Abstract
The protein universe is the set of all proteins of all organisms. Here, all currently known sequences are analyzed in terms of families that have single-domain or multidomain architectures and whether they have a known three-dimensional structure. Growth of new single-domain families is very slow: Almost all growth comes from new multidomain architectures that are combinations of domains characterized by ≈15,000 sequence profiles. Single-domain families are mostly shared by the major groups of organisms, whereas multidomain architectures are specific and account for species diversity. There are known structures for a quarter of the single-domain families, and >70% of all sequences can be partially modeled thanks to their membership in these families.
Keywords: domain architecture, protein sequence, protein structure, structural genomics
The protein universe, a concept first mentioned in 1992 (1), is the collection of all proteins of every biological species that lives or has lived on earth. It is a very large, poorly defined, even mysterious entity, which also happens to be an essential underpinning of all biology. Studies of the protein universe as it exists today began with the first determination of a protein sequence by Sanger in 1952 (2). Now, there are almost 8 million sequences in a nonredundant (NR) database of protein sequences, including the complete genomes of ≈1,800 different species. This large body of data is doubling in size every 28 months. The sequences are very different, with polypeptide chain lengths that range from 6 to almost 37,000 amino acid residues. Biological knowledge on sequences also varies enormously. For some proteins, we know their three-dimensional structure and how and where they function and at what kinetic rate. For most, we know just the sequence deduced from the DNA sequence.
Coming to grips with the protein universe is unarguably central, given its importance to biology and the consequent devotion of large resources to accumulate all of this experimental data. In this endeavor, we are aided by the evolutionary relatedness of all life on earth, which provides a shortcut that speeds analysis of the protein universe. Many sequences show detectable levels of similarity (measured, say, by the percentage of identical amino acids when suitably aligned). Appreciable levels of similarity generally imply homology or descent from a common ancestor, which allows related sequences to be grouped into families (3). The number of families is much smaller than the number of sequences, making the entire task more manageable.
To reveal the nature of the protein universe, we ask: How many protein sequences are there? How many sequences are novel vs. repetitious? How many sequences are characterized at structural and functional levels? Are sequences of prokaryotes, eukaryotes, and viruses different? Is the number of sequence families saturating or is it still expanding rapidly?
An obvious way to cluster sequences into families is by pairwise comparison (4) of all sequences preceded by indexing (5) to eliminate close pairs. Such a combination led to massive clustering of millions of protein sequences from both known species and environmental samples by Yooseph et al. (6). Their remarkable conclusion was that the number of protein families as measured by the number of sequence clusters showed no sign of saturation. Indeed, the cluster count was increasing at the same rate as new sequences were being determined. This result featured in a recent report on the Protein Structure Initiative (7) that expressed concern that because the number of new families is expanding rapidly determining three-dimensional structures for a representative of each family may not be possible (8).
Here, we approach the problem differently. Instead of clustering entire protein sequences (6), we rely on the occurrence of protein sequence patterns termed “sequence profiles.” These patterns can be derived from a few members of the family and then used to add new members that match the same pattern. They are related to structural domains, the independent globular parts of the polypeptide chain found in protein structures, but the correspondence is not exact (9).
The first major set of sequence profiles, PFAM (Protein FAMilies), is curated as a consortium (10), which has grown from 100 to >10,000 different sequence profiles. Our analysis uses the Conserved Domain Architecture Retrieval Tool (CDART) resource at the National Center for Biotechnology Information (NCBI) (11), which includes >30,000 sequence profiles from seven different databases and searches sequences using RPS-BLAST (12), which is based on PSI-BLAST (13). Methods such as PFAM, RPS-BLAST, and others (14, 15) build a profile from a multiple sequence alignment and use it to search for any protein sequence. RPS-BLAST uses heuristics for efficiency and is almost as sensitive as the probabilistic hidden Markov models (HMMs) (16, 17) used by PFAM, with a matching threshold of ≈20% sequence identity (18).
On the basis of early work of Chothia (19), Holm and Sander (20), and Koonin et al. (21), the present work provides a concise description of the protein universe: (i) The number of single-domain architecture families (SDAs; with one region matched by a sequence profile) is increasing very slowly. (ii) Multidomain architecture families (MDAs; with more than one region matched by a sequence profile) continue to grow rapidly and at the same exponential rate as deposited sequences. (iii) Almost all novelty comes from the arrangement of known SDA domains along an MDA sequence. (iv) Structural information is known for a quarter of sequence profiles, with one-fifth of these structures coming from structural genomics. (v) Evolution proceeds by creating new MDA families, particularly for eukaryotes. (vi) Less than 25% of the sequences do not match any sequence profile (referred to as the “dark matter”) and likely contain additional sequence profiles. (vii) The distribution of SDA family sizes does not follow a simple power law, preventing an estimate of the effective total number of SDAs.
Different Growth of SDAs and MDAs
The growth in the number of SDA and MDA families is very different (Fig. 1). Although the number of MDA families is growing rapidly with time, the number of SDA families appears to be saturating. In 1980, there were 8,000 sequences in the NR database, with 4,500 different SDA families and 400 different MDA families. By mid-2000, the numbers of MDA families and SDA families were equal. In the past 9 years, the number of deposited sequences has increased 13.6-fold, the number of MDA families has increased 5.6-fold, but the number of SDA families has increased only 21%. The vast majority of sequence profiles found in MDAs (98.2%) also occur independently in SDAs.
Fig. 1.
As the NR database grows, the number of different multidomain architecture (MDA) families found by CDART is increasing rapidly with year (Left) or added sequence (Right). In contrast, the number of single-domain architecture (SDA) families is increasing much more slowly. Because the number of sequences is growing exponentially, fractional sequence coverage (number of sequences in a SDA or MDA family divided by the total number of NR sequences) has dropped slightly from 0.88 to 0.76; more than three-quarters of current sequences still contain a domain recognized by a known sequence profile. Merged CDART sequence profiles are used here. Corresponding results with unmerged CDART sequence profiles are given in Fig. S1. The solid curves marked “2008” were made with a release of CDART from February 9, 2008, which contained fewer sequence profiles (24,083 compared with 27,036). This gave rise to smaller numbers of SDA and MDA families and lower coverage. During this time, the number of sequences in the NR database increased by 2 million.
Part of the slow growth of the numbers of SDAs is due to the time needed to define a new sequence profile (Fig. S2). A year ago, the number of merged sequence profiles was smaller (11,678 vs. 14,119), resulting in lower values of the number of SDAs and MDAs. Although almost 2,000,000 new sequences were added to the NR database in this period, the fractional sequence coverage actually increased from 0.766 to 0.774 to reduce the dark matter fraction by >3% (from 0.234 to 0.226). Even a few additional sequence profiles allow characterization of a larger fraction of sequences.
The NCBI's NR database of sequences used here is large, with almost 8 million sequences and 2.6 billion residues (Table S1), and includes approximately equal amounts of data from prokaryotes and eukaryotes. To ensure that the radically different growth seen for SDA and MDA families is not an artifact of the definition of sequence profiles and my method to merge duplicated sequence profiles (see Materials and Methods), I repeated this analysis with all of the CDART sequence profiles and CDARTPFAM, the subset of sequence profiles from PFAM. I also tested the role of the sequence matching algorithm using the actual matches found by PFAM in version 23.0 (PFAM23). The results (Fig. S1) show the same slow growth of SDA families and rapid growth of MDA families seen with CDART. In CDART and indeed in PFAM (Table S2 and Table S3), there is the duplication of sequence profiles, characterized by more than one sequence profile matching the same region of a particular sequence. Such duplication directly affects the number of different domain architectures found.
Structural Coverage Is High
Fig. 2Left shows the percentage of different SDA families that have a sequence of known structure in the family (unique coverage); it has grown from 17% in 1980 to 26% today. Recent growth is very dependent on structures solved by structural genomics programs: Without these structures, the coverage would have peaked at 21% and be on the decline. A similar picture is seen when coverage is plotted against the total number of sequences in the NR database (Fig. 2 Center) and emphasizes the dramatic increase in percentage coverage achieved by structural genomics even though the number of NR sequences has increased 4-fold. Chandonia and Brenner (22) also used PFAM to assess progress of structural genomics efforts.
Fig. 2.
Unique and repetitious structural coverage as a function of year and size of the sequence database. Coverage is the percentage of single-domain architecture (SDA) families containing at least one sequence of known three-dimensional structure (in the PDB). For unique coverage, we count each family once, whereas for repetitious coverage we count every sequence in the family. If all of the known structures belonging to a particular family are determined by structural genomics, then that family is counted in structural genomics coverage. (If any structure of a family is not from structural genomics, then the entire family is not.). (Left) Unique coverage with merged CDART sequence profiles increasing from 17% in 1980 to 26% now, with a 5% increase since 2004 due to structural genomics. (Right) This increase in coverage occurred during a period when the number of sequences increased 900-fold (from 8,600 to 7.6 million) The upper curves show corresponding data for repetitious coverage that are higher at 71%; this is expected because larger families are more likely to contain a member with a known structure. It is an indication of the maximum number of sequences (4.2 million) that could be modeled by homology. (Center) Coverage with unmerged sequence profiles is significantly lower (22% and 54% for unique and repetitious coverage, respectively); this is expected because families are smaller with unmerged sequence profiles and less likely to contain a member with a known structure.
Knowing the structure of one member of a family allows one to extrapolate (at least partially) to all members of the family. When every sequence in a family is counted (repetitious coverage, Fig. 2 Right), 50% of all characterized sequences had some structural information in 1980; now this number is 71%.
An unexpected consequence of merging duplicated sequence profiles is to increase repetitious coverage (Fig. 2 Right): Merged SDA families are larger in size and have a higher chance of including a member with a known three-dimensional structure. The effect is surprisingly large: The structural coverage of repetitious sequence falls from 71% to 54%.
I also quantified the structural coverage of the MDA families by checking if the individual domains were in a SDA family with a known structure. The results were surprising in that 42% of the unique MDA families had known structures for all domains, 46% had known structures for some domains, and only 12% had no known structure for any domain. The corresponding numbers allowing for sequence repetition in the family (repetitious coverage) are very similar at 49%, 37%, and 14%, respectively.
Evolution via MDA Families
The Venn diagrams in Fig. 3 show that most SDA families occur in more than one major organism group (prokaryotes, eukaryotes, or viruses). Such commonality disappears when one considers MDA families, which are much more organism specific. Sharing drops from 61% to 6% in going from SDA families to MDA families. Prokaryotes have more SDA families, and eukaryotes have more MDA families, in accordance with the finding that domain combinations give rise to new function (23). A simpler reason for more MDA sequences in eukaryotes is to ensure that certain proteins are coexpressed and colocalized in these multicellular organisms.
Fig. 3.
Scaled Venn diagrams of the numbers of single-domain architecture (SDA) and multidomain architecture (MDA) families for the three major organism groups of life: prokaryotes, eukaryotes, and viruses. (Upper Left) For SDA families, there is a good deal of commonality, with 64% of SDAs shared between two or more groups. (Upper Right) For MDA families, the situation is very different, with 96% of MDAs unique to a particular group. The larger eukaryote disk in Upper Left compared with Upper Right shows that although prokaryotes have the highest fraction of SDA families (88%), eukaryotes have the highest fraction of MDA families (68%). The very small number of shared MDAs in Upper Right (4%) shows the relationship that MDAs have to evolutionary diversity. Results with merged sequence profiles are very similar in that Lower Left and Lower Right have corresponding percentages of 61%, 94%, 85%, 68%, and 6%, respectively. The MDA panels are drawn on a different scale from the SDA panels; the area of the prokaryote disk is kept fixed to facilitate comparison.
Although most MDA families consist of a few domains, a few families consist of many domains to give the power-law number_of_cases = 400,000/(number_of_domains)2.9. Repeats of the same domain in a particular MDA family are very common: A power law is also found for the number times that a particular sequence profile repeats: number_of_cases = 2,000/(number_of_repeats)1.7. Repeating domains often have known structures with 17 of the top 20 most frequent repeat domains in the Protein Data Bank (PDB), partially explaining MDA structural coverage. Study of the evolution of domain architectures is an active field (24–27) beyond the scope of this work.
Relation to Earlier Work
Previous work (6) suggests that the protein universe is growing rapidly and without bounds; we find that only the MDA sequences are growing linearly with added sequences. The new MDA families are almost always combinations of a smaller number of existing domains found in SDA sequences. This discrepancy arises from the different ways that sequences are matched: Previous work (6) matched entire sequences without concern for domain structure. Earlier analysis (28) also concluded that the number of protein families is growing rapidly.
The slow growth that we find for SDAs is also consistent with the Structural Classification of Proteins (SCOP) classification of protein structural domains into family, superfamily, or fold categories (29). I showed earlier (30) that each category is becoming saturated. Although the correspondence between CDART families and SCOP categories is not straightforward, interestingly, SDA families are growing slowly.
Role of Homology Modeling
The limited number of different SDA families found here has implications for structural genomics. If almost all novelty in newly discovered sequences is coming from new MDA families that are combinations of domains already found in SDA families, then the aim of determining structural representatives for each sequence profile is achievable.
Structural coverage of SDA families has increased linearly for the past 5 years thanks to structural genomics. Continuing at the same rate for another 40 years would lead to coverage of 70% (Fig. S3). Although the current level of repetitious coverage of SDA families is much higher at 71%, it is growing more slowly (new families have fewer members than existing families). Continued linear growth of repetitious coverage would lead to 85% coverage in 2050. Extrapolations like this are fraught with uncertainty, but researchers should be heartened that achieving 70% coverage would require just 60,000 additional X-ray or NMR structures. In this same period, the number of protein sequences is expected to grow 100,000-fold to 1 trillion (1012).
Currently, 4.2 million sequences have some relationship to structural data: They are matched to one or more sequence profiles that are in the same family as a sequence of known structure (Table S1). The value of this relationship depends on being able to make a useful homology model for the particular sequence. For a sequence to be modelable in this way, it must align well to a known structure, and this is likely to be achieved because RPS-BLAST is a fairly conservative alignment method (31). The field of homology modeling has been active for almost 40 years (32, 33) and is improving rapidly as we accumulate more structural data. Aligning part of the sequence generally only allows that part to be modeled. The average lengths are 336 and 716 aa for SDA and MDA sequences, respectively (Table S1). The average lengths of the unmatched regions are 77 and 294 aa, respectively, showing that known domains cover 59% and 77% of the SDA and MDA protein lengths, respectively. Modeling MDA sequences may require assembly of the individual domains, an area of considerable activity (34). In modeling, every residue counts: A billion residues are matched to a PDB structure (39.3% of all 2.7 billion residues in the NR database) with higher structural coverage for SDAs than MDAs (57.2% vs. 42%).
Dark Matter of the Sequence Universe
Our analysis has been able to characterize 78% of all known sequences longer than 50 aa by matching all or part of the sequence to a sequence profile. The remaining 22% is uncharacterized and considered as dark matter. Dark matter contains equal numbers of prokaryote and eukaryote sequences, but there are more eukaryote residues.
Uncharacterized sequences could exist for four reasons: (i) the DNA-deduced protein sequences are not real; (ii) these are low-complexity, nonglobular protein sequences; (iii) many of the dark matter sequences belong in known families but pattern matching methods are not sensitive enough to detect them; (iv) discovery of new sequence profiles lags so far behind the increase in the number of sequences that very many sequence profiles remain to be discovered in the dark matter. Support for i comes from Sammut et al. (35), who find that UniProt sequences marked as having little evidence of existence have a much higher chance of being identified as dark matter. Support for ii comes from the shorter length of dark matter sequences (median length of 155 aa, half that of other sequences). These sequences are also 50% more likely to be from eukaryotes, whereas new sequence profiles are expected to be more common in prokaryotes (Fig. 3).
Reason iii is supported by the dependence of the percentage of dark matter on the definitions of the sequence profiles and the methods used for matching. The subset of CDART sequence profiles found in PFAM reduces sequence coverage from 78% to 72% and increases the dark matter percentage from 22% to 28%. Improved recognition could be obtained by using matching methods more sensitive than those of RPS-BLAST. For example, on a common set of almost 3 million sequences, HMMs used in PFAM give 5% more sequence coverage than the PSI-BLAST method used in CDART (see Materials and Methods). Using the HMM method on all of the sequence profiles in CDART would be expected to reduce the dark matter percentage to 18%. The PSI-BLAST method was taken as the default method for 7th Critical Assessment of Structure Prediction (CASP7) assessment (31); better sequence coverage is obtained by methods found to work best for CASP7 (36–38).
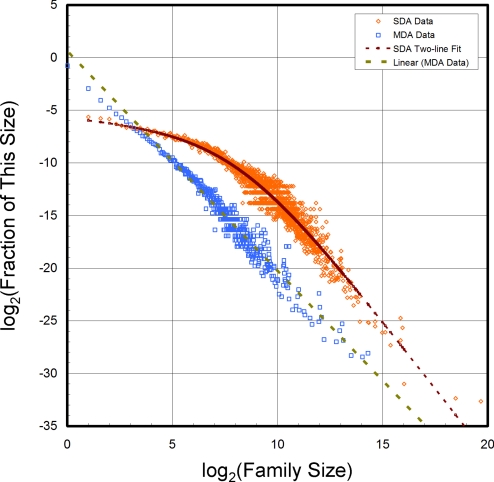
Reason iv is the hardest to assess, because it depends on the activity of scientists defining new sequence profiles. The definition of what constitutes a new sequence profile is arbitrary and will depend on the particular sequence profile database. We have known for some time that the number of SDA families of a given size does not follow a linear power law, whereas the number of MDAs does (39) (Fig. 4). Specifically, there seem to be too few SDA families with small numbers of members (<128). Does this result from the greater sensitivity of methods such as PSI-BLAST and HMM, where the sequence profile is derived from large families, or could it be a reluctance to define a new SDA until it has been seen many times? Fig. S2 shows that new sequence profiles defined in the last year characterize sequences deposited in the NR database decades ago.
Fig. 4.
Although the fraction of MDA families with a particular number of members has a power-law dependence on the family size (as shown by the linear log–log plots), the fraction of SDA families with a particular number of members does not. For MDA families, the fraction of families with m members varies as m−2.09. For small SDA families, the fraction drops much more slowly than that for large SDA families (varies as m−0.18 for m < 32 and then as m−2.57 for m > 64).
Nature of the Sequence Universe
I provide two illustrations of the protein universe (Fig. 5): The repetitious protein universe counts each sequence once and shows current sequence holdings; the unique protein universe counts each domain architecture once and shows novelty or diversity. In the repetitious universe, SDA sequences dominate (88% of 5.9 million sequences), because SDA families are much larger than MDA families. Most of the SDA sequences (71%) are in a family with at least one member of known structure, and 5% of these 3.7 million sequences come from structural genomics structures.
Fig. 5.
Illustrations of sequence space in which area is proportional to the number of sequences or sequence families in that region. Sequences not characterized by any merged CDART sequence profile are the dark matter of the protein universe (23% of 7,500,000, the gray core). (Top Left) The unique sequence universe contains all sequence families. Eighty-six percent of the families are MDAs, and the other 14% are SDAs. Thirty-two percent of SDA sequence families have a known structure, with one-fifth of these from structural genomics. For 49% of the MDAs, all domains have a known structure (hatched), and another 42% have at least one domain with a known structure (part PDB). (Top Right) The repetitious sequence universe contains all sequences. Most characterized sequences (88%, orange area) have single domain architectures (SDAs), where one region of the sequence is matched by a sequence profile (colored bar on black line). The remainder (12%, blue area) have multidomain architectures (MDAs), with more than one region of the sequence matched (several colored bars on sequence). Over three-quarters (76%) of the SDA sequences are matched by a sequence profile family that has a known three-dimensional structure, and 4% of the SDA sequences were solved by structural genomics (brown area, hatching indicates domain of known structure). (Middle) Numbers of sequences in the corresponding regions of Top Right. (Bottom) Numbers of families in the corresponding regions of Top Left.
The number of different MDA families, which are different combinations of SDAs, can clearly expand with the number of sequences. The slow growth of SDAs and the leveling off seen in Fig. 1 would seem to imply saturation in the number of SDAs, but as new sequence profiles are discovered, the saturation level increases. Given the limited sensitivity of methods to recognize homology in sequences, those already at the limits of detection easily can be imagined to drift apart further to give rise to a new SDA family not recognized by an existing sequence profile. Is the number of SDAs going to continue increasing more slowly than the number of sequences? Fig. 4 shows that for large SDA family sizes, the value of the power is less than −2.0; this means that the number of SDA families will eventually increase linearly with the number of sequences (6). This is contrary to the very slow increase in the number of SDAs observed in Fig. 1 and remains to be resolved.
Limitations of This Study
Any study that predicts the future from the past is fraught with uncertainty. The NCBI NR database used is large and representative, with almost 8 million sequences and 2.7 billion amino acids. It contains the complete genomes of >1,800 organisms and partial genome sequences of many more. Particularly uncertain is whether the uncharacterized dark matter and metagenomic sequences that are omitted from the NR database (6) contain large numbers of new sequence profiles. PFAM23 does analyze metagenomic sequences and finds them to have a lot more dark matter (54% vs. 34% for PFAM in its version of NCBI's GenBank, which differs from the NR database).
Implications
Beyond conclusions coming directly from the data, this work suggests that attention be focused on three areas: (i) Improved ability to recognize and model sequence would reduce the amount of additional experimental structure determination. (ii) Dark matter needs to be analyzed for new sequence profiles. (iii) Frequent updates of sequence profile databases are needed to keep up with the rapid growth in the number of protein sequences, doubling in 28 months.
Materials and Methods
Databases Used.
This work depends on a database of sequence profiles that are matched to all known sequences; I used CDART because it contains all sequence profiles and is matched several times per month to the NCBI's NR database.
The NCBI provided us with the first deposit dates of NR sequences to February 7, 2009. In all (Table S1), there were almost 8 million nonredundant (nonidentical) protein sequences (7,624,220). That same day, I downloaded the CDART data from ftp://ftp.ncbi.nih.gov/pub/mmdb/cdart/ and the NR sequences from ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz.
The sequence profiles used in CDART are taken from the NCBI's Conserved Domain Database (13). This database includes all of the sequence profiles from four external resources: (i) PFAM, (ii) SMART (Simple Modular Architecture Research Tool), (iii) COGs (Clusters of Orthologous Groups of proteins), and (iv) PRK (PRotein Klusters). In addition, there are entries from three other sources: (i) KOGs (eukaryotic counterpart to COGs), (ii) CHL (Chloroplast and organelle proteins, a subset of PRK), and (iii) cd (a database curated at NCBI). There were no hits to any of the KOG sequence profiles: the effective number of profiles in CDART is 27,036 (Table S2).
PFAM23 (July 2008) was downloaded from http://pfam.janelia.org/, PDB entries solved by structural genomics from http://targetdb.rcsb.org/target_files, and protein taxonomy from ftp://ftp.ncbi.nih.gov/pub/taxonomy/gi_taxid_prot.dmp.gz.
Data Processing.
The lengths of the sequence profiles in CDART vary greatly from a minimum of 5 to a maximum of 5,019 aa. There are 1,182 sequence profiles that are shorter than 50 aa (74% from PFAM). Almost 250,000 of the NR sequences are shorter than 50 aa, and these were omitted. Sequence profiles shorter than 50 aa were included. The results and conclusions of this work are not sensitive to these choices.
The CDART files (cdart_hits1.txt.gz to cdart_hits2.txt.gz) list all of the matches of each of the 27,036 sequence profiles to each NR sequence that is below the expectation value (E-Value) threshold of 0.01. Many different sequence profiles may overlap a given region of the sequence under consideration, and I used a greedy method to select just one arrangement of sequence profiles. All of the sequence profiles that match a particular sequence are given a score that is a combination of its eval, which is defined as 10 × loge(E-value) plus the length of the sequence matched, SCORE = eval × 0.01 + (S2 − S1 + 1), where S1 is the hit start and S2 is the hit stop. Matches are sorted by decreasing SCORE, the first match that does not overlap with any other already included sequence profile is accepted, and this is repeated until no more matches can be added. This method weights the match length more strongly than its CDART E-Value. Different scoring schemes are possible by changing the relative weights of match length and E-value. Thresholds also can be set for length and E-value. Tests of different schemes gave only small differences.
As a further test, I compared the hits found with CDARTPFAM and those found in PFAM23. For the almost 3 million sequences in common, CDART found 2,859,558 hits; PFAM found these and 129,921 more (4.5%). PFAM residue coverage is also higher than that of CDARTPFAM by a similar margin (4.9%).
Once matches are found, the domain architecture is defined by the type and order of sequence profiles along the particular sequence. The position and length of nonmatched sequence are ignored.
Merged CDART Subset.
Use of different names for essentially the same sequence profile could give rise to different domain architectures that are really equivalent. Here, the CDART sequence profiles were clustered to get a merged subset of sequence profiles. This was done by looking at particular sequences on which different sequence profiles matched well (E-value better than 0.0001) and overlapped so extensively as to be identical. Specifically, I use a stringent overlap criterion, ensuring that the lengths of the two sequence profiles and their extent of overlap on a particular sequence are within 10%. The 16,099 sequence profiles that overlapped in this way were clustered by single-linkage clustering to give 4,049 sequence profiles. These were added to the 10,937 sequence profiles without overlap to give a total of 14,986 merged sequence profiles. All of the sequence profiles in a particular cluster are given the name of the central member. PFAM provides 57% of the merged profiles, much more than COG (19%) and PRK (13%, Table S2). Some of the sequence profiles in both CDART and the merged subset are never matched (1,276 and 863, respectively).
In all, there are 112,804 overlapping sequence profiles. Surprisingly, more than half of the overlap pairs (71,705 or 64%) are between sequence profiles within the same CDART subset (Table S3). The overlaps also occur within well-curated databases such as PFAM and SMART and are unavoidable if one is to maximize sensitivity. Overlaps include PF00106 and PF08659 in PFAM (short chain dehydrogenase and KR domain) and sm00406 and sm00409 in smart (IGv and IG).
Supplementary Material
Acknowledgments.
I thank students, postdocs, and colleagues for critical discussion and encouragement. I am grateful to the National Center for Biotechnology Information for providing the earliest dates associated with each NR sequence. This work was supported by National Institutes of Health Grant GM63817.
Footnotes
The author declares no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905029106/DCSupplemental.
References
- 1.Ladunga I. Phylogenetic continuum indicates galaxies in the protein universe: Preliminary results on the natural group structures of proteins. J Mol Evol. 1992;4:358–375. doi: 10.1007/BF00160244. [DOI] [PubMed] [Google Scholar]
- 2.Sanger F. Arrangement of amino acids in proteins. Adv Protein Chem. 1952;7:1–66. doi: 10.1016/s0065-3233(08)60017-0. [DOI] [PubMed] [Google Scholar]
- 3.Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 4.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 6.Yooseph D, et al. The Sorcerer II global ocean sampling expedition: Expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith J, et al. Report of the Protein Structure Initiative Assessment Panel. Bethesda: Natl Inst Health; 2007. available at www.nigms.nih.gov/News/Reports/PSIAssessmentPanel2007.htm. [Google Scholar]
- 8.Service RF. Structural biology. Protein structure initiative: Phase 3 or phase out. Science. 2008;319:1610–1613. doi: 10.1126/science.319.5870.1610. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Chandonia J-M, Ding C, Holbrook SR. Comparative mapping of sequence-based and structure-based protein domains. BMC Bioinformatics. 2005;6:77. doi: 10.1186/1471-2105-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonnhammer ELL, Eddy SR, Birney E, Bateman A, Durbin R. Pfam: Multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geer Y, Domrachev M, Lipman DL, Bryant SH. CDART: Protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchler-Bauer A, et al. CDD: A curated Entrez database of conserved domain alignments. Nucleic Acids Res. 2002;31:383–387. doi: 10.1093/nar/gkg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul F, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5964. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnamurthy N, Brown D, Sjölander DK. FlowerPower: Clustering proteins into domain architecture classes for phylogenomic inference of protein function. BMC Evol Biol. 2007;7:S1. doi: 10.1186/1471-2148-7-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krogh A, Brown M, Mian IS, Sjölander K, Haussler D. Hidden Markov models in computational biology: Applications to protein modeling. J Mol Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 17.Eddy S. Hidden Markov models. Curr Opin Struct Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 18.Park J, et al. Sequence comparisons using multiple sequences detect three times as many remote homologues as pairwise methods. J Mol Biol. 1998;284:1201–1210. doi: 10.1006/jmbi.1998.2221. [DOI] [PubMed] [Google Scholar]
- 19.Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 20.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 21.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 22.Chandonia JM, Brenner SE. The impact of structural genomics: Expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 23.Bashton M, Chothia C. The generation of new protein functions by the combination of domains. Structure. 2007;15:85–99. doi: 10.1016/j.str.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Fong JH, Geer LY, Panchenko AR, Bryant SH. Modeling the evolution of protein domain architectures using maximum parsimony. J Mol Biol. 2007;366:307–315. doi: 10.1016/j.jmb.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forslund K, Henricson A, Hollich V, Sonnhammer ELL. Domain tree-based analysis of protein architecture evolution. Mol Biol Evol. 2008;25:254–264. doi: 10.1093/molbev/msm254. [DOI] [PubMed] [Google Scholar]
- 26.Vogel C, Berzuini C, Bashton M, Gough J, Teichmann SA. Supra-domains: Evolutionary units larger than single protein domains. J Mol Biol. 2004;336:809–823. doi: 10.1016/j.jmb.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 27.Bjorklund AK, Ekman D, Light S, Frey-Skott J, Elofsson A. Domain rearrangements in protein evolution. J Mol Biol. 2005;353:911–923. doi: 10.1016/j.jmb.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 28.Kunin V, Cases I, Enright AJ, de Lorenzo V, Ouzounis CA. Myriads of protein families, and still counting. Genome Biol. 2003;4:401. doi: 10.1186/gb-2003-4-2-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 30.Levitt M. Growth of novel protein structural data. Proc Natl Acad Sci USA. 2007;104:3183–3188. doi: 10.1073/pnas.0611678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battey JDN, Kopp J, Bordoli L, Read RJ, Clarke ND, Schwede T. Automated server predictions in CASP7. Proteins. 2007;69(Suppl 8):68–82. doi: 10.1002/prot.21761. [DOI] [PubMed] [Google Scholar]
- 32.Browne WJ, North ACT, Phillips DC. A possible three-dimensional structure of bovine α-lactalbumin based on that of hen's egg-white lysozyme. J Mol Biol. 1969;42:65–86. doi: 10.1016/0022-2836(69)90487-2. [DOI] [PubMed] [Google Scholar]
- 33.Warme PK, Momany FA, Rumball SV, Tuttle RW, Scheraga HA. Computation of structures of homologous proteins. α-Lactalbumin from lysozyme. Biochemistry. 1974;13:768–782. doi: 10.1021/bi00701a020. [DOI] [PubMed] [Google Scholar]
- 34.Lensink MF, Méndez R, Wodak SJ. Docking and scoring protein complexes: CAPRI 3rd edition. Proteins. 2007;69:704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 35.Sammut SJ, Finn RD, Bateman A. Pfam 10 years on: 10,000 families and still growing. Brief Bioinform. 2008;9:210–219. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- 36.Zhou H, et al. Analysis of TASSER-based CASP7 protein structure prediction results. Proteins. 2007;69(Suppl 8):90–97. doi: 10.1002/prot.21649. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(Suppl 8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 38.Das R, et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69(Suppl 8):118–128. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- 39.Unger R, Uriel S, Havlin S. Scaling law in sizes of protein sequence families: From super-families to orphan genes. Proteins. 2003;51:569–576. doi: 10.1002/prot.10347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.