Abstract
CTXφ is a filamentous, temperate bacteriophage whose genome includes ctxAB, the genes that encode cholera toxin. In toxigenic isolates of Vibrio cholerae, tandem arrays of prophage DNA, usually interspersed with the related genetic element RS1, are integrated site-specifically within the chromosome. We have discovered that these arrays routinely yield hybrid virions, composed of DNA from two adjacent prophages or from a prophage and a downstream RS1. Coding sequences are always derived from the 5′ prophage whereas most of an intergenic sequence, intergenic region 1, is always derived from the 3′ element. The presence of tandem elements is required for production of virions: V. cholerae strains that contain a solitary prophage rarely yield CTX virions, and the few virions detected result from imprecise excision of prophage DNA. Thus, generation of the replicative form of CTXφ, pCTX, a step that precedes production of virions, does not depend on reversal of the process for site-specific integration of CTXφ DNA into the V. cholerae chromosome. Production of pCTX also does not depend on RecA-mediated homologous recombination between adjacent prophages. We hypothesize that the CTXφ-specific proteins required for replication of pCTX can also function on a chromosomal substrate, and that, unlike the processes used by other integrating phages, production of pCTX and CTXφ does not require excision of the prophage from the chromosome. Use of this replication strategy maximizes vertical transmission of prophage DNA while still enabling dissemination of CTXφ to new hosts.
CTXφ is a filamentous phage whose single-stranded DNA genome includes ctxAB, the genes that encode cholera toxin (CT) (1), the primary virulence factor produced by the choleragenic bacterium Vibrio cholerae (2). Unlike the filamentous phages that infect Escherichia coli, CTXφ can give rise to lysogens, which contain the phage genome integrated into the bacterial chromosome (1, 3, 4). Integration of CTXφ DNA into the V. cholerae chromosome is a site-specific process that does not require RecA (5). Both O1 El Tor and O139 strains of V. cholerae, which have caused virtually all recent cholera epidemics, have just one chromosomal locus within which the phage genome is integrated (3, 6). The core of this integration site (attB) is a 17-bp sequence that is almost identical to an 18-bp sequence within the phage genome. Integration of phage DNA into the genome of an attB+, CTXφ− El Tor strain of V. cholerae yields single or tandem prophages flanked by these 17/18-bp sequences, which are known as end repeats (ERs) (5). If such a lysogen is infected by an additional phage or transformed with the replicative form (RF) (a plasmid) of phage DNA, the new phage DNA will integrate at an ER between tandem prophages or between a 3′ end and chromosomal DNA; the ER between the 5′ end of a prophage and adjacent chromosomal DNA is not used (7).
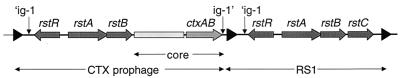
Toxigenic strains of V. cholerae frequently also contain a genetic element related to CTXφ, known as RS1 (4). This element is found on the chromosome adjacent (5′ and/or 3′) to CTX prophages (Fig. 1). Its genes include rstR, which encodes the phage repressor, and rstA and rstB, which are the phage genes needed for autonomous replication of phage DNA and for phage DNA integration. RS1 also contains an additional gene, rstC, not found in the CTXφ genome; however, it lacks ctxAB and all other genes of the phage “core” region, which are thought to encode virion coat and assembly proteins (1, 4). Many different CTXφ/RS1 arrays have been observed among epidemic V. cholerae isolates (3). However, all of the arrangements are similar in that prophages and RS1s are immediately adjacent to each other; they are never separated by chromosomal loci. Furthermore, to our knowledge, natural O1 El Tor and O139 isolates harboring a solitary prophage, not flanked by either an RS1 or another CTX prophage, have not been identified.
Figure 1.
RS1 is a genetic element frequently found adjacent to CTX prophages; it contains some but not all of the genes encoded within CTXφ. Black triangles represent end repeat (ER) sequences. The core region is not drawn to scale. Within the RF of CTXφ, intergenic region-1 (ig-1; composed of ig-1′ and ′ig-1) is a continuous sequence between ctxAB and rstR.
Most prophage/RS1 arrays in O1 El Tor and O139 strains yield infectious virions at a low but measurable rate, even in the absence of inducing stimuli such as mitomycin C (7, 8). These virions are secreted, rather than released through bacterial lysis, and virion production is not known to limit growth of the host bacterium. It is likely that virion production by lysogens is preceded by formation of the RF of the phage genome (also known as pCTX). pCTX can be detected at low levels (fewer than 1 copy per 10 cells) within plasmid DNA prepared from most CTX lysogens (B.M.D. and H. H. Kimsey, unpublished data). This plasmid is apparently a preferred template for synthesis of viral proteins and for further rounds of extrachromosomal CTX replication, because strains engineered to contain at least one copy of the RF per bacterium produce a relatively high number of virions. In contrast, strains that harbor mutant RFs whose packaging depends on chromosome-encoded viral proteins yield far fewer virions (B.M.D., K. E. Moyer, E. F. Boyd, and M.K.W., unpublished work).
Based on studies of the excision of lambda and other temperate phages that integrate into the chromosome in a site-specific fashion, we initially presumed that the CTXφ RF arises in lysogens through excision of the prophage via site-specific recombination between the flanking ER sequences. That is, we believed that the processes for generating CTX prophages and pCTX would be inversely related, as has been observed for integration/excision of lambdoid phages and the phage P2 (9). However, we have discovered that a distinct process, apparently unrelated to phage DNA integration, usually underlies formation of CTXφ virions. This process, which does not require homologous recombination, depends on the presence of tandem prophages or of a prophage followed by an RS1. Engineered strains harboring solitary prophages rarely give rise to pCTX or to infectious virions. The RFs and virions produced from phage/RS1 arrays are hybrids, derived from the coding sequences of the 5′ prophage and an intergenic region from the 3′ prophage or RS1. We hypothesize that replication of phage DNA from a chromosomal template, rather than prophage excision, is the first step in generating pCTX and ultimately CTXφ.
Materials and Methods
Bacterial Strains and Culture Conditions.
All bacteria were cultured in Luria–Bertani broth at 37°C unless otherwise noted. Antibiotics were used at the following concentrations: ampicillin (Ap), 50 μg/ml (V. cholerae); Ap, 100 μg/ml (E. coli); kanamycin (Kn), 50 μg/ml. Sucrose selection was performed as described (10).
Phage Transduction Assays.
Donor cultures were grown to an OD600 of ≈1. Both marked and unmarked CTX phage transduction assays were performed as described (7).
Molecular Biology Methods.
Southern hybridization was carried out by using horseradish peroxidase-labeled DNA probes, which were prepared and hybridized by using the ECl direct nucleic acid labeling and detection system (Amersham Pharmacia) according to the manufacturer's instructions. PCR products were cloned into the TA cloning vector pCRII-TOPO (Invitrogen) according to the manufacturer's instructions. Other techniques were performed by using standard protocols (11).
Plasmid and Strain Construction.
Plasmid derivatives of pCTX and RS1 used in this study are described in Table 1. Strains are described in Fig. 4, Fig. 6, and/or below. AS207 CTXET∷Kn was generated with the allele exchange vector pHK260 as described (7). CTXφ lysogens were generated by transforming 2740-80 [an O1 El Tor, attB+, CTXφ− strain (5)] with the appropriate pCTX; prophage copy numbers were determined by Southern blotting. BD206 was generated with a derivative of the ΔrecA∷Kn targeting vector pGP84 (12) in which the XbaI fragment containing the KnR cassette was deleted. Mutants made with this new ApR, dominant streptomycin-sensitive, counterselectable vector (pBD176) lack a 2.8-kb XbaI chromosomal segment that includes most of recA. ΔrecA clones were identified by their sensitivity to methyl methanesulfonate (Sigma) as described (13). The targeting vector (pBD570) used to generate BD578 contains a 1.09-kb HpaI-XbaI fragment of 2740-80 DNA that spans most of recA. The recA643 mutation was introduced on PCR primers (K215QF: 5′GGTAACGCACTGCAATTCTACGC and K215QR: 5′CAGAAGCGTAGAATTGCAGTGCG3′) by using standard protocols, and the mutant fragment was cloned into the counterselectable (sacB) suicide vector pCVD442 (10). This allele was crossed onto the chromosome as described (10). recA643 clones were identified by their inability to integrate, via homologous recombination, an unrelated ApR suicide vector. The presence of the A643C mutation in BD578 recA (which results in the amino acid change K215Q) was confirmed by DNA sequence analysis. The insert for pBD275 was amplified from a circularly permuted form of RS1 (B.M.D., unpublished results) by using rstC3 (5′GCTTTATATCTGCGTTCAGGCG3′) and rig1-1 (5′CACGCTACGTCGCTTATGT3′). The insert for pBD276 was similarly amplified with rstC3 and rstR9 (5′CTTCGACATCAAATGGCA3′). The related plasmids pBD286 and pBD287 were introduced into V. cholerae lysogens by mating; they could integrate into the V. cholerae chromosome by ER-mediated site specific recombination because the necessary proteins (RstB, perhaps RstA) were encoded by the resident prophage in the recipient strains. Strains with a single plasmid integrant downstream of the prophage were identified by Southern blotting.
Table 1.
pCTX and RS1-derived plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pCTXET-Kn | Replicative form of CTXET-Knφ | Ref. 1 |
| pCTXET-Ap | Xba I fragment of pCTXET-Kn (contains all CTX phage genes), inserted into pGP704 | Ref. 7 |
| pCTXETNF-Kn | pCTXET-Kn with NotI site filled in and religated | This study |
| pCTXETH1-Kn | Hybrid pCTXET-Kn from KnR transductants of AS207 CTXET∷Kn | This study |
| pGP704 | oriR6K mobRP4 suicide vector, ApR | Ref. 21 |
| pBD275 | PCR-amplified fragment (rstC3-rig1-1) of circular permutation of RS1 cloned into pCRII-Topo | This study |
| pBD276 | PCR-amplified fragment (rstC3-rstR9) of circular permutation of RS1 cloned into pCRII-Topo | This study |
| pBD286 | EcoRI insert of pBD275 in pGP704 | This study |
| pBD287 | EcoRI insert of pBD276 in pGP704 | This study |
Figure 4.
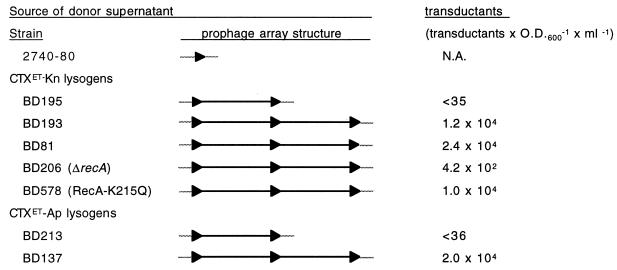
Production of CTX virions by lysogens with tandem arrays of prophages vs. solitary prophages. All strains were derived from strain 2740-80. BD 213, BD137, BD81, and its derivatives BD206 and BD578 are based on a lac− 2740-80, BD22. Black triangles represent the ERs that flank each prophage after integration into attB. Gray lines represent chromosomal DNA. Transductants × OD600−1 × ml−1 is normalized to the OD600−1 of the donor culture; the titer is the average result obtained in at least three experiments. Detection of one transductant/assay corresponds to a titer of ≈35; the exact value depends on the donor OD600−1. When no transductants were obtained, the titer was, thus, less than the calculated threshold value. N.A., not applicable.
Figure 6.
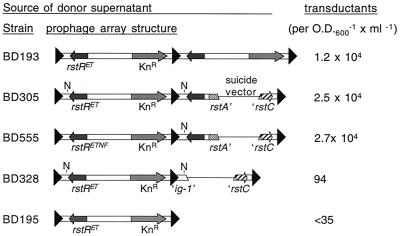
A solitary prophage with a fragment of RS1 inserted downstream can efficiently produce CTX virions. BD555 is identical to BD305 except that it contains a CTXETNF-Kn prophage (derived from pCTXETNF-Kn), not a CTXET-Kn prophage. KnR transductants derived from BD555 always contain pCTXET-Kn. The inserted fragment in BD328 is too small to rescue the 5′ prophage.
Results
“Hybrid” CTXφ Virions Produced by a Clinical Isolate of V. cholerae.
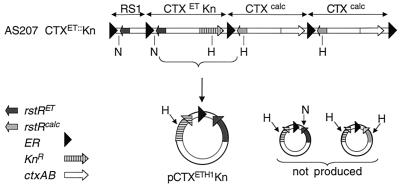
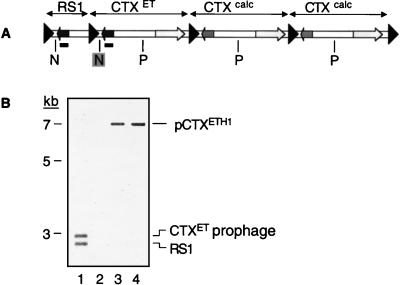
Studies of V. cholerae strain AS207 (14), a recent O139 clinical isolate from Calcutta, India, provided the first indication that CTXφ is not generated via ER-mediated excision of a CTX prophage. AS207 carries two distinct forms of CTX prophages: the prophage found in O1 El Tor strains of V. cholerae, CTXET, which was the first prophage to be identified as a producer of infectious virions, and a new prophage, CTXcalc, which also yields phage particles (7). These two prophages contain distinct rstR genes and distinct intergenic sequences flanking these genes (7, 15). To study production of virions by AS207, we created derivatives of this strain in which the various copies of ctxAB were replaced by a kanamycin resistance (KnR) cassette (7). Filtered supernatants from cultures of such marked strains can be used to transduce an O1 classical recipient strain, O395, to KnR. The CTX prophages in O395 encode a third form of RstR (RstRclass) and therefore do not interfere with replication of CTXET or CTXcalc. Copies of transduced DNA can subsequently be isolated by preparing plasmid DNA from the KnR clones because O395, like other classical strains (e.g., RV508, CA401) lacks a functional integration site for newly introduced CTXφ DNA (ref. 1; data not shown). Unexpectedly, AS207 CTXET∷Kn (depicted in Fig. 2) yielded Kn-transducing virions that did not exactly correspond to the CTXETKn prophage. Instead, AS207 CTXET∷Kn generated virions that contained coding sequences, including rstR, derived from the El Tor prophage, but also included part of an intergenic sequence (′ig-1; see Fig. 1 and Fig. 2) derived from the downstream CTXcalc prophage. This “hybrid” phage, CTXETH1-Knφ, was initially recognized because of the absence of an expected NotI restriction site in the corresponding pCTXETH1-Kn plasmid, and its structure was subsequently confirmed by DNA sequencing. The plasmid contains all of the CTXET prophage sequences except the first 357–473 bp, which have been replaced by the first 479–595 bp of the CTXcalc prophage. It is not currently possible to identify the junction between CTXET and CTXcalc-derived sequences more precisely because bp 358–473 of CTXET are identical to bp 480–595 of CTXcalc. (ig-1 of the CTXcalc prophage is slightly larger than the corresponding region of CTXETφ; hence, the numerical boundaries for the regions of sequence identity differ between the two prophages. For this analysis, the first base pair after the 5′ ER has been considered to be bp1.) Sequence analysis of AS207 CTXET∷Kn chromosomal DNA confirmed that the Kn-marked prophage in the chromosome still contained El Tor ′ig-1 sequences; these sequences had not been disrupted during generation of the Kn-marked AS207 derivative.
Figure 2.
KnR transductants derived from supernatants of AS207 CTXET∷Kn contain a plasmid with sequences from both the CTXET∷Kn prophage and the downstream CTXcalc prophage. A specific hybrid plasmid (pCTXETH1-Kn) is always produced; neither alternate hybrids nor the CTXET-Kn RF are ever detected. H, HindIII; N, NotI.
Analysis of four independently derived pools of KnR transductants, composed of >20 transductants, revealed that all contained plasmids with restriction maps corresponding to pCTXETH1Kn (described above and in Fig. 2). Alternate hybrid plasmids (e.g., containing the HindIII site within rstRcalc as well as the ′ig-1 sequence of the CTXcalc prophage) were not detected. These data strongly suggest that homologous recombination between adjacent prophages to release a KnR-marked plasmid does not underlie formation of the CTXφ RF and virions, because such a mechanism would be expected to yield KnR-transducing virions containing rstRcalc at least some of the time. Consistent formation of a specific hybrid is more likely to be the outcome of a replication and/or excision process with some sequence specificity.
We confirmed that formation of the hybrid phage was not an aberrant outcome induced by the presence of the KnR cassette by analyzing unmarked virions produced by AS207. Supernatants of AS207 were mixed with O395 and grown overnight at 30°C to maximize infection of the recipient, as described (7). Plasmid DNA was then prepared from these unselected potential transductants, and the DNA was analyzed by Southern blotting (Fig. 3). Hybridization of the blot with an rstRET probe revealed that all transduced DNA that included rstRET did not contain the NotI site found 5′ to this gene in the prophage. Thus, AS207 appeared to produce unmarked, RstRET-encoding CTXETH1φs with structures matching the marked hybrid virions produced by AS207 CTXET∷Kn and differing from the CTXET prophage from which they are principally derived. This finding is consistent with the hypothesis that virions regularly incorporate DNA from two, rather than just one, CTX prophage. This hypothesis was also supported by analyses of virions produced by additional lysogens containing tandem prophages that could be distinguished by their restriction maps. As with AS207 and AS207 CTXET∷Kn, these lysogens produced virions in which ′ig-1 sequences were derived from the second prophage in the array whereas core sequences were derived from the upstream prophage (data not shown).
Figure 3.
The restriction map of the CTXET prophage in AS207 differs from the map of pCTXET found in transductants derived from AS207. (A) Restriction map of AS207 chromosomal DNA. N, NotI; P, PstI. (B) A Southern blot of donor chromosomal DNA and plasmid DNA from potential transductants probed with rstRET (underlined in A). This probe detected a 2.9-kb band in a NotI/PstI digest of AS207 chromosomal DNA (lane 1) but a 7-kb band in a NotI/PstI digest (lane 3) of plasmid DNA from a pool of potential transductants. Digestion of the plasmid DNA with PstI alone (lane 4) revealed that the NotI site (shaded in A), rather than the PstI site, was absent from the plasmid. No hybridization was observed to NotI/PstI-digested plasmid DNA isolated from the transduction recipient O395 grown without donor supernatant (lane 2).
Negligible Production of Virions by Lone CTX Prophages.
A corollary of the hypothesis that virions regularly incorporate DNA from two CTX prophages, rather than just one prophage, is that lysogens that contain only a single CTX prophage should not yield CTX virions. To test whether this is true, we constructed an isogenic set of strains that contain either a single or tandem antibiotic-marked prophage(s) by transforming V. cholerae 2740-80 with pCTXET-Kn. The resulting transformed strains were analyzed by Southern blotting to determine the number of prophages present in each (data not shown). Culture supernatants of these strains were then tested for the presence of KnR transducing particles (Fig. 4). Supernatants from strains containing tandem CTXET-Kn prophages (e.g., BD193 and BD81) yielded more than 104 KnR transductants × OD600−1 × ml−1. In contrast, supernatants from a strain with a single CTXET-Kn prophage (BD195) rarely yielded any transductants. Only one trial of BD195 supernatant yielded a single KnR colony. Similar data were obtained with single and tandem CTXET-Ap prophages, which transduce recipients to ampicillin (Ap) resistance (Fig. 4). As expected, based on this data, pCTX could be detected within plasmid DNA isolated from cultures of lyosgens with tandem prophages but not within DNA from lysogens with a single prophage (data not shown). These data indicate that production of virions from a solitary prophage is a very infrequent event.
Imprecise Excision of Solitary CTX Prophages.
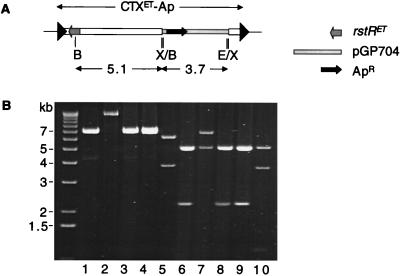
To gain insight into the mechanism by which virions occasionally arise from solitary CTX prophages, we used restriction mapping to analyze the plasmids within ApR transductants generated from supernatants of BD213, a strain with a single CTXET-Ap prophage. In four of four transductants studied, the ApR plasmid did not correspond precisely to pCTXET-Ap (Fig. 5, compare lanes 1–4 to lane 5 and lanes 6–9 to lane 10). None of the four contained sequences from the 3′ end of the CTX prophage. Instead, restriction mapping and PCR analyses indicated that the plasmids all contained some chromosomal sequences from upstream of the CTXET-Ap prophage in BD213 as well as a portion of the CTXET-Apφ genome (Fig. 5; data not shown). These data suggest that BD213, and presumably other strains with a solitary prophage as well, only produces virions after imprecise excision of the prophage from the chromosome. Thus, it appears that ER-mediated site specific recombination, which was initially believed to underlie all CTXφ virion production, is not even used to generate the rare subset of virions produced from solitary CTX prophages.
Figure 5.
Plasmids isolated from ApR transductants derived from the supernatant of BD213, which contains a solitary CTXET-Ap prophage, are not equivalent to pCTXET-Ap. (A) Restriction map of BD213 chromosomal DNA. B, BglII; X, XbaI; E, EcoRI. (B) pCTXET-Ap (lanes 5 and 10) yielded different restriction fragments than did plasmid DNA from four transductants (lanes 1–4 and 6–9) after digestion with XbaI (lanes 1–5) and EcoRI/BglII (lanes 6–10). All of the new ApR plasmids contained the 5.1-kb BglII fragment and the adjacent ApR cassette; all lacked the EcoRI and XbaI sites at the 3′ end of the prophage.
Production of CTXφ from Prophage Arrays Does Not Depend On Homologous Recombination.
The elevated phage yields from lysogens with tandem prophages (relative to single prophage strains; Fig. 4) is consistent with the model that virions incorporate genetic material from two distinct prophages. However, the uniformity of hybrids produced by AS207 suggests that unrestricted homologous recombination between adjacent CTX prophages is not the dominant mechanism for producing pCTX from such arrays. Consequently, we assessed whether homologous recombination is necessary for production of CTX virions by strains with tandem prophages, using two recA− derivatives of BD81. One derivative, BD206, has a 2.8-kb deletion encompassing most of recA whereas the other, BD578, has a point mutation (K215Q) expected to impair homologous recombination but not RecA-mediated protease activity (16). We found that supernatants from BD206 yielded significantly fewer transductants than the isogenic wild type strain, although KnR colonies were still obtained with an average frequency of 420 transductants × OD600−1 × ml−1 (Fig. 4). In contrast, BD578 yielded 104 transductants × OD600−1 × ml−1, similar to the wild-type strain BD81, despite the fact that its capacity for homologous recombination was equivalent to that of the recA deletion strain (data not shown). pCTX DNA was detectable within plasmid DNA prepared from cultures of both strains (data not shown). These data suggest that homologous recombination between adjacent prophages is not required for efficient production of virions but that RecA-mediated proteolysis does contribute to formation of CTXφ. As with other temperate phages, RecA may mediate cleavage of the CTXφ-specific repressor, RstR, and thereby facilitate viral replication. Interestingly, CTXφ lysogens still yield a low level of virions even in the absence of RecA (i.e., BD206), indicating that the generation of CTXφ does not entirely depend on RecA-mediated cleavage of RstR.
“Rescue” of a Solitary Prophage by Downstream Insertion of RS1 Derivatives.
Because the CTXETH1-Knφ produced by AS207 CTXET∷Kn consistently contain only a short sequence derived from the 3′ prophage in the array, we speculated that a complete prophage followed by just a fragment of a prophage might still efficiently give rise to virions. We therefore generated derivatives of 2740-80 in which fragments of RS1 were inserted downstream of a solitary prophage. Like the sequentially integrated prophages in BD315 and BD550, the new downstream fragments were introduced into the chromosomal DNA by ER-mediated recombination. The fragments do not encode the proteins needed for CTXφ replication and integration (RstA and RstB), but these were provided by the prophage already present on the chromosome. The new prophage arrays generated by using this strategy are diagrammed in Fig. 6; the structure of each was confirmed by Southern blotting (data not shown).
Supernatants from these new strains were used in transduction assays to determine whether insertion of a prophage fragment could rescue a solitary prophage (i.e., enhance its capacity to yield virions). We found that insertion of a 1,280-bp sequence (from ER through the beginning of rstA) downstream of a single prophage was as effective as insertion of a complete prophage in rescuing virion production by the 5′ prophage. Supernatants from strains containing the 1,280-bp insertion (e.g., BD305, BD555) yielded at least 104 transductants × OD600−1 × ml−1, a titer equivalent to the tandem prophage strain BD193 (Fig. 6). Furthermore, the RF of virions produced by BD555 always contained a NotI site (pool of 15 assayed), although the 5′ prophage in BD555 lacks this site, suggesting that virions had again incorporated sequences from the 3′ element within the array. These data also demonstrated that a defect in the solitary prophage in BD195 (which was used to construct BD305) was not responsible for its failure to yield virions.
Insertion of a shorter fragment of RS1 was much less effective at rescuing virion production by a solitary prophage. Introduction of a 340-bp fragment (which included all but the final 17 bp of the region known to be derived from the 3′ prophage; e.g., in BD328) resulted in production of 96 transductants × OD600−1 × ml−1. Thus, the presence of the 340-bp fragment downstream of the solitary prophage enhanced virion production relative to the yield from the prophage alone; however, the effect from this fragment was far below that of a larger fragment or a complete prophage.
Discussion
We have shown that the “unit prophage” from which CTXφ virions are generally derived is in fact a hybrid, composed from two adjacent prophages or prophage-related elements. Virions derive all of their coding sequence from the 5′ prophage in an array, but their DNA for an intergenic region (′ig-1) is primarily from the 3′ genetic element. The 3′ element can be a second prophage, an RS1, or a derivative of RS1. Formation of virions from tandem elements does not depend on RecA-mediated homologous recombination between adjacent prophages.
Virions are rarely produced by CTX lysogens that contain only a solitary prophage, even if such strains are treated with Mitomycin C (Fig. 4; data not shown). Analysis of the few tranductants derived from these strains suggests that virions were produced after imprecise excision of the single CTX prophage from the chromosome. Similar imprecise excision of a CTX precursor phage is thought to have enabled a ctxAB− precursor phage to acquire the genes encoding cholera toxin (E. F. Boyd and M.K.W., unpublished work). We have never detected virions that appear to have arisen from site specific recombination between the ERs flanking the prophage, a process that could theoretically also result in excision of prophage DNA. Many mobile genetic elements, including lambdoid phages, phage P2, and Tn916, use such a process to generate extrachromosomal forms of DNA (9, 17). However, these elements depend on an element-encoded excisionase (e.g., Xis) to reverse the integration pathway, and such a protein is not known to be encoded within CTXφ. The absence of an Xis could account for the failure of solitary CTX prophages to yield virions with intact genomes.
Interestingly, clinical isolates of O1 El Tor and O139 V. cholerae rarely, if ever, harbor solitary CTX prophages. In fact, all of our strains with solitary prophages were engineered in the laboratory. Some O1 El Tor and O139 clinical isolates do contain a single prophage, but to our knowledge all of these prophages are flanked by at least one RS1. This raises the possibility that virion-producing lysogens cause more disease and thus are more frequently detected in patients than lysogens that produce CT but few, if any, virions. Selection for production of virions might account for the widespread distribution of RS1 among O1 El Tor lysogens. RS1 is found in most epidemic strains along with CTX prophages, despite the fact that it has not been shown to play a direct role in bacterial survival or pathogenesis. Selection for virion production might also account for the fact that O1 strains of the classical biotype, whose prophages do not yield virions, have been supplanted by O1 El Tor and O139 strains as the dominant cause of epidemic disease.
The precise mechanism by which prophage arrays yield CTXφ has not been determined, and it could be very difficult, using currently available reagents, to analyze this process and characterize bacterial chromosomal DNA after virion production. Virion yields from lysogens are so low (maximum 105 × OD600−1 × ml−1, which corresponds to approximately 1 virion per 1,000 cells) that most cells in a culture have not produced virions. Consequently, identifying individual virion producers within a population is very challenging. Currently, there is no evidence that production of virions by a lysogen induces any changes within chromosomal DNA; indeed, Southern blot analysis suggests that the structures of prophage/RS1 arrays are quite stable, at least in the absence of extreme selective pressures. Furthermore, our data indicates that homologous recombination between adjacent prophages does not underlie formation of pCTX. We therefore suspect that production of pCTX and subsequently CTXφ relies on the mechanism described below, which is consistent with the results presented in this paper, the observed stability of prophage arrays, and previous analyses of plasmid and virion replication.
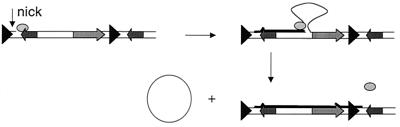
We hypothesize that the phage-replicative machinery, which probably comprises RstA and several V. cholerae encoded proteins (4), can use chromosomal as well as plasmid templates for DNA synthesis. Thus, instead of commencing phage DNA replication at a nicked site within pCTX [a site possibly produced by RstA, which shows some similarity to rolling circle initiator proteins (4)], phage replication could originate at an RstA-induced chromosomal nick within the prophage (Fig. 7). As in rolling circle replication (18), DNA synthesis could then proceed along the prophage, displacing the original DNA, until a sequence and/or secondary structure equivalent to the initial nick site was encountered. In plasmid replication, this would in fact be the site of initiation; in a prophage array, this site would be the corresponding site in the 3′ prophage (or RS1). In either case, this site could then prompt production of a new nick and circularization of the DNA displaced by the synthesis, which would be equivalent to a single strand of the CTX RF. Second strand synthesis, presumably identical to that after CTXφ infection of a new host, would then ensue to yield pCTX.
Figure 7.
Model for generation of the replicative form of CTXφ from a prophage array. The phage replication protein RstA (oval) introduces a nick into ′ig-1 of the 5′ prophage. DNA synthesis (thick black line) originates at the nick and proceeds along the prophage, displacing one strand of DNA but keeping it tethered to the replication complex, until the replication machinery encounters the origin within the 3′ prophage. A new nick is introduced, generating a 3′ end that can be joined to the 5′ end of the displaced single strand to yield a circular, single stranded copy of the phage genome. Second strand synthesis is equivalent to synthesis after infection by CTXφ.
Site-specific nicking/initiation within ig-1 of the CTX prophage would account for the uniform composition of the hybrids produced by each of the clinical and lab-derived lysogens that we studied. Furthermore, this model is consistent with independent analyses of the origin of pCTX replication, which suggest that the double-stranded origin is near the 3′ end of ig-1 (K. Moyer, personal communication). Thus, we propose that production of pCTX from an array of prophages occurs without excision of a prophage from the chromosome. In this respect, replication of the CTX genome may resemble propagation of replicative transposons or phage Mu more closely than it resembles that of phages such as λ or P2. It may be particularly similar to transposition of IS91, which also depends on a protein with homology to rolling circle replication proteins (19). However, although we favor the replicative process outlined above as the explanation for generation of pCTX from a prophage array, our current data does not exclude the possibility that pCTX production could result from a conservative process in which hybrid genomes are excised from the chromosome. Still, regardless of whether the process yielding pCTX is replicative or conservative, it is clearly distinct from the typical excision mechanisms of other prophages, because it depends on sequences within tandem elements rather than a solitary prophage. Tandem lambdoid prophages can also yield hybrid virions (20); however, lambdoid prophages are generally found as solitary, rather than tandem, elements.
A replicative strategy in which production of virions and maintenance of proviral DNA are not mutually exclusive pathways could have significant evolutionary advantages for the prophage. If virions can be produced for horizontal transmission of phage DNA (transduction) without disrupting the vertical transmission of the prophage (via chromosomal replication), then virion production does not need to be solely an escape mechanism used to release prophage DNA from a damaged or replication-deficient host. Instead, constitutive (albeit at low levels) virion production under normal growth conditions is feasible. The fact that CTXφ is secreted, rather than released by host lysis, is a complementary feature of CTXφ dissemination. In summary, the CTX genome appears to replicate by a strategy that maximizes stable propagation of the lysogen while still enabling spread of the phage to new hosts.
Acknowledgments
We are grateful to our colleagues, Drs. A. Camilli, A. Kane, and B. Hochhut, for helpful suggestions and critical reading of the manuscript. We thank A. Kane and the New England Medical Center GRASP Center for preparation of plates and media. This work was supported by National Institutes of Health Grant AI-42347 to M.K.W., Grant P30DK-34928 for the NEMC GRASP Digestive Center, Grant GM20483-01 to B.M.D., and the Howard Hughes Medical Institute. M.K.W. is a Pew Scholar in the Biomedical Sciences.
Abbreviations
- Kn
kanamycin
- Ap
ampicillin
- RF
replicative form
- CT
cholera toxin
- ER
end repeat
- ig-1 intergenic region 1.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140109997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140109997
References
- 1.Waldor M K, Mekalanos J J. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 2.Kaper J B, Morris J G, Jr, Levine M M. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mekalanos J J. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 4.Waldor M K, Rubin E J, Pearson G D, Kimsey H, Mekalanos J J. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 5.Pearson G D, Woods A, Chiang S L, Mekalanos J J. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waldor M K, Mekalanos J J. J Infect Dis. 1994;170:278–283. doi: 10.1093/infdis/170.2.278. [DOI] [PubMed] [Google Scholar]
- 7.Davis B M, Kimsey H H, Chang W, Waldor M K. J Bacteriol. 1999;181:6779–6787. doi: 10.1128/jb.181.21.6779-6787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimsey H H, Waldor M K. Infect Immun. 1998;66:4025–4029. doi: 10.1128/iai.66.9.4025-4029.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neidhardt F C, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: Am. Soc. Microbiol.; 1996. [Google Scholar]
- 10.Donnenberg M S, Kaper J B. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidmann J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Greene and Wiley; 1990. [Google Scholar]
- 12.Pearson G D N. Ph.D. thesis. Cambridge, MA: Harvard Univ. Medical School; 1989. [Google Scholar]
- 13.Goldberg I, Mekalanos J J. J Bacteriol. 1986;165:715–722. doi: 10.1128/jb.165.3.715-722.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma C, Maiti S, Mukhopadhyay A K, Basu A, Basu I, Nair G B, Mukhopadhyaya R, Das B, Kar S, Ghosh R K, Ghosh A. J Clin Microbiol. 1997;35:3348–3350. doi: 10.1128/jcm.35.12.3348-3350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimsey H H, Nair G B, Ghosh A, Waldor M K. Lancet. 1998;352:457–458. doi: 10.1016/S0140-6736(05)79193-5. [DOI] [PubMed] [Google Scholar]
- 16.Larminat F, Cazaux C, Germanier M, Defais M. J Bacteriol. 1992;174:6264–6269. doi: 10.1128/jb.174.19.6264-6269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poyart-Salmeron C, Trieu-Cuot P, Carlier C, Courvalin P. EMBO J. 1989;8:2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan S A. Microbiol Mol Biol Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendiola M V, Bernales I, de la Cruz F. Proc Natl Acad Sci USA. 1994;91:1922–1926. doi: 10.1073/pnas.91.5.1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottesman M E, Yarmolinsky M B. J Mol Biol. 1968;31:487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- 21.Miller V L, Mekalanos J J. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]