Abstract
The xCR1 protein is a maternal determinant and cofactor for nodal signaling in vertebrate embryos. The xCR1 protein accumulates specifically in the animal cells of Xenopus embryos, but maternal xCR1 mRNA is distributed equally throughout all embryonic cells. Here, we show that vegetal cell-specific translational repression of xCR1 mRNA contributes to this spatially restricted accumulation of the xCR1 protein in Xenopus embryos. xCR1 mRNA was associated with polyribosomes in animal cells but not vegetal cells. A 351-nucleotide region of xCR1 mRNA's 3′ untranslated region was sufficient to confer a spatially restricted pattern of translation to a luciferase reporter mRNA by repressing translation in vegetal cells. Repression depended upon the mRNA's 5′ cap but not its 3′ poly(A) tail. Furthermore, the region of xCR1 mRNA sufficient to confer vegetal cell-specific repression contained both Pumilio binding elements (PBEs) and binding sites for the CUG-BP1 protein. The PBEs and the CUG-BP1 sites were necessary but not sufficient for translation repression. Our studies of xCR1 mRNA document the first example of spatially regulated translation in controlling the asymmetric distribution of a maternal determinant in vertebrates.
In vertebrate organisms, temporally and spatially regulated posttranscriptional events direct development during the first hours following fertilization, when there is an absence of zygotic transcription (5, 24, 49). These posttranscriptional events are critical for controlling the expression and/or the activity of maternally derived determinants, unique maternal proteins that control embryonic cell fate decisions (18). While studies of Xenopus laevis embryos have revealed that a diverse set of mechanisms control the expression of maternal determinants, including mRNA localization (23) and protein stability (54), the role of regulated maternal mRNA translation during early embryonic development is poorly understood. Translational regulation is a well-known mechanism that activates stored quiescent cell cycle mRNAs during oocyte maturation (5, 6, 49). However, the mechanisms that control the translation of mRNAs that encode maternal determinants during early vertebrate embryogenesis are likely to have unique features distinct from those that activate mRNAs encoding cell cycle proteins. In particular, mRNAs encoding cell cycle proteins must be translationally activated to produce copious amounts of uniformly distributed cell cycle proteins to drive the rapid early cell divisions. In contrast, the expression of maternal determinants must be tightly controlled in terms of absolute dosage as well as in terms of spatial and temporal location within the developing embryo. In this study, we analyze the control of the xCR1 mRNA encoding the nodal signaling protein Cripto, as a model for translational regulation of a maternal determinant in Xenopus embryos.
The nodal signaling pathway is a maternally derived pathway important for the development of vertebrate organisms (41). Cripto proteins are critical coreceptors of this pathway. In Xenopus, the Cripto protein xCR1 (Xenopus Cripto 1) is an important maternal determinant required for nodal signaling. Perturbing the expression of xCR1 affects anterior/posterior patterning (45, 53). Significantly, accumulation of the xCR1 protein during Xenopus development is both temporally and spatially regulated (9). The xCR1 protein accumulates in embryonic cells only after fertilization, and then, only in animal and marginal zone cells; the xCR1 protein is absent from vegetal cells. RNA localization mechanisms cannot explain the asymmetric distribution of the xCR1 protein since the xCR1 maternal mRNA is present during all stages of development and in all embryonic cells. Thus, regulated translation and differential protein stability are mechanisms that could explain the restricted localization of the xCR1 protein to animal cells.
In this study, we examined the contribution of regulated xCR1 mRNA translation to the spatially restricted accumulation of the xCR1 protein in animal cells of Xenopus embryos. We provide evidence that translation of xCR1 mRNA was activated after fertilization but was specifically repressed within the vegetal cells of embryos through the action of Pumilio binding elements (PBEs) and CUG-BP1 binding sites contained within xCR1 mRNA's 3′ untranslated region (3′ UTR). Thus, the correct temporal and spatial distribution of the maternal determinant xCR1 resulted from a combination of temporally controlled translational activation and spatially restricted repression mechanisms mediated by xCR1 mRNA's 3′ UTR.
MATERIALS AND METHODS
Polyribosome isolation and analysis.
Polyribosomes were isolated, and polyribosome-associated mRNAs were analyzed as described previously (10).
Luciferase reporter mRNA plasmids and mRNA synthesis.
xCR1 3′ UTRs were generated from the xCR1 cDNA by using PCR (9). The first nucleotide of the 3′ UTR is designated +1, and the last is designated +941. mRNAs were located as follows: wild-type xCR1-3′UTR-WT, nucleotides 1 to 941; xCR1-3′UTR-Mut1, nucleotides 1 to 308; xCR1-3′UTR-Mut2, nucleotides 286 to 637; xCR1-3′UTR-Mut3, nucleotides 615 to 941; xCR1 MUT2-PBE-Mut, nucleotides 286 to 637, where the UGU sequences of PBEs 1 to 3 were changed to ACA (see Fig. 6); 3′UTR-Mut4, nucleotides 286 to 464; xCR1-SH-Mut, nucleotides 469 to 941; xCR1-3′UTR-MUT2-CSFV-IRES, which contains the classical swine fever virus (CSFV) internal ribosome entry site (IRES) (35) and the Mut2 xCR1 3′ UTR; xCR1 MUT2-CUG-BP1-Mut, nucleotides 286 to 637, where the UGU sequences of the CUG-BP1 sites were changed to CGC (see Fig. 7). The 3′ UTRs were cloned via BglII and BamHI sites into the pT7LucBglII plasmid to generate luciferase reporter plasmids (11). The luciferase reporter plasmid containing the Xenopus cyclin B1 3′ UTR (pT7Luc/Xen CyclinB1 WT 3′UTR) has been described previously (10, 42). All plasmids were linearized with BamHI, and mRNAs were generated as described previously (10).
FIG. 6.
PBEs in the 3′ UTR of the xCR1 mRNA were necessary but not sufficient for vegetal cell-specific translational repression. (A) Sequence of the central 351 nucleotides of the xCR1 mRNA's 3′ UTR (nucleotides [nt] 286 to 637) present in the Mut2 reporter mRNA. The core sequences of three putative PBEs are highlighted in black. The sequences contained in the PBE-1/2 and PBE-3 RNAs used for Pumilio binding experiments are underlined. Nucleotide 464, which is the 3′ extent of the xCR1 3′UTR contained in Mut4, is indicated with a black triangle. (B) Xenopus Pumilio-1 protein binds the PBEs in the xCR1 3′ UTR. Radiolabeled RNAs were mixed with 0, 50, and 150 pmol of GST-Xen-pum1 protein, and the formation of protein/RNA complexes was analyzed by native gel electrophoresis. Lanes 1 to 3, PBE-1/2 RNA; lanes 4 to 6, PBE-3 RNA; lanes 7 to 9, negative control A30 RNA; lanes 10 to 12, NRE RNA, a positive control RNA derived from the Drosophila hunchback mRNA. (C) Pumilio binding to the xCR1 3′ UTR is specific. Radiolabeled RNAs were mixed with 0, 50, and 150 pmol of GST-Xen-pum1 protein, and binding was analyzed by native gel electrophoresis. Lanes 1 to 3, PBE-1/2 RNA; lanes 4 to 6, PBE-Mut RNA (UGU sequences of PBEs 1 and 2 were replaced by ACA); lanes 7 to 9, NRE positive control RNA. (D) Diagram of reporter mRNAs with xCR1 3′ UTRs containing wild-type and mutant PBEs. (E) Reporter mRNAs were injected and analyzed as described in the legends to Fig. 3 and 4. (F) Reporter mRNAs were equally stable in animal and vegetal cells after injection. Reporter RNA from injected cells was analyzed by RT-PCR, and the products were visualized by agarose gel electrophoresis.
FIG. 7.
CUG-BP1 binding sites in the 3′ UTR of the xCR1 mRNA were necessary but not sufficient for vegetal cell-specific translational repression. (A) Sequence of the central 351 nucleotides of the xCR1 mRNA's 3′ UTR (nucleotides [nt] 286 to 637) present in the Mut2 reporter mRNA. Two putative CUG-BP1 binding sites are boxed. (B) Diagram of reporter mRNAs with xCR1 3′ UTRs containing wild-type and mutant CUG-BP1 sites. (C) Reporter mRNAs were injected and analyzed as described in legends for Fig. 3 and 4. (D) Reporter mRNAs were equally stable in animal and vegetal cells after injection. Reporter RNA from injected cells was analyzed by RT-PCR, and the products were visualized by agarose gel electrophoresis.
mRNA injections and luciferase assays.
Each reporter mRNA was diluted to a concentration of 2.5 nM, and 5 nanoliters (12.5 amol) was injected into either the animal pole cells or the vegetal pole cell of eight-cell embryos. Injected embryos were cultured until stage 7, and extracts prepared from samples of 10 embryos were analyzed for luciferase activity.
Ligation-coupled reverse transcription-PCR (RT-PCR) to detect poly(A).
RNA was isolated from oocytes, eggs, embryos, and embryo fragments using Trizol. To remove poly(A) tails, oligo(dT) was annealed to the total RNA treated with RNase H (42). Four micrograms of RNA was ligated to 50 pmol of primer P1 by using T4 RNA ligase. The ligated RNA was used for cDNA synthesis, utilizing a primer complementary to P1 (P1′) and reverse transcriptase. The resulting cDNA products served as the template for PCR with a gene-specific sense primer 1280-1305, GCAGCAATGTAAGTGCTAGCCTGTGG, and the P1′ antisense primer in the presence of [32P]dCTP. Products were fractionated on 8% native polyacrylamide gels and visualized by autoradiography.
RT-PCR.
Equal amounts of total RNA extracted from luciferase reporter mRNA-injected embryos were used to synthesize cDNAs. The cDNA was used as a template to perform PCR using the following primers for luciferase: for-primer, 5′-GCTGTTTTTACGATCCCTTCAGG-3′, and rev-primer, 5′-CGGTCAACTATGAAGAAGTGTTCG-3′ (498-nucleotide product).
Electrophoretic mobility assays.
Recombinant glutathione S-transferase (GST)-Xpum1 protein was generated as described previously (40). RNA substrates were chemically synthesized and radiolabeled using T4 polynucleotide kinase. The PBE-1/2 RNA (nucleotides 323 to 369 of the xCR1 3′ UTR) contains PBEs 1 and 2 (see Fig. 5A). The PBE-3 RNA (nucleotides 364 to 389 of the xCR1 3′ UTR) contains PBE-3 (see Fig. 5A). The PBE-Mut RNA (nucleotides 323 to 369 of the xCR1 3′ UTR) contains ACA substitutions for the UGU sequences in PBEs 1 and 2 (see Fig. 5A). The A30 RNA is a 30-nucleotide poly(A) substrate that serves as a negative control. The positive control is the Nanos response element (NRE) RNA (from the Drosophila hunchback mRNA). Binding reaction mixtures (20 μl) were assembled with GST-Xenopus Pumilio-1 protein (either 0, 120, or 500 nM), 10 mM HEPES (pH 8.0), 1 mM EDTA, 50 mM KCl, 0.02% Tween 20, 1 μg/ml Saccharomyces cerevisiae tRNA, 50 μg/ml bovine serum albumin, 1 mM dithiothreitol, and 5 nM radiolabeled RNA substrate. Reaction mixtures were supplemented with loading buffer containing 35% Ficoll, and the products were separated on 6% (1× Tris-borate-EDTA) native polyacrylamide gels at 4°C.
FIG. 5.
The 5′ cap was required for vegetal cell-specific translational repression. (A) Schematic of mutant xCR1 3′ UTRs contained in different luciferase reporter mRNAs. nt, nucleotide. (B) Reporter mRNAs were injected into animal cells or vegetal cells of eight-cell embryos and the injected embryos were cultured until stage 7. Lysates prepared from each sample were assayed for luciferase activity. The VG/AN luciferase ratio was calculated for each reporter. (C) Reporter mRNAs were equally stable in animal (An) and vegetal (Vg) cells after injection. Reporter RNA from injected cells was analyzed by RT-PCR, and the products were visualized by agarose gel electrophoresis.
Synthesis and analysis of 32P-labeled 3′ UTRs.
32P-labeled 3′ UTRs were generated by in vitro transcription as previously described (10). Ten nanoliters of radiolabeled RNA was injected into animal or vegetal cells of eight-cell embryos. RNAs from injected embryos were isolated and analyzed by denaturing polyacrylamide gel electrophoresis as previously described (10).
RESULTS
The association of xCR1 mRNA with polyribosomes was temporally and spatially regulated.
The Xenopus xCR1 protein is not present in eggs, but it accumulates in animal cells following fertilization (9). To determine whether regulated translation contributed to the temporally regulated accumulation of the xCR1 protein, the association of xCR1 mRNA with polyribosomes from eggs and different embryonic stages was analyzed (Fig. 1A) (10). xCR1 mRNA was not associated with polyribosomes in eggs, but an increasing fraction of the xCR1 mRNA was associated with polyribosomes in embryos as development proceeded (Fig. 1B, compare lanes 2 and 3). The xCR1 mRNA-polyribosome association was disrupted by the presence of EDTA, a treatment that dissociates polyribosomes (Fig. 1B, compare lanes 2 and 4). Analysis of the same fractions for the cyclin B1 mRNA showed a robust association of this mRNA with polyribosomes in both eggs and embryos (Fig. 1B, compared lanes 7 and 8), consistent with the robust translation of cyclin B1 mRNA previously observed in eggs and embryos (10, 42). Thus, translational activation of maternal xCR1 mRNA was delayed until the 16- to 32-cell stage, providing evidence that the temporal accumulation of the xCR1 protein was controlled at the level of xCR1 mRNA translation.
FIG. 1.
Translation (polyribosome association) of the xCR1 mRNA is temporally and spatially regulated. (A) Polyribosomes were isolated from eggs, from embryos at different stages, or from animal or vegetal halves dissected from stage 7 embryos. The mRNAs in the supernatant (non-polyribosome-associated) and polyribosomal pellets were analyzed by blot hybridization using radiolabeled probes. (B) xCR1 mRNA temporal polyribosome association during development (egg to stage 7 [St.7]) (left panel). Cyclin B1 mRNA temporal polyribosome association during development (egg to stage 7) (right panel). (C and D) xCR1 mRNA polyribosome association in animal (An) vegetal (Veg) halves from stage 7 embryos. For each experimental sample, RNA was isolated from unfractionated samples (total RNA [T]), the polyribosome fraction (P), and the nonpolyribosome fraction (S). Samples were fractionated in the presence of EDTA where indicated.
To test whether translational regulation also contributed to the differential spatial distribution of the xCR1 protein in embryos, polyribosome analysis was repeated using isolated animal and vegetal embryo fragments (Fig. 1C). Polyribosomes from animal pole cells contained xCR1 mRNA (Fig. 1D, lane 2), whereas polyribosomes from vegetal cells were depleted of xCR1 mRNA (Fig. 1D, lane 5). In contrast, the cyclin B1 mRNA was associated with polyribosomes from both the animal and vegetal cells (Fig. 1D, compare lanes 2 and 5). These data provided evidence that translation of maternal xCR1 mRNA was restricted to cells of the animal pole.
Changes in poly(A) correlated with temporal but not spatial regulation of xCR1 mRNA translation.
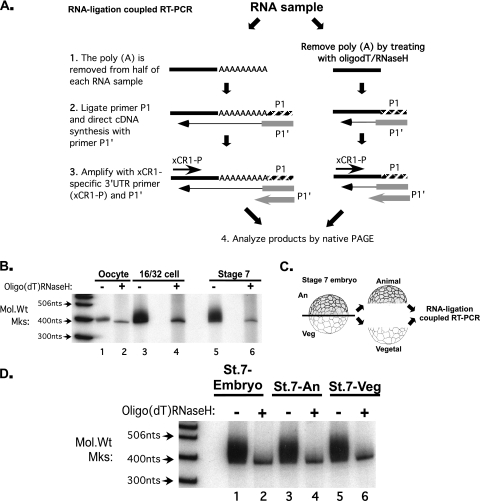
The addition and removal of poly(A) are mechanisms commonly used to regulate maternal mRNA translation (48, 49). In general, mRNAs with long poly(A) tails are translated more efficiently than are mRNAs with short tails. To determine whether regulated xCR1 mRNA translation could be explained by changes in polyadenylation, the poly(A) tails of the xCR1 mRNAs from eggs and embryos were analyzed by RNA ligation-coupled RT-PCR (Fig. 2A) (3). Prior to analysis, half of each RNA sample was treated with oligo(dT)/RNase H to remove any poly(A) present. In this assay, the presence of 3′ poly(A) generates a heterogeneous distribution of high-molecular-weight RT-PCR products, whose size in comparison to poly(A)-deficient controls indicates the lengths of the poly(A) tails on the mRNAs in a sample.
FIG. 2.
The xCR1 mRNA is polyadenylated after fertilization in both animal and vegetal cells. (A) RNA ligation-mediated poly(A) test (RL-PAT). Primer P1 is ligated to the 3′ end of the RNA. cDNA is primed using primer P1′ (complementary to P1). PCR amplification of the cDNA using a gene-specific primer and primer P1′ generates a PCR product whose size depends on the length of the poly(A) tail. Poly(A) can be removed prior to RL-PAT by treatment with oligo(dT)/RNase H. PAGE, polyacrylamide gel electrophoresis. (B) The xCR1 mRNA has a short poly(A) tail in oocytes that is extended by ∼60 adenosines in 16- to 32-cell and stage 7 embryos. RL-PAT performed on RNA from oocytes, 16- to 32-cell embryos, and intact stage 7 embryos. (C and D) The xCR1 mRNA is polyadenylated in animal and vegetal cells. RL-PAT was performed on RNA from the animal and vegetal halves of stage 7 embryos. nts, nucleotides; Mol.Wt Mks, molecular weight markers; An, animal; Veg, vegetal; St.7, stage 7. +, present; −, absent.
The xCR1 mRNA in oocytes contained a short (<20 nucleotides) poly(A) tail (Fig. 2B, lanes 1 to 2), as the RT-PCR products with and without RNase H/oligo(dT) treatment were similar in size and the poly(A) did not increase in length during oocyte maturation (data not shown). However, the xCR1 mRNAs from 16- to 32-cell and stage 7 embryos possessed poly(A) tails of ∼60 nucleotides in length, as indicated by higher-molecular-weight RT-PCR products that were sensitive to RNase H/oligo(dT) treatment (Fig. 2B, lanes 3 to 6). Thus, the poly(A) tail of the xCR1 mRNA was elongated during embryogenesis, coincident with the temporal polyribosome association of the xCR1 mRNA (Fig. 1B) and consistent with the established role of poly(A) tail lengthening in translational activation.
If polyadenylation were important for the spatial differences in translation, then the xCR1 mRNA should be polyadenylated in animal cells but not in vegetal cells. However, analysis of the xCR1 mRNA isolated from either isolated animal or vegetal halves of stage 7 embryos revealed that the xCR1 mRNA contained a poly(A) tail of similar length regardless of its source (Fig. 2C and D). Thus, although polyadenylation might contribute to the temporally regulated translational activation of xCR1 mRNA, it was not sufficient to explain the spatial distribution of translationally active xCR1 mRNA.
The 3′ UTR of the Xenopus xCR1 mRNA was sufficient to confer vegetal cell-specific translational repression to a luciferase reporter mRNA.
To further investigate the mechanisms of spatially restricted translation, a reporter mRNA containing the 3′ UTR of xCR1 mRNA (Luc/xCR1-3′UTR-WT) was created (Fig. 3A). For comparison, a second reporter mRNA was used, which contained the 3′ UTR of cyclin B1 mRNA (10, 42). The cyclin B1 mRNA 3′ UTR was chosen as our control because, in contrast to xCR1 mRNA, cyclin B1 mRNA was recruited to polyribosomes in both animal and vegetal cells of embryos (Fig. 1D). Each reporter was injected into either the animal or vegetal cells of eight-cell embryos (Fig. 3B). Extracts prepared from injected embryos when they reached stage 7 were assayed for luciferase activity.
FIG. 3.
The 3′ UTR of the Xenopus xCR1 mRNA represses translation in vegetal cells. (A) xCR1 3′ UTR reporter mRNAs. nt, nucleotides. (B) Embryo injections for analyzing spatially regulated translation. Reporter mRNAs injected into animal cells or vegetal cells of eight-cell Xenopus embryos were cultured until stage 7, and lysates were assayed for luciferase activity. (C) The 3′ UTR of the Xenopus xCR1 mRNA represses translation in vegetal cells. The absolute luciferase values observed in animal (AN) and vegetal (VG) cells with each reporter from three independent experiments are shown in the top graphs. The VG/AN luciferase ratios calculated from the same experiments are shown in the graphs below. (D) Reporter mRNAs were equally stable in animal (An) and vegetal (Vg) cells after injection. Reporter RNA from injected cells was analyzed by RT-PCR, and the products were visualized by agarose gel electrophoresis.
In three independent experiments, the absolute levels of luciferase produced by the Luc/xCR1-3′UTR-WT reporter mRNA were greater in animal cells than in vegetal cells (Fig. 3C, top panels). In contrast, the levels of luciferase produced by the Luc/cyclin B1-3′UTR reporter mRNA were similar in animal and vegetal cells. The consistency of these data between experiments was particularly obvious when ratios of luciferase activity produced by vegetal cell-injected embryos to animal cell-injected embryos (vegetal/animal luciferase ratio [VG/AN ratio]) were presented (Fig. 3C, bottom panel). These effects were due to translational differences rather than differences in mRNA stability, as the levels of each reporter mRNA were similarly stable in animal and vegetal cells (Fig. 3D). Together, these data indicated that the Luc/xCR1-3′UTR-WT and Luc/cyclin B1-3′UTR reporter mRNAs produced spatial translation patterns within embryos that recapitulated the behavior of the native xCR1 and cyclin B1 mRNAs on polyribosomes (Fig. 1D). Therefore, the xCR1 mRNA 3′ UTR contained elements that restricted the efficient translation of xCR1 mRNA to animal cells. In particular, based on the absolute luciferase values produced by the reporter mRNAs in animal cell- and vegetal cell-injected embryos, the xCR1 mRNA 3′UTR contained elements that repressed translation of xCR1 mRNA in vegetal cells.
The central region of the xCR1 mRNA 3′UTR was sufficient to direct vegetal cell-specific translational repression.
To define the sequences responsible for vegetal cell-specific translational repression, reporter mRNAs containing different regions of the xCR1 3′ UTR (Fig. 4) were analyzed. The Luc/xCR1-3′UTR-Mut2 reporter that contained only the central 351 nucleotides of the xCR1 3′UTR was translated less efficiently by vegetal cells than it was by animal cells (VG/AN ratio of 0.12). This ratio was comparable to that produced by the reporter mRNA containing the entire xCR1 3′ UTR (Fig. 4B, compare Mut2 to xCR1-WT). In contrast, the reporter mRNAs containing the Mut1, Mut3, or Mut-SH xCR1 3′ UTRs produced similar levels of luciferase in both vegetal and animal cells (Fig. 4B), suggesting that these reporter mRNAs lacked sequences necessary for efficient vegetal cell-specific translational repression. Control experiments (Fig. 4D) indicated that the reporter mRNAs were equally stable in animal and vegetal cells. Thus, the central 351 nucleotides of the xCR1 3′ UTR (nucleotides 286 to 637) contained sequences sufficient for vegetal cell-specific translational repression of the xCR1 mRNA.
FIG. 4.
The central 351 nucleotides of the xCR1 3′ UTR were sufficient to direct vegetal cell-specific translational repression. (A) Schematic of mutant xCR1 3′ UTRs contained in different luciferase reporter mRNAs. nt, nucleotide. (B) Reporter mRNAs were injected into animal cells or vegetal cells of eight-cell embryos, and the injected embryos were cultured until stage 7. Lysates prepared from each sample were assayed for luciferase activity. The vegetal/animal luciferase ratio was calculated for each reporter. (C) Reporter mRNAs were equally stable in animal (An) and vegetal (Vg) cells after injection. Reporter RNA from injected cells was analyzed by RT-PCR, and the products were visualized by agarose gel electrophoresis. (D) The xCR1 Mut2 3′ UTR is not polyadenylated during embryogenesis. 32P-labeled xCR1-Mut2 RNA or cyclin B1 3′UTR was injected into animal or vegetal cells of eight-cell embryos. The RNAs from injected cells were analyzed by denaturing gel electrophoresis. +, present; −, absent.
The xCR1 mRNA is polyadenylated in both animal and vegetal cells (Fig. 2), suggesting that differential polyadenylation cannot explain the differences in translation. To further address this issue, we analyzed the polyadenylation of an RNA consisting of the xCR1 Mut2 3′ UTR. Radiolabeled versions of this RNA and the cyclin B1 3′ UTR were injected into animal or vegetal cells of eight-cell embryos. Once the embryos reached stage 7, the RNAs were isolated and analyzed by denaturing gel electrophoresis. The xCR1 Mut2 RNA was not polyadenylated in either animal or vegetal cells (Fig. 4D). In contrast, the cyclin B1 3′ UTR was efficiently polyadenylated in both cell types (Fig. 4D). These results provided additional evidence that vegetal cell-specific repression occurs independently of polyadenylation.
The 5′ cap was required for vegetal cell-specific translational repression.
Some repression mechanisms that function on maternal mRNAs interfere with the binding of translational initiation factors to the mRNA 5′ cap and such repression is cap dependent (5, 48). To test whether vegetal cell-specific repression of the xCR1 mRNA was cap dependent, a reporter mRNA that contained the IRES from the CSFV was generated (Luc/xCR1-3′UTR-MUT2-CSFV-IRES [Fig. 5A]) (35). The CSFV IRES allows translation to occur without a 5′ cap. To protect the stability of this reporter mRNA, it was synthesized with an ApppG structure in place of the normal GpppG 5′ cap. ApppG cannot direct translation but ensures that the mRNA is not degraded by 5′ exonucleases. The reporter mRNA containing 5′ ApppG, the CSFV IRES and the Mut2 region of the xCR1 mRNA's 3′ UTR (Luc/xCR1-3′UTR-MUT2-CSFV-IRES) was injected into animal and vegetal cells, and the ratios of luciferase activities were compared to those of the controls. The CSFV IRES-Mut2 reporter mRNA was not repressed in vegetal cells, in contrast to the control reporter containing the Mut2 region and a normal 5′ cap (Fig. 5B). Control experiments indicated that reporter mRNAs were equally stable in animal and vegetal cells (Fig. 5C). Therefore, vegetal cell-specific repression by the Mut2 region of the xCR1 3′ UTR required a 5′ cap and suggested that repression of xCR1 mRNA translation occurred by interfering with an aspect of translation initiation that required the 5′ cap binding complex.
PBEs in the 3′ UTR of the xCR1 mRNA were necessary but not sufficient for vegetal cell-specific translational repression.
The xCR1 Mut2 3′ UTR (nucleotides 286 to 637) that was sufficient to confer translational repression on the luciferase reporter mRNA contained three potential PBEs (UGUANAUA) (Fig. 6A) (36, 40, 51, 52). PBEs are binding sites for Pumilio proteins that function as translational repressors of specific mRNAs, such as the cyclin B1 and ringo mRNAs in Xenopus oocytes. Therefore, we postulated that these potential PBEs contributed to the translational repression of xCR1 mRNA in vegetal cells of embryos. To test whether these sequences were PBEs, we performed RNA binding experiments, using purified recombinant Pumilio-1 protein (GST-Xpum1) (Fig. 6B) (40). In these electrophoretic mobility shift assays, short radiolabeled RNAs were tested for their ability to be bound by GST-Xpum1, which would retard migration through the gel compared to free RNA. A short RNA containing the potential PBEs 1 and 2 (PBE-1/2; nucleotides 323 to 369) (Fig. 6A) was bound as efficiently by GST-Xpum1 as was the positive control RNA that contained a known PBE (NRE, from the Drosophila hunchback mRNA) (Fig. 6B, compare lanes 1 to 3 with lanes 10 to 12). In contrast, a negative control RNA (A30 RNA) of similar size was not bound by GST-Xpum1 (Fig. 6B, lanes 7 to 9). An RNA containing the third potential PBE (Fig. 6A, PBE-3, nucleotides 364 to 389) was bound weakly by GST-Xpum1 (Fig. 6B, lanes 4 to 6). This third potential PBE contained a weaker match to the PBE consensus, which probably explained its diminished binding.
To further test the binding specificity of the PBE-1/2 sites, another radiolabeled RNA, PBE1/2-Mut, was generated, in which the conserved UGU nucleotides known to be critical for Pumilio binding were replaced by AGA (Fig. 6A and C). GST-Xpum1 failed to bind the PBE-Mut RNA, whereas it bound the PBE-1/2 RNA efficiently (Fig. 6C, compare lanes 1 to 3 to lanes 4 to 6). Thus, the Mut2 region of xCR1 3′ UTR contained functional PBEs.
To test the importance of the PBEs for translational repression of xCR1 mRNA within vegetal cells, reporter mRNAs were microinjected into animal or vegetal cells of embryos as in Fig. 4 and 5 and luciferase activity was measured (Fig. 6D). The Luc/xCR1-3′UTR-MUT2 reporter mRNA previously shown to be sufficient for translational repression in vegetal cells (Fig. 4) contained wild-type PBEs (Fig. 6D). A second reporter RNA, Luc/xCR1-3′UTR-Mut2-PBE-mut, was the same except that it contained UGU to ACA nucleotide substitutions in all three of the PBEs noted in Fig. 6A (Fig. 6D). Strikingly, Luc/xCR1-3′UTR-Mut2-PBE-Mut RNA produced similar levels of luciferase in animal or vegetal cells and behaved similarly to the control cyclin B1 mRNA (Fig. 6E, compare Mut2-PBE-Mut to cyclin B1). In contrast, and as expected (Fig. 4), the corresponding wild-type RNA that contained functional PBEs, Luc/xCR1-3′UTR-MUT2, produced significantly greater luciferase activity when injected into animal cells of embryos compared to vegetal cells (Fig. 6E, Mut2). Control experiments indicated that the reporter mRNAs were equally stable in animal and vegetal cells (Fig. 6F).
However, although the PBEs were necessary for vegetal cell-specific repression, additional experiments revealed that they were not sufficient. Specifically, the Mut4 reporter mRNA (Luc/xCR1-3′UTR-MUT4) that contained the three PBEs (nucleotides 286 to 413) but lacked sequences at positions 414 to 637 within the Mut2 region was not efficiently repressed in vegetal cells (Fig. 6E). Thus, these data indicated that PBEs within the 3′ UTR of xCR1 mRNA were necessary but not sufficient to repress translation of xCR1 mRNA specifically in the vegetal cells. These data suggested that Pumilio functions together with other RNA binding proteins to mediate translational repression of xCR1 mRNAs in vegetal cells.
CUG-BP1 protein binding sites were also necessary but not sufficient for vegetal cell-specific repression.
The Mut2 region of the xCR1 3′ UTR also contains two binding sites for the CUG-BP1 protein (Fig. 7A) (16, 27). The Xenopus CUG-BP1 protein is also referred to as EDEN-BP (16, 37). The binding of CUG-BP1 proteins to mRNA 3′ UTRs can negatively regulate mRNA translation in Xenopus embryos (37). Recent experiments with Xenopus tropicalis (16), using CUG-BP1 immunoprecipitation, identified the xCR1 mRNA as a CUG-BP1 target and demonstrated that the 3′ UTR contained CUG-BP1 binding sites. Therefore, we postulated that the CUG-BP1 sites in the Mut2 region of the xCR1 3′ UTR contributed to the translational repression of xCR1 mRNA in vegetal cells. To test this hypothesis, a reporter RNA containing mutant CUG-BP1 sites (Luc/xCR1-3′UTR-MUT2-CUGBP1-Mut) (Fig. 7B) was assayed for vegetal cell-specific translation repression. The Luc/xCR1-3′UTR-Mut2-CUGBP1-Mut RNA produced similar levels of luciferase in animal and vegetal cells (Fig. 7C, compare Mut2-CUGBP1-Mut to cyclin B1). In contrast, the corresponding wild-type RNA that contained functional CUG-BP1 binding sites (Luc/xCR1-3′UTR-MUT2) produced significantly greater luciferase activity when injected into animal cells of embryos than when injected into vegetal cells (Fig. 7C). Furthermore, the Mut-SH reporter RNA (Luc/xCR1-3′UTR-MUT-SH) analyzed in the experiments shown in Fig. 4 contained the CUG-BP1 sites but lacked sequences at positions 286 to 468 present in the Mut2 region, including the PBEs. The Mut-SH reporter was not capable of efficiently repressing translation in vegetal cells (Fig. 4). Together, these data indicated that CUG-BP1 sites within the 3′ UTR of xCR1 mRNA were necessary but not sufficient to repress translation of xCR1 mRNA specifically in the vegetal cells.
DISCUSSION
This report provides evidence that regulated translation of the xCR1 mRNA contributes to the restricted spatial accumulation of the xCR1 protein in the animal cells of Xenopus embryos. Specifically, translational repression of xCR1 mRNA within vegetal cells of embryos was mediated by PBEs and CUG-BP1 sites in the xCR1 mRNAs 3′ UTR. To the best of our knowledge, this is the first example of a role for spatially regulated translation in the asymmetric distribution of a key maternal determinant within a vertebrate embryo.
Animal cell-specific accumulation of the xCR1 protein was due to vegetal cell-specific translational repression of the xCR1 mRNA. Two lines of evidence supported the conclusion that the xCR1 mRNA was subjected to translational repression within the vegetal cells of Xenopus embryos. First, although the xCR1 mRNA was uniformly distributed and polyadenylated throughout the cells of stage 7 embryos, only the xCR1 mRNA present in animal cells was associated with polyribosomes. These data provided evidence that some mechanism prevented the recruitment of xCR1 mRNA for translation in vegetal cells. Second, a reporter mRNA containing the 3′ UTR of xCR1 mRNA was efficiently translated in the animal cells of embryos compared to the vegetal cells. In contrast, a reporter mRNA containing the 3′ UTR of cyclin B1 mRNA was translated equally well in animal and vegetal cells. Thus, the 3′ UTR of xCR1 mRNA contained sequence elements that repressed mRNA translation in vegetal cells but not animal cells. Together, these data provided evidence that vegetal cell-specific translational repression of xCR1 mRNA contributed to the accumulation of the xCR1 protein specifically within animal cells of embryos (9).
Pumilio and CUG-BP1 binding elements within the xCR1 3′ UTR contributed to vegetal cell-specific translational repression.
The central 351 nucleotides of the xCR1 mRNA 3′ UTR were sufficient to direct vegetal cell-specific translational repression. This region of the 3′ UTR contained PBEs and CUB-BP1 binding sites. Mutations within either the PBE or the CUG-BP1 sequences eliminated the vegetal cell-specific translational repression of the reporter mRNA in vivo. However, although both sequence elements are required for repression, neither the PBEs nor the CUG-BP1 sites were sufficient. These results suggest that efficient vegetal cell-specific repression requires the combined actions of Pumilio and CUG-BP1 proteins potentially functioning with other components yet to be defined.
It is interesting to note that the 3′ UTR of cyclin B1 contains both a PBE and a CUG-BP1 site and yet its translation is not repressed in vegetal cells (28, 32, 33). Perhaps efficient vegetal cell translation repression exhibited by the xCR1 mRNA requires the actions of the multiple PBEs and CUG-BP1 sites present in the xCR1 mRNA's 3′ UTR. Such multiple sites, arranged in a particular context, may be capable of translation repression, whereas single sites are not. Interestingly, the 3′ UTR of the xCR1 mRNA contains putative cytoplasmic polyadenylation elements (CPEs) situated between the PBEs and CUG-BP1 sites (39). Perhaps these CPEs contribute to translational repression in vegetal cells via CPE binding protein and Maskin proteins (7, 44). This seems unlikely, since the 3′ UTR of the cyclin B1 mRNA contains CPEs that can clearly repress translation in oocytes, but not in embryos. Another possibility is that the CPEs in the 3′ UTR of the cyclin B1 are optimized for polyadenylation and translational activation, while the CPEs in the 3′ UTR of the xCR1 mRNA are not. Such optimal CPEs in the cyclin B1 3′ UTR and/or the polyadenylation that they direct may counteract the repressive effects of the adjacent PBE and CUG-BP1 site (28, 32, 33). Systematic and comprehensive mutational analyses are necessary to sort out the balances between translational activation and repression signals that achieve remarkably different modes of mRNA behavior while presumably using similar sequence elements.
Pumilio and vegetal cell-specific translational repression.
A role for Pumilio proteins in the negative regulation of mRNA function is well established and occurs by the repression of translation or the promotion of mRNA degradation (51, 52). In Drosophila oocytes and embryos, Pumilio represses the translation of mRNAs encoding proteins that regulate embryonic patterning. For example, the Drosophila hunchback mRNA encodes a transcription factor critical for embryonic patterning, and hunchback mRNA translation is repressed in posterior regions of embryos (2, 21, 26, 31, 43). In the case of the hunchback mRNA, Pumilio functions within a complex of proteins through Pumilio binding sequences (called NREs) in the hunchback mRNA's 3′ UTR. This spatial translational repression ensures that hunchback protein is produced only by anterior cells and is thus is a critical mechanism for fly development. xCR1 is important for embryonic patterning by controlling the activity of the nodal signaling pathway (41). It is interesting that although the molecular functions of the hunchback and xCR1 maternal determinants differ so fundamentally, the need for their spatial control relies on a conserved regulatory mechanism—translational repression via Pumilio proteins. Thus, despite the fundamental differences in the pathways that underlie the early patterning of embryos in invertebrate and vertebrate organisms, spatial regulation of translation via Pumilio is a conserved mechanism for controlling the patterning molecules of both types of organisms.
The 5′ cap and repression.
Vegetal cell-specific repression was disrupted when translation initiation was mediated by the CSFV IRES, providing evidence that a 5′ cap was required for repression. This requirement raises the possibility that the messenger RNP that forms on the Mut2 region may mediate repression by affecting the binding of initiation factors to the 5′ cap. Such mechanisms function with other maternal mRNAs (5, 48). For example, the CPE binding protein in Xenopus oocytes represses the translation of mRNAs, such as the cyclin B1 mRNA, through interactions with the Maskin protein (1, 44). Maskin binds to eIF4E and prevents eIF4E's association with eIF4G to block cyclin B1 mRNA translation. Also, in yeast, the Puf6 protein related to Pumilio binds to the eIF5b initiation factor and blocks translation of the Ash1 mRNA (8, 17).
Pumilio and deadenylation complexes.
In many cases, Pumilio proteins negatively regulate mRNAs as components of deadenylation complexes that contain the POP2 and the CCR4 deadenylases (15). These complexes are multifunctional and can negatively regulate mRNAs by mechanisms other than deadenylation. For example, in yeast, the PUF5 protein binds to particular target mRNAs and directs the formation of a POP2/CCR4 deadenylation complex (13, 14, 20). This complex is sufficient to repress target mRNA translation without deadenylation. By analogy, Pumilio binding to the 3′ UTR of the xCR1 mRNA could direct the formation of a unique deadenylation complex that includes CUG-BP1 and represses translation in vegetal cells, independent of poly(A) removal.
CUG-BP1 proteins as negative regulators of mRNA function.
In Drosophila, Bruno, a protein similar to CUB-BP1, represses the translation of specific maternal mRNAs, such as the oskar mRNA (48). This repression requires Bruno response elements (BREs) in the 3′ UTR of target mRNAs (22). Bruno binds to the BREs and recruits the CUP protein (34). CUP inhibits eIF4G binding to eIF4E to blocks the translation of BRE-containing mRNAs. In addition, the BREs bound by Bruno also negatively affect translation by promoting the oligomerization of target mRNAs (4). This oligomerization disrupts productive interactions of the BRE-containing mRNAs with translation factors. Further studies will be necessary to determine whether similar mechanisms involving Xenopus CUG-BP1 are used to repress translation of the xCR1 mRNA in the vegetal cells of embryos.
In Xenopus embryos, CUG-BP1 negatively regulates the translation of specific mRNAs, such as the c-mos mRNA by directing mRNA deadenylation (16, 37). However, such a mechanism cannot account for repression of the xCR1 mRNA, as the xCR1 mRNA is polyadenylated in both animal and vegetal cells. This suggests that CUG-BP1 represses translation of the xCR1 mRNA via a different mechanism, such as disrupting one of the 5′ cap-dependent steps of translational initiation. Thus, our results with the xCR1 mRNA suggest that CUB-BP1 has a broader role in regulating maternal mRNAs in Xenopus embryos than previously suspected. This idea is consistent with studies of mammalian cells in which CUG-BP1 regulates pre-mRNA splicing (19, 25, 38), translation (46, 47), and mRNA degradation via deadenylation (29, 50, 55).
A critical key to understanding the spatially regulated repression of xCR1 mRNA translation will be to identify the proteins that form distinct messenger RNP complexes on the xCR1 3′ UTR in vegetal cells. One possibility is that the Pumilio or CUG-BP1 proteins themselves are restricted to the vegetal cells of embryos. At present, there is no information available concerning the expression of these proteins in Xenopus embryos. An alternative possibility is that either Pumilio or CUG-BP1 is present in all embryonic cells but an important functional cofactor is confined to vegetal cells. This type of regulation governs Pumilio function in Drosophila embryos, where Pumilio is controlled by spatially restricting the Nanos protein, a Pumilio cofactor (12, 21). Interestingly, the Xenopus Xcat-2 mRNA that encodes a protein related to Drosophila Nanos is localized to the vegetal cortex of oocytes (30). After fertilization, both the Xcat-2 mRNA and the Xcat-2 protein are restricted to vegetal cells of embryos. Thus, the possibility that Xcat-2 could fulfill the role of a Pumilio cofactor and restrict Pumilio's function to vegetal cells of Xenopus embryos is intriguing.
Acknowledgments
We thank Marv Wickens and Labib Rouhana for the GST-Xpum1 plasmid and advice on RNA binding experiments and Catherine Fox, Tricia Kiley, and Marv Wickens for comments on the manuscript. We thank Justin Spanier for assistance with the RNA binding experiments. We thank Caroline Hill for reagents and Betty Craig for use of the luminometer.
This work was supported by grants from the NIH (no. HD43996 to M.D.S.) and the James and Dorothy Shaw Fund of the Greater Milwaukee Foundation.
Footnotes
Published ahead of print on 13 April 2009.
REFERENCES
- 1.Cao, Q., and J. D. Richter. 2002. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 213852-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chagnovich, D., and R. Lehmann. 2001. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 9811359-11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlesworth, A., L. L. Cox, and A. M. MacNicol. 2004. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J. Biol. Chem. 27917650-17659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chekulaeva, M., M. W. Hentze, and A. Ephrussi. 2006. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124521-533. [DOI] [PubMed] [Google Scholar]
- 5.Colegrove-Otero, L. J., N. Minshall, and N. Standart. 2005. RNA-binding proteins in early development. Crit. Rev. Biochem. Mol. Biol. 4021-73. [DOI] [PubMed] [Google Scholar]
- 6.de Moor, C. H., H. Meijer, and S. Lissenden. 2005. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin. Cell Dev. Biol. 1649-58. [DOI] [PubMed] [Google Scholar]
- 7.de Moor, C. H., and J. D. Richter. 1999. Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 182294-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, Y., R. H. Singer, and W. Gu. 2008. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 221037-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorey, K., and C. S. Hill. 2006. A novel Cripto-related protein reveals an essential role for EGF-CFCs in nodal signalling in Xenopus embryos. Dev. Biol. 292303-316. [DOI] [PubMed] [Google Scholar]
- 10.Fritz, B. R., and M. D. Sheets. 2001. Regulation of the mRNAs encoding proteins of the BMP signaling pathway during the maternal stages of Xenopus development. Dev. Biol. 236230-243. [DOI] [PubMed] [Google Scholar]
- 11.Gallie, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 52108-2116. [DOI] [PubMed] [Google Scholar]
- 12.Gavis, E. R., and R. Lehmann. 1994. Translational regulation of nanos by RNA localization. Nature 369315-318. [DOI] [PubMed] [Google Scholar]
- 13.Goldstrohm, A. C., B. A. Hook, D. J. Seay, and M. Wickens. 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13533-539. [DOI] [PubMed] [Google Scholar]
- 14.Goldstrohm, A. C., D. J. Seay, B. A. Hook, and M. Wickens. 2007. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 282109-114. [DOI] [PubMed] [Google Scholar]
- 15.Goldstrohm, A. C., and M. Wickens. 2008. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 9337-344. [DOI] [PubMed] [Google Scholar]
- 16.Graindorge, A., O. Le Tonqueze, R. Thuret, N. Pollet, H. B. Osborne, and Y. Audic. 2008. Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis. Nucleic Acids Res. 361861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu, W., Y. Deng, D. Zenklusen, and R. H. Singer. 2004. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 181452-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heasman, J. 2006. Maternal determinants of embryonic cell fate. Semin. Cell Dev. Biol. 1793-98. [DOI] [PubMed] [Google Scholar]
- 19.Ho, T. H., D. Bundman, D. L. Armstrong, and T. A. Cooper. 2005. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 141539-1547. [DOI] [PubMed] [Google Scholar]
- 20.Hook, B. A., A. C. Goldstrohm, D. J. Seay, and M. Wickens. 2007. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 28215430-15438. [DOI] [PubMed] [Google Scholar]
- 21.Irish, V., R. Lehmann, and M. Akam. 1989. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature 338646-648. [DOI] [PubMed] [Google Scholar]
- 22.Kim-Ha, J., K. Kerr, and P. M. Macdonald. 1995. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81403-412. [DOI] [PubMed] [Google Scholar]
- 23.King, M. L., T. J. Messitt, and K. L. Mowry. 2005. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell 9719-33. [DOI] [PubMed] [Google Scholar]
- 24.Kuersten, S., and E. B. Goodwin. 2003. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 4626-637. [DOI] [PubMed] [Google Scholar]
- 25.Ladd, A. N., N. Charlet, and T. A. Cooper. 2001. The CELF family of RNA binding proteins is implicated in cell specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 211285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehmann, R., and C. Nusslein-Volhard. 1987. hunchback, a gene required for segmentation of an anterior and posterior region of the Drosophila embryo. Dev. Biol. 119402-417. [DOI] [PubMed] [Google Scholar]
- 27.Marquis, J., L. Paillard, Y. Audic, B. Cosson, O. Danos, C. Le Bec, and H. B. Osborne. 2006. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem. J. 400291-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meijer, H. A., H. E. Radford, L. S. Wilson, S. Lissenden, and C. H. de Moor. 2007. Translational control of maskin mRNA by its 3′ untranslated region. Biol. Cell 99239-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moraes, K. C., C. J. Wilusz, and J. Wilusz. 2006. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA 121084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosquera, L., C. Forristall, Y. Zhou, and M. L. King. 1993. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development 117377-386. [DOI] [PubMed] [Google Scholar]
- 31.Murata, Y., and R. P. Wharton. 1995. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80747-756. [DOI] [PubMed] [Google Scholar]
- 32.Nakahata, S., Y. Katsu, K. Mita, K. Inoue, Y. Nagahama, and M. Yamashita. 2001. Biochemical identification of Xenopus pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 27620945-20953. [DOI] [PubMed] [Google Scholar]
- 33.Nakahata, S., T. Kotani, K. Mita, T. Kawasaki, Y. Katsu, Y. Nagahama, and M. Yamashita. 2003. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev. 120865-880. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, A., K. Sato, and K. Hanyu-Nakamura. 2004. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell 669-78. [DOI] [PubMed] [Google Scholar]
- 35.Otero, L. J., A. Devaux, and N. Standart. 2001. A 250-nucleotide UA-rich element in the 3′ untranslated region of Xenopus laevis Vg1 mRNA represses translation both in vivo and in vitro. RNA 71753-1767. [PMC free article] [PubMed] [Google Scholar]
- 36.Padmanabhan, K., and J. D. Richter. 2006. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 20199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paillard, L., F. Omilli, V. Legagneux, T. Bassez, D. Maniey, and H. B. Osborne. 1998. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 17278-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philips, A. V., L. T. Timchenko, and T. A. Cooper. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280737-741. [DOI] [PubMed] [Google Scholar]
- 39.Piqué, M., J. M. Lopez, S. Foissac, R. Guigo, and R. Mendez. 2008. A combinatorial code for CPE-mediated translational control. Cell 132434-448. [DOI] [PubMed] [Google Scholar]
- 40.Rouhana, L., and M. Wickens. 2007. Autoregulation of GLD-2 cytoplasmic poly(A) polymerase. RNA 13188-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schier, A. F. 2003. Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 19589-621. [DOI] [PubMed] [Google Scholar]
- 42.Sheets, M. D., C. A. Fox, T. Hunt, G. Vande Woude, and M. Wickens. 1994. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 8926-938. [DOI] [PubMed] [Google Scholar]
- 43.Sonoda, J., and R. P. Wharton. 1999. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 132704-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stebbins-Boaz, B., Q. Cao, C. H. de Moor, R. Mendez, and J. D. Richter. 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell 41017-1027. (Erratum, 5:766.) [DOI] [PubMed] [Google Scholar]
- 45.Tao, Q., C. Yokota, H. Puck, M. Kofron, B. Birsoy, D. Yan, M. Asashima, C. C. Wylie, X. Lin, and J. Heasman. 2005. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell 120857-871. [DOI] [PubMed] [Google Scholar]
- 46.Timchenko, L. T., E. Salisbury, G. L. Wang, H. Nguyen, J. H. Albrecht, J. W. Hershey, and N. A. Timchenko. 2006. Age-specific CUGBP1-eIF2 complex increases translation of CCAAT/enhancer-binding protein beta in old liver. J. Biol. Chem. 28132806-32819. [DOI] [PubMed] [Google Scholar]
- 47.Timchenko, N. A., A. L. Welm, X. Lu, and L. T. Timchenko. 1999. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res. 274517-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vardy, L., and T. L. Orr-Weaver. 2007. Regulating translation of maternal messages: multiple repression mechanisms. Trends Cell Biol. 17547-554. [DOI] [PubMed] [Google Scholar]
- 49.Vasudevan, S., E. Seli, and J. A. Steitz. 2006. Metazoan oocyte and early embryo development program: a progression through translation regulatory cascades. Genes Dev. 20138-146. [DOI] [PubMed] [Google Scholar]
- 50.Vlasova, I. A., N. M. Tahoe, D. Fan, O. Larsson, B. Rattenbacher, J. R. Sternjohn, J. Vasdewani, G. Karypis, C. S. Reilly, P. B. Bitterman, and P. R. Bohjanen. 2008. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell 29263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wharton, R. P., and A. K. Aggarwal. 2006. mRNA regulation by Puf domain proteins. Sci. STKE 2006pe37. [DOI] [PubMed] [Google Scholar]
- 52.Wickens, M., D. S. Bernstein, J. Kimble, and R. Parker. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18150-157. [DOI] [PubMed] [Google Scholar]
- 53.Yabe, S., K. Tanegashima, Y. Haramoto, S. Takahashi, T. Fujii, S. Kozuma, Y. Taketani, and M. Asashima. 2003. FRL-1, a member of the EGF-CFC family, is essential for neural differentiation in Xenopus early development. Development 1302071-2081. [DOI] [PubMed] [Google Scholar]
- 54.Yost, C., M. Torres, J. R. Miller, E. Huang, D. Kimelman, and R. T. Moon. 1996. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 101443-1454. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, L., J. E. Lee, J. Wilusz, and C. J. Wilusz. 2008. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J. Biol. Chem. 28322457-22463. [DOI] [PMC free article] [PubMed] [Google Scholar]