Abstract
The protein modifier ubiquitin is a signal for proteasome-mediated degradation in eukaryotes. Proteasome-bearing prokaryotes have been thought to degrade proteins via a ubiquitin-independent pathway. We have identified a prokaryotic ubiquitin-like protein, Pup (Rv2111c), which was specifically conjugated to proteasome substrates in the pathogen Mycobacterium tuberculosis. Pupylation occurred on lysines and required proteasome accessory factor A (PafA). In a pafA mutant, pupylated proteins were absent and substrates accumulated, thereby connecting pupylation with degradation. Although analogous to ubiquitylation, pupylation appears to proceed by a different chemistry. Thus, like eukaryotes, bacteria may use a small-protein modifier to control protein stability.
Similar to the eukaryotic 20S proteasome, the Mycobacterium tuberculosis (Mtb) proteasome is a multisubunit barrel-shaped protease composed of two rings of catalytic β subunits sandwiched by rings of α subunits (1–5). The eukaryotic 26S proteasome is composed of a 20S core particle and one or two 19S regulatory caps, which include adenosine triphosphatases (ATPases) that recognize, unfold, and translocate substrates into the core for degradation [reviewed in (6)]. In Mtb, Mpa (Mycobacterium proteasome ATPase) shares homology with regulatory cap ATPases. Substrates of the Mtb proteasome have been identified (7), but it remains unclear how they were targeted for degradation. Proteins delivered to the eukaryotic proteasome are usually conjugated with ubiquitin, which covalently attaches to substrate lysines (Lys) as well as onto ubiquitin itself [reviewed in (8)]. Genes encoding ubiquitin-like proteins (Ubls) have not been identified in the Mtb genome.
To further define the Mtb proteasome system, we looked for proteins that interacted with Mpa using an Escherichia coli bacterial two-hybrid system (9, 10). A fusion protein that encoded the last 26 amino acids of Rv2111c (here referred to as “Pup”) interacted with the Mpa bait fusion [Fig. 1A (10)]. Full-length Pup also specifically interacted with Mpa (Fig. 1A). The pup gene has been identified (11, 12), but the function of Pup was unknown. pup homologs have so far only been identified in Actino-bacteria by BLAST search (13). In Mtb, pup is part of a putative operon with the proteasome core genes prcB and prcA (fig. S2). pup is predicted to encode a 64–amino acid protein with a molecular size of 6.9 kD (GenBank accession number EU914921). Recombinant Pup purified from E. coli migrated to a position around 15 kD in a denaturing polyacrylamide gel (Fig. 1B); however, certain Ubls, like SUMO-1, migrate more slowly than expected (14, 15).
Fig. 1.
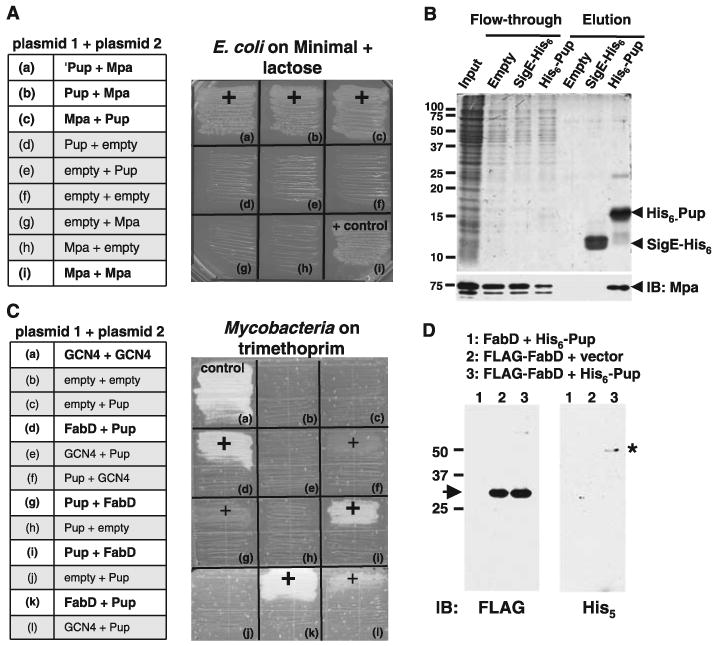
Pup interacts with the ATPase Mpa and the proteasome substrate FabD. (A) Mpa interacted with Pup in an E. coli two-hybrid system. E. coli (cya) was transformed with combinations of plasmids encoding either of the two domains of Bordetella pertussis Cya, T25 (“plasmid 1”) or T18 (“plasmid 2”), fused to test proteins (for plasmid details, see fig. S1A and table S1). ‘Pup represents the 26–amino acid fragment identified from an Mtb genomic T25 library with T18C-Mpa as bait (a). Interactions that reconstituted functional Cya permitted growth on minimal lactose agar (“+”). All strains grew on minimal glucose agar (fig. S1A). (B) Mpa interacted with Pup in vitro. His6-Pup, SigE-His6, or E. coli “vector only” lysate on Ni-NTA agarose was incubated with recombinant Mpa (“input”). Fractions were separated by 15% SDS–polyacrylamide gel electrophoresis (PAGE) and visualized with Coomassie Brilliant Blue (CBB). The same samples were analyzed by anti-Mpa immunoblot (IB, below). (C) Pup interacted with FabD in an Msm two-hybrid system. Msm was transformed with combinations of plasmids encoding either of the two domains of murine dihydrofolate reductase, F(1,2) (“plasmid 1”) or F(3) (“plasmid 2”), fused to Pup, FabD, GCN4 (a Saccharomyces cerevisiae leucine zipper domain), or no other protein (for plasmid details, see fig. S1B and table S1). Positive interactions permitted growth on trimethoprim (Trim) (“+”). Pup had weak interactions with GCN4 (f, l). All strains grew on media lacking Trim (fig. S1B). (D) Pup formed a stable complex with FabD in Msm. FLAG-tagged proteins were enriched from equal amounts of lysates of Msm with plasmids encoding FLAG-FabD and either empty vector or His6-Pup. Untagged FabD was the negative control. Samples were separated by 12% SDS-PAGE, and analyzed by anti-FLAG or anti-His5 immunoblotting. FLAG-FabD migrated at the predicted size (arrow, left) and at a higher molecular size (fig. S3A); the ∼45-kD anti-His5–reactive protein (asterisk, right) is only seen in mycobacteria producing FLAG-FabD and His6-Pup.
We then tested the Pup/Mpa interaction in vitro using nickel-nitrilotriacetic acid (Ni-NTA) agarose bound with purified His6-Pup, and Pup was able to bind Mpa (Fig. 1B) (10). Mpa was not retained by agarose that had first been incubated with E. coli lysate or with SigE-His6, a Salmonella typhimurium protein that is similar in size and charge to Pup (16). Thus, Pup specifically and noncovalently interacted with Mpa in an E. coli lysate under native conditions.
Additional genetic and biochemical experiments with E. coli to test for interactions between Pup and other Mtb proteasome components were unsuccessful. Thus, we hypothesized that E. coli lacked cofactors that were necessary to promote certain Mtb protein-protein interactions. We therefore used a mycobacterial protein fragment complementation assay (17) to test for interactions between various Mtb proteasome components and substrates in Mycobacterium smegmatis (Msm). Surprisingly, we observed a strong positive interaction between Pup and the proteasome substrate FabD [mal-onyl coenzyme A acyl carrier protein] (Fig. 1C). To confirm the interaction, we expressed constructs encoding FLAG-FabD and His6-Pup in Msm. Antibodies to FLAG (anti-FLAG) detected purified FLAG-FabD at the predicted size of ∼30 kD (Fig. 1D). Unexpectedly, His5-specific antibodies detected a purified ∼45-kD species when FLAG-FabD and His6-Pup were coproduced in mycobacteria (Fig. 1D). We also observed the ∼45-kD band upon a longer exposure with anti-FLAG (fig. S3A). This ∼45-kD species, probably representing a Pup∼FabD complex, was highly stable because it was maintained under reducing and denaturing conditions. When FLAG-FabD was purified from an E. coli strain making His6-Pup, we were unable to detect the ∼45-kD species (fig. S3B). Thus, Pup interacts with an Mtb proteasome substrate in a manner that is not supported in E. coli, and requires Mycobacterium-specific factors.
The formation of a stable complex between our model substrate FabD and Pup was reminiscent of the covalent attachment of ubiquitin to proteasome substrates in eukaryotes. Sequence and structural prediction comparisons between Pup and ubiquitin showed no overall homology. However, we noticed conservation of either of the basic amino acids arginine (Arg) or Lys, followed by two glycines (Gly) at the C termini (Fig. 2A). This di-Gly motif is conserved in most members of the ubiquitin-like protein family, and is usually followed by one or more amino acids [reviewed in (18)]. The C termini of Ubls are generally processed to expose the di-Gly and then activated for conjugation to substrate proteins through a series of enzyme-catalyzed reactions [reviewed in (19)]. The terminal Gly of ubiquitin is essential for the formation of an isopeptide bond with the Lys of a substrate [reviewed in (8)].
Fig. 2.
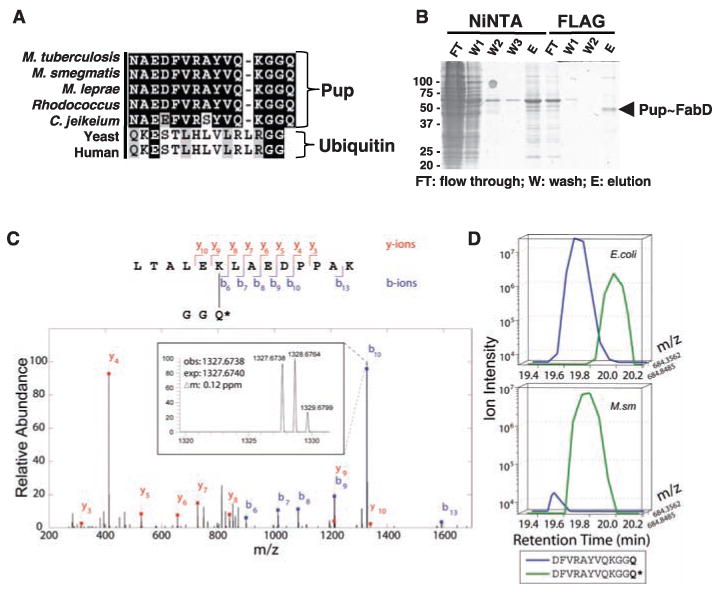
The C terminus of Mtb Pup covalently attaches to Lys173 of Mtb FabD. (A) Alignment of the C terminus of Pup to that of Pup or ubiquitin from representative Actinomycetes or eukaryotes, respectively. Identical amino acids are shaded black. Sequences were compiled from the National Center for Biotechnology Information server and aligned by means of ClustalW (23). (B) Purification of the FabD∼Pup complex. Msm was cotransformed with plasmids encoding FLAG-FabD and His6-Pup. FLAG-FabD∼His6-Pup was purified sequentially with Ni-NTA agarose and anti-FLAG M2 affinity matrix. Proteins from each purification step were analyzed by 12% SDS-PAGE and visualized with CBB. (C) Tandem mass (MS/MS) spectrum of a FabD tryptic peptide derived by collision-induced dissociation of the (M + 2H)2+ precursor, mass/charge ratio (m/z) 869.963 [1.55 parts per million (ppm)]. Singly charged fragment ions marked in the spectrum represent peptide bond cleavage resulting in the sequence information recorded from both the N and C termini (b- and y-type ions, respectively). This spectrum, searched with the SEQUEST program, matched to the peptide shown with a mass shift corresponding to a deamidation event, converting the Pup C-terminal Gln to Glu (Q*). High mass accuracy MS/MS unambiguously confirms covalent modification of lysine in FLAG-FabD by His6-Pup, with multiple matching b- and y-type ions. Additional detailed fragment ion information and additional spectra are presented in fig. S4. (D) Extracted ion chromatograms of the C-terminal peptide of Asp-N–digested His6-Pup. The traces correspond to the m/z of MH22+ precursors ±3 ppm). In E. coli, deamidated Gln was detected at a low abundance (∼10%), whereas in Msm, the C-terminal Gln deamidation predominated. Q* denotes a deamidated Gln, equivalent to Glu. See fig. S5 and (10) for additional details. Amino acid residues: A, Ala; D, Asp; E, Glu; F, Phe; G, Gly; H, His; K, Lys; L, Leu; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; and Y, Tyr.
Consequently, we used tandem affinity chro-matography to purify FLAG-FabD∼His6-Pup (Fig. 2B) and characterized the interaction using mass spectrometry (MS) (20). MS analysis of ubiquitylated substrates typically identifies substrate peptides with the tryptic Gly-Gly ubiquitin fragment covalently attached to a lysine (20). Our MS analysis confirmed the presence of both Mtb proteins and, given the Pup C-terminal sequence (Gly-Gly-Gln; Gln, glutamine), we performed a high-resolution tandem MS/MS search allowing for either a Gly-Gly or Gly-Gly-Gln modification of FabD (Fig. 2C). This analysis revealed several spectral matches to a FabD tryptic peptide with the Pup C-terminal sequence attached through an isopeptide bond to Lys173 of FabD. The precursor mass deviation (ΔM), however, suggested a deamidation event (ΔM = +0.984), pointing to a probable C-terminal Gln→Glu conversion. This result showed that the Gln following the di-Gly of Pup was not removed. We then purified unconjugated His6-Pup from E. coli and Msm, digested the protein with Asp-N protease, and analyzed peptides by MS/MS. Using the raw intensity data, we estimated roughly a 1:10 ratio of deamidated Gln:Gln at the Pup C terminus when purified from E. coli (Fig. 2D). In contrast, the deamidated form dominated by two orders of magnitude in Msm, strongly suggesting that enzymatic activity was responsible for the conversion of Pup into its active form. Thus, deamidated Pup was covalently bound to a specific Lys residue of an Mtb proteasome substrate in a manner analogous to the conjugation of ubiquitin to eukaryotic proteasome substrates.
FabD and other Mtb proteasome substrates accumulate in mpa and pafA mutants (7). If Pup, like ubiquitin, targets proteins for degradation, pupylated FabD should also accumulate in these mutants. FLAG-tagged FabD abundance was increased in the mpa and pafA strains compared to wild-type (WT) Mtb (Fig. 3A). We detected Pup∼FabD in WT Mtb, and an accumulation of this species in the mpa mutant (Fig. 3A). We also observed Pup∼FabD in WT samples using FLAG-specific antibodies (Fig. 3A). Pup∼FabD is present at extremely low steady-state amounts, suggesting that the transition from an unpupylated to a pupylated state is a tightly regulated process, like that of Ubl conjugation (19, 21). Unexpectedly, Pup∼FabD was undetectable in the pafA strain (Fig. 3A), despite the accumulation of unpupylated FabD. Similar observations were made for another Mtb proteasome substrate, PanB (ketopantoate hydroxymethyltransferase) (fig. S6A), but not for DlaT (dihydrolipoamide acyltransferase) (Fig. 3A), which is not a substrate (7). Therefore, PafA is involved in pupylation, a process that seems to be specific for Mtb proteasome substrates.
Fig. 3.
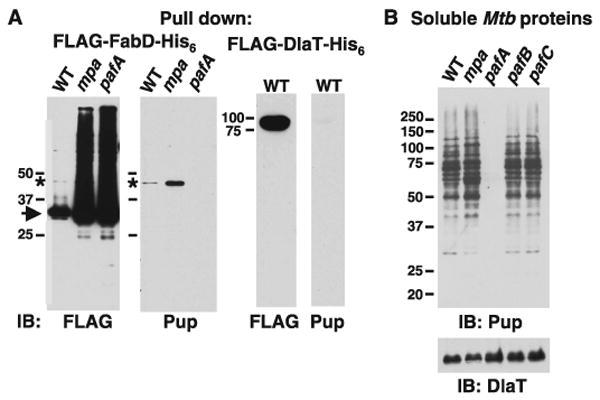
Pupylation is associated with Mtb proteasome substrates. (A) Aberrant amounts of pupylation correlated with proteasome-defective states. Equal amounts of soluble Mtb lysates from WT, mpa and pafA strains were incubated with Ni-NTA agarose for enrichment of FLAG-FabD-His6. Samples were deliberately overloaded to detect pupylated protein and observe the relative amounts of unpupylated versus pupylated FabD. Anti-FLAG immunoblots of Ni-NTA eluates detected both unpupylated (arrow) and pupylated (asterisk) FLAG-FabD-His6. Anti-Pup immunoblots of the same samples detected Pup∼FLAG-FabD-His6 (asterisk) in WT and mpa Mtb but not in the pafA strain. As a control, FLAG-DlaT-His6 was purified from WT Mtb. Anti-FLAG immunoblots detected a protein at the predicted size of FLAG-DlaT-His6, but no pupylated species was detected. Ponceau S staining shows that protein is present on this membrane (fig. S6B). (B) Multiple pupylated proteins were present in Mtb, but not in a pafA mutant. Anti-Pup immunoblots of Mtb lysates from WT, mpa, pafA, pafB, and pafC strains. Equivalent cell numbers were analyzed and the same blot was used for detection of endogenous DlaT. All samples were separated by 10% SDS-PAGE.
If Pup acts like ubiquitin, then multiple pupylated proteins could exist in Mtb. Immunoblot analysis with a Pup-specific antibody against soluble proteins from WT and mpa Mtb strains revealed a ladder of proteins (Fig. 3B). Again, no anti-Pup reactive bands were observed in the pafA sample (Fig. 3B), implying that this phenomenon extends to all targets of pupylation within the limits of detection. We were unable to detect the unconjugated form of Pup, suggesting that most Pup molecules are conjugated to substrates at steady state, or are rapidly degraded by an unidentified protease. Because pafA is in an operon with pafBC, we also tested pafB and pafC mutants for substrate pupylation (Fig. 3B). The extent of pupylation did not differ between the WT strain and the pafBC mutants, confirming that PafB and PafC do not seem to be involved in substrate degradation (22).
Our data suggest that PafA-dependent pupylation of Lys173 leads to the degradation of FabD. To test this hypothesis, we followed the stability of purified 35S-labeled FLAG-FabD- K173A-His6 from WT Msm. The K173A mutant was markedly more stable than WT FabD (Fig. 4, A and B), providing further evidence that pupylation is a signal for degradation. We then purified radiolabeled His6-pupylated proteins from WT and mpa-deficient Msm and observed the disappearance of these proteins over time in WT but not mpa-deficient bacteria (Fig. 4C and fig. S7). Thus, Pup covalently conjugates to a specific Lys of an Mtb proteasome substrate, and pupylated proteins are degraded in an Mpa/proteasome-dependent manner (fig. S8).
Fig. 4.
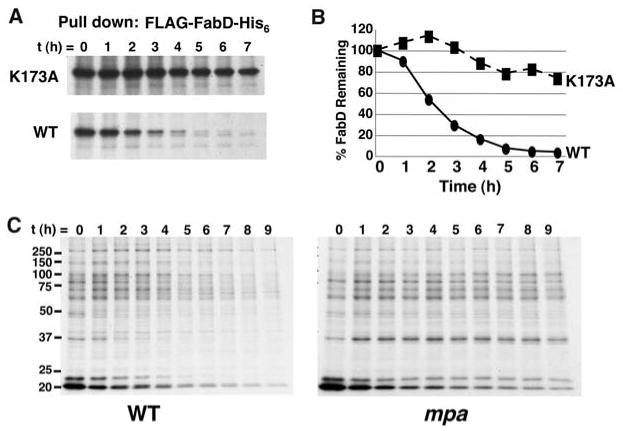
Pupylation is required for Mpa-dependent protein degradation. (A) K173A mutation stabilized FabD. WT Msm expressing WT fabD or fabD with the K173 codon mutated to alanine was pulse labeled with 35S-methionine and cysteine. Samples were collected over time and FLAG-FabD-His6 (WT or K173A mutant) was purified and analyzed by 10% SDS-PAGE (10). This image represents a 12-hour exposure. A 6-hour exposure of the same gel is shown in fig. S7A. Immunoblot analysis showed that the K173A mutant was also not efficiently pupylated (fig. S7B). (B) Quantification of labeled protein in (A). (C) Pupylated proteins were degraded in an Mpa-dependent manner. WT and mpa mutant Msm were treated as in (A) and His6-pupylated proteins were purified and analyzed. Total 35S protein labeling is shown in fig. S7C. All data are representative of at least two independent experiments.
There are similarities between the ubiquitin and Pup systems, but there are also notable differences. Unique aspects of pupylation may include the mechanism of Pup activation and conjugation to substrates, the chemistry involved in the linkage of Pup to Lys, and the involvement of PafA. We speculate that PafA plays a part in conjugating Pup to substrates, but this idea requires further investigation. Additionally, it remains to be determined if proteins can be poly-pupylated in Mtb.
Aside from a role in protein degradation, Pup and other small protein modifiers may have important implications for other cellular processes in bacteria. Considering the multitude of activities coordinated by ubiquitylation or SUMOylation in eukaryotes (19, 21), prokary-otes may also use posttranslational protein modifiers for functions ranging from subcellular sorting to secretion.
Supplementary Material
Acknowledgments
We thank S. Ehrt, D. Ladant, V. Miller, and A. Steyn for plasmids used in this study. We are grateful to C. Arias and C. Perez for advice on pulse-labeling experiments and to T. Huang, I. Mohr, and M. Pagano for helpful discussions. We thank A. Darwin and M. Pagano for critical review of this manuscript. This work was supported by NIH grants AI065437 and HL092774 (to K.H.D.) and GM67945, HG3456, and HG3616 (to S.P.G.). M.J.P. was supported by grant 5T32AI07189-25.
Footnotes
References and Notes
- 1.Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL. Mol Cell. 2003;11:69. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 2.Groll M, et al. Nature. 1997;386:463. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 3.Hu G, et al. Mol Microbiol. 2006;59:1417. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- 4.Lin G, et al. Mol Microbiol. 2006;59:1405. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- 5.Unno M, et al. Structure. 2002;10:609. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 6.Baumeister W, Walz J, Zühl F, Seemüller E. Cell. 1998;92:367. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 7.Pearce MJ, et al. EMBO J. 2006;25:5423. doi: 10.1038/sj.emboj.7601405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 9.Karimova G, Pidoux J, Ullmann A, Ladant D. Proc Natl Acad Sci USA. 1998;95:5752. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Materials and methods are available as supporting material on Science Online.
- 11.Knipfer N, Shrader TE. Mol Microbiol. 1997;25:375. doi: 10.1046/j.1365-2958.1997.4721837.x. [DOI] [PubMed] [Google Scholar]
- 12.Tamura T, et al. Curr Biol. 1995;5:766. doi: 10.1016/s0960-9822(95)00153-9. [DOI] [PubMed] [Google Scholar]
- 13.Altschul SF, et al. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. Cell. 1997;88:97. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 15.Matunis MJ, Coutavas E, Blobel G. J Cell Biol. 1996;135:1457. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darwin KH, Robinson LS, Miller VL. J Bacteriol. 2001;183:1452. doi: 10.1128/JB.183.4.1452-1454.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, Mai D, Kumar A, Steyn AJ. Proc Natl Acad Sci USA. 2006;103:11346. doi: 10.1073/pnas.0602817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hochstrasser M. Nat Cell Biol. 2000;2:E153. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- 19.Kerscher O, Felberbaum R, Hochstrasser M. Annu Rev Cell Dev Biol. 2006;22:159. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 20.Kirkpatrick DS, Denison C, Gygi SP. Nat Cell Biol. 2005;7:750. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiss-Friedlander R, Melchior F. Nat Rev Mol Cell Biol. 2007;8:947. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 22.Festa RA, Pearce MJ, Darwin KH. J Bacteriol. 2007;189:3044. doi: 10.1128/JB.01597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chenna R, et al. Nucleic Acids Res. 2003;31:3497. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


