Abstract
During the S phase of the cell cycle, the entire genome is replicated. There is a high level of orderliness to this process through the temporally and topologically coordinated activation of many replication origins situated along chromosomes. We investigated the program of replication from origins initiating in early S phase by labeling synchronized normal human fibroblasts (NHF1) with nucleotide analogs for various pulse times and measuring labeled tracks in combed DNA fibers. Our analysis showed that replication forks progress 9–35 kilobases from newly initiated origins, followed by a pause in synthesis before replication resumes. Pausing was not observed near origins that initiated in the middle of S phase. No evidence for pausing near origins was found at the beginning of the S phase in glioblastoma T98G cells. Treatment with the S phase checkpoint inhibitor caffeine abrogated pausing in NHF1 cells in early S phase. This suggests that pausing may comprise a novel aspect of the intra-S phase checkpoint pathway or a related new early S checkpoint. Further, it is possible that the loss of this regulatory process in cancer cells such as T98G could be a contributing factor in the genetic instability that typifies cancers.
Introduction
Progress on our understanding of nuclear DNA replication in eukaryotic cells has involved studies of the molecular mechanisms of initiation, the enzymatic process of chain elongation, and nucleotide analog labeling studies to investigate DNA replication dynamics. Early studies demonstrated that DNA consists of long fibers that replicate in separate units that are tandemly joined. 1, 2 Subsequently, Huberman and Riggs evaluated different grain densities produced by incorporation of tritiated thymidine followed by a chase with non-radioactive thymidine to demonstrate the directionality of replication and reveal that replication typically diverges bidirectionally from origin sites. 3 More recent studies, such as those by Diffley and by Jackson and Pombo, used sequential incorporation of two different thymidine analogs that could be identified using separate fluorescent antibody probes to reveal directionality of replication and the comparable timing of replication of genomic sites in sequential cell cycles. 4, 5 The introduction of techniques for aligning DNA fibers on slides by “combing” 6 has also permitted the investigation of replication events to be examined at particular sites in the genome.
We have been studying the fraction of DNA replicated in the early S phase following our earlier observations that cells in the early S phase are exquisitely vulnerable to transformation by treatments with chemical carcinogens. 7–9 Synchronization studies with cultured fibroblasts released from confluence were improved by using aphidicolin to transiently block the cycling cells in early S phase. 10, 11 We found that in addition to improving synchronization of populations of cells through the S phase, fibroblasts held in aphidicolin leaked into S phase and replicated DNA very slowly, which allowed for improved precursor labeling of DNA replicating in early S phase. Using this labeling approach, previous observations of the localization of DNA precursor incorporation have shown that at the very beginning of the S phase, replication occurs in only a small number of chromosomal sites. 12 Subsequent efforts involving the cloning of DNA replicating in early S phase and mapping the sequenced clones to the human genome revealed about 1700 regions of early replication. 13, 14 Replication timing analysis of these regions and comparison with replication timing studies performed with human lymphoblasts confirmed the early replication of the majority of these regions and the similarities of replication timing between the two different human cell types. 14–16
The coordinated expression of cyclin-dependent kinases promotes the orderly entry into S phase and progression through the cell cycle. Pre-replication complexes assembled at potential origins must be activated at the appropriate time and only once per cell cycle. Once DNA replication has begun, the integrity of DNA replication is monitored through the intra-S phase checkpoint. This checkpoint is invoked in response to DNA damage during S phase and acts to suppress firing of new origins as well as to inhibit elongation and stabilize stalled replication forks. The intra-S phase checkpoint pathway involves activation of transducers and effector kinases that phosphorylate downstream substrates leading to inhibition of cell cycle progression. 17–24 There is also evidence that constituents of the intra-S phase checkpoint function to coordinate origin firing during an unperturbed S phase. For instance, Chk1 kinase has been shown to be required for normal cell proliferation as well as the survival of mouse embryos and ES cells, suggesting that checkpoint proteins are essential for normal S phase progression. 18, 25, 26
In the current study, we have employed DNA fiber analysis to evaluate DNA replication in the early S phase and at other times in S phase. We show an apparent pause in replication fork progression within 9–35 kilobases from origins that initiate early in S phase in normal human fibroblasts (NHF1). This pausing is absent in mid S phase in NHF1 cells as well as the early S phase of glioblastoma T98G cells. Treatment of NHF1 cells with the S phase checkpoint inhibitor caffeine abrogated pausing in early S phase NHF1 cells. This suggests that pausing may constitute a regulatory mechanism contributing to the orderly progression of early S phase that may be overridden during oncogenesis.
Materials and Methods
Cell lines and synchronization
Normal human diploid fibroblasts (NHF1) 27 immortalized by ectopic expression of the catalytic subunit of telomerase 24 and T98G glioblastoma cells were cultured in minimum essential medium (MEM) (Invitrogen) supplemented with 2 mM L-glutamine (Invitrogen) and 10% fetal bovine serum (FBS) (Hyclone Laboratories, Inc.). For synchronization, NHF1 cells were arrested at confluence as described previously. 28, 29 Briefly, NHF1 cells were plated at 1 × 106 cells/100 mm plate and were grown for seven days with medium changes on the third and fifth days. The confluence-arrested cells were presence or absence of 2 µg/ml aphidicolin (A.G. Scientific, Inc.) at 374,000 cells/60 mm plate for pulse labeling and fiber spreading, or 600,000–800,000 cells/100 mm plate for flow cytometry. T98G cells were serum starved for three days in MEM containing only 0.2% FBS followed by release into MEM with 10% FBS in the presence or absence of aphidicolin. Aphidicolin was removed through three washes with Hanks’ balanced salt solution (HBSS) (HyClone), and cells were incubated in complete medium for 15 min to remove any effects of the aphidicolin block. Cells were then labeled with nucleotide analogs as described below.
Caffeine treatment
Thirty minutes prior to labeling, caffeine (Sigma Aldrich) was added at a concentration of 2 mM to the culture medium of cells synchronized by confluence arrest only. Labeling was performed either in the presence of or 2 mM caffeine or Phosphate Buffered Saline (PBS, Sigma Aldrich) as control.
Labeling of cells with thymidine analogs
Actively replicating cells were pulsed first with the thymidine analog IdU (50 µM) (Sigma Aldrich) for 10, 20 or 45 min followed by a pulse with another thymidine analog CldU (100 µM) (Sigma Aldrich) for 20 min.
DNA fiber spreading
DNA was spread on glass slides as described previously 30, 31 with modifications. After labeling, the cells were trypsinized and resuspended in PBS at 200 cells/µl. Two microliters of cell suspension was streaked on a silane-prep slide (Sigma Aldrich), and the cell suspension was allowed to air-dry almost completely. Eight microliters of spreading buffer (0.5% SDS in 200 mM Tris-HCl, pH 7.4, 50 mM EDTA) was then added over the cells. After 10 min, the slides were tilted at 15°, an additional 8 µl was added and the buffer was allowed to run down the slide. The resulting DNA spreads were air-dried, fixed in 3:1 methanol/acetic acid, and frozen overnight.
Immunostaining
Slides were treated with 2.5 M HCl for 30 min, washed once in PBS/0.1% Tween 20 and twice in PBS and then incubated in 2% bovine serum albumin in PBS for 40 min to decrease non-specific binding of antibodies. The slides were then incubated with the primary antibody mixture at room temperature in a humid chamber: 1 hour in 1:250 rat anti-bromodeoxyuridine (detects CldU) (OBT0030, Accurate) plus 1:250 mouse anti-bromodeoxyuridine (detects IdU) (Becton Dickinson). After incubation, slides were incubated for 10 min in a buffer containing 10 mM Tris HCl pH 7.4, 400 mM NaCl, 0.2% Nonidet 40 (NP40) to eliminate any non-specifically bound primary antibodies. Slides were then washed twice in PBS. The slides were then incubated in the secondary antibody mixture in a humid chamber at room temperature: 30 min in 1:250 Alexafluor 488-conjugated chicken anti-rat (Molecular Probes) plus 1:333 Alexafluor 594-conjugated rabbit anti-mouse (Molecular Probes). Slides were washed once in PBS/0.1% Tween 20 followed by two washes in PBS. Before incubation in the third antibody mixture, slides were blocked in 2% Normal Goat Serum (Gibco) in PBS for 15 min. The third antibody mixture was added in a humid chamber at room temperature: 30 min in 1:250 Alexafluor 488-conjugated goat anti-chicken (Molecular Probes) plus 1:333 Alexafluor 594-conjugated goat anti-rabbit (Molecular Probes). The slides were then washed as described for the secondary antibody mixture. The slides were mounted in antifade (UNC Microscopy Core). Microscopy was carried out using an Olympus FV500 confocal microscope using the sequential scanning mode. A single-blind evaluation was performed to measure the lengths of continuously stained red tracks using Image J software. To demonstrate the presence or absence of pausing across many origins, during the scanning and selection process the images taken were not limited to any one area of the slide; instead, pictures were taken of all possible origins on the entire slide and on multiple slides, reducing the chance of over- or under-representing certain origins or the genomes of individual cells.
Conversion from arbitrary units to kilobases
To determine the conversion factor from arbitrary units to kilobases, FISH was used to probe a region of 282984 base pairs in the DNA fiber spreads, and the hybridized region was measured in arbitrary units. The average of 13 measurements (326.2 arbitrary units) was divided into the length of the probed region to give a conversion of 867.5 bp per arbitrary unit. This analysis was done on fibers spread by the same person at the time the experimental slides were prepared to ensure a similar stretching of the DNA.
Statistical analysis
Levene’s test showed that the group variances were not equal in many of the data sets even after log transformation of the original data. As such, the F test in the traditional one-way ANOVA will be affected as the group sample sizes differ greatly in many data sets. Consequently we adopted a general linear model approach with unequal group variances on the log transformed data (SAS, PROC MIXED). We compared the means of the 10 and 20 min and 10 and 45 min data sets and the p-values were adjusted for multiple comparison using Tukey’s method.
Flow cytometry
Cells for flow cytometric analysis were labeled for 10 min with 50 µM IdU. The cells were then trypsinized, collected, washed twice with PBS and fixed in 70% cold ethanol. Staining was performed following a published protocol. 32 Cells were incubated in primary antibody, 1:10 mouse anti-bromodeoxyuridine (Becton Dickinson), for 30 min, followed by incubation in the secondary antibody, 1:500 goat anti-mouse FITC (Sigma Aldrich), for 30 min in the dark. The cells were analyzed and data was collected using a CyAn flow cytometer.
Results
Pausing of replication forks near origins occurs in early S phase in NHF1 cells
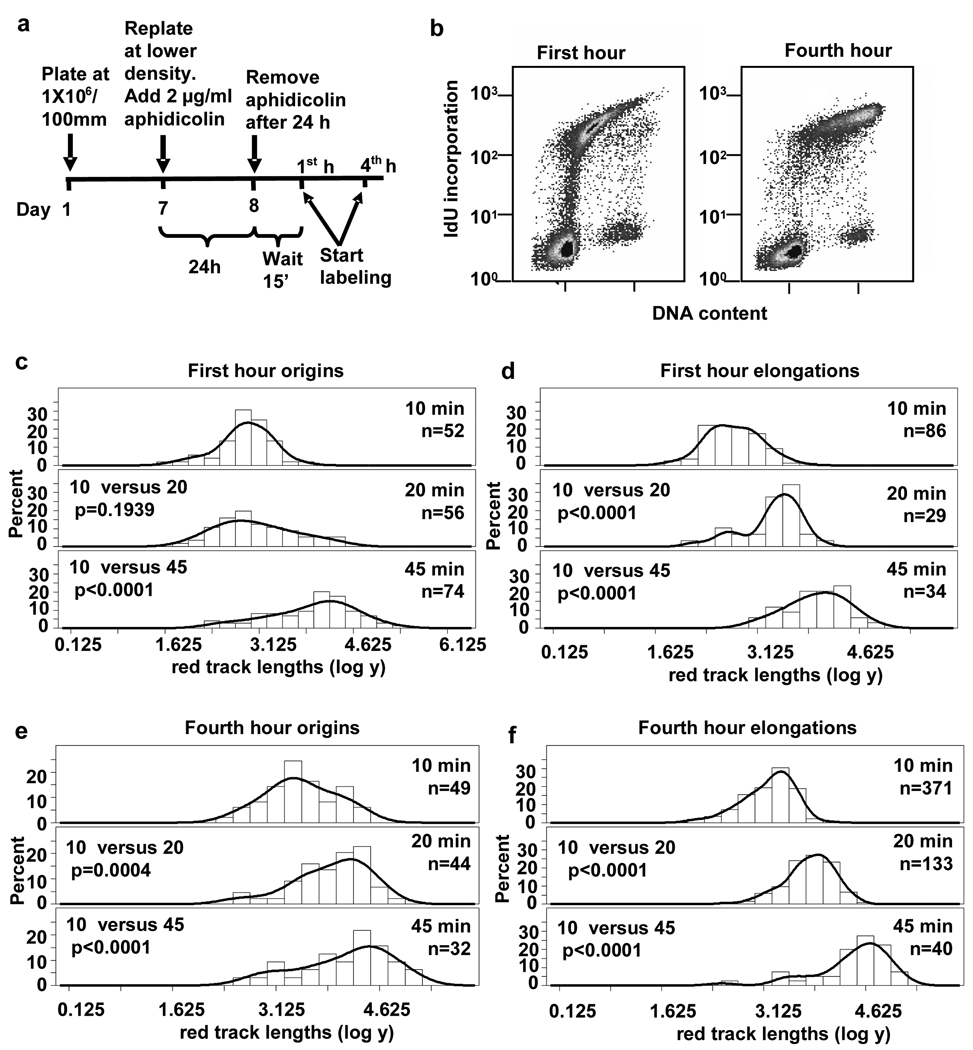
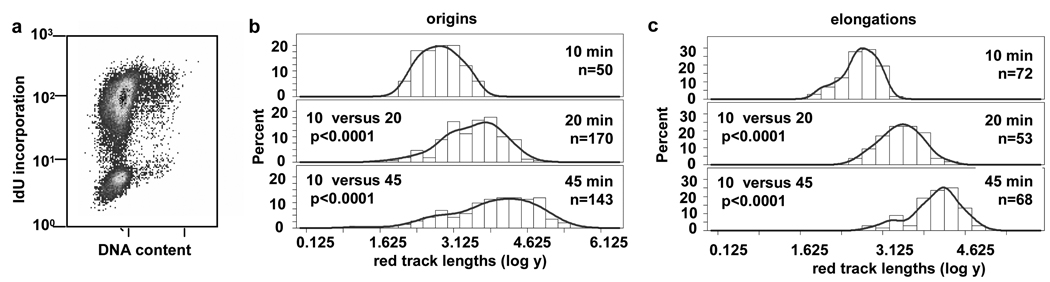
To characterize the replication program from origins that initiate at the onset of the S phase, we initially synchronized normal human fibroblasts (NHF1) by confluence arrest followed by replating in the presence of 2 µg/ml aphidicolin for 24 h. Fifteen minutes after removal of aphidicolin, the synchronized population of early S phase cells was sequentially pulse-labeled with the thymidine analogs IdU and CldU (Fig. 2a). These cells then were harvested and lysed, and their genomic DNA straightened and aligned by fiber spreading.5, 6, 30, 31, 33, 34 Incorporated IdU and CldU in the DNA fibers were detected using red or green fluorescently-tagged antibodies, respectively, and visualized by confocal microscopy. During pulse-labeling, cells were incubated with IdU (red) for 10, 20 or 45 min followed by a 20 min pulse with CldU (green) to track replication patterns and directionality of fork movement (Fig. 1). Newly initiated origins are indicated by a red track flanked on both sides by green, since these patterns can only be obtained as a consequence of replication origin initiating during the first pulse (red) and replication forks still moving bidirectionally during the second pulse (green). Flow cytometric analysis of IdU incorporation during a 10 min pulse at the start of this time point demonstrated the early S phase cell cycle profile at the time of the first nucleotide analog pulse (Fig.2b, left panel). For comparison, a flow cytometric profile of an asynchronous population of NHF1 cells is shown in Supplemental Fig. S1b. The profile of confluence arrested NHF1 cells incubated with IdU and aphidicolin for 24 h is shown in Supplemental Fig. S1a. This showed that 33% of cells were already in early S phase at the end of the incubation with aphidicolin; once aphidicolin was removed, cells moved synchronously through S phase (40% IdU-positive cells in Fig. 2b, left panel).
Fig. 2.
DNA replication forks pause near origins in early but not mid S phase in NHF1 cells. (a) Schematic of the synchronization protocol with aphidicolin. (b) Flow cytometric analysis of NHF1 cells during the first and fourth hour after release from aphidicolin arrest. (c to f) Frequency distributions of red track lengths (log of arbitrary units) during the first hour after release from aphidicolin arrest at (c) origins and (d) elongations and during the fourth hour at (e) origins and (f) elongations.
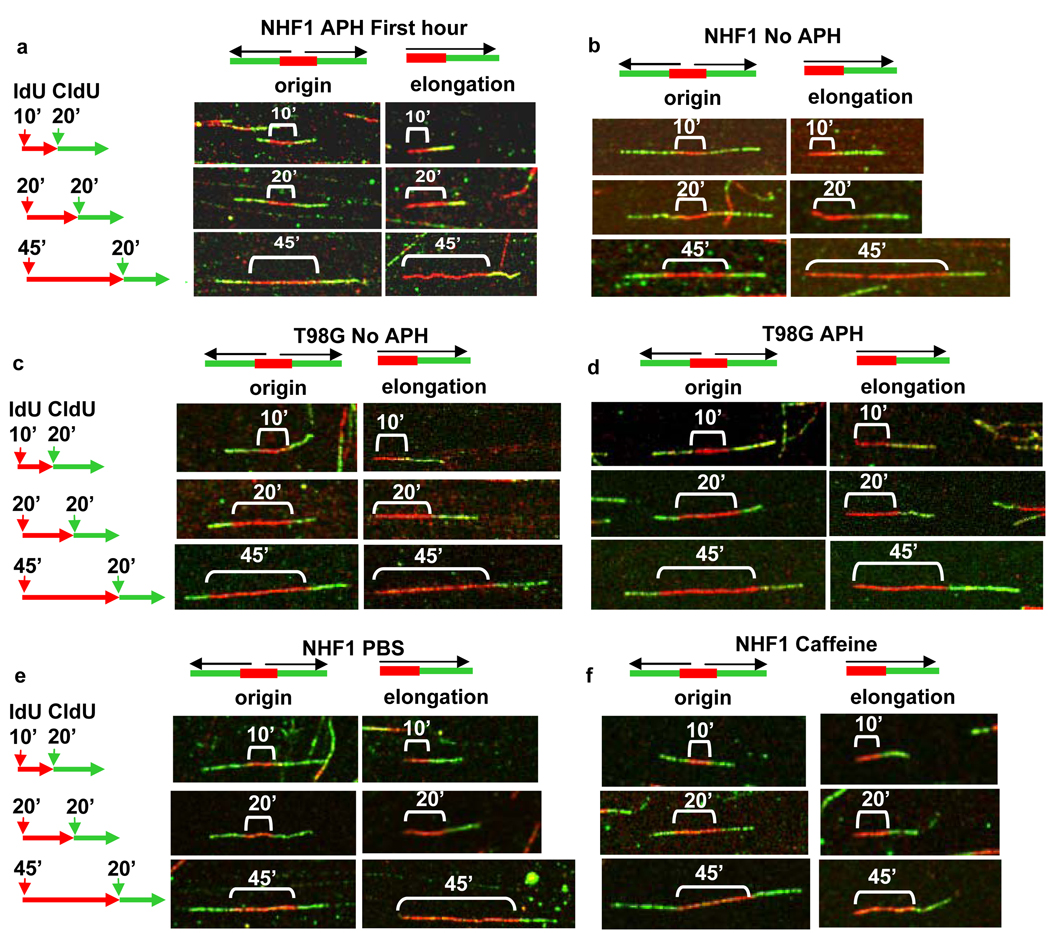
Fig. 1.
Detection of origin and unidirectional elongation fork patterns by DNA fiber spreading. Early S phase cells were pulsed with IdU for 10, 20 or 45 min followed by a pulse with CldU for 20 min. The origin and elongation patterns for representative fibers near the median length of red tracks produced by this labeling strategy are shown for early S phase cells in the following populations: (a) NHF1 synchronized by confluence arrest followed by treatment with aphidicolin (APH); (b) NHF1 synchronized by confluence arrest in the absence of aphidicolin; (c) T98G synchronized by serum starvation; (d) T98G synchronized by serum starvation followed by treatment with aphidicolin; (e and f) NHF1 synchronized by confluence arrest in the absence of aphidicolin and pre-treated for 30 min and labeled in the presence of (e) PBS (control) or caffeine (f).
We expected the red track lengths at origins to increase proportionally to labeling time with IdU and the distribution of the lengths of the red tracks at origins to be different at the increasing pulse times. A frequency curve was generated for the red track lengths at origins, and a statistical analysis was performed on the different populations. The statistical analysis assumed that red track lengths (after log transformation) are normally distributed with different means and variances. The means of the red track lengths at 10, 20 and 45 min pulses were compared for origins fired in early S phase.
To our surprise, we found that replication tracks near origins did not increase in direct proportion to labeling time in NHF1 cells during early S phase (Fig. 1a and Fig. 2c). Replication forks near origins synthesized DNA in a range measured in arbitrary units during the 10 min incubation, but the range did not increase significantly during the next 10 min of incorporation. During the following 25 min, the red track lengths at origins increased significantly from that at the 10 min pulse (Fig. 1a and Fig. 2c). The distributions of lengths for the different pulse times are shown as logarithms of the arbitrary units in Fig. 2c, as we used for the statistical analysis. Since taking the logarithm helps to give the data a more normal distribution, the data is shown as log to better illustrate pausing visually. The distributions of red tracks after conversion from arbitrary units to kilobases are shown in Fig. S2 (see Materials and Methods). This analysis suggests that during the beginning of S phase, replication initiation near single origins proceeded within 9 to 35 kb and was followed by a decrease in replication incorporation, or a “pause” in DNA synthesis before replication resumed.
Pausing does not occur at unidirectional elongating forks
To determine whether pausing is specific for replication forks near origins or affects all replication sites globally, we measured the red tracks from unidirectionally elongating forks distinguished as a single red track adjoined to a single green track (henceforth referred to as “unidirectional elongation forks”) (Fig. 1a). Statistical analysis of the red track lengths suggested that the rate of displacement of single forks away from the origins led to different red track lengths after 10 min and 20 min of synthesis, as well as after 10 and 45 min (Fig. 2d). This finding indicates that pausing does not occur at all replication sites but only proximal to the origin initiation site. Also, since the range of lengths of unidirectional forks increased with time, fork pausing near origins cannot be due to changes in nucleotide pools that would affect replication globally. Histograms comparing the distributions of red tracks converted to kilobases are shown for origins and elongations in Supplemental Fig. S2a and S2b.
Pausing does not occur during mid S phase in NHF1 cells
We next determined whether pausing near origins is specific for replication in early S phase. Cells synchronized by confluence arrest and aphidicolin treatment were pulsed with IdU and CldU during the fourth hour after release from aphidicolin. The flow cytometric profile of these mid S to late S phase cells, with 47% of the cells incorporating IdU, is shown in Fig. 2b (right panel). During this time point, the mean track length of IdU incorporation at origins (red tracks) labeled for 10, 20 or 45 min increased in proportion with time, suggesting that forks near origins do not pause during mid S phase (Fig. 2e). Red track lengths at unidirectional elongation sites also increased progressively with pulse times during this mid S phase time interval (Fig. 2f). The distributions of red tracks for origins and elongations, in lengths converted to kb, are shown in Supplemental Figs. S2c and S2d.
Pausing occurs in early S phase of confluence arrested NHF1 cells in the absence of aphidicolin treatment
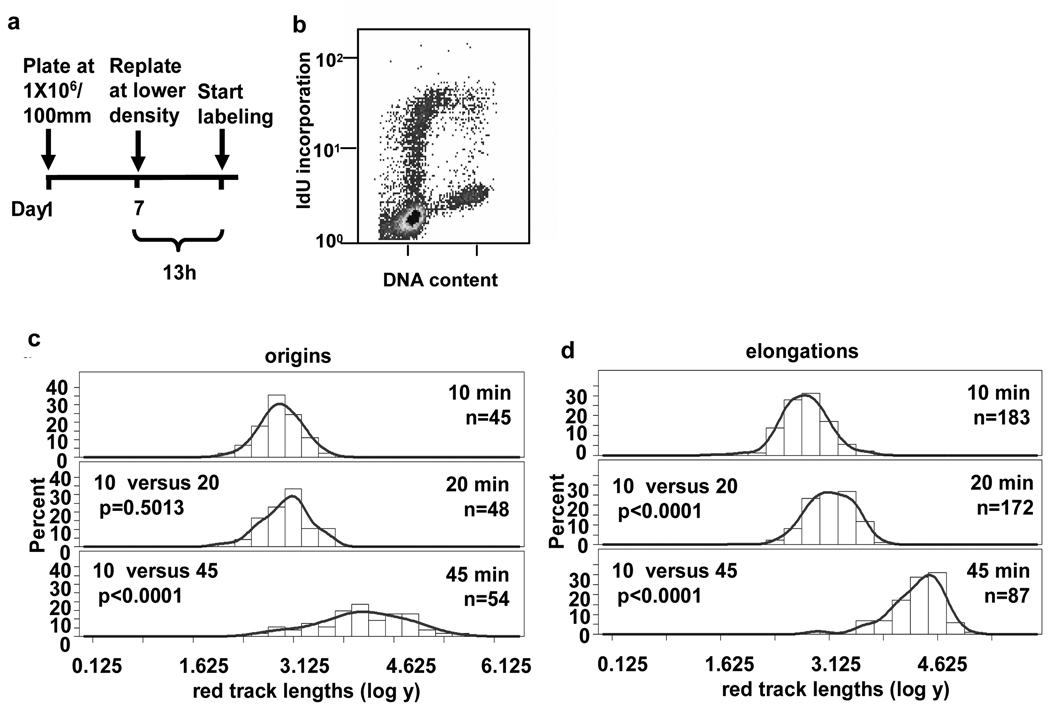
To demonstrate that synchronization with aphidicolin did not activate a signal that was responsible for the pause seen near origins, we labeled normal human fibroblasts synchronized by confluence arrest in the absence of aphidicolin. A schematic of the labeling protocol used in this study is shown in Fig. 3a. Analysis by flow cytometry confirmed that 17% of the cells labeled during the thirteenth hour after release from confluence arrest were replicating and had an early S phase flow profile similar to the profile of NHF1 cells synchronized using aphidicolin in Fig. 2b, although a smaller percent of the cell population entered S phase in the absence of aphidicolin at the thirteenth hour after release compared to cells released from confluence arrest followed by incubation in aphidicolin for 24 h. Once again we found that in these early S phase cells the lengths of DNA tracks did not increase significantly between the 10 and 20 min pulses near origins but replication was resumed between the 20 and 45 min pulses (Fig. 1b, left panels, and Fig. 3c). Individual elongation forks continue synthesis without pausing between the 10, 20 and 45 min pulses (Fig. 1b, and Fig. 3d). The lengths of red tracks at origins and elongations converted to kb are shown in Supplemental Figs. S3a and S3b. These data suggest that pausing of forks near origins is a process inherent to the normal replication program and is not a consequence of aphidicolin treatment.
Fig. 3.
Replication forks near early origins pause in NHF1 cells. (a) NHF1 cells were synchronized by confluence arrest. (b) Thirteen hours after release, a portion of the cells are in early S phase. (c and d) Frequency distributions of red track lengths (log of arbitrary units) measured blind are shown for (c) origins and (d) elongations.
Pausing of newly initiated forks near origins is absent in early S phase in T98G glioblastoma cells
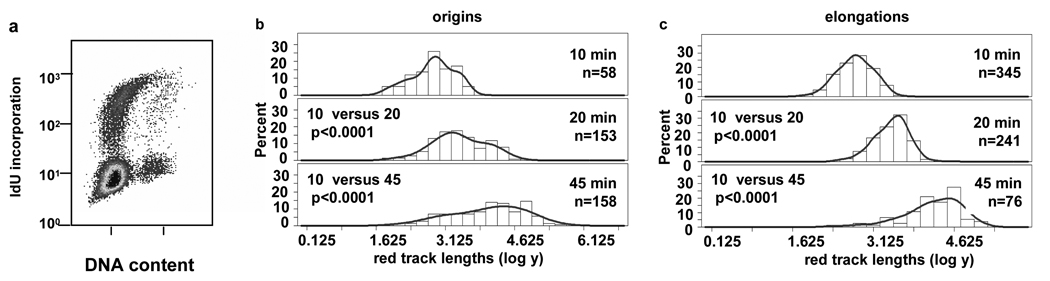
Since the orderly progression of DNA replication is presumed to be necessary to preserve genomic integrity and prevent the genetic instability typical of cancer, we hypothesized that the regulation of replication during early S phase provided by pausing near origins might be lost in cancer cells. To test this hypothesis, we analyzed replication fork progression near origins in early S phase T98G glioblastoma cells, a cancer cell line that can be efficiently synchronized by serum starvation. The cell cycle profile of the T98G cells labeled following serum starvation (Fig. 4a) had 33% of the cells IdU-positive and a distribution of early S phase cells comparable to the profile of NHF1 cells that exhibit origin pausing. A profile of asynchronous T98G cells is shown in Supplemental Fig. S1c. In contrast to our observation with NHF1 cells, labeling of T98G cells with IdU for 10, 20 or 45 min during the fourteenth hour after release from serum starvation resulted in mean red track lengths at origins that increased in proportion to pulse time (Fig. 1c, and Fig. 4b). Their distribution appeared similar to that observed for individual elongating forks (Fig. 1c, and Fig. 4c, and Supplemental Figs. S4a and S4b). This finding suggests that pausing of replication forks near origins that are activated in early S phase does not occur in T98G cancer cells.
Fig. 4.
Forks near early origins do not pause in T98G cells synchronized by serum starvation. (a) Fourteen hours after release from serum starvation, T98G cells exhibited a flow cytometric profile comparable to that of NHF1 cells during which pausing occurs (Fig. 2b). Frequency distributions of red track lengths (log of arbitrary units) measured blind are shown for (b) origins and (c) elongations.
Pausing cannot be induced near early S phase origins in T98G cells by treatment with aphidicolin
To confirm that pausing in NHF1 cells is independent of aphidicolin treatment, we treated T98G cells with aphidicolin for 24 h after release from serum starvation. If pausing is an effect of aphidicolin, treatment with the drug should induce pausing in T98G cells. Fifteen minutes after release from aphidicolin, the T98G cells showed a similar, though enriched (81% IdU-positive), entry of cells into early S phase cells at the time of labeling (Fig. 5a). Although the enriched population of early S phase cells obtained with the aphidicolin treatment suggests that aphidicolin functions in these cells, analysis of labeled tracks near origins showed that pausing was still absent near origins (Fig. 1d, Fig 5b and Supplemental Fig. S5a), and replication also proceeded in proportion to time at individual elongating forks (Fig. 1d, Fig. 5c and Supplemental Fig. S5b). This demonstrates that pausing is not an artifact of aphidicolin treatment.
Fig. 5.
DNA replication forks associated with early S phase origins do not pause in T98G cells synchronized by serum starvation and aphidicolin treatment. (a) Flow cytometric analysis of T98G cells at early S phase after release from aphidicolin treatment. (b and c) Frequency distributions of red track lengths (log of arbitrary units) measured blind are shown for (b) origins and (c) elongations.
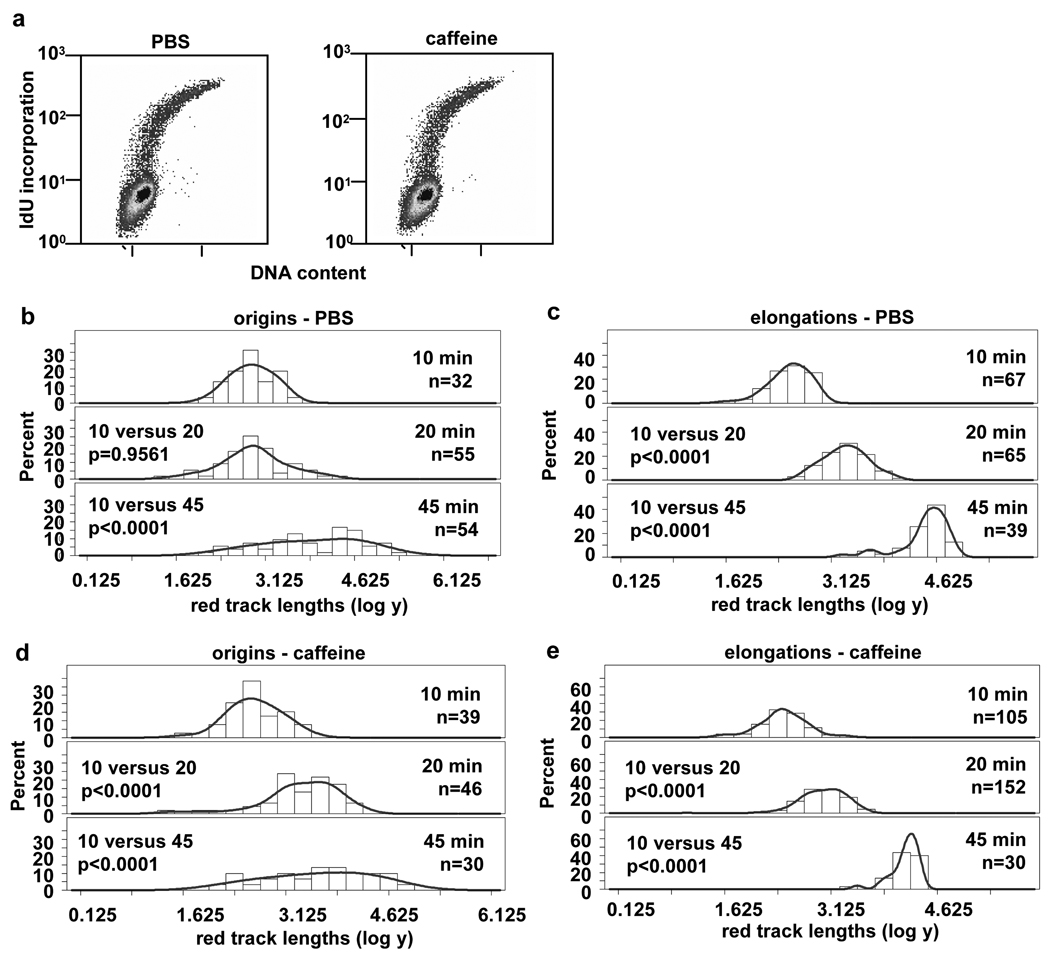
Fork pausing near origins is ablated in early S phase NHF1 cells by caffeine treatment
We hypothesized that since pausing does not occur in T98G cells, it may be due to a defective checkpoint pathway. To test this hypothesis, we synchronized NHF1 cells by confluence arrest to avoid possible complicating effects of aphidicolin treatment. At 12.5 h after replating to release cells into the cell cycle, cells were treated either with PBS (control) or 2 mM caffeine for 30 min prior to labeling. Treatment with caffeine at this concentration has been shown to effectively abrogate ATM- and ATR-dependent checkpoint responses. 35 Cells were then labeled at 13 h after release as before, either in the presence of PBS or 2 mM caffeine. The cell cycle profiles of PBS- (14% IdU-positive) and caffeine- (13% IdU-positive) treated cells at the start of the labeling time point (Fig. 6a) shows their comparable early S phase status. The measurements of red tracks at origins again showed the 10 and 20 min populations were statistically the same in the PBS (control) cells, suggesting pausing occured near the origins (Fig. 1e, and Fig. 6b). However, treatment with caffeine resulted in the red track length distributions at origins for the 10 and 20 min pulses that were statistically different (Fig. 1f and Fig. 6d) Elongations between 10, 20 and 45 min in both conditions were statistically different (Fig. 1e and 1f, Fig. 6c and Fig. 6e). The distributions of red track lengths converted to kb for the PBS- and caffeine-treated cells are shown in Supplemental Fig. S6. This data suggests that the process of pausing near early S phase origins can be abrogated in NHF1 cells by treatment with caffeine.
Fig. 6.
Pausing of DNA replication forks in early S phase of NHF1 cells is abrogated by caffeine treatment . (a) Cell cycle profile of NHF1 cells released for 13 h from confluence arrest, after pre-treatment with PBS (left panel) or caffeine (right panel) for 30 min prior to the start of labeling. (b to e) Frequency distributions of red track lengths (log of arbitrary units) for origins (b and d) and elongations (c and e) with treatment with PBS (b and c) or caffeine (d and e).
Discussion
We have used DNA fiber spreading to study the pattern of replication near origins in early and mid S phase. We show that pausing of forks occurs near most if not all of the early S phase origins analyzed in NHF1 cells, but is absent from origins that fire during mid S phase as well as early S phase T98G glioblastoma cells. The whole genome approach in analyzing origins by fiber spreading suggests the widespread occurrence of pausing across many origins may be a manifestation of a general control mechanism to regulate fork progression near origins in early S phase. We have demonstrated that pausing occurs in the early S phase of NHF1 cells whether they are synchronized in the presence or absence of aphidicolin following release from confluence arrest. In our experience, NHF1 cells can be synchronized to G0 by confluence arrest, while the synchrony of cells that enter S phase is increased after release from treatment with aphidicolin following confluence arrest. Despite the total percent of cells that enter S phase during the labeling period with aphidicolin (40%, Fig. 2) or without (17%, Fig. 3), the early S phase profile of cells replicating during labeling is similar between the two conditions, as well as the synchronized T98G cells in which pausing does not occur, suggesting the cells are at a comparable point in the cell cycle when the labeling is performed.
We have previously determined that it takes at least 12 hours for cells released from confluence arrest to traverse G1 and enter S phase; thus the incorporation of IdU in the presence of aphidicolin for 24 h before release should proceed during the 12–24th h as cells enter S phase. Aphidicolin reduces the rate of replication to 2.6% of the normal rate, 12 suggesting that for the normal ~8 h S phase of NHF1 cells, incubation in aphidicolin is equivalent to approximately 20 min of the normal S phase. Once aphidicolin is removed from the media and the cells are allowed to recover from the drug for 15 min, this suggests that the “early” S phase timepoint in which pausing is observed corresponds to a time within approximately the first hour of a normal S phase. The cell cycle profile of NHF1 cells released from confluence arrest into aphidicolin and IdU for 24 h is shown in Supplemental Fig. S1a.
We have shown that pausing occurs in early S phase in NHF1 cells after replication proceeds within a range of 9 to 35 kb. We have not yet determined the reason for pausing at a distance of 9 to 35 kb from the origin, whether for example it is determined by sequence or if it is related to chromatin structure. During the labeling time when pausing is observed, we continue to see incorporation of IdU at individual forks although incorporation near origins slowed or ceased. This suggests that pausing is not due to alterations in nucleotide pools that globally affect replication. The rates of replication measured for the red tracks of individual forks appeared to be similar to replication rates reported for other cell lines 36, 37 (Supplemental Fig. S7).
The observation of pausing of forks near origins prompted us to speculate about processes that would only involve replication near the first cohort of DNA replication origins, that would engage after several kb of DNA had been replicated and would require a brief interval of cessation of replication before resuming. Since pausing is absent in the early S phase of T98G glioblastoma cells, this suggests that pausing in early S phase may contribute to the orderly progression of replication in normal cells that is disrupted to give a growth advantage characteristic of cancer. It is possible that pausing near origins in early S phase may be part of a checkpoint that helps to ensure the integrity of DNA as replication initiates at the start of S phase. It has been demonstrated that the ATR and ATM checkpoint responses, which mediate their effects through proteins such as Chk1 and p53, can be reversed by treatment with caffeine. 35 To determine whether pausing is an effect of these pathways, we have analyzed early S phase NHF1 cells treated with PBS (control) or caffeine. Our data shows that whereas PBS-treated control cells demonstrate pausing near origins in early S phase, pausing can be abrogated in these cells by treatment with caffeine. This suggests that pausing may constitute a novel S phase checkpoint function that facilitates surveillance of the earliest replicated DNA for the presence of damage near origins at the start of S phase. Our data suggests that since pausing can be ablated by caffeine treatment, this checkpoint function may involve the functioning of ATR. It is also possible that pausing is mediated through ATM, which phosphorylates p53 during the DNA damage response. Since T98G cells express mutant p53, it is possible that pausing is dependent on functional p53.
Our data suggests that replication from origins that initiate in early S phase may occur in a highly-controlled, coordinated manner important for protecting the orderly process of replication. Since faithful duplication of DNA from cell to daughter cell is critical for proper regulation of cell growth, the lack of pausing of forks near origins in early S phase in glioblastoma T98G cells may reflect or explain the abnormal DNA content of these cells and the disrupted organization of their chromosomes.
Supplementary Material
The log phase cell cycle distribution is shown for (a) NHF1 cells incubated for 24 h in IdU and aphidicolin after replating from confluence arrest. The log cell cycle profile of NHF1 is shown in (b) and T98G cells is shown in (c).
The distribution of red track lengths converted to kb for NHF1 cells is shown for first hour (a) origins and (b) elongations and fourth hour (c) origins and (d) elongations after release from aphidicolin block.
The distribution of red track lengths converted to kb for NHF1 cells is shown for first hour (a) origins and (b) elongations 13 h after release from confluence arrest.
The distribution of red track lengths converted to kb for T98G cells is shown for first hour (a) origins and (b) elongations 14 h after release from serum starvation.
The distribution of red track lengths converted to kb for T98G cells is shown for first hour (a) origins and (b) elongations after release from aphidicolin block.
The distribution of red track lengths converted to kb for NHF1 cells treated with PBS (control) (a and b) or caffeine (c and d) is shown for early (a and c) origins and (b and d) elongations 13 h after release from confluence arrest.
The overall rate of unidirectional elongating forks was determined for each cell line and condition. For each experimental condition, the lengths of all red tracks at unidirectional elongation forks were plotted for the 10, 20 and 45 min pulses, and a best fit line between the time points was drawn, as shown in (a) for the first hour after release from synchronization using aphidicolin in NHF1 cells. The slope of the best fit line determined the overall rate of synthesis for the cell lines and conditions as shown in (b).
Acknowledgments
We thank Bruna Brylawski for technical advice, Jean Cook for providing T98G cells, Kathleen Nevis for help with flow cytometry, Marila Cordeiro-Stone for constructive discussions and suggestions, and Mahesh Ramamoorthy for encouragement. This research was supported by a research grant from the National Cancer Institute (R01-CA084493) (DGK), a pilot grant from the University of North Carolina CEHS Program (P30ES10126) (PDC) and a Postdoctoral Fellowship from the National Institute of Environmental Health Sciences (T32-ES07017) (RAF).
References
- 1.Cairns J. Autoradiography of HeLa cell DNA. J Mol Biol. 1966;15:372–373. doi: 10.1016/s0022-2836(66)80233-4. [DOI] [PubMed] [Google Scholar]
- 2.Huberman J, Riggs AD. Autoradiography of chromosomal DNA fibers from Chinese hamster cells. Proc Natl Acad Sci USA. 1966;55:599–606. doi: 10.1073/pnas.55.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huberman J, Riggs AD. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- 4.Merrick CJ, Jackson D, Diffley JF. Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem. 2004;279:20067–20075. doi: 10.1074/jbc.M400022200. [DOI] [PubMed] [Google Scholar]
- 5.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 7.Grisham JW, Greenberg DS, Kaufman DG, Smith GJ. Cycle-related toxicity and transformation in 10T1/2 cells treated with N-methyl-N'-nitro-N-nitrosoguanidine. Proc Natl Acad Sci USA. 1980;77:4813–4817. doi: 10.1073/pnas.77.8.4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaufmann WK, Rice JM, Wenk ML, Devor D, Kaufman DG. Cell cycle-dependent initiation of hepatocarcinogenesis in rats by methyl(acetoxymethyl)nitrosamine. Cancer Res. 1987;47:1263–1266. [PubMed] [Google Scholar]
- 9.Kaufmann WK, Rahija RJ, MacKenzie SA, Kaufman DG. Cell cycle-dependent initiation of hepatocarcinogenesis in rats by (+/−)-7r,8t-dihydroxy-9t-,10t-epoxy-7,8,9,10-tetrahydrobenzo(a)pyrene. Cancer Res. 1987;47:3771–3775. [PubMed] [Google Scholar]
- 10.Pedrali-Noy G, Spadari S, Miller-Faures A, Miller AO, Kruppa J, Koch G. Synchronization of HeLa cell cultures by inhibition of DNA ploymerase alpha with aphidicolin. Nucleic Acids Res. 1980;8:377–387. doi: 10.1093/nar/8.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordeiro-Stone M, Kaufman DG. Kinetics of DNA replication in C3 H 10T1/2 cells synhcronized by aphidicolin. Biochemistry. 1985;24:4815–4822. doi: 10.1021/bi00339a015. [DOI] [PubMed] [Google Scholar]
- 12.Cohen SM, Cobb ER, Cordeiro-Stone M, Kaufman DG. Identification of chromosomal bands replicating early in the S phase of normal human fibroblasts. Exp Cell Res. 1998;245:321–329. doi: 10.1006/excr.1998.4258. [DOI] [PubMed] [Google Scholar]
- 13.Brylawski BP, Cohen SM, Longmire JL, Doggett NA, Cordeiro-Stone M, Kaufman DG. Construction of a cosmid library of DNA replicated early in the S phase of normal human fibroblasts. J Cell Biochem. 2000;78:509–517. doi: 10.1002/1097-4644(20000901)78:3<509::aid-jcb15>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 14.Cohen SM, Furey TS, Doggett NA, Kaufman DG. Genome-wide sequence and functional analysis of early replicating DNA in normal human fibroblasts. BMC Genomics. 2006;7:301. doi: 10.1186/1471-2164-7-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP. Replication timing of the human genome. Hum Mol Genet. 2004;13:191–202. doi: 10.1093/hmg/ddh016. [DOI] [PubMed] [Google Scholar]
- 16.Woodfine K, Beare DM, Ichimura K, Debernardi S, Mungall AJ, Fiegler H, Collins VP, Carter NP, Dunham I. Replication timing of human chromosome 6. Cell Cycle. 2005;4:172–176. doi: 10.4161/cc.4.1.1350. [DOI] [PubMed] [Google Scholar]
- 17.Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: Linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H, Watkins JL, Piwnica-Worms H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci USA. 2002;99:14795–14800. doi: 10.1073/pnas.182557299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorensen CS, Syljuasen RG, Falck J, Schroeder T, Ronnstrand L, Khanna KK, Zhou BB, Bartek J, Lukas J. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3:247–258. doi: 10.1016/s1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson-Rosenthal C, Millar JB. Mechanisms of checkpoint inhibition and recovery. Trends Cell Biol. 2006;16:285–292. doi: 10.1016/j.tcb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, Cordeiro-Stone M, Kaufmann WK. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol. 2002;22:8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M, Nakayama K. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 2004;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 26.Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6:45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 27.Boyer JC, Kaufmann WK, Cordeiro-Stone M. Role of postreplication repair in transformation of human fibroblasts to anchorage independence. Cancer Research. 1991;51:2960–2964. [PubMed] [Google Scholar]
- 28.Cordeiro-Stone M, Boyer JC, Smith BA, Kaufmann WK. Effect of benzo[a]pyrene-diolepoxide- I on growth of nascent DNA in synchronized human fibroblasts. Carcinogenesis. 1986;7:1775–1781. doi: 10.1093/carcin/7.10.1775. [DOI] [PubMed] [Google Scholar]
- 29.Brylawski BP, Cohen SM, Norne H, Irani N, Cordeiro-Stone M, Kaufman DG. Transitions in replication timing in a 340 kb region of human chromosomal R-Band 1p36.1. Journal of Cellular Biochemistry. 2004;92:755–769. doi: 10.1002/jcb.20101. [DOI] [PubMed] [Google Scholar]
- 30.Chastain PD, Heffernan TP, Nevis KR, Lin L, Kaufmann WK, Kaufman DG, Cordeiro-Stone M. Checkpoint regulation of replication dynamics in UV-irradiated human cells. Cell Cycle. 2006;5:2160–2167. doi: 10.4161/cc.5.18.3236. [DOI] [PubMed] [Google Scholar]
- 31.Unsal-Kacmaz K, Chastain PD, Qu P, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White AE, Livanos EM, Tlsty D. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 33.Hand R, Tamm I. DNA replication: direction and rate of chain growth in mammalian cells. J Cell Biol. 1973;58:410–418. doi: 10.1083/jcb.58.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herrick J, Bensimon A. Single molecule analysis of DNA replication. Biochimie. 1999;81:859–871. doi: 10.1016/s0300-9084(99)00210-2. [DOI] [PubMed] [Google Scholar]
- 35.Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A. DNA replication origin interference increases the spacing between initiation events in human cells. Molecular Biology of the Cell. 2006;17:5337–5345. doi: 10.1091/mbc.E06-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daboussi R, Courbet S, Benhamou S, Kannouche P, Zdzienicka MZ, Debatisse M, Lopez BS. A homologous recombination defect affects replication-fork progression in mammalian cells. J Cell Sci. 2008;121:162–166. doi: 10.1242/jcs.010330. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The log phase cell cycle distribution is shown for (a) NHF1 cells incubated for 24 h in IdU and aphidicolin after replating from confluence arrest. The log cell cycle profile of NHF1 is shown in (b) and T98G cells is shown in (c).
The distribution of red track lengths converted to kb for NHF1 cells is shown for first hour (a) origins and (b) elongations and fourth hour (c) origins and (d) elongations after release from aphidicolin block.
The distribution of red track lengths converted to kb for NHF1 cells is shown for first hour (a) origins and (b) elongations 13 h after release from confluence arrest.
The distribution of red track lengths converted to kb for T98G cells is shown for first hour (a) origins and (b) elongations 14 h after release from serum starvation.
The distribution of red track lengths converted to kb for T98G cells is shown for first hour (a) origins and (b) elongations after release from aphidicolin block.
The distribution of red track lengths converted to kb for NHF1 cells treated with PBS (control) (a and b) or caffeine (c and d) is shown for early (a and c) origins and (b and d) elongations 13 h after release from confluence arrest.
The overall rate of unidirectional elongating forks was determined for each cell line and condition. For each experimental condition, the lengths of all red tracks at unidirectional elongation forks were plotted for the 10, 20 and 45 min pulses, and a best fit line between the time points was drawn, as shown in (a) for the first hour after release from synchronization using aphidicolin in NHF1 cells. The slope of the best fit line determined the overall rate of synthesis for the cell lines and conditions as shown in (b).