no cross-reaction with NPAS2, a paralog of CLOCK (data not shown). On the other hand, the BH domain was selected for immunization (Fig. 1A), and three anti-BH antibodies, termed B1BH1, -2, and -8, were established. These MAbs recognized Arg83-Val99 of BMAL1, and we verified that B1BH2 showed no cross-reaction with BMAL2 by immunoblotting (data not shown).
In vivo phosphorylation rhythms of CLOCK and BMAL1.
Because BMAL1 undergoes nucleocytoplasmic shuttling (31, 50), we first examined daily variations of subcellular localization of CLOCK in the mouse liver by using the anti-CLOCK MAb CLSP3 (Fig. 1D). Although it was reported that CLOCK is present in both the nucleus and the cytoplasm in the mouse liver (28, 32), CLOCK protein was detected only in the nuclear extracts at ZT6 and ZT18, and no CLOCK band was detected in the cytoplasmic fraction. Similar results were obtained with other anti-CLOCK MAbs (CLNT1 and CLSP1, -4, -10, and -12), the epitopes of which are spread over the CLOCK molecule (Fig. 1A and B). These data are supportive of the nuclear localization of CLOCK protein in mouse liver cells.
The electrophoretic mobility of CLOCK in the liver nuclei at ZT18 appeared slower than that at ZT6 (Fig. 1D). The mobility change of CLOCK was investigated in detail by purifying CLOCK protein from the mouse liver nuclear extract by IP with an anti-CLOCK MAb. When the amount of CLSP4 was increased, constant amounts of CLOCK and BMAL1 proteins were detected in the IP pellet (Fig. 1E, lanes 7 and 8), whereas no protein band was detected in the supernatant (Fig. 1E, lanes 3 and 4), indicating that almost all CLOCK and BMAL1 proteins were immunoprecipitated quantitatively from the nuclear extract and that they associate with each other in the nuclei. Parallel immunoblot analyses of the anti-CLOCK immunoprecipitates demonstrated coprecipitation of CRY2 (Fig. 1E, bottom). Similar results were obtained when CLSP3 or CLNT1 MAb was used (data not shown). The results indicate that these anti-CLOCK MAbs are applicable to quantitative IP of CLOCK protein interacting with its partners, such as BMAL1 and CRY2.
The liver nuclear extracts prepared from the entrained mice over 1 day were subjected to quantitative IP. CLOCK and BMAL1 proteins thus purified exhibited daily and circadian changes in their electrophoretic mobilities (Fig. 2A and B). CLOCK and BMAL1 had three (Fig. 2, black, gray, and open arrowheads) and two (Fig. 2, black and gray arrowheads) visible bands, respectively, among which the slowest-migrating species of CLOCK and BMAL1 had their peaks at ZT/CT18 and at CT10 to CT14, respectively. Incubation of the immunoprecipitates with lambda protein phosphatase resulted in a down-shift of the CLOCK and BMAL1 bands in the SDS-PAGE gel (Fig. 2D, black arrowheads), indicating that the temporal changes of the electrophoretic mobilities represent circadian variations in phosphorylation states of CLOCK and BMAL1. Importantly, the up-shifted form of phosphorylated BMAL1 in the CLOCK-containing complex had its peak in abundance at around dusk (Fig. 2B, bottom). On the other hand, the hyperphosphorylated form of CLOCK (Fig. 2A and B, open arrowheads) had its peak at midnight (ZT/CT18), while the hypophosphorylated form was detected to be abundant throughout the day (Fig. 2A and B, gray arrowheads). The peak time of CLOCK hyperphosphorylation matched the timing of the trough level of Dbp (Fig. 2C), which is regulated by E-box elements in the Dbp promoter region (54). Obviously, the circadian phosphorylation events of CLOCK and BMAL1 occur at different times of the day in the mouse liver nuclei, most probably via distinct mechanisms.
Negative regulators control phosphorylation states of CLOCK and BMAL1.
Molecular circadian changes in the liver are generated not only by the cell-autonomous clock but also by receipt of extracellular signals (45). To investigate which mechanism regulates the phosphorylation rhythms of CLOCK and BMAL1, the temporal changes were examined using cultured cells, in which the molecular clocks can be synchronized by a variety of stimuli, such as serum and dexamethasone (3, 4, 36). When NIH 3T3 cells were synchronized by a pulse treatment with dexamethasone, endogenous CLOCK and BMAL1 exhibited clear phosphorylation rhythms (Fig. 3A) . Accumulation of hyperphosphorylated CLOCK peaked 30 h after the dexamethasone pulse, while the level of phosphorylated BMAL1 peaked 22 to 26 h after the pulse. The phase angles of these phosphorylation rhythms (Fig. 3A) relative to the cycling of the Dbp transcript levels (Fig. 3B) were very similar to those observed in the mouse liver (Fig. 2B and C). The phosphorylation mechanism of CLOCK and BMAL1 in the mouse liver is most likely conserved in cultured NIH 3T3 cells. These data indicate that the phosphorylation rhythms are generated by the cell-autonomous molecular clock and suggest that they are important for intracellular events, such as the transactivation ability rhythm of the CLOCK-BMAL1 complex.
We next explored the contribution of interactions among the clock proteins to the circadian phosphorylation of CLOCK and BMAL1 by transient transfection of the components to the cultured cells. When CLOCK or BMAL1 was expressed individually in NIH 3T3 cells, no significant up-shift of the immunoreactive band was detected (Fig. 3C, lanes 1 and 2). On the other hand, coexpression of CLOCK and BMAL1 resulted in the generation of phosphorylated species of these proteins (Fig. 3C, lane 3 and right), as previously reported (28). Coexpression of CLOCK not only promoted phosphorylation of BMAL1 but also reduced the amount of BMAL1 (Fig. 3C, left). Then, the phosphorylation states of CLOCK and BMAL1 were examined in the presence of negative regulators. Coexpression of CRY2 remarkably suppressed phosphorylation of both CLOCK and BMAL1 and increased the amount of BMAL1 (Fig. 3D) as if they had returned to the states when they were expressed individually (Fig. 3C, left). In contrast, PER2 coexpression had little effect on CLOCK phosphorylation (Fig. 3E). We found that coexpression of a negative regulator, CIPC (CLOCK-interacting proteins, circadian) (63), not only stimulated CLOCK phosphorylation but also increased the protein levels of CLOCK and BMAL1 (Fig. 3F, lanes 5 and 6, and G). The CIPC-dependent stabilization of BMAL1 was no longer observed in the absence of coexpressed CLOCK (Fig. 3F, lanes 3 and 4), suggesting that CIPC impinged on the BMAL1 state only in the CLOCK-BMAL1 complex. Although CLOCK interacts directly with CIPC (63), coexpression of CIPC with CLOCK alone (without BMAL1 expression) had only a marginal effect on CLOCK protein in terms of the phosphorylation level (Fig. 3F, lanes 1 and 2). In short, CIPC stimulates phosphorylation of coexpressed CLOCK probably only in the complex state and increases the stability of the complex. Thus, these negative regulators, CRY2 and CIPC, have their own unique effects on the phosphorylation states of CLOCK and BMAL1, and a combination of negative regulators may underpin fine-tuned regulation of the phosphorylation states of CLOCK and BMAL1.
Phosphorylation sites of CLOCK.
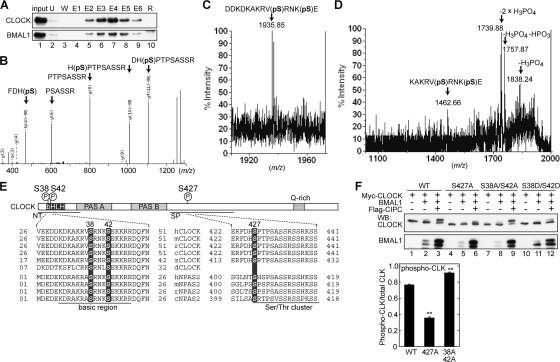
To investigate the biological significance of CLOCK phosphorylation, we searched for the residues of CLOCK to be phosphorylated in vivo by MS analysis. The mouse liver nuclear extract was prepared at ZT18, when hyperphosphorylated CLOCK was detected (Fig. 2A). Then, the CLOCK-BMAL1 complex was purified through DEAE-Sepharose column chromatography and subsequent CLSP3-conjugated affinity column chromatography (Fig. 4A). The eluates containing CLOCK/BMAL1 immunoreactivities were subjected to SDS-PAGE, and proteins in the sliced gels were digested with trypsin for analysis by nanoflow liquid chromatography-electrospray ionization-MS-MS. This analysis revealed that Ser427 of CLOCK is phosphorylated in the mouse liver (Fig. 4B). Ser427 is located in a region that is highly conserved among chicken, mouse, and human CLOCK proteins, and the Ser residue is also conserved in the corresponding regions of chicken, mouse, and human NPAS2 proteins (Fig. 4E). By this MS analysis, the peptide sequence coverage of the CLOCK protein was 34.3%, and we were unable to detect any fragments in the Lys- and Arg-rich regions, including the basic region of the DNA-binding domain. To analyze the basic region(s), endoproteinase Glu-C, instead of trypsin, was used for the in-gel digestion, and the digests were analyzed similarly. Then, we detected a peptide showing a molecular ion peak at m/z 1,935.85 (Fig. 4C), a value identical to the protonated molecular mass (MH+ 1,935.87) calculated from the CLOCK sequence Asp29-Glu43 (DDKDKAKRVpSRNKpSE), which is phosphorylated at two residues, possibly Ser residues (pS). The MS-MS fragment ions, observed to occur intensely at m/z 1,838.24, 1,757.87, and 1,739.88 (Fig. 4D), were attributable to the specific cleavage of phosphoester bonds at phospho-Ser, which, as a result, gave the signature for the double phosphorylation in the sequence. The analyzed peptide was identical to a synthetic one phosphorylated both at Ser38 and at Ser42 by reversed-phase liquid chromatography and MS-MS analysis (data not shown). These data demonstrate that Ser38 and Ser42 are phosphorylated in the mouse liver isolated at ZT18. Ser38 and Ser42 are located in the basic region of the bHLH DNA-binding domain and are conserved in not only CLOCK from Drosophila to humans but also NPAS2 from chickens to humans (Fig. 4E).
FIG. 4.
Identification of in vivo phosphorylation sites of CLOCK. (A) Purification of the CLOCK-containing protein complex by immunoaffinity column chromatography. The mouse liver nuclear extract prepared at ZT18 was subjected to DEAE-Sepharose column chromatography, and the eluates (lane 1, input) were loaded onto the CLSP3-coupled column. The fractions obtained by immunoaffinity column chromatography were resolved by SDS-PAGE and immunoblotted with CLSP3 anti-CLOCK MAb (top) and B1BH2 anti-BMAL1 MAb (bottom). U, unbound fraction; W, washing fractions with IP buffer; E1 to E6, eluates with SPN5 epitope peptide; R, resin collected from the column after the elution. (B) Electrospray ionization-MS-MS spectrum of the phosphorylated peptide (Phe424-Arg435). The position of the phosphorylated residue was read out based on b- and y-series ions, which were produced upon cleavage of peptide bonds during MS-MS. The b-98 and y-98 ions denote the fragment ions derived from β elimination of the phosphoric acid (H3PO4), most probably from pSer427. (C) MALDI-MS spectrum of the phosphorylated peptide (Asp29-Glu43) derived from the endoproteinase Glu-C digest of CLOCK. (D) MALDI-MS-MS spectrum from the ion at m/z 1,935.85 in panel C. The fragment ions, observed to occur predominantly at m/z 1,838.24, 1,757.87, and 1,739.88, could be assigned to those lacking H3PO4, H3PO4 plus HPO3, and 2× H3PO4, respectively, which were produced by cleavage of the phosphoester bonds at the phosphorylation sites, most probably at Ser38 and/or Ser42. (E) Sequence alignment of CLOCK and NPAS2 around phosphorylated Ser from various species. h, human; m, mouse; r, rat; c, chicken; z, zebrafish; d, Drosophila. (F) (Left) CLOCK mutants were expressed with BMAL1 and CIPC as indicated and subjected to immunoblot analysis as described in the legend for Fig. 3C to F. (Right) Quantitative analysis of the upshifted CLOCK protein compared with the total CLOCK protein (phospho-CLK) by densitometry of the blots shown at the top of the left panel, lanes 3, 6, and 9. Data are means with standard errors of the means from four experiments. Double asterisks indicate a P value of <0.01 (Student's t test). WB, Western blotting; WT, wild type.
To investigate whether the mobility shift of the CLOCK protein is due to the phosphorylation of Ser38, Ser42, and/or Ser427, a Ser-to-Ala mutation was introduced at these sites, and the mutants were expressed with BMAL1 and CIPC in NIH 3T3 cells. The Ser427Ala mutation significantly decreased the intensity of the slower-migrating band of CLOCK (Fig. 4F), indicating that Ser427 of wild-type CLOCK is phosphorylated in NIH 3T3 cells. On the other hand, a paired mutation of Ser38Ala and Ser42Ala (Fig. 4F) caused no reduction of CLOCK phosphorylation (upshifted band).
Ser38Asp and Ser42Asp mutations of CLOCK inhibit its transactivation activity, with reduction of its nuclear amount and the DNA-binding activity.
The CLOCK-BMAL1 heterodimer is known to bind rhythmically to E-box elements in the promoter region of the Dbp gene (41). To test the hypothesis that phosphorylation of Ser38 and Ser42 affects CLOCK-BMAL1-mediated transactivation, we prepared additional CLOCK mutants, in which either Ser38 or Ser42 (or both) was mutated to Asp to mimic the phosphorylation. A dual luciferase reporter assay with HEK293T cells showed that a single Asp mutation at either Ser38 or Ser42 significantly decreased CLOCK-BMAL1-mediated transactivation (Fig. 5A). The double mutation suppressed the activity much more strongly, due to the additive inhibitory effect of the Asp mutations (Fig. 5A and B), with no significant change in total protein levels (data not shown). The additive inhibition by Asp mutations at Ser38 and Ser42 of CLOCK was also observed in the dual luciferase reporter assay with NIH 3T3 cells (data not shown).
A possible mechanism that accounts for the reduction in the transactivation ability of the Ser38Asp/Ser42Asp mutant could be the phosphorylation-dependent regulation of the subcellular localization of CLOCK. The nuclear localization is tightly coupled with the transactivation ability of CLOCK-BMAL1, and the nuclear localization signal (NLS) of BMAL1 has been identified at a position near the bHLH region (31). To investigate the domain(s) responsible for the nuclear translocation of CLOCK, we performed a computer-based sequence analysis, which predicted two putative NLSs, K33AKR36 and K44KRR47, which are located very close to each other and to the phosphorylation sites Ser38 and Ser42, respectively. To determine whether the bipartite NLS motif was functional, we constructed a plasmid expressing a CLOCK hexadecapeptide, Asp32-Arg47, that is N-terminally tagged with green fluorescent protein (GFP) (termed GFP-basic) and investigated its subcellular localization in NIH 3T3 cells. While GFP was localized in the nucleus and the cytoplasm with similar intensities (class III), GFP-basic was localized exclusively in the nucleus (class I), indicating that Asp32-Arg47 of CLOCK functions as the NLS (Fig. 5C and D). To investigate the possibility that the bipartite NLS function is regulated by phosphorylation of Ser38 and Ser42, we constructed an additional plasmid expressing “GFP-mut-basic,” in which Ser38 and Ser42 were both mutated to Asp. When GFP-mut-basic was expressed in NIH 3T3 cells, the distribution pattern of the GFP signals in the major population (∼79%) of the cells shifted to class II (localized strongly in the nucleus and weakly in the cytoplasm), indicating that Ser38Asp and Ser42Asp mutations weaken the bipartite NLS function (Fig. 5C and D). To determine the significance of the NLS function in the native CLOCK-BMAL1 heterodimer, we investigated the subcellular localization of wild-type CLOCK or Ser38Asp/Ser42Asp mutant CLOCK coexpressed with BMAL1 in HEK293T cells. Quantification of the nuclear abundance showed that the Ser38Asp and Ser42Asp mutations reduced the nuclear abundance of CLOCK to ∼64% of that of wild-type CLOCK (Fig. 5E), while no significant change in the total CLOCK protein levels was detected (data not shown). The decrease in nuclear abundance of the Ser38Asp/Ser42Asp mutant was also detected when CLOCK and BMAL1 were coexpressed with CIPC in NIH 3T3 cells (data not shown).
Ser38 and Ser42 of CLOCK are located in the basic region of the bHLH DNA-binding domain (Fig. 4E), and the DNA-binding activity of CLOCK-BMAL1 to the E-box sequence is known to exhibit circadian changes (41), raising the possibility that phosphorylation of Ser38 and Ser42 may also reduce the DNA-binding activity of CLOCK. To test the hypothesis, the binding activity of CLOCK-BMAL1 with a DNA probe that includes the E-box sequence of the Per1 promoter was investigated by EMSA. Under this condition, wild-type CLOCK and BMAL1 formed a complex with the DNA probe (Fig. 5F), and Asp mutations at Ser38 and Ser42 additively and significantly reduced the DNA-binding activity of CLOCK-BMAL1 (Fig. 5G). Collectively, these results show that CLOCK phosphorylation, especially at Ser38 and Ser42, appears to suppress the transactivation activity of CLOCK through regulation of the bHLH functions: inhibition of the NLS function and decrease of DNA binding.
Phosphorylation of CLOCK leads to its protein degradation.
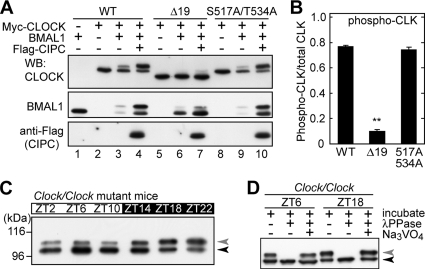
The physiological significance of CLOCK phosphorylation was studied in vivo by using Clock mutant mice (58). The CLOCKΔ19 protein, encoded by the mutated Clock gene, lacks 51 amino acids (2, 24) corresponding to exon 19, which constitutes the CIPC-binding domain (63). We predicted that CIPC-stimulated phosphorylation of CLOCK (Fig. 3F and G) should be impaired in CLOCKΔ19. We first investigated the phosphorylation level of CLOCKΔ19 expressed in NIH 3T3 cells. When CLOCKΔ19, instead of wild-type CLOCK, was coexpressed with BMAL1 and CIPC, not only the phosphorylation level of CLOCK but also the CLOCK-dependent up-shift (phosphorylation) of BMAL1 was significantly reduced (Fig. 6A, compare lane 4 with lane 7, and B). It should be noted, however, that BMAL1 was again stabilized by coexpression with CLOCKΔ19 and CIPC (Fig. 6A, compare lane 6 with lane 7), as it was by CLOCK and CIPC coexpression (Fig. 6A, compare lane 3 with lane 4). In this way, CIPC influenced BMAL1 stability even without efficient interaction with CLOCKΔ19. It is probable that CIPC also interacts with CLOCKΔ19-BMAL1 in a manner divergent from that with CLOCK-BMAL1 (see Fig. 7F).
FIG. 6.
The CLOCKΔ19 mutation reduces CLOCK phosphorylation. (A) CLOCK mutants were expressed with BMAL1 and CIPC as indicated and subjected to immunoblot analysis, as described in the legend for Fig. 3C to F. (B) Quantitative analysis of up-shifted CLOCK protein (phospho-CLK) compared with total CLOCK protein by densitometry of the blots shown in panel A, lanes 4, 7, and 10. Data are means with standard errors of the means from four experiments. A double asterisk indicates a P value of <0.01 (Student's t test). (C) Daily variation of CLOCK phosphorylation in the livers of Clock/Clock mutant mice. Nuclear extracts (100 μg of proteins) prepared at the indicated time points were immunoprecipitated with CLNT1 anti-CLOCK MAb. The immunoprecipitates were subjected to immunoblot analysis with CLSP3 anti-CLOCK MAb. (D) The immunoprecipitates were incubated at 30°C for 30 min with lambda protein phosphatase (λPPase) and 25 mM sodium vanadate as indicated. After the incubation, the samples were immunoblotted with CLSP3 anti-CLOCK MAb. WB, Western blotting; WT, wild type.
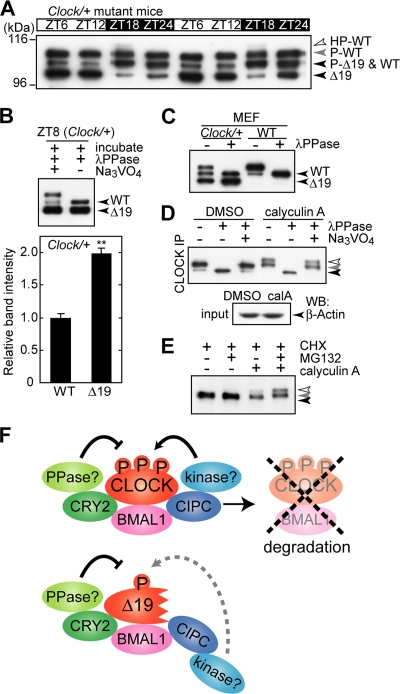
FIG. 7.
CLOCK phosphorylation leads to proteasomal degradation. (A) Daily variations of CLOCK and CLOCKΔ19 proteins in Clock/+ heterozygous mice. Nuclear extracts (100 μg of proteins) prepared from the mouse liver at the indicated time points were immunoprecipitated with anti-CLOCK MAb. The immunoprecipitates were subjected to immunoblot analysis with CLSP3 anti-CLOCK MAb. (B) (Top) Nuclear extracts were prepared from Clock/+ heterozygous mice and subjected to IP, followed by phosphatase treatment and immunoblot analysis with CLSP3 anti-CLOCK MAb. (Bottom) Quantitative analysis of amounts of CLOCK and CLOCKΔ19 proteins by densitometry of the blots shown in the top panel, lane 2. The mean value of the band intensity of wild-type (WT) CLOCK was set to 1. Data are means with standard errors of the means from four experiments. A double asterisk indicates a P value of <0.01 (Student's t test). (C) Mouse embryonic fibroblasts (MEF) (WT and Clock/+) cultured in a 10-cm dish were collected, and total protein extracts were immunoprecipitated with CLNT1 anti-CLOCK MAb, followed by phosphatase treatment and immunoblot analysis with CLSP3 anti-CLOCK MAb. (D) NIH 3T3 cells were cultured in a 10-cm dish, and 20 nM calyculin A was added. After a 90-min incubation, the cells were collected and immunoprecipitated with CLNT1 anti-CLOCK MAb. The immunoprecipitates were incubated at 30°C for 30 min with lambda protein phosphatase (λPPase) and 25 mM sodium vanadate as indicated. After the incubation, the samples were immunoblotted with CLSP3 anti-CLOCK MAb (top). Total protein extracts were immunoblotted with anti-β-actin antibody as a sampling control (bottom). (E) NIH 3T3 cells were cultured with 100 μM cycloheximide (CHX) and 25 μM MG132 as indicated. After a 1-h incubation, 20 nM calyculin A was added. After a 1-h incubation, the cells were collected and immunoprecipitated with CLNT1 anti-CLOCK MAb. The immunoprecipitates were subjected to immunoblot analysis with CLSP3 anti-CLOCK MAb. (F) Model of the difference between CLOCKΔ19 and WT CLOCK. Translated CLOCK and BMAL1 form a heterodimer, and the dimerization causes their phosphorylation. CRY2 suppresses their phosphorylation possibly by recruiting protein phos-phatase (PPase), while CIPC stimulates CLOCK phosphorylation possibly by recruiting protein kinase. Hyperphosphorylation of CLOCK leads to proteasomal degradation. In the case of Clock mutant mice, CLOCKΔ19 and BMAL1 can dimerize but are hypophosphorylated because of a deficiency in the reception of phosphorylation signals for degradation. HP-, hyperphosphorylated; P-, phosphorylated; DMSO, dimethyl sulfoxide; calA, calyculin A.
Based on these observations, we positioned the Clock mutant mouse as the mouse model harboring a deficiency in receiving normal signals for CIPC-dependent phosphorylation of the CLOCK-BMAL1 complex. As expected, in Clock/Clock mutant mice, the Clock gene product CLOCKΔ19 was poorly phosphorylated in the liver, as visualized by the relative abundance of the unphosphorylated band (Fig. 6C and D, black arrowhead), which corresponds to the extremely minor species of CLOCK in the wild-type mouse liver (Fig. 2A, black arrowhead). In the immunoblot analysis, we noticed that the signal intensity of CLOCKΔ19 in the liver nuclear extract prepared from the Clock/Clock mutant mouse was remarkably higher than that of CLOCK in the wild-type mouse when the same amounts of nuclear proteins were loaded on the gel (data not shown). These data suggest the phosphorylation-dependent destabilization of wild-type CLOCK. This possibility was examined in the Clock/+ heterozygous mouse in vivo by comparing the protein amount of CLOCKΔ19 encoded by the mutated allele with that of CLOCK encoded by the wild-type allele. For easier comparison between the proteins, the liver lysate prepared at ZT8 was treated with lambda protein phosphatase in order to integrate the shifted and nonshifted bands into a single band for each of the wild-type CLOCK and the CLOCKΔ19 proteins. Then, it was demonstrated that the amount of CLOCKΔ19 was twofold higher than that of wild-type CLOCK (Fig. 7B). Furthermore, the stabilization of CLOCKΔ19 was observed in Clock/+ heterozygous mouse embryonic fibroblasts (Fig. 7C). Again, even in heterozygous (Clock/+) mice, the wild-type CLOCK protein was kept mostly in the phosphorylated form throughout the day, whereas the major species of CLOCKΔ19 at ZT6 to ZT12 was the unphosphorylated band (Fig. 7A). It is considered that a reduction in CLOCK phosphorylation caused the protein stabilization in Clock mutant mice.
Finally, we examined the effect of the promotion of CLOCK phosphorylation on protein stability in NIH 3T3 cells. We found that the hyperphosphorylated form of CLOCK accumulated by treatment of cultured NIH 3T3 cells with calyculin A (Fig. 7D, open arrowhead), which is a cell-permeable inhibitor of protein phosphatase (types 1 and 2A) and is known to promote PER2 phosphorylation (11). These results substantiate the equilibrium between phosphorylation and dephosphorylation of CLOCK protein in NIH 3T3 cells. Importantly, the calyculin A treatment reduced the amount of CLOCK protein (Fig. 7E), and the calyculin A-induced decrease of CLOCK protein was inhibited in the presence of MG132, a 26S proteasome inhibitor (Fig. 7E). These observations demonstrate that phosphorylated CLOCK, most probably in the hyperphosphorylated form, is directed toward the 26S proteasome-mediated degradation pathway (Fig. 7F). Together, these results show that the phosphorylation rhythm of the CLOCK protein should play an important role in the temporal regulation of not only the protein level but also the function of CLOCK.
DISCUSSION
CLOCKΔ19 has less ability than wild-type CLOCK to receive a phosphorylation signal that is important for its degradation.
Clock (circadian locomotor output cycles kaput) was identified as the first vertebrate clock gene mutated in Clock mutant mice (2, 24), which exhibit a free-running period of locomotor activity that is ∼1 h longer (Clock/+ mice) or arrhythmic/∼4 h longer (Clock/Clock mice) than that exhibited by wild-type mice (58). The mutant mice express CLOCKΔ19, which has an internal deletion of 51 amino acids (2, 24), and the mutation was shown to reduce the transactivation ability of CLOCK without affecting its dimerization with BMAL1 or its binding to E-box elements (14). In the present study, we found that CLOCKΔ19 is far less phosphorylated and much more stabilized than wild-type CLOCK (Fig. 7F). These molecular phenotypes of the mutant protein can be explained by the black widow model (35, 51), which illustrates properties of some transcription factors that limit temporally their own activities through destabilization of their active forms so as to fine-tune the transactivation of the downstream genes. In this sense, CLOCKΔ19 may be characterized by a deficiency in its conversion into the active form that can timely receive the phosphorylation signals for degradation. However, this model does not explain our data that MG132 treatment of HEK293T cells reduced the transactivation of the CLOCK-BMAL1 complex in the dual luciferase reporter assay (data not shown), raising another possibility that the degradation of CLOCK itself is required for the enhancement of its transactivation activity. Transcription factors, which bind to DNA elements for transactivation of the downstream genes, need to be removed from the elements in order to proceed to the next round of the “transcriptional clock” (34). CLOCKΔ19 may take a longer time than wild-type CLOCK to be removed from E-box elements, due to its lesser ability to receive the phosphorylation signals for degradation, and hence CLOCKΔ19 should have been unable to keep its level of transactivation activity as high as that of wild-type CLOCK.
Phosphorylations of Ser38/Ser42 located in the bHLH of CLOCK reduce the transactivation ability of CLOCK.
In this study, we identified Ser38, Ser42, and Ser427 as in vivo phosphorylation sites of CLOCK (Fig. 4), but additional phosphorylation sites of CLOCK are likely left undetermined in the present analysis, considering the sequence coverage in the MS-MS analysis. We found an important role of CLOCK phosphorylation in the degradation of CLOCK, whereas no significant change in protein stability was caused by any mutation of the phosphorylation sites, indicating that additional phosphorylation sites of CLOCK might contribute to the promotion of its protein degradation. On the other hand, Ser-to-Asp mutations of Ser38 and Ser42 markedly suppressed the transactivation ability of CLOCK in cultured cells (Fig. 5A and B). A subcellular-distribution analysis of GFP fusion proteins revealed that the basic region (Asp32-Arg47) of CLOCK retains an NLS function and that the Ser38Asp and Ser42Asp mutations inhibit the NLS function (Fig. 5C and D). In fact, the Ser38Asp and Ser42Asp mutations decreased nuclear amounts of the CLOCK protein expressed in HEK293T cells (Fig. 5E) and NIH 3T3 cells (data not shown). A bipartite NLS has been found in the basic region of SRC-3/ACTR, a bHLH transcription factor having a HAT domain, as CLOCK does (8, 33). This similarity suggests that the basic region of some bHLH transcription factors may function as an NLS, and our data imply that the NLS of CLOCK may be regulated by phosphorylation.
In addition to inhibiting NLS function, the Ser38Asp and Ser42Asp mutations reduced the DNA-binding activity of CLOCK (Fig. 5F and G). Ser38 and Ser42 are located in the basic region of the bHLH DNA-binding domain of CLOCK (Fig. 4E), and the DNA-binding activity of CLOCK-BMAL1 to the E-box sequence is known to exhibit circadian changes (41), raising the possibility that the phosphorylation of Ser38 and Ser42 may reduce the DNA-binding activity of CLOCK. With the Neurospora clock, the DNA binding of WC1 is thought to be inhibited by phosphorylation of five Ser residues that are located very close to the DNA-binding region (16, 17, 21, 44). Circadian phosphorylation-dependent regulation of the DNA-binding ability of the positive regulators may be a key mechanism commonly underlying the clockwork operating in a variety of organisms.
CLOCK phosphorylation is regulated by both protein kinase and phosphatase.
The protein kinase and phosphatase that regulate the phosphorylation states of CLOCK and BMAL1 in vivo are left undetermined. In vitro experiments revealed that CLOCK and BMAL1 are phosphorylated by multiple kinases: PKG (52) and PKC (47) phosphorylate CLOCK, while MAPK (42) and CKIɛ (10) phosphorylate BMAL1. In the present study, we found that Ser38, Ser42, and Ser427 of CLOCK are phosphorylated in the mouse liver (Fig. 4). It appears that CLOCK has multiple phosphorylation sites and that the total phosphorylation levels of CLOCK and BMAL1 have their peaks at different circadian phases in the mouse liver (Fig. 2). It is most probable that the phosphorylation state of the CLOCK-BMAL1 complex is regulated by multiple protein kinases at different phases of the circadian rhythm and that each of the phosphorylation sites contributes to multiple aspects of CLOCK-BMAL1 function in order to fine-tune the negative-feedback loop oscillating with 24-h periodicity.
A variety of clock proteins have been shown to undergo dephosphorylation by protein phosphatase in vivo (reviewed in reference 13). For example, protein phosphatase PP1 negatively regulates PER2 phosphorylation, which leads to the degradation of PER2 in mammals (12). In Drosophila, the PP2A regulatory subunits TWS and WDB were shown to target PER and stabilize it (43). In mammalian cells, CRY1 and CRY2 interact with PP5, and knockdown of PP5 reduces the circadian change of PER1/2 protein abundance (39). In the present study, calyculin A treatment of NIH 3T3 cells promoted the hyperphosphorylation and degradation of CLOCK (Fig. 7D and E), indicating that CLOCK phosphorylation is regulated by protein phosphatase in the cells. It is intriguing to note that coexpression of CRY2 with CLOCK and BMAL1 caused significant inhibition of CLOCK-BMAL1 phosphorylation in NIH 3T3 cells (Fig. 3D). Consistently, coexpression of either CRY1 or CRY2 was shown to increase the amount of nonphosphorylated BMAL1 in HEK293 cells (29) and COS7 cells (6). The CRY (and PER) protein may serve as an adaptor protein that recruits protein phosphatase to the CLOCK-BMAL1 complex (Fig. 7F).
Novel role of CIPC as a regulator of CLOCK phosphorylation and function.
In the mouse liver, the mRNA level of Clock shows a robust circadian rhythm, with a high peak/trough ratio peaking at the suppression phase of E-box-dependent transcription (Fig. 2C), while the total protein level of CLOCK is kept almost constant throughout the day (Fig. 2A and B). Considering that the hyperphosphorylated form of CLOCK was detected only at the suppression phase (ZT/CT18) (Fig. 2) and that CLOCK phosphorylation plays a key role in triggering the degradation of CLOCK (Fig. 7F), we speculate that the CLOCK protein may undergo temporally regulated turnover, with an extremely labile period in the suppression phase. The acceleration of CLOCK degradation near the peak time of its mRNA level may help to keep its protein level apparently constant throughout the day. Recent studies of Drosophila have demonstrated the regulatory mechanism of the phosphorylation-dependent degradation of the dCLK protein (22, 23, 37, 62). The mechanism appears to be partially conserved between mammals and flies; the total protein level of dCLK is constant despite the circadian oscillation of the dClk mRNA level, peaking at the suppression phase, a time period when PER recruits Doubletime kinase (DBT) to the dCLK-CYC complex to yield hyperphosphorylated dCLK. In contrast to the clockwork in Drosophila, however, the coexpression of PER2 with CLOCK in NIH 3T3 cells resulted in only slight upregulation of CLOCK phosphorylation (Fig. 3E). Instead, we found that BMAL1-dependent phosphorylation of CLOCK is stimulated by the coexpression of CIPC (Fig. 3F and G), a protein that has no invertebrate homolog. The Cipc gene has E-box elements, and not only its transcript level but also its protein level shows circadian rhythms (63). Furthermore, CLOCKΔ19, lacking the CIPC-binding domain, is far less phosphorylated than wild-type CLOCK in the mouse liver and NIH 3T3 cells (Fig. 6). In vertebrates, CIPC may regulate CLOCK phosphorylation in a manner similar to that by which invertebrate PER regulates dCLK phosphorylation.
The 51 amino acids encoded by exon 19 of Clock include two phosphorylatable residues, Ser517 and Thr537, but a paired mutation of Ser517Ala and Thr537Ala had no significant effect on the mobility shifts of CLOCK protein when it was coexpressed with BMAL1 and CIPC in NIH 3T3 cells (Fig. 6A and B). Therefore, Ser517 and Thr534 of CLOCK are unlikely to contribute to the CIPC-mediated regulation of CLOCK phosphorylation. Rather, a functional interaction of the 51-amino-acid region with clock proteins, such as CIPC, should be important for the regulation of CLOCK phosphorylation (Fig. 7F). We showed that the CLOCKΔ19 protein was far less phosphorylated and much more stabilized than wild-type CLOCK in the mouse liver (Fig. 7E). The in vivo importance of the molecular phenotypes of the mutant protein is strengthened by the fact that Clock/Clock mutant mice exhibit a free-running period of the locomotor activity as much as ∼4 h longer than that of wild-type mice (58). Collectively, these results show that CIPC/CRY2-modifiable CLOCK/BMAL1 phosphorylation should play a key role in triggering the suppression of the E-box-dependent transactivation and contribute to the 24-h cycling of the molecular clock.
Acknowledgments
We thank Joseph S. Takahashi (Northwestern University, Evanston, IL) for the generous gifts of Clock mutant mice and Takeshi Todo (Osaka University) for the anti-CRY2 antibody. We also thank Munenori Komori, Shumpei Yamauchi, and Hiroshi Kiyota for technical assistance and Naohiro Kon for critical reading of the manuscript.
This work was supported in part by grants-in-aid and by the Global COE program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from MEXT, Japan. This work was performed partly under the Cooperative Research Program of the Institute for Protein Research, Osaka University. H.Y. is supported by JSPS Research Fellowships for Young Scientists.
Footnotes
Published ahead of print on 4 May 2009.
REFERENCES
- 1.Akashi, M., Y. Tsuchiya, T. Yoshino, and E. Nishida. 2002. Control of intracellular dynamics of mammalian period proteins by casein kinase I ɛ (CKIɛ) and CKIδ in cultured cells. Mol. Cell. Biol. 221693-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoch, M., E. Song, A. Chang, M. Vitaterna, Y. Zhao, L. Wilsbacher, A. Sangoram, D. King, L. Pinto, and J. Takahashi. 1997. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell 89655-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsalobre, A., S. Brown, L. Marcacci, F. Tronche, C. Kellendonk, H. Reichardt, G. Schütz, and U. Schibler. 2000. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 2892344-2347. [DOI] [PubMed] [Google Scholar]
- 4.Balsalobre, A., F. Damiola, and U. Schibler. 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93929-937. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S., J. Ripperger, S. Kadener, F. Fleury-Olela, F. Vilbois, M. Rosbash, and U. Schibler. 2005. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308693-696. [DOI] [PubMed] [Google Scholar]
- 6.Dardente, H., E. Fortier, V. Martineau, and N. Cermakian. 2007. Cryptochromes impair phosphorylation of transcriptional activators in the clock: a general mechanism for circadian repression. Biochem. J. 402525-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dignam, J., R. Lebovitz, and R. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doi, M., J. Hirayama, and P. Sassone-Corsi. 2006. Circadian regulator CLOCK is a histone acetyltransferase. Cell 125497-508. [DOI] [PubMed] [Google Scholar]
- 9.Dunlap, J. 1999. Molecular bases for circadian clocks. Cell 96271-290. [DOI] [PubMed] [Google Scholar]
- 10.Eide, E., E. Vielhaber, W. Hinz, and D. Virshup. 2002. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J. Biol. Chem. 27717248-17254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eide, E., M. Woolf, H. Kang, P. Woolf, W. Hurst, F. Camacho, E. Vielhaber, A. Giovanni, and D. Virshup. 2005. Control of mammalian circadian rhythm by CKIɛ-regulated proteasome-mediated PER2 degradation. Mol. Cell. Biol. 252795-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallego, M., H. Kang, and D. Virshup. 2006. Protein phosphatase 1 regulates the stability of the circadian protein PER2. Biochem. J. 399169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallego, M., and D. Virshup. 2007. Post-translational modifications regulate the ticking of the circadian clock. Nat. Rev. Mol. Cell Biol. 8139-148. [DOI] [PubMed] [Google Scholar]
- 14.Gekakis, N., D. Staknis, H. Nguyen, F. Davis, L. Wilsbacher, D. King, J. Takahashi, and C. Weitz. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 2801564-1569. [DOI] [PubMed] [Google Scholar]
- 15.Harada, Y., M. Sakai, N. Kurabayashi, T. Hirota, and Y. Fukada. 2005. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J. Biol. Chem. 28031714-31721. [DOI] [PubMed] [Google Scholar]
- 16.He, Q., J. Cha, H. Lee, Y. Yang, and Y. Liu. 2006. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 202552-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, Q., H. Shu, P. Cheng, S. Chen, L. Wang, and Y. Liu. 2005. Light-independent phosphorylation of WHITE COLLAR-1 regulates its function in the Neurospora circadian negative feedback loop. J. Biol. Chem. 28017526-17532. [DOI] [PubMed] [Google Scholar]
- 18.Hirota, T., and Y. Fukada. 2004. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog. Sci. 21359-368. [DOI] [PubMed] [Google Scholar]
- 19.Hirota, T., T. Okano, K. Kokame, H. Shirotani-Ikejima, T. Miyata, and Y. Fukada. 2002. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured Rat-1 fibroblasts. J. Biol. Chem. 27744244-44251. [DOI] [PubMed] [Google Scholar]
- 20.Honma, S., T. Kawamoto, Y. Takagi, K. Fujimoto, F. Sato, M. Noshiro, Y. Kato, and K. Honma. 2002. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419841-844. [DOI] [PubMed] [Google Scholar]
- 21.Huang, G., S. Chen, S. Li, J. Cha, C. Long, L. Li, Q. He, and Y. Liu. 2007. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 213283-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, E., and I. Edery. 2006. Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc. Natl. Acad. Sci. USA 1036178-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, E., H. Ko, W. Yu, P. Hardin, and I. Edery. 2007. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol. Cell. Biol. 275014-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King, D., Y. Zhao, A. Sangoram, L. Wilsbacher, M. Tanaka, M. Antoch, T. Steeves, M. Vitaterna, J. Kornhauser, P. Lowrey, F. Turek, and J. Takahashi. 1997. Positional cloning of the mouse circadian clock gene. Cell 89641-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiyohara, Y., K. Nishii, M. Ukai-Tadenuma, H. Ueda, Y. Uchiyama, and K. Yagita. 2008. Detection of a circadian enhancer in the mDbp promoter using prokaryotic transposon vector-based strategy. Nucleic Acids Res. 36e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Köhler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256495-497. [DOI] [PubMed] [Google Scholar]
- 27.Kon, N., T. Hirota, T. Kawamoto, Y. Kato, T. Tsubota, and Y. Fukada. 2008. Activation of TGF-beta/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat. Cell Biol. 101463-1469. [DOI] [PubMed] [Google Scholar]
- 28.Kondratov, R., M. Chernov, A. Kondratova, V. Gorbacheva, A. Gudkov, and M. Antoch. 2003. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 171921-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondratov, R., A. Kondratova, C. Lee, V. Gorbacheva, M. Chernov, and M. Antoch. 2006. Post-translational regulation of circadian transcriptional CLOCK(NPAS2)/BMAL1 complex by CRYPTOCHROMES. Cell Cycle 5890-895. [DOI] [PubMed] [Google Scholar]
- 30.Kume, K., M. Zylka, S. Sriram, L. Shearman, D. Weaver, X. Jin, E. Maywood, M. Hastings, and S. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98193-205. [DOI] [PubMed] [Google Scholar]
- 31.Kwon, I., J. Lee, S. Chang, N. Jung, B. Lee, G. Son, K. Kim, and K. Lee. 2006. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol. Cell. Biol. 267318-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, C., J. Etchegaray, F. Cagampang, A. Loudon, and S. Reppert. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107855-867. [DOI] [PubMed] [Google Scholar]
- 33.Li, C., R. Wu, L. Amazit, S. Tsai, M. Tsai, and B. O'Malley. 2007. Specific amino acid residues in the basic helix-loop-helix domain of SRC-3 are essential for its nuclear localization and proteasome-dependent turnover. Mol. Cell. Biol. 271296-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Métivier, R., G. Penot, M. Hübner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115751-763. [DOI] [PubMed] [Google Scholar]
- 35.Muratani, M., and W. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4192-201. [DOI] [PubMed] [Google Scholar]
- 36.Nagoshi, E., C. Saini, C. Bauer, T. Laroche, F. Naef, and U. Schibler. 2004. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119693-705. [DOI] [PubMed] [Google Scholar]
- 37.Nawathean, P., D. Stoleru, and M. Rosbash. 2007. A small conserved domain of Drosophila PERIOD is important for circadian phosphorylation, nuclear localization, and transcriptional repressor activity. Mol. Cell. Biol. 275002-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okano, T., K. Yamamoto, K. Okano, T. Hirota, T. Kasahara, M. Sasaki, Y. Takanaka, and Y. Fukada. 2001. Chicken pineal clock genes: implication of BMAL2 as a bidirectional regulator in circadian clock oscillation. Genes Cells 6825-836. [DOI] [PubMed] [Google Scholar]
- 39.Partch, C., K. Shields, C. Thompson, C. Selby, and A. Sancar. 2006. Posttranslational regulation of the mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc. Natl. Acad. Sci. USA 10310467-10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reppert, S., and D. Weaver. 2002. Coordination of circadian timing in mammals. Nature 418935-941. [DOI] [PubMed] [Google Scholar]
- 41.Ripperger, J., and U. Schibler. 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38369-374. [DOI] [PubMed] [Google Scholar]
- 42.Sanada, K., T. Okano, and Y. Fukada. 2002. Mitogen-activated protein kinase phosphorylates and negatively regulates basic helix-loop-helix-PAS transcription factor BMAL1. J. Biol. Chem. 277267-271. [DOI] [PubMed] [Google Scholar]
- 43.Sathyanarayanan, S., X. Zheng, R. Xiao, and A. Sehgal. 2004. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 116603-615. [DOI] [PubMed] [Google Scholar]
- 44.Schafmeier, T., A. Haase, K. Káldi, J. Scholz, M. Fuchs, and M. Brunner. 2005. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122235-246. [DOI] [PubMed] [Google Scholar]
- 45.Schibler, U., and P. Sassone-Corsi. 2002. A web of circadian pacemakers. Cell 111919-922. [DOI] [PubMed] [Google Scholar]
- 46.Shearman, L., X. Jin, C. Lee, S. Reppert, and D. Weaver. 2000. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol. Cell. Biol. 206269-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shim, H., H. Kim, J. Lee, G. Son, S. Cho, T. Oh, S. Kang, D. Seen, K. Lee, and K. Kim. 2007. Rapid activation of CLOCK by Ca2+-dependent protein kinase C mediates resetting of the mammalian circadian clock. EMBO Rep. 8366-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takano, A., Y. Isojima, and K. Nagai. 2004. Identification of mPer1 phosphorylation sites responsible for the nuclear entry. J. Biol. Chem. 27932578-32585. [DOI] [PubMed] [Google Scholar]
- 49.Takano, A., K. Shimizu, S. Kani, R. Buijs, M. Okada, and K. Nagai. 2000. Cloning and characterization of rat casein kinase 1epsilon. FEBS Lett. 477106-112. [DOI] [PubMed] [Google Scholar]
- 50.Tamaru, T., Y. Isojima, G. van der Horst, K. Takei, K. Nagai, and K. Takamatsu. 2003. Nucleocytoplasmic shuttling and phosphorylation of BMAL1 are regulated by circadian clock in cultured fibroblasts. Genes Cells 8973-983. [DOI] [PubMed] [Google Scholar]
- 51.Tansey, W. 2001. Transcriptional activation: risky business. Genes Dev. 151045-1050. [DOI] [PubMed] [Google Scholar]
- 52.Tischkau, S., J. Mitchell, L. Pace, J. Barnes, J. Barnes, and M. Gillette. 2004. Protein kinase G type II is required for night-to-day progression of the mammalian circadian clock. Neuron 43539-549. [DOI] [PubMed] [Google Scholar]
- 53.Toh, K., C. Jones, Y. He, E. Eide, W. Hinz, D. Virshup, L. Ptácek, and Y. Fu. 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 2911040-1043. [DOI] [PubMed] [Google Scholar]
- 54.Ueda, H., S. Hayashi, W. Chen, M. Sano, M. Machida, Y. Shigeyoshi, M. Iino, and S. Hashimoto. 2005. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 37187-192. [DOI] [PubMed] [Google Scholar]
- 55.Vanselow, K., J. Vanselow, P. Westermark, S. Reischl, B. Maier, T. Korte, A. Herrmann, H. Herzel, A. Schlosser, and A. Kramer. 2006. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 202660-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vielhaber, E., D. Duricka, K. Ullman, and D. Virshup. 2001. Nuclear export of mammalian PERIOD proteins. J. Biol. Chem. 27645921-45927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vielhaber, E., E. Eide, A. Rivers, Z. Gao, and D. Virshup. 2000. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I ɛ. Mol. Cell. Biol. 204888-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vitaterna, M., D. King, A. Chang, J. Kornhauser, P. Lowrey, J. McDonald, W. Dove, L. Pinto, F. Turek, and J. Takahashi. 1994. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yagita, K., F. Tamanini, M. Yasuda, J. Hoeijmakers, G. van der Horst, and H. Okamura. 2002. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 211301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yagita, K., S. Yamaguchi, F. Tamanini, G. van der Horst, J. Hoeijmakers, A. Yasui, J. Loros, J. Dunlap, and H. Okamura. 2000. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 141353-1363. [PMC free article] [PubMed] [Google Scholar]
- 61.Yoo, S., C. Ko, P. Lowrey, E. Buhr, E. Song, S. Chang, O. Yoo, S. Yamazaki, C. Lee, and J. Takahashi. 2005. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. USA 1022608-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu, W., H. Zheng, J. Houl, B. Dauwalder, and P. Hardin. 2006. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 20723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao, W., N. Malinin, F. Yang, D. Staknis, N. Gekakis, B. Maier, S. Reischl, A. Kramer, and C. Weitz. 2007. CIPC is a mammalian circadian clock protein without invertebrate homologues. Nat. Cell Biol. 9268-275. [DOI] [PubMed] [Google Scholar]