Summary
Ribosomes and spliceosomes are ribonucleoprotein nanomachines that catalyze translation of mRNA to synthesize proteins and splicing of introns from pre-mRNAs, respectively. Assembly of ribosomes involves more than 300 proteins and RNAs, and that of spliceosomes over 100 proteins and RNAs. Construction of these enormous ribonucleoprotein particles (RNPs) is a dynamic process, in which the nascent RNPs undergo numerous ordered rearrangements of RNA-RNA, RNA-protein, and protein-protein interactions. Here we outline similar principles that have emerged from studies of ribosome and spliceosome assembly. Constituents of both RNPs form subassembly complexes, which can simplify the task of assembly and segregate functions of assembly factors. Reorganization of RNP topology, and proofreading of proper assembly, are catalyzed by protein- or RNA- dependent ATPases or GTPases. Dynamics of intermolecular interactions may be facilitated or regulated by cycles of posttranslational modifications. Despite this repertoire of tools, mistakes occur in RNP assembly or in processing of RNA substrates. Quality control mechanisms recognize and turnover misassembled RNPs and reject improper substrates.
Introduction
Ribosomes and spliceosomes are two of the best characterized large RNPs, multimolecular complexes that contain both RNA and protein constituents. Mature ribosomes in eukaryotes consist of two RNP subunits, the large subunit containing 47 different proteins and three rRNAs, and the small subunit containing 32 proteins and one rRNA. Assembly of these molecules into ribosomes begins with synthesis of rRNA in the nucleolus, followed by proper folding of nascent rRNA, to enable its modification (methylation or pseudouridylation), processing by exo- and endonucleases, and binding to ribosomal proteins. More than 180 assembly factors and 100 small nucleolar RNPs (snoRNPs) associate with pre-rRNA to catalyze ribosome assembly [1]. The snoRNPs catalyze posttranscriptional modifications of pre-rRNA, primarily at sequences that ultimately form or surround the active site of mature ribosomes. Each snoRNP contains four core proteins (including the modifying enzyme) plus a unique snoRNA, which targets the modifying enzyme via snoRNA-pre-rRNA basepairing [2]. After each assembly factor and snoRNP completes its function, it must somehow dissociate from pre-rRNPs, to be recycled for construction of other ribosomes. Ribosomal proteins also play a role in assembly of ribosomes, and remain integral components of ribosomes, together with rRNA, to also function in the dynamics of protein synthesis.
Preribosomes traffic from the nucleolus through the nucleoplasm to the cytoplasm, undergoing additional steps in maturation at each point, to finally assemble into functional subunits (Figure 1). In order to be exported to the cytoplasm, preribosomes must be sufficiently small to fit through nuclear pores. Presumably this is achieved by release of most of the assembly factors prior to nuclear export, as well as changes in pre-rRNP conformation [3,4]. Both pre-40S and pre-60S particles are directed specifically to and through nuclear pores by multiple export receptors, including Crm1/Xpo1 in concert with Ran-GTP [5–8]. Successful navigation through the hydrophobic environment of FG repeats of nucleoporins, by preribosomes likely to contain a hydrophilic exterior, may be aided by binding of the alpha-helical HEAT repeat protein Rrp12 to the surface of pre-rRNPs [9].
Fig. 1.
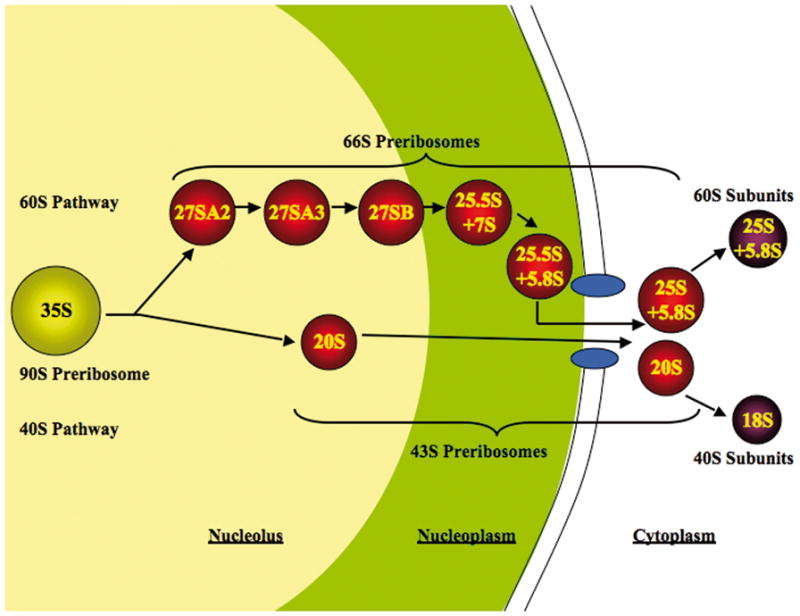
Maturation of preribosomes in Saccharomyces cerevisiae. Ribosome biogenesis begins in the nucleolus, where pre-rRNA is transcribed and packaged into the 90S pre-rRNP, together with a subset of ribosomal proteins and ribosome assembly factors. Subsequent steps of maturation occur in the nucleolus, nucleoplasm and cytoplasm. The 90S pre-rRNP is converted into the 66S and 43S particles by cleavage within the pre-rRNA. There are at least six consecutive 66S precursors to mature 60S ribosomal subunits, distinguished by the consecutive pre-rRNA processing intermediates contained within them. The 43S pre-rRNP containing 20S pre-rRNA is exported to the cytoplasm where mature 40S subunits are formed.
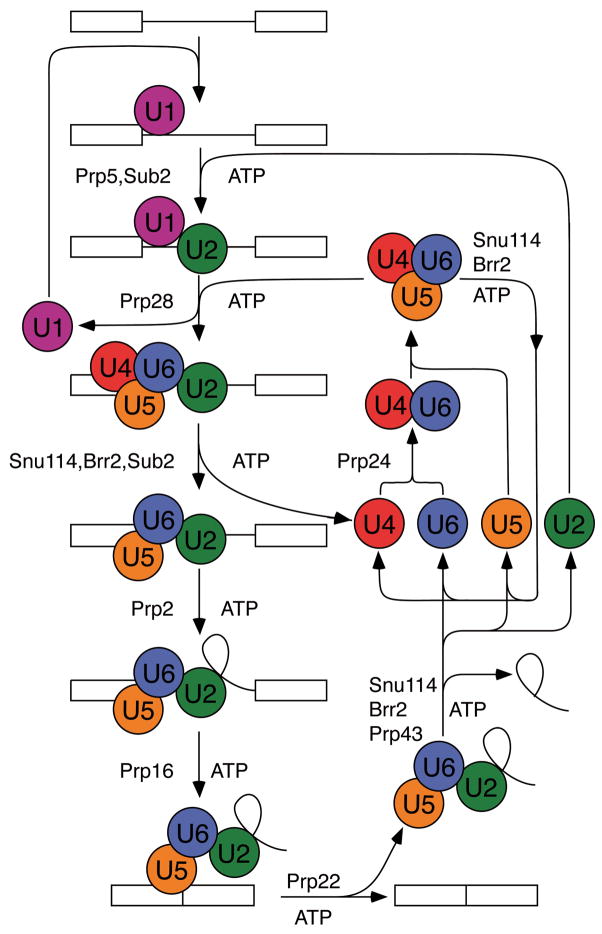
Spliceosomes contain five small nuclear ribonucleoprotein particles (snRNPs) plus more than 100 other splicing factors [for review, see 10 and references therein]. During each cycle of splicing, these factors assemble de novo on the pre-mRNA substrate. An intron is defined by sequences at the 5′ splice site, the branch site, and the 3′ splice site. These sequences participate in the two transesterification reactions in splicing—5′ splice site cleavage and exon ligation. After catalysis, the spliceosome dissociates the products and disassembles, so that the spliceosome can be reused in splicing of another pre-mRNA (Figure 2).
Fig. 2.
The spliceosome cycle. The spliceosome assembles de novo on a pre-mRNA transcript, catalyzes intron removal, dissociates the products and disassembles to permit recycling for subsequent rounds of splicing. Numerous ATP-dependent steps require factors belonging to the DExD/H box family of proteins. Revised from Cell 1998 92:315–326, with permission from Elsevier.
Here we outline common principles underlying construction of these complex RNPs, revealed by studies of the assembly of ribosomes and the assembly and function of spliceosomes.
Coupling transcription to assembly of RNPs and processing of RNAs
Pre-rRNA processing and ribosome assembly, as well as pre-mRNA processing and spliceosome assembly, occur co-transcriptionally and are physically linked to transcription of the respective RNAs. Proteins involved in 5′ mRNA capping, pre-mRNA splicing, and 3′-end formation of mRNA bind to RNA polymerase II, transcription factors or chromatin. Certain ribosome assembly factors bind not only to pre-rRNA but also to rDNA chromatin. These physical interactions between the transcription and RNA processing machinery enable reciprocal, functional coupling of transcription, RNA processing, and RNP assembly. These concepts have been reviewed recently [11,12], and will not be discussed further in this review.
Subcomplexes and hierarchical assembly of RNPs
The complexity of assembling the enormous ribosome and spliceosome RNPs is reduced in part by preassembling subcomplexes. More than a dozen subassembly complexes of preribosomal molecules have been discovered [reviewed in 1,13]. Five of these subcomplexes, t-UTP/UTP-A, UTP-B, UTP-C, the Mpp10 complex, and the U3 snoRNP, form independently of each other, then associate with the pre-rRNA in a hierarchical, stepwise fashion to complete the 90S preribosome [14••]. The tUTP complex proteins, which bind to rDNA and are necessary for transcription of pre-rRNA [15], are the first assembly factors to bind to pre-rRNA. The remaining complexes assemble via two independent pathways: the U3 snoRNP, UTP-B complex, and Mpp10 complex bind to rRNA after tUTP, to form a stable intermediate. The UTP-C complex assembles in a parallel pathway.
Many of the proteins in these SSU processome subcomplexes contain predicted protein-protein interaction domains or RNA binding motifs, and none contain predicted enzymatic motifs. Thus, interactions established through these motifs may enable formation of an RNP scaffold upon which GTPases, ATPases, and nucleases can act to reconfigure the preribosome to catalyze rRNA modification and processing and particle maturation. Stepwise assembly of such large and dynamic RNPs may also provide the advantage of temporally and spatially separating different steps in their biogenesis [14••].
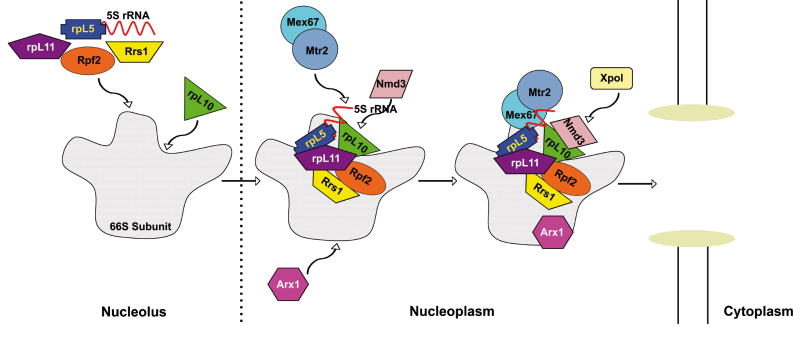
Identification of subcomplexes has provided clues about cofactors of assembly factors and substrates upon which they might act. For example, the discovery of the Rpf2 subcomplex containing the assembly factors Rpf2, Rrs1, ribosomal proteins rpL5 and rpL11, and 5S rRNA helped uncover the mechanism for assembly of 5S rRNA into preribosomes [16•]. Three of the four rRNAs within mature ribosomes are derived by processing of a single primary transcript, within the assembling particles. However, 5S rRNA is transcribed separately. Rpf2 and Rrs1 were shown to recruit 5S rRNA as well as ribosomal proteins rpL5 and rpL11 into 90S preribosomes. Failure to recruit these molecules blocks maturation of preribosomes several steps later in the assembly pathway. Because the abortive assembly intermediates lack 5S rRNA and rpL10, they cannot bind to nuclear export receptors Mex67-Mtr2 [8] or Nmd3 [5], respectively, and therefore accumulate in the nucleus (Figure 3). This example reminds us how assembly and export are coupled by creation of binding sites for receptors during biogenesis of the pre-rRNPs.
Fig. 3.
Coupling assembly of ribosomes with their export to the cytoplasm. Assembly factors Rpf2 and Rrs1 are necessary for incorporation of 5S rRNA and ribosomal proteins rpL5, rpL10, and rpL11 into 90S preribosomes in the nucleolus. Subsequently, nuclear export receptor Mex67-Mtr2 can bind to 5S rRNA in preribosomes, Nmd3 can bind to rpL10 in pre-rRNPs, and Arx1 can associate with nascent ribosomes, in the nucleoplasm. Mex67-Mtr2, Nmd3, and Arx1 then function to direct preribosomes to and through nuclear pores into the cytoplasm. Nmd3 does so via binding to Xpo1/Crm1.
As with ribosome assembly, spliceosome assembly is reduced in complexity through the formation of subcomplexes. For example, the five snRNPs are observed as independent particles. These particles can be reduced even further; the SF3a and SF3b components of the U2 snRNP form stable subcomplexes. Conversely, the snRNPs also form higher order structures. Before interacting with pre-mRNA, the U4 snRNP interacts with the U6 snRNP via extensive base pairing between the snRNAs and then this complex binds to the U5 snRNP to form the U4/U6.U5 snRNP. Protein complexes also feature prominently. For example, a large protein complex, termed the Prp19p complex or NTC, has recently been found to be a major salt-stable component of purified, catalytically active spliceosomes [17••]. Experimentally, the U1, U2, U4/U6.U5 snRNPs and NTC were first observed to bind a pre-mRNA sequentially, but subsequently a U1.U2.U4/U6.U5 snRNP, or penta-snRNP, that includes the NTC was discovered, potentially a “holospliceosome” [18]. While compelling, the penta-snRNP has not yet been observed in vivo [19,20] and is not essential in vitro for early spliceosome assembly steps [21]. Regardless, preassembly of subcomplexes simplifies spliceosome assembly.
Several core splicing factors likely function as scaffolds both to recruit other splicing factors and to time their recruitment. For example, the C-terminal domain of the U5 snRNP protein and DExD/H box ATPase Brr2 interacts with at least five splicing factors that bind to the spliceosome at different stages [22]. Because some of these interactions with Brr2 are overlapping, Brr2 may necessitate sequential interactions with these factors. Additionally, Brr2 as an ATPase may assume alternative conformations during the splicing cycle that favor one interaction over another, and thereby time the recruitment and function of specific splicing factors. While assembly and processing of a genuine pre-mRNA appears to be a largely ordered process, the spliceosome can also traverse alternative pathways – especially in rejecting and discarding incorrect substrates (see below).
NTPases and dynamic rearrangements involving RNA and protein
A number of GTPases and ATPases are required for assembly of ribosomes and spliceosomes [23•,24]. Cycles of NTP binding and hydrolysis could drive assembly forward by several different mechanisms to recruit or release factors [23•]: (1) by direct binding to a protein to stabilize or destabilize its association with preribosomes, (2) by direct catalysis of conformational switches of RNA or RNP structures in preribosomes, or (3) by functioning as timers for assembly or acting as placeholders to prevent premature association of factors.
One example is the GTPase Bms1, which binds to the putative endonuclease Rcl1, and is required to deliver Rcl1 into preribosomes [23•,25,26]. Studies of the binding of Bms1 to its ligands in the presence of GTP or GDP led to the following model: binding of Bms1-GDP to Rcl1 could lead to exchange of GDP for GTP and subsequent association of Bms1-GTP-Rcl1 with pre-rRNPs containing U3 snoRNA. Entry into preribosomes or subsequent rearrangements of the pre-rRNPs, including dissociation of U3 snoRNA, may trigger the intramolecular GAP of Bms1 to activate GTP hydrolysis. This would enable release of Bms1 from Rcl1, strengthen association of Rcl1 with preribosomes, and trigger subsequent pre-rRNA processing.
A second example is Rea1, one of three AAA ATPases that catalyze release of assembly factors from preribosomes [27–29]. Rea1 is found in late nucleoplasmic pre-60S particles, and functions in late steps of pre-rRNA processing and subsequent nuclear export of pre-rRNPs [29]. Rea1 may remodel pre-rRNPs to expose the 3′ ends of 7S pre-rRNA to the exosome complex of nucleases for 3′ trimming. An important breakthrough was the demonstration of factor release activity by an ATPase in vitro [29]. When pre-60S particles were purified in the presence of ATP, Rea1 and another assembly factor Nug2 dissociated from preribosomes, but not when nonhydrolyzable analogues of ATP were used or ATP was omitted.
Nineteen DExD/H-box proteins (DBPs) are involved in ribosome biogenesis in yeast [24]. Likely substrates for these potential RNA helicases are the snoRNAs. Indeed, two DEAD-box proteins, Has1 and Dbp4, have been implicated in releasing snoRNAs from preribosomes [30,31]. Inactivation of Dbp4 or Has1 by mutation of residues necessary for ATP binding or hydrolysis prevents release of several different snoRNAs from pre-rRNPs. Depletion of each of the remaining 17 preribosomal DBPs results in defects at different steps in pre-rRNA processing. An important next step will be to discover the specific substrates of these 17 DBPs, either RNA helices to be unwound, RNP substructures to be remodeled, or proteins to be recruited into or released from pre-rRNPs.
At least eight DExD/H box ATPases are also required for spliceosome assembly and pre-mRNA processing. During splicing, the spliceosome dramatically rearranges RNA-RNA and RNA-protein interactions within the spliceosome or involving the substrate. The DExD/H box ATPases are strong candidates for, or known catalysts of, these key transitions. The DEAD box ATPase Prp5 promotes an intramolecular rearrangement of U2 stem IIc to stem IIa that is required for binding of the U2 snRNP to the pre-mRNA [32•]. After the first cleavage event, the DEAH box ATPase Prp16 has been implicated in toggling this switch again from the U2 stem IIc state to the stem IIa state, suggesting a role for Prp16 in modulating substrate-spliceosome interactions during the catalytic phase of splicing [32•,33•]. After exon ligation, the DEAH box ATPase Prp22, a 3′→5′ unwindase, promotes release of the mRNA product and appears to do so by binding downstream of the exon junction and then translocating upstream along the mRNA [34••]. Each of these ATPases has been implicated in promoting the fidelity of pre-mRNA splicing (see below). Suggesting an explicit parallel with ribosome assembly, the DEAH box ATPase Prp43 is required for both pre-mRNA splicing and pre-rRNA processing [35–37]. In splicing, the G-patch protein Ntr1 recruits and catalytically activates Prp43 to promote intron release [38,39•,40]. The G-patch, a short glycine-rich sequence, is found in a number of RNA binding proteins, including several that interact with DBPs [41]. In ribosome assembly, a related G-patch protein may similarly recruit and activate Prp43 to promote pre-rRNA processing and ribosome assembly. The dual role for Prp43 in splicing and ribosome assembly could provide a mechanism for coupling ribosome biogenesis with gene expression.
The only integral spliceosomal DExD/H box ATPase, Brr2, is regulated by the sole spliceosomal GTPase, Snu114. This ATPase promotes both spliceosome assembly, by unwinding base-paired U4/U6, and spliceosome disassembly, perhaps by unwinding the mutually exclusive and catalytically essential U2/U6 interaction, and thereby likely necessitates tight regulation. Functioning as a classic G-protein switch, Snu114 in the GTP state promotes Brr2 function while in the GDP state, represses Brr2 function [42•]. While it is currently unclear what signals control this switch, they likely include the splicing substrate and the snRNAs, which together can specify the stage in the splicing cycle and thereby dictate a requirement for activating Brr2. In the future, it will be interesting to determine whether Snu114 also responds to signals outside of the spliceosome and/or whether Snu114 sends signals beyond the spliceosome. Intriguingly, the close paralog of Snu114 corresponds to the translation elongation factor EF-2, suggesting another connection between the spliceosome and the ribosome [43,44]. While EF-2 utilizes GTP hydrolysis to promote work, it is not yet clear if Snu114 also functions in this way.
Posttranslational modification: another mode to power dynamics
Cycles of posttranslational modifications of preribosomal or spliceosomal proteins add an extra layer of flexibility and complexity to the dynamics of RNP biogenesis. For example, an isoform of casein kinase I, Hrr25, is present in pre-40S and pre-60S ribosomal particles and is necessary for their maturation. Phosphorylation of assembly factor Tif6 by Hrr25 is required for production of 60S subunits [45]. It remains to be determined whether function of Tif6, or its association with or dissociation from preribosomes, depends on its state of phosphorylation. Hrr25-dependent phosphorylation followed by dephosphorylation of ribosomal protein S3, is necessary for proper integration of rpS3 into preribosomes, and induces remodeling of the structure of pre-40S particles, perhaps to enable export through the nuclear pore [3••].
Analysis of the SUMO proteome revealed that a number of ribosome assembly factors, especially those functioning in the nucleolus or nucleoplasm, are modified by sumoylation [reviewed in 46•]. In addition, screens for mutants defective in ribosome biogenesis, including those impaired in nuclear export of pre-rRNPs, identified mutants in the SUMO modification pathway [46•]. Interestingly, the upl1- mutant, defective in the SUMO deconjugating enzyme that is associated with nuclear pores, exhibited genetic interactions with the mtr2-33 mutant defective in pre-60S subunit export. By preventing incorrect protein-protein interactions, sumoylation may enable orderly arrangement of molecules within assembling RNPs. Desumoylation may be important for efficient nuclear export of nascent ribosomes.
As with ribosome assembly and pre-rRNA processing, efficient spliceosome assembly and pre-mRNA processing require orderly progression through the splicing pathway. Posttranslational modifications are emerging as a mechanism to establish order. Cycles of phosphorylation and dephosphorylation have long been implicated in controlling the splicing cycle. Recently, direct evidence has revealed a requirement for PP1/PP2A phosphatases in the exon ligation step of splicing [47•]. Intriguingly, this requirement correlates with dephosphorylation of the GTPase U5-116 kDa (hSnu114) and the HEAT-repeat protein SAP155. In each case, dephosphorylation may regulate conformational rearrangements, given the switch like nature of G-proteins and the conformational flexibility of HEAT-repeat proteins. Ubiquitylation has also been implicated in splicing and the first direct evidence revealed conjugation of ubiquitin to Prp8 [48•], a central U5 snRNP component that interacts with all consensus sequences in the pre-mRNA [reviewed in 49]. The ubiquitylation state of Prp8, like the guanine nucleotide bound state of Snu114, regulates Brr2-dependent unwinding, suggesting that ubiquitylation also regulates conformation rearrangements. Ultimately, it will be important to determine what regulates these regulators, to understand how order is established in ribosome biogenesis and spliceosome assembly and function.
Quality control
In many mutants defective in ribosome assembly, pre-rRNAs do not accumulate to levels predicted if assembly intermediates were completely stable. Rather, these pre-rRNAs undergo significant turnover [reviewed in 1]. These results indicated the existence of a surveillance machinery that could recognize and destroy misassembled preribosomes. Indeed, the nuclear exosome, a complex of ten nucleases that can process or degrade nuclear RNAs, turns over pre-rRNAs when ribosome assembly is blocked [50,51••]. These RNAs targeted for turnover include aberrant processing intermediates that form when pre-rRNA cleavage is blocked [50]. Likewise, normal 27S pre-rRNAs are degraded by the exosome in mutants blocked in late steps of pre-60S maturation and nuclear export [51••], or in cells treated with 5-FU, an inhibitor of pre-60S subunit maturation [52].
The TRAMP nuclear polyadenylation complex, which includes the polyA polymerase Trf4, the zinc knuckle protein Air2, and the DEAD-box protein Mtr4, also is required for pre-rRNA degradation in vivo [53]. This purified complex can add polyA to RNA in vitro, perhaps to make it a better substrate for the exosome, and it also activates the processive activity of the exosome. Consistent with this role of TRAMP in pre-rRNA turnover, pre-rRNAs destined for destruction are polyadenylated in vivo [54].
Turnover of RNA in abortive ribosome assembly intermediates may occur in a discrete locale within the nucleolus. Pre-rRNPs destined for demolition, as well as components of the exosome and the TRAMP complex, are localized to subnucleolar foci called “No-bodies”[51••]. Formation of No-bodies requires the exosome and TRAMP complex. However, a distinct nucleolar body containing polyA+ RNA is observed in the absence of the exosome enzyme Rrp6 or Mtr4, but requires the presence of Trf4 [55]. It remains to be determined how abortive intermediates are recognized for destruction. Dez and collaborators speculate that assembly factors that fail to be released in a timely manner when assembly is aborted could recruit TRAMP or the exosome [51••].
As in pre-rRNA processing, exonucleases appear to promote resolution of stalled spliceosomes through turnover of the substrate; in addition, exonucleases turnover incorrect substrates. Substrates stalled by cis or trans mutations can be targeted for turnover by nuclear exonucleases, particularly the exosome [56]. Additionally, incorrect pre-mRNA substrates that fail to engage the spliceosome are exported to the cytoplasm where they are subject to nonsense-mediated decay. Remarkably, incorrect intermediates are also turned over by cytoplasmic nucleases, particularly Xrn1, implicating an energy-dependent mechanism for rejecting, dissociating and discarding incorrect substrates [57].
Indeed, the spliceosomal DExD/H box ATPases have been implicated in establishing fidelity through kinetic proofreading in which the DExD/H box protein utilizes the energy of ATP to compete with the pathway to splicing products (Figure 4, reviewed in 58). In this model, the spliceosome establishes specificity by favoring a correct substrate on the path to products and/or favoring incorrect substrates on the competitive branch, which leads to rejection. While the spliceosomal DExD/H box ATPases promote splicing of a correct substrate, they can antagonize splicing of an incorrect substrate if they act prematurely. In this way, Prp5 proofreads formation of the U2/branch site interaction [59•], Prp16 proofreads lariat intermediate formation [60,61] and Prp22 proofreads exon ligation [62••]. Perhaps the numerous ribosomal DExD/H box ATPases function similarly to proofread rearrangements and RNA processing steps in ribosome biogenesis. Given the role of spliceosomal DExD/H box ATPases in repressing incorrect splice sites, it will be interesting to determine whether these factors can also regulate splice site choice to control alternative splicing.
Fig. 4.
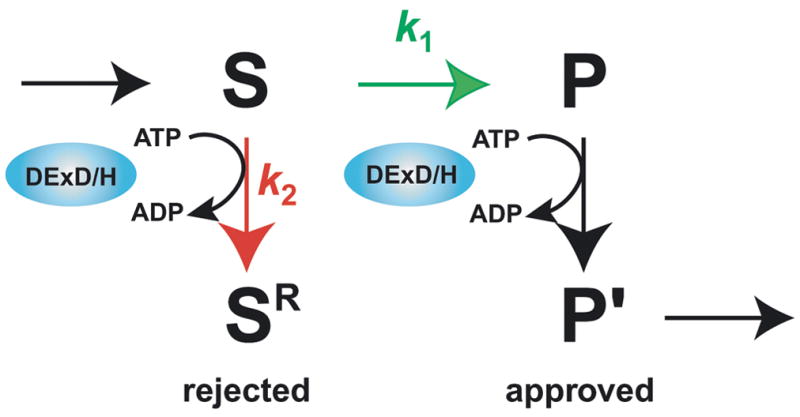
A general mechanism for proofreading RNP transitions by DExD/H box ATPases. In the kinetic proofreading scheme, k1 (for S→P, shown in green) represents the rate of a chemical reaction, such as exon ligation, a binding event, such as binding of U2 to the branch site consensus, or potentially a conformation change; k2 (for S→SR, shown in red) represents the rate of rejecting a substrate. Specific discrimination against an incorrect substrate can be established by a slower k1 and/or a faster k2. Note that the DExD/H box ATPase expends energy to reject an incorrect substrate but also expends energy to promote a genuine product (P→P′), if the DExD/H box ATPase functions after, rather than before, the step under inspection. It is currently unclear what determines whether the DExD/H box ATPase acts before or after the proofread step and how the ATPase antagonizes splicing before while promoting splicing after the proofread step.
Neither Prp16 nor Prp22 reject incorrect substrates by dissociating the substrates, which requires additional factors [61,62••]. Indeed, the spliceosome appears to reject substrates in part by rearranging and thereby sequestering the substrates in effectively inactive conformations [33,61]. After rejecting substrates at the stage of 5′ splice site cleavage, the spliceosome rearranges to a conformation that resembles the exon ligation conformation [61]. Conversely, after rejecting substrates at the stage of exon ligation, the spliceosome can rearrange back to an intermediate state or further to the 5′ splice site cleavage conformation [33,61]. Moreover, consistent with the conservation of phosphodiester bonds in splicing, the spliceosome can reverse both steps of splicing [63•], suggesting that splicing could improve fidelity through self-correction. Unless pre-rRNA processing similarly conserves phosphodiester bonds, errors in pre-rRNA processing are likely catastrophic.
An evolutionary connection between the spliceosome and the ribosome?
The evidence that the catalytic function of the spliceosome evolved from self-splicing group II introns is compelling [64 and references therein]. In particular, there are strong similarities of group II intron domains V and VI with U6 and U2 snRNAs, respectively. However, the origins of the other snRNAs and the spliceosomal cofactors is less clear. An attractive hypothesis is that the evolving spliceosome borrowed activities already established to support the assembly and function of a mature ribonucleoprotein machine – the ribosome. For example, U4 snRNA shares intriguing similarities with the box C/D snoRNAs. Specifically, both bind Snu13 and while box C/D snoRNAs bind Nop56 and Nop58, U4 binds the highly homologous Prp31 [65 and references therein]. Perhaps in the evolution of the spliceosome, a snoRNA that functioned to modify rRNA evolved a separate function in base pairing with the catalytic domain of a group II intron to downregulate its activity, giving rise to base pairing of U4 with the catalytically central U6 snRNA. Given the role of Prp43 at the earliest stages of pre-rRNA processing when snoRNPs modify pre-rRNA [35–37], the recruitment of Prp43 to the evolving spliceosome may also be rationalized by an evolutionary connection between U4 snRNA and box C/D snoRNAs. Additionally, U5 snRNA shares similarities with tRNA [66], the GTPase Snu114 shares similarities with EF-2 [43], and the DExD/H box ATPase Brr2 shares similarities with Slh1, a protein involved in repressing translation of mRNAs lacking a polyA tail [67]. Deeper studies will reveal whether such parallels hold at a mechanistic level and provide support for a role for ribosomal factors in taming group II introns and evolving a mature spliceosome.
Conclusions
Many challenges lie ahead to develop a higher resolution view of the dynamics of RNP assembly. Further experiments will reveal additional interactions between components of assembling ribosomes and spliceosomes and the order in which these encounters occur. A critical next step is to determine how the order is defined – whether by obligatory, sequential steps and/or by the relative rates of different reactions that define a landscape of kinetically accessible pathways. To better understand how NTPases drive assembly or monitor substrate transitions, we will need to identify their cofactors and targets and define their enzymology, both outside of an RNP and within an RNP. We will need to define which DBPs are RNPases and which are helicases and determine if any function simply as ATP-dependent RNA-binding proteins. We will need to discover which GTPases perform work and which act as molecular switches and then elucidate their mechanisms. We are only beginning to appreciate the impact of posttranslational modifications on RNP assembly and we need to reveal their frequency, timing, and mechanistic roles. Studies of splicing have largely preceded those of ribosome biogenesis and in many cases will inform future studies. As research on ribosome biogenesis blossoms, it should in turn guide future studies in splicing.
Acknowledgments
We apologize to the many colleagues whose work we have been unable to cite because of space limitations. We thank Prakash Koodathingal and Jason Talkish for assistance with figures. JS is supported by grants from the National Institutes of Health (GM62264) and from the American Cancer Society Illinois Division (Stephen F. Sener, M.D.-Research Scholar Award; RSG-06-099-01-GMC), and JW is supported by grants from the National Institutes of Health (GM28301) and the National Science Foundation (MCB081854).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decatur W, Fournier MJ. RNA guided nucleotide modification of ribosomal and other RNAs. J Biol Chem. 2003;278:695–698. doi: 10.1074/jbc.R200023200. [DOI] [PubMed] [Google Scholar]
- 3••.Schafer T, Maco B, Petfalski E, Tollervey D, Bottcher B, Aebi M, Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. In vitro assays with purified preribosomes revealed that phosphorylation then dephosphorylation of ribosomal protein rpS3 caused it to be more stably associated with preribosomes. This cycle of modifications of rpS3, required for subunit biogenesis, was also correlated with structural rearrangements of the pre-40S particle, visible by cryo-EM. Mature 40S particles contain a “beak” structure, caused by protruding helix 33 of the 18S rRNA, whereas pre-40S particles lack the structure. The protein kinase Hrr25 was found to be associated with these preribosomes and required in vivo for this phosphorylation of rpS3 and maturation of pre-40S particles. [DOI] [PubMed] [Google Scholar]
- 4.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Nuclear export of 60S ribosomal subunits depends on Xpo1 and requires a nuclear export sequence-containing factor, Nmd3, that associates with the large subunit protein RpL10p. Mol Cell Biol. 2002;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung N-J, Lo K-Y, Patel SS, Helmke K, Johnson AW. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol Biol of the Cell. 2008;19:735–744. doi: 10.1091/mbc.E07-09-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradatsch B, Katahira J, Kowalinski E, Bange G, Yao W, Sekimoto T, Baumgartel V, Boese G, Bassler J, Wild K, Peters R, Yoneda Y, Sinning I, Hurt E. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol Cell. 2007;27:767–779. doi: 10.1016/j.molcel.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 8.Yao W, Roser D, Kohler A, Bradatsch B, Bassler J, Hurt E. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol Cell. 2007;26:51–62. doi: 10.1016/j.molcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Oeffinger M, Dlakic M, Tollervey D. A preribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes and Dev. 2004;18:196–209. doi: 10.1101/gad.285604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith DJ, Query CC, Konarska MM. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granneman S, Baserga SJ. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr Opin Cell Biol. 2005;17:281–286. doi: 10.1016/j.ceb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang L, Sahasranaman A, Jakovljevic J, Schleifman E, Woolford JL., Jr Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes. Mol Biol Cell. 2008;19:2844–2856. doi: 10.1091/mbc.E07-12-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Pérez-Fernández J, RománÁ Rivas JDL, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. Assays of subcomplexes and ribosome assembly in wildtype cells and in mutants defective for each subcomplex demonstrate a stepwise pathway for the association of discrete subcomplexes of ribosome assembly factors with pre-rRNA to form the 90S preribosome. The t-UTP subcomplex assembles first with pre-rRNA, and is required for subsequent association of subcomplexes in two independent pathways: UTP-B, the U3 snoRNP, and the Mpp10 complex in one pathway, and UTP-C in the other. These results are complemented by bioinformatic analysis of the network of protein-protein interactions among these assembly factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher JEG, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL., Jr Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 2007;21:2580–2592. doi: 10.1101/gad.1569307. Genetic and biochemical approaches used to disrupt assembling ribosomes enabled identification and purification of the Rpf2 subcomplex containing ribosome assembly factors Rpf2 and Rrs1, plus ribosomal proteins rpL5 and rpL11 and 5S rRNA. Depletion of each of these four proteins demonstrated their mutual interdependence for assembly into preribosomes and their requirement to recruit 5S rRNA into nascent ribosomes. In the absence of the Rpf2 complex, 66S preribosomes are otherwise largely intact, but cannot carry out processing of 27SB pre-rRNA and fail to be exported from the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. Isolation of an active step I spliceosome and composition of its RNP core. Nature. 2008;452:846–850. doi: 10.1038/nature06842. In this manuscript, the authors purify assembled spliceosomes to homogeneity and show that these spliceosomes are catalytically active in the absence of additional factors. Further, they show that a salt-resistance core of the spliceosomes includes, in addition to the U2, U5 and U6 snRNAs, the Prp19 complex and U5 snRNP proteins, including the human orthologs of the GTPase Snu114, the post-translationally modified Prp8 and the DExD/H box ATPase Brr2. [DOI] [PubMed] [Google Scholar]
- 18.Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 19.Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Tardiff DF, Rosbash M. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA. 2006;12:968–979. doi: 10.1261/rna.50506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behzadnia N, Hartmuth K, Will CL, Luhrmann R. Functional spliceosomal A complexes can be assembled in vitro in the absence of a penta-snRNP. RNA. 2006;12:1738–1746. doi: 10.1261/rna.120606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23•.Karbstein K. Role of GTPases in ribosome assembly. Biopolymers. 2007;87:1–11. doi: 10.1002/bip.20762. This review provides a number of detailed models for the functions of GTPases in the assembly of ribosomes in prokaryotes and eukaryotes, with very useful case studies to illustrate mechanisms driven by GDP or GTP binding and hydrolysis. [DOI] [PubMed] [Google Scholar]
- 24.Bleichert F, Baserga S. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Karbstein K, Jones S, Doudna J. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol Cell. 2005;20:633–643. doi: 10.1016/j.molcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Karbstein K, Doudna JA. GTP-dependent formation of a ribonucleoprotein subcomplex required for ribosome biogenesis. J Mol Biol. 2006;356:432–443. doi: 10.1016/j.jmb.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 27.Pertschy B, Saveanu C. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol Cell Biol. 2007;27:6581–6592. doi: 10.1128/MCB.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kressler D, Roser D, Pertschy B, Hurt E. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J Cell Biol. 2008;181:935–944. doi: 10.1083/jcb.200801181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nissan TAS, Galani K, Maco B, Tollervey D, Aebi M, Hurt E. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol Cell. 2004;15:295–301. doi: 10.1016/j.molcel.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 30.Kos M, Tollervey D. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae. Mol Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 31.Liang X, Fournier M. The helicase Has1p is required for snoRNA release from pre-rRNA. Mol Cell Biol. 2006;26:7437–7450. doi: 10.1128/MCB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Perriman RJ, Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. In this and the following manuscript, the authors provide evidence that U2 snRNA switches between two mutually exclusive states at least four times during the splicing cycle and that the DExD/H box ATPases Prp5 and Prp16 promote two of these transitions. Because these two conformations of the U2 snRNP interact differentially with the substrate, these transitions may modulate snRNP-substrate interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. The author provides our deepest understanding of spliceosomal DExD/H box ATPase function by showing that Prp22 interacts with the mRNA just downstream of the exon junction, in a manner that requires exon ligation, positioning Prp22 to translocate upstream to disrupt interactions between the spliceosome and the upstream exon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Combs DJ, Nagel RJ, Ares M, Jr, Stevens SW. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai RT, Tseng CK, Lee PJ, Chen HC, Fu RH, Chang KJ, Yeh FL, Cheng SC. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol Cell Biol. 2007;27:8027–8037. doi: 10.1128/MCB.01213-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. This manuscript demonstrates for the first time in splicing that a cofactor activates unwinding by a spliceosomal DExD/H box ATPase, Prp43 in this case, and that this activation is required for Prp43-mediated intron release. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boon KL, Auchynnikava T, Edwalds-Gilbert G, Barrass JD, Droop AP, Dez C, Beggs JD. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol Cell Biol. 2006;26:6016–6023. doi: 10.1128/MCB.02347-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aravind L, Koonin EV. G-patch; a new conserved domain in RNA-processing proteins and type D retroviral polyproteins. Trends in Biochem Sci. 1999;24:342–344. doi: 10.1016/s0968-0004(99)01437-1. [DOI] [PubMed] [Google Scholar]
- 42•.Small EC, Leggett SR, Winans AA, Staley JP. The EF-G–like GTPase Snu114 regulates spliceosome dynamics mediated by Brr2p, a DExD/H-box ATPase. Mol Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. The authors show that Snu114 regulates Brr2 and that Snu114 does so by acting as a classical regulatory G-protein. This finding was suprising given the high similarity between Snu114 and the GTPase EF-2, the ribosomal translocation factor, which acts as a chemomechanical motor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabrizio P, Laggerbauer B, Lauber J, Lane WS, Lührmann R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 1997;16:4092–4106. doi: 10.1093/emboj/16.13.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenner TJ, Guthrie C. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics. 2005;170:1063–1080. doi: 10.1534/genetics.105.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray P, Basu U, Ray A, Majumdar R, Deng H, Maitra U. The Saccharomyces cerevisiae 60S ribosome biogenesis factor Tif6p is regulated by Hrr25p-mediated phosphorylation. J Biol Chem. 2008;283:9681–9691. doi: 10.1074/jbc.M710294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Panse VP, Kressler D, Pauli A, Petfalski E, Gnadig M, Tollervey D, Hurt E. Formation and nuclear export of preribosomes are functionally linked to the small-ubiquitin-related modifier pathway. Traffic. 2006;7:1311–1321. doi: 10.1111/j.1600-0854.2006.00471.x. This work reveals possible roles of sumoylation and desumoylation in ribosome biogenesis. Screens for mutants defective in nuclear export of preribosomes identified the RIX16/UBA2 gene, encoding a component of the SUMO conjugation pathway. Subsequent analysis revealed that other SUMO pathway mutants are also blocked in pre-rRNA processing and nuclear export of pre-rRNPs. A number of proteins required for assembly of either the 40S or the 60S ribosomal subunit were found to be sumoylated. -Double mutants containing a mutation in the SUMO deconjugation enzyme Upl1 and ribosome export factor Mtr2 exhibited stronger defects in nuclear export of preribosomes than either single mutant, suggesting that desumoylation is important for nuclear export of pre-rRNPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Shi Y, Reddy B, Manley JL. PP1/PP2A phosphatases are required for the second step of Pre-mRNA splicing and target specific snRNP proteins. Mol Cell. 2006;23:819–829. doi: 10.1016/j.molcel.2006.07.022. In this manuscript, the authors present the first direct evidence of a requirement for dephosphorylation. They demonstrate that dephosphorylation is required for exon ligation and they implicate dephosphorylation of the human ortholog of the GTPase Snu114 and SAP155, a HEAT-repeat U2 snRNP component. [DOI] [PubMed] [Google Scholar]
- 48•.Bellare P, Small EC, Huang X, Wohlschlegel JA, Staley JP, Sontheimer EJ. A role for ubiquitin in the spliceosome assembly pathway. Nat Struct Mol Biol. 2008;15:444–451. doi: 10.1038/nsmb.1401. In this manuscript, the authors provide the first direct evidence of a role for ubiquitin in splicing. They identify the central splicing factor Prp8 as a ubiquitin conjugate and they show that Brr2-dependent U4/U6 unwinding is repressed by ubiquitin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucl Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Dez C, Houseley J, Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. Ribosome assembly factor Sda1 is required for late steps in pre-60S subunit biogenesis, before nuclear export of the nascent particles. In sda1- mutants, RNA constituents of preribosomes are polyadenylated and undergo rapid turnover. Under these conditions, protein and RNA reporters of preribosomes, and protein constituents of the TRAMP polyadenylation complex and the exosome complex of nucleases, localize to discrete foci within the nucleolus, called No-bodies. Knockouts of the TRAMP protein Trf4 or the exosome protein Rrp6 prevent polyadenylation and turnover of the RNA and prevent abortive assembly intermediates from localizing to Nobodies. These results suggest that quality control of misassembled ribosomes is mediated by TRAMP and the exosome in Nobodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang F, Hoskins J, Butler JS. 5-Fluorouracil enhances exosome-dependent accumulation of polyadenylated RNA. Mol Cell Biol. 2004;24:10766–10776. doi: 10.1128/MCB.24.24.10766-10776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lacava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 54.Kuai L, Fang F, Butler JS, Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carneiro T, Carvalho C, Braga J, Rino J, Milligan L, Tollervey D, Carmo-Fonseca M. Depletion of yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleus. Mol Cell Biol. 2007;27:4157–4165. doi: 10.1128/MCB.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 57.Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- 58.Query CC, Konarska MM. Splicing fidelity revisited. Nat Struct Mol Biol. 2006;13:472–474. doi: 10.1038/nsmb0606-472. [DOI] [PubMed] [Google Scholar]
- 59•.Xu YZ, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. The authors identify Prp5 as the third example of a DExD/H box ATPase that proofreads splicing and implicate for the first time proofreading of a specific interaction - the U2/branchsite interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- 61.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 62••.Mayas RM, Maita H, Staley JP. Exon ligation is proofread by Prp22p, a DExD/H-box ATPase. Nature Structural and Molecular Biology. 2006;13:482–490. doi: 10.1038/nsmb1093. While Prp22 promotes genuine substrates after exon ligation, the authors show in vitro that Prp22 also antagonizes aberrant substrates before exon ligation, providing the first biochemical evidence for kinetic proofreading in splicing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63••.Tseng CK, Cheng SC. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science. 2008;320:1782–1784. doi: 10.1126/science.1158993. The authors extend parallels with group II introns by showing that 5′ splice site cleavage and exon ligation are reversible and that the spliceosome can cleave an mRNA by hydrolysis at the exon-exon junction. Each of these reactions could be exploited to enhance fidelity. [DOI] [PubMed] [Google Scholar]
- 64••.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. The authors reveal that that the catalytic domain V of a group II intron participates in a “catalytic triplex” that binds two divalent metal ions separated by 3.9 Å, the ideal distance for such metal ions to stabilize the build up of negative charge in catalyzing a phosphoryl transfer reaction. The structure supports, establishes and predicts striking parallels between the catalytic domain V of group II introns and the spliceosomal snRNA U6, consistent with a common catalytic mechanism and evolutionary origin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Luhrmann R, Carlomagno T, Wahl MC. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science. 2007;316:115–120. doi: 10.1126/science.1137924. [DOI] [PubMed] [Google Scholar]
- 66.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 67.Searfoss A, Dever TE, Wickner R. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol Cell Biol. 2001;21:4900–4908. doi: 10.1128/MCB.21.15.4900-4908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]