Abstract
Delta-like 3 (Dll3) is a Delta family member expressed broadly in the developing nervous system as neural progenitor cells initiate differentiation. A proximal promoter sequence for Dll3 is conserved across multiple species and is sufficient to direct GFP expression in a Dll3-like pattern in the neural tube of transgenic mice. This promoter contains multiple E-boxes, the consensus binding site for bHLH factors. Dll3 expression and the activity of the Dll3-promoter in the dorsal neural tube depends on the basic helix-loop-helix (bHLH) transcription factors Ascl1 (Mash1) and Neurog2 (Ngn2). Mutations in each E-box identified in the Dll3-promoter allowed distinct enhancer or repressor properties to be assigned to each site individually or in combination. In addition, each E-box has distinct characteristics relative to binding of bHLH factors Ascl1, Neurog1, and Neurog2. Surprisingly, novel Ascl1 containing DNA binding complexes are identified that interact with specific E-box sites within the Dll3-promoter in vitro. These complexes include Ascl1/Ascl1 homodimers and Ascl1/Neurog2 heterodimers, complexes that in some cases require additional undefined factors for efficient DNA binding. Thus, a complex interplay of E-box binding proteins spatially and temporally regulate Dll3 levels during neural tube development.
Keywords: Mash1, Neurogenin, bHLH transcription factors, dorsal spinal cord development, gene regulation, Delta, Notch ligands
Introduction
The choice within neural progenitor populations to differentiate or be maintained as a progenitor relies on the activity of bHLH transcription factors in balance with the Notch signaling pathway. The ability of proneural bHLH factors to induce differentiation is at least partially accounted for by their transcriptional regulation of Notch ligands, such as Dll1 and Dll3 (for reviews see Bertrand et al., 2002; Kageyama and Ohtsuka, 1999; Louvi and Artavanis-Tsakonas, 2006). Since Notch signaling results in down regulation of neural bHLH factor activity, the regulation of Notch ligands as downstream targets of neural bHLH factors is important in understanding the balance between the progenitor state and initiation of differentiation.
Mammals have multiple Notch receptors (Notch 1-4), and multiple types of Notch ligands, most notably those of the Delta and Serrate families (Lendahl, 1998; Lindsell et al., 1996). Of the Delta family including Dll1, Dll3, and Dll4 (Bettenhausen et al., 1995; Dunwoodie et al., 1997; Shutter et al., 2000), Dll3 is the smallest and most divergent as it shares only 36% overall amino acid homology with Dll1. Dll3 is expressed along the entire dorsal-ventral axis of the neural tube in cells that have recently exited the cell-cycle and have begun to differentiate (Dunwoodie et al., 1997). Dll3 has a unique function in that unlike Dll1, which functions as a classic Delta protein by stimulating Notch signaling and inhibiting neural differentiation, Dll3 does not stimulate Notch signaling (Geffers et al., 2007; Ladi et al., 2005). Indeed, in some instances it may actually function cell autonomously to attenuate Notch signaling (Ladi et al., 2005). Here we examine the regulation of Dll3 as a downstream target of the bHLH factors Ascl1 (previously Mash1) (Johnson et al., 1990) and Neurog2 (previously Ngn2, Math4A) (Gradwohl et al., 1996) in the dorsal neural tube.
Proliferating neural progenitor cells reside in the ventricular zone of the neural tube. As these cells initiate their differentiation program they exit the cell cycle and move laterally out of this zone. Neural bHLH factors such as Ascl1 and Neurog2 are expressed transiently in neural progenitor cells in the ventricular zone in specific regions and with distinct temporal characteristics throughout the developing nervous system (Bertrand et al., 2002; Helms et al., 2005; Ma et al., 1997; Sommer et al., 1996). They belong to a subclass of bHLH family proteins that form heterodimers with E-protein bHLH factors (such as Tcfe2a-E12/E47), bind DNA at E-box consensus sites, and activate transcription (Gradwohl et al., 1996; Johnson et al., 1992; Massari and Murre, 2000). Although recent advances have begun to identify target genes regulated by Ascl1 and Neurog2, particularly in telencephalon development (Castro et al., 2006), there is still much to be learned about the molecular strategies used by these transcription factors to control neural differentiation.
The importance of Ascl1 and Neurog2 in regulating Dll3 expression is evident by the dramatic reduction in Dll3 levels in embryos mutant for these bHLH factors (this report and Casarosa et al., 1999). Here we show that Ascl1 and Neurog2 regulate Dll3 transcription in the mouse dorsal neural tube through a complex series of E-box consensus binding sites. Furthermore, we identify novel DNA binding complexes containing Ascl1 homodimers or Ascl11/Neurog2 heterodimers that likely contribute to this regulation. Thus, as neural progenitor cells differentiate into neurons, Dll3 levels are regulated by the integrated activity of the different bHLH factor complexes and specific E-boxes within the Dll3 promoter. This integrated mode of regulation through sequences proximal to the Dll3 gene is distinct from that reported for the related Notch ligand Dll1, which appears to have discrete enhancers responding either to Ascl1 or Neurog2 regulation (Castro et al., 2006).
Materials and Methods
DNA Constructs
PCR was used to amplify the Dll3 proximal promoter from the mouse genome. Using primers 5’-aaggatccTAATTTCCTGTCCGTTTG-3’ and 5’-aaccatggCTTTGGGGGACAGGATG-3’, a 640 bp promoter from -640 to +3 was obtained and cloned into BamHI/NcoI sites of the BgEGFP expression vector (Timmer et al., 2001) to generate Dll3wt-GFP. This cloning replaces the basal promoter of BgEGFP with that of the Dll3 gene. A PCR based site directed mutagenesis strategy was used to generate each mutant construct. For E-box mutations the CANNTG was mutated to CANNAT and the N-box from CACACGAG to ATCACGTA. All constructs were sequenced to establish their integrity.
Transgenic mice
Transgenic mice were generated by standard procedures (Brinster et al., 1985) using fertilized eggs from B6D2F1 (C57BL/6×DBA) or B6SJLF1 (C57BL/6J×SJL) crosses. The Dll3-GFP fragments were isolated from the vector with BamHI and XhoI following separation on a standard 1% agarose gel. The DNA in the excised band was placed at -20°C for 10 minutes atop a 45 μm microfiltration column. After spinning at top speed in a microfuge, the DNA in the flow through was purified using the QIAquick PCR Purification Kit (Qiagen) according to the manufacturer protocol. Each Dll3-GFP transgene was injected into the pronucleus of fertilized mouse eggs at 1-3 ng/μl in 10 mM Tris (pH 7.5), 0.1 mM EDTA. Transgenic embryos were identified by PCR analysis using yolk sac DNA. Ascl1, Neurog2, and Neurog1 null mouse strains were previously published (Fode et al., 1998; Guillemot et al., 1993; Ma et al., 1998). Embryos were staged based on assumed copulation at E0, halfway through the dark cycle.
mRNA in situ hybridization and GFP Visualization
For mRNA in situ hybridization, embryos were harvested at E11.5, fixed in PBS containing 4% formaldehyde for 2 hours, rinsed in cold PBS, incubated overnight in a 30% sucrose/PBS solution, embedded in OCT and cryosectioned. In situ hybridizations were performed as previously described (Gowan et al., 2001) using probes specific for Dll3, GFP, Ascl1, Neurog2, or Neurog1. Probes are available upon request.
Embryos were harvested at E9.5, E11.5, or E13.5 and directly imaged for GFP fluorescence. After whole mount images were taken, embryos were fixed in PBS containing 4% formaldehyde for 30 minutes at room temperature, and processed for cyrosection as above. Whole mount images were all taken at the same exposure times for comparative purposes using a Magna Fire imaging system attached to a stereomicroscope with fluorescence capabilities (Olympus). Cross sections were imaged on a BioRad MRC 1024 confocal microscope keeping all imaging parameters constant.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed similar to (Castro et al. 2006) but using mouse E12.5 neural tube as tissue source. Briefly, E12.5 neural tubes dissected from wild-type or Ascl1 null embryos were dissociated in cold PBS and fixed in 1% formaldehyde/PBS for 10 minutes at room temperature. Isolated chromatin was sonicated to ~0.5-1.2 kb. 15 μg of chromatin was incubated with 2.5 μg antibody in 20mM Hepes, 20mM NaCl, 2mM EDTA, 0.1% Na-DOC, 1% Triton X-100, 1 mg/ml BSA, and protease inhibitors overnight at 4° C. Antibodies used were mouse anti-Ascl1 (BD Pharmingen anti-Mash1, 85103) and mouse anti-RNA Polymerase II (Active Motif, 101307). Chromatin complexes were captured using sheep anti-mouse IgG magnetic beads (Invitrogen Dynabeads M-280, H54700). A detailed protocol for the ChIP assay is available upon request. Target DNA was quantified by real time PCR with Fast SYBR Green Master Mix (ABI) using an ABI 7500. % ChIP efficiency was calculated as (2(Threshold Cycle Input – Threshold Cycle ChIP)) × 1/dilution factor × 100. The following primers were used: Dll3 (fw) TGCCCGAAGACTGAAGACTAATT, (rev) TGGGCTCAGGAAGGTGTGA; Gapdh (fw) CACAGATGTCCAGCTGGTGACA, (rev) ATGATTCCAGGGATGGGTCTTGG; and from (Castro et al., 2006) Dll1-M (fw) GCGTGGCTGTCATTAAGG, (rev) GGTGCTGTCTGCATTACC; Dll1 ORF (fw) GTCTCAGGACCTTCACAGTAG, (rev) GAGCAACCTTCTCCGTAGTAG.
Electromobility shift assays (EMSA)
In vitro translated proteins were synthesized with the TNT kit from Promega, Inc. Translations were quantified using 35S Met according to the manufacturers directions. Nuclear extracts were prepared from mouse E10.5 neural tube using the CelLytic™ NuCLEAR™ Extraction Kit from Sigma-Aldrich. TNT lysates or nuclear extracts were first added to binding buffer (20 mM Hepes pH 7.9, 10mM MgCl2, 50 mM KCl, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, and 5 μg/ml Poly dI/dC) and incubated at 30°C for 15 minutes. (γ-32P)ATP end-labeled oligo probe (50,000 cpm) was added and allowed to incubate for an additional 15 minutes. Complexes were separated on a 5.5% polyacrylamide matrix gel run at 4° C in 0.5% TBE under constant voltage at a rate of 10V/cm gel length. The amount of cold competitor probe was always in 100-fold excess with respect to the labeled probe. Mutant competitors used the same nucleotide changes found in their corresponding mutant transgene constructs. Competitor oligonucleotides and antibodies used for supershifts were added during the initial 15 minute incubation at 30° C prior to the addition of the labeled probe. Rabbit polyclonal antibodies used in the super-shift assays were against Ascl1 (Chemicon, AB5696), Neurog2 (CeMines, AB/HLH2), GFP (Clontech, 8367-1), and E2a (Santa Cruz, SC-763X). For EMSA experiments summarized in Fig. 5, probes were labeled and adjusted to the same specific activity for comparison. EMSAs were quantified using a Storm imaging system.
Figure 5. EMSA using in vitro translated proteins reveal differences in bHLH complexes binding Dll3 promoter E-boxes.
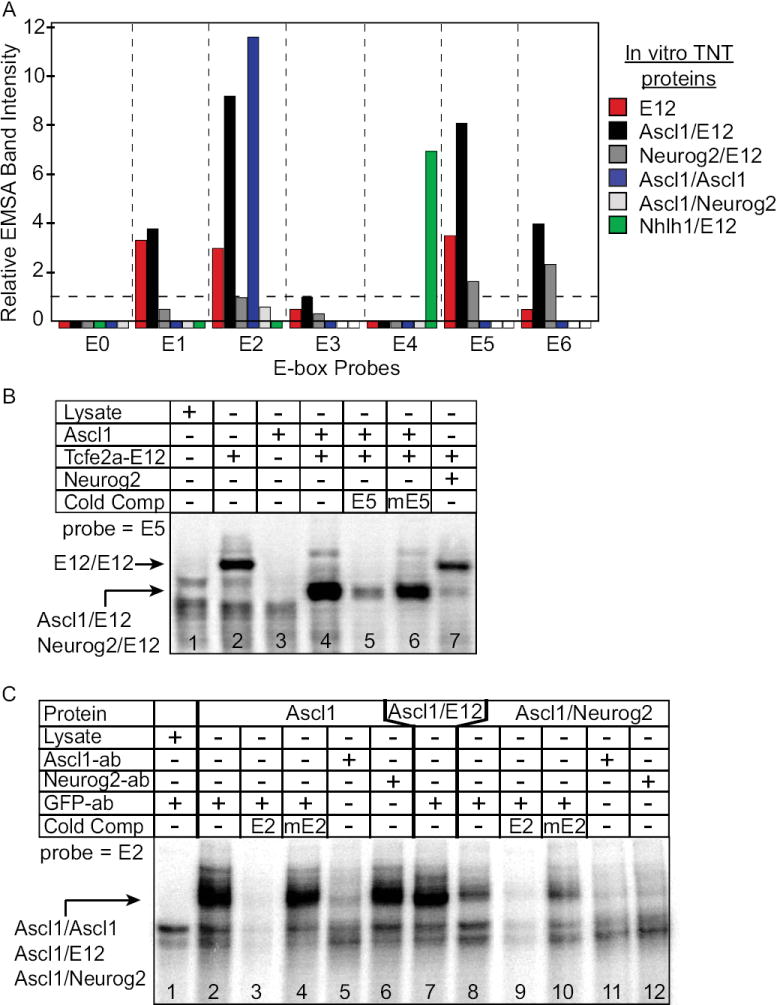
(A) A summary of multiple EMSA experiments with different in vitro translated proteins showing band intensities for each E-box probe (E0-E6) normalized to the lowest measurable Ascl1/E12 heterodimer band (probe E3). No detectable gel shifted band is shown as a box below the X-axis. A white box indicates that condition was not tested. (B) A representative EMSA with E5 probe that generated the data summarized in (A). In each case cold competitor oligonucleotides with wild-type or mutant E-box sequences demonstrate the requirement for the E-box. (C) EMSA demonstrating Ascl1/Ascl1 homodimers (lanes 2-6) and Ascl1/Neurog2 heterodimers (lanes 8-12) can bind E2 probe in an E-box dependent manner. Pretreating lysates with antibodies (ab) to Ascl1 and Neurog2, but not control GFP disrupted formation of the Ascl1 containing complexes.
Oligonucleotides used in EMSA are indicated below with the E-box underlined and the mutated nucleotides in lowercase:
E0-TGAGCCCAAATGGGAGGC; mE0-TGAGCCatAATGGGAGGC;
E1-GAGGCCCAGCTGCGGCCC; mE1-GAGGCCCAGCatCGGCCC;
E2-GCCCGGCAGATGGCGACA; mE2-GCCCGGatGATGGCGACA;
E3-TACATACAGCTGGGAGGC; mE3- TACATAatGCTGGGAGGC;
E4-AAGAAGGGGAAACACACATGCATACA;
mE4-AAGAAGGGGAAACACACaatCATACA; E5-GGAATGCACCTGTATTTC;
mE5-GGAATGCACCatTATTTC; E6-ATCCAGCAGCTGCGACTC;
mE6-ATCCAGCAGCatCGACTC
Transcription Activation Assay
Myc-tagged Ascl1, Neurog2, and Tcfe2a proteins were expressed from pMiWIII derived vectors (Matsunaga et al., 2001; Suemori et al., 1990). The tether peptide in the Ascl1 tethered proteins Ascl1tAscl1, Ascl1tTcfe2a, and Ascl1tNeurog2 is AAAGTSAGGAAAGTSASAATGA. These vectors are also myc-tagged and in pMiWIII expression cassettes. The Firefly luciferase reporters were constructed by cloning a tandem repeat of six E2 or mE2 (sequences as above) E-box sites upstream of the Ela1 basal promoter of EIp.Luc (Beres et al., 2006). Firefly luciferase reporters and phRL-TK (Renilla transfection control) were co-transfected using Fugene (Roche) reagent into 293 human embryonic kidney cells (ATCC CRL-1573) grown in 12 well tissue culture plates. Cells were assayed 48 hours post-transfection using a Dual luciferase system kit (Promega). Firefly Luciferase activity was normalized to the Renilla activity for both the pE2Luc and pmE2Luc reporters. Fold activation was calculated by comparing the activity of the wild-type to mutant reporter (pE2Luc/pmE2Luc).
Results
A 640 bp proximal promoter in the Dll3 gene directs neural tube specific expression
In order to discover regulatory sequences important in controlling the expression of Dll3, we examined the sequence surrounding the open reading frame for regions of conservation between the mouse and human genomes. The Dll3 gene has a high degree of evolutionary divergence that exposed a region of 640 bp strongly conserved between the mouse and human genomes (Evoprinter, Odenwald et al., 2005). This 640 bp region is the only sequence outside of the Dll3 open reading frame that showed significant conservation within 20 kb surrounding the gene. The 640 bp sequence is located immediately upstream of the start codon and extends beyond the predicted transcriptional initiation site (Fig. 1A). Within the 640 bp are three blocks of homology: Homology A is 78% identical over 250 bp, Homology B is 77 % identical over 13 bp, and Homology C is 70% identical over 100 bp. To test the activity of this 640 bp promoter, we assayed its ability to drive GFP expression in transgenic mouse embryos. Six of six embryos expressing the (Dll3wt-GFP) transgene had strong, consistent GFP signal at E11.5 in the neural tube, dorsal root ganglia, hindbrain, ventral telencephalon, somites, and limbs (Fig. 1B, Dll3-WT). These domains accurately reflect the expression pattern reported for Dll3 (Dunwoodie et al., 1997; Fig. 1), although GFP mRNA and protein persist in more differentiated cells likely owing to differences in stability relative to Dll3. These differences are visualized by comparing Dll3 and GFP mRNA in the neural tube of E11.5 Dll3wt-GFP embryos (Fig. 1C,D). Importantly, the 640 bp Dll3wt promoter retains activity for many aspects of Dll3 expression including initiation of expression in the ventricular zone and enriched expression at the lateral edges of the ventricular zone (Fig. 1C’,D’), as well as restriction to neural tissue.
Figure 1. A proximal Dll3 promoter conserved between mouse and human directs Dll3 like expression in transgenic mice.
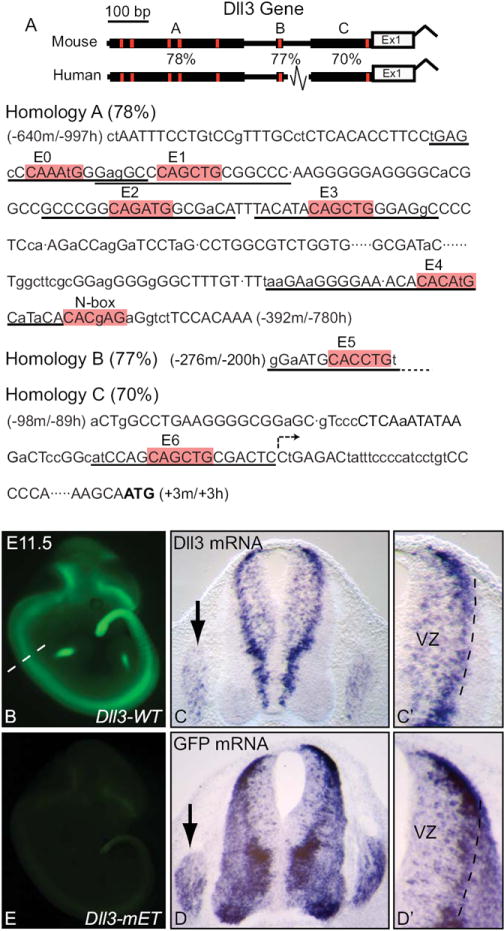
(A) Diagram illustrating sequence homology regions A, B, and C in the 5’ proximal sequence of Dll3. Sequence from mouse for each homology region is shown with capital letters indicating conserved nucleotides between human and mouse sequences. Location relative to the start codon (ATG in bold) in mouse and human is given. E0-E6 E-boxes and an N-box are highlighted in red. The predicted transcription start site is indicated by the arrow in Homology C. Oligonucleotide probes used for EMSA experiments are underlined. (B) whole mount GFP fluorescence in a transgenic embryo expressing GFP under the control of the Dll3 proximal regulatory sequence (Dll3-WT). Dashed line indicates location of sections shown in (C,D). (C,D) cross sections of the neural tube with flanking dorsal root ganglia (arrows) showing mRNA in situ hybridization for Dll3 and GFP. (C’,D’) are higher magnification images of the dorsal neural tube highlighting the ventricular zone (VZ). (E) whole mount GFP fluorescence in a transgenic embryo expressing GFP from the Dll3 promoter that has been mutated at all seven E-box sites (Dll3-mET, see Fig. 4 for diagram).
This analysis was extended to multiple embryonic stages by generating a stable transgenic line and assaying for GFP expression (Supplemental Fig. 1). Expression of GFP in the somites first appears at E9.5. At E13.5, expression was seen in the distal lateral muscle in the limbs, telencephalon (striatum) and diencephalon (hypothalamus), dorsal spinal cord, and retina. This spatial and temporal pattern of expression for the GFP reporter also mimics expression of the endogenous Dll3 gene (Dunwoodie et al., 1997). These observations demonstrate that the 640 bp promoter contains sufficient information for transcriptional regulation of Dll3.
Efficient activity of the 640 bp Dll3 promoter requires E-box sites
Contained within the conserved 640 bp sequence block are seven E-boxes (Fig. 1A), the consensus binding site for the Class II neural bHLH transcription factors (Murre et al., 1994). The functional significance of the E-box sequences was assessed by mutating all seven of the E-box sites in the Dll3wt-GFP transgene and assaying GFP expression at E11.5 in transgenic embryos (Fig. 1E, Dll3-mET). In the absence of all E-boxes, there was a dramatic reduction in the activity of the promoter (3 transgenic embryos had detectable but low expression). The low level GFP expression remaining was restricted to the neural tube largely in the wild-type pattern (see Fig. 4, Dll3-mET, inset). This result clearly establishes an important role for E-box sequences in Dll3 regulation; however, it also indicates that the cell-type specific activity of the promoter is not solely dependent on these E-box sequences.
Figure 4. E-box sites are required for activity of the 640 bp Dll3 promoter.
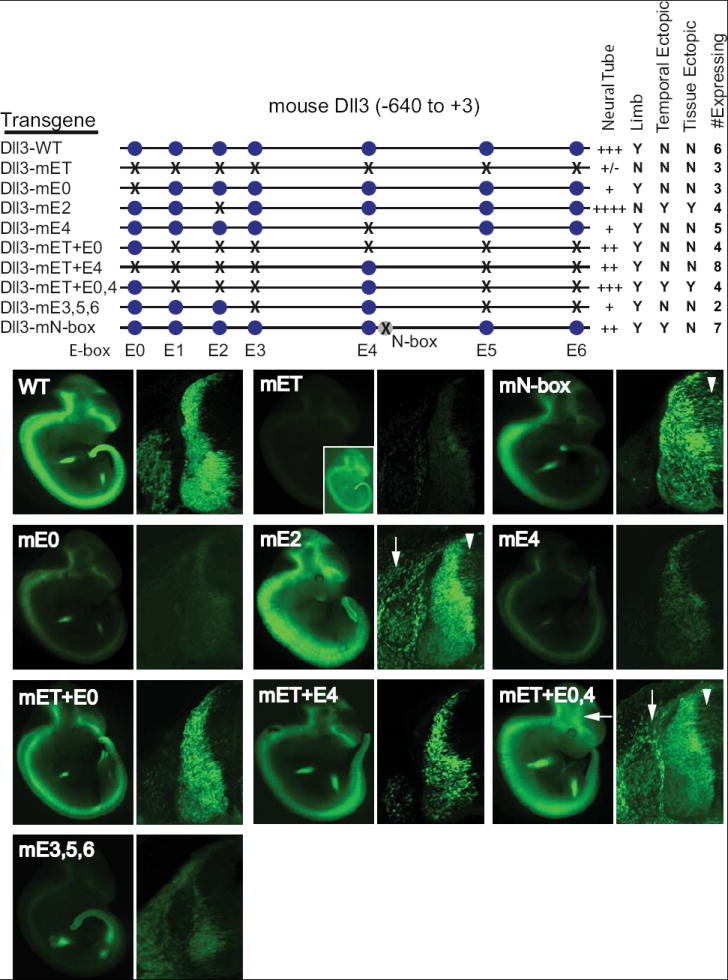
Transient transgenic embryos with a wild-type or E-box mutant Dll3 promoter driving GFP expression at E11.5 are shown in whole mount or as one half of a cross section through the neural tube. The blue circles represent each E-box, and the X indicates a mutation of the site. The relative expression in the neural tube is indicated by +, and expression in limb or in ectopic locations is indicated by Y (expression seen) or N (no expression seen). Temporal ectopic expression is early expression in the ventricular zone (arrowheads), and tissue ectopic expression is expression detected aberrantly outside the neural tube (arrows). The number of expressing embryos analyzed for each transgene is indicated (# Expressing). The images shown were obtained using identical exposure time for the whole mount embryos and identical imaging parameters on the confocal for the cross sections. Each embryo is representative of those obtained for each transgene. The inset in Dll3-mET was imaged at a higher gain to illustrate the low level GFP expression detected is restricted to Dll3 domains.
Ascl1 and Neurog2 regulate Dll3 expression in the developing dorsal neural tube
The identification of a Dll3 promoter whose activity is dependent on E-box sequences suggested that the E-box binding bHLH factors present in the developing neural tube may directly regulate Dll3 levels through these sequences. To begin to address this possibility we examined Dll3 expression in embryos mutant for the bHLH factors Ascl1, Neurog2 and Neurog1. In wild-type mouse embryonic neural tube at E11.5, Dll3 is expressed strongly at the lateral edge of the ventricular zone (VZ) and in scattered cells in the dorsal VZ along the entire dorsal/ventral axis (Fig. 2A). mRNA in situ hybridization for Dll3 in null mutants of Ascl1 (Guillemot et al., 1993), Neurog2 (Fode et al., 1998), and Neurog1 (Ma et al., 1998) was assessed (Fig. 2C,E,G). Dll3 was most dramatically affected in the Ascl1 mutant. In this mutant, Dll3 was not detected specifically within and adjacent to the normal expression domain of Ascl1 (Fig. 2B,C). A subset of the Dll3 expression pattern was also lost in the Neurog2 mutant. In this case, the strong lateral expression seen in the dorsal neural tube is clearly lost (compare Fig. 2E with 2A). This is consistent with Neurog2 expression in the dorsal neural tube being enriched in the lateral, more differentiated cells (Fig. 2D; Helms et al., 2005). No perturbation in Dll3 was detected in the Neurog1 mutant (Fig. 2F,G). Thus, the activity of Ascl1 and Neurog2, but not Neurog1, is required for proper expression of Dll3 specifically in the dorsal neural tube. Furthermore, the sequential nature of Ascl1 and Neurog2 expression in the dorsal neural tube (Fig. 2B,D; Helms et al., 2005), and the discrete pattern of Dll3 perturbation in the two mutants, suggests Dll3 is regulated by integrating activities of multiple bHLH transcription factors.
Figure 2. Dll3 expression and Dll3 promoter activity in the dorsal neural tube requires Ascl1 and Neurog2.
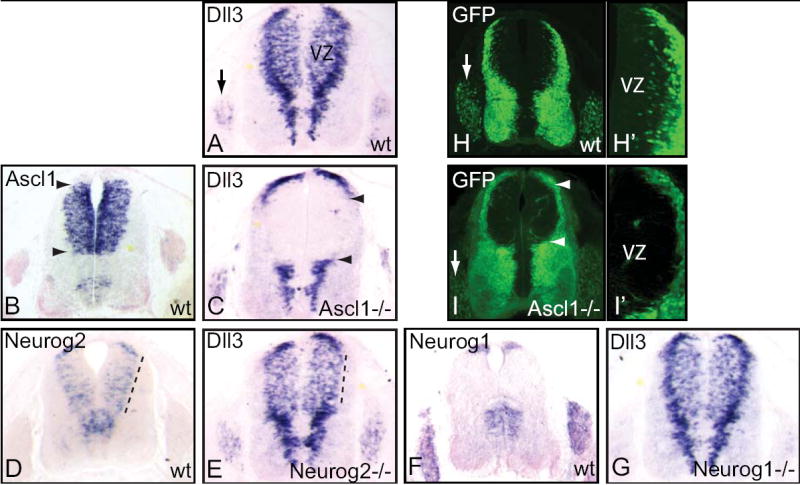
(A-G) mRNA in situ hybridization on transverse sections of E11.5 mouse neural tube. Dll3 expression in wild-type (A), Ascl1-/- (C), Neurog2-/- (E), and Neurog1-/- (G) embryos showing the requirement for Ascl1 for much of the dorsal Dll3 expression (between arrowheads in C), and Neurog2 for Dll3 expression in dorsolateral domains (dashed line in E compared to A). For reference, mRNA expression domains for Ascl1 (B), Neurog2 (D), and Neurog1 (F) in wild-type embryos are shown. (H-I) GFP fluorescence in Dll3-GFP transgenic embryos at E11.5 in the presence (H,H’) or absence (I,I’) of Ascl1. (H’,I’) are higher magnification images of (H,I) to highlight the loss of GFP cells in the ventricular zone (VZ) of the Ascl1 mutants. Arrows indicate the dorsal root ganglia and the arrowheads indicate the normal dorsal domain of Ascl1 expression.
The requirement for Ascl1 in the activity of the 640 bp Dll3 promoter was also tested in transgenic mice. Dll3wt-GFP transgenic mice were bred onto the Ascl1 mutant background. In the presence of normal levels of Ascl1, Dll3wt-GFP expresses GFP in scattered cells within the dorsal VZ, with intense GFP in more differentiated cells at the lateral edges of the neural tube (Fig. 2H,H’). In the absence of Ascl1, the scattered GFP cells in the VZ are absent and there are fewer differentiated cells at the lateral edges of the dorsal neural tube (Fig. 2I,I’). It is likely that much of the remaining GFP containing cells at the lateral edge are from dI1 and dI2 interneurons streaming ventrally from their origin in more dorsal regions. These populations do not require Ascl1, rather they require the other bHLH factors Atoh1 and Neurog1 (Gowan et al., 2001). The lack of GFP signal in the dorsal VZ is consistent with a role for Ascl1 in activation of expression through the Dll3 promoter. Similar experiments were attempted with Neurog2 mutant mice but no Dll3wt-GFP+;Neurog2-/- embryos were obtained from over 10 litters suggesting the transgene randomly inserted into the genome near the Neurog2 locus.
Ascl1 binds the Dll3 promoter in vivo
The dramatic loss of Dll3 expression in the Ascl1 mutant neural tubes and the presence of binding consensus sites for bHLH factors in the Dll3 promoter suggested that Ascl1 functions directly through at least some of these sites. We used Chromatin Immuno-Precipitation (ChIP) analysis to determine whether Ascl1 is localized to the Dll3 promoter in vivo. Chromatin was immunoprecipitated from formaldehyde cross-linked E12.5 neural tubes with antibodies specific to Ascl1. To determine if Ascl1 localized to the Dll3 promoter, qPCR analysis was performed using primers to this region. DNA immunoprecipitated with Ascl1 antibodies showed significant enrichment for the Dll3 promoter target similar to a regulatory region for Dll1 (Dll1-M), an enhancer that was previously shown to be directly regulated by Ascl1 (Castro et al. 2006). Negative controls including the Dll1 open reading frame (ORF) or Gapdh had no enrichment. In addition, no enrichment was seen with chromatin isolated from Ascl1 null neural tubes (Fig. 3, top panel). Chromatin from wild-type and Ascl1 mutant neural tubes was immunoprecipitated similarly with antibodies to RNA polymerase II, demonstrating the chromatin from the mutant neural tubes was competent in this assay (Fig. 3, bottom panel). These results demonstrate Ascl1 directly binds to the Dll3 promoter in vivo in E12.5 neural tubes.
Figure 3. Ascl1 occupies the regulatory regions of Dll1 and Dll3 in embryonic neural tube.
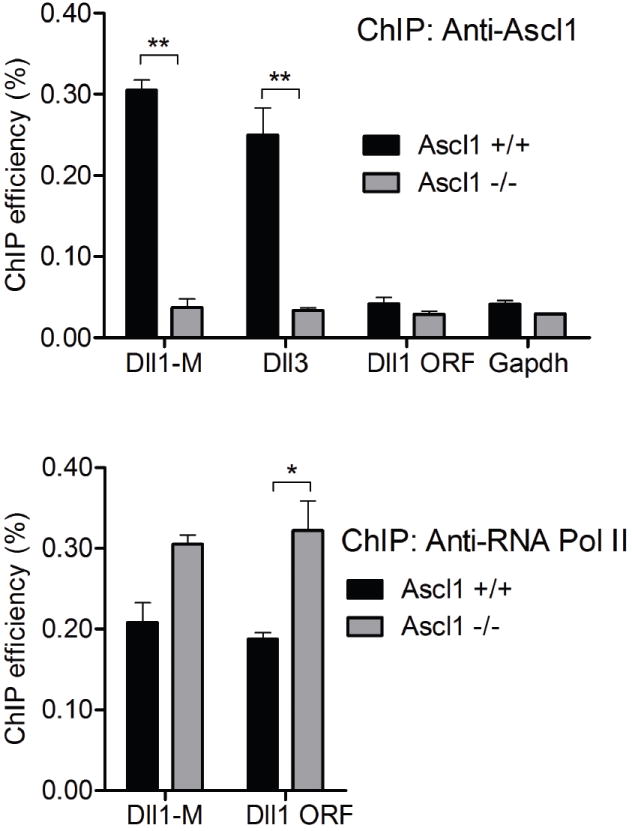
Chromatin from E12.5 neural tubes immunoprecipitated using Ascl1 antibodies (top) is enriched for the Dll3 promoter (Dll3) and a previously identified enhancer in Dll1 (Dll1-M) (Castro et al., 2006). Control DNA regions including the open reading frame of the Dll1 gene (Dll1 ORF) and Gapdh are not enriched. Chromatin immunoprecipitated with Ascl1 antibodies from Ascl1 mutant neural tubes was not enriched for any regions tested. The ChIP efficiency with chromatin immunoprecipitated using antibodies to RNA polymerase II (bottom panel) is comparable or higher from the Ascl1 null tissue than from wild-type tissue, confirming the competence of the Ascl1 null tissue in this assay. ** p value <0.001, * p value <0.05.
In a similar set of experiments we utilized ChIP assays to test whether Neurog2 directly binds to the Dll3 promoter in vivo. Although the Dll3 promoter was enriched after ChIP with Neurog2 antibodies relative to negative controls, the efficiency of the pull-downs from embryonic neural tube was low, and thus, these experiments were not definitive (data not shown).
Individual E-box sites have distinct properties with respect to Dll3-promoter activity
The results above demonstrate that at least Ascl1 is directly regulating the expression of Dll3 through the 640 bp Dll3 promoter. Furthermore, the activity of the promoter requires intact E-box sites. To more precisely define the contribution of each E-box to the activity of the Dll3 promoter, the requirement for each individual E-box was assayed in transgenic mice. The results reveal a complex use of the E-box sequences for both activation and suppression of transgene expression. Mutation of four of the E-boxes, E1, E3, E5, and E6, had minor, if any, detectable effects on promoter activity when mutated individually (Supplementary Fig. S2). However, when mutated in combination, such as in Dll3-m3,5,6, enhancer activity was markedly decreased (Fig. 4, Dll3-mE3,5,6). These data suggest a model in which multiple, redundant E-box sites are important for Dll3 expression. In contrast, individual mutations of E0, E2, and E4 revealed their individual importance to the activity of the Dll3 promoter. The following sections detail the properties of each of these sites for Dll3 promoter activity.
E-boxes E0 and E4 serve major activator function in the Dll3 promoter
Of the seven E-boxes present in the promoter, only E-boxes E0 and E4 were required to maintain activity of the Dll3 promoter when tested individually. With each single mutation, a profound loss of expression was seen in all embryos assayed (Fig. 4, Dll3-mE0 and Dll3-mE4). To test whether E0 and E4 are sufficient within the context of the Dll3 promoter to drive the wild-type Dll3 pattern, a reconstructive approach was taken. Starting with the E-box null mutant (Dll3-mET), E0 and/or E4 were mutated back to wild-type creating three new constructs-- Dll3-mET+E0 (E0 only), Dll3-mET+E4 (E4 only), and Dll3-mET+E0,4 (E0 and E4 only) (Fig. 4). When tested in transgenic mice, E0 or E4 alone could rescue efficient GFP expression throughout the neural tube in all embryos expressing the transgene in a pattern consistent with wild-type, albeit at a consistently reduced intensity compared to the wild-type promoter (Fig. 4, compare Dll3-mET+E0 and Dll3-mET+E4 with Dll3-WT). The inability of E0 or E4 individually to restore the high level of GFP seen with the wild-type promoter suggests their function may be additive. This was directly tested by assaying Dll3-mET+E0,4. This construct directed efficient expression of GFP at levels exceeding the constructs with the individual E0 or E4 and approaching those seen with the wild-type promoter (Fig. 4, Dll3-mET+E0,4). However, relative to wild-type, this construct also showed expanded expression within the brain and ectopic expression in the mesenchyme, suggesting at least one of the other E-boxes has repressor activity. Thus, in the wild-type promoter, the combined activator activity of E0 and E4 must be attenuated by the presence of the other E-boxes.
E-box E2 serves major repressor function in the Dll3 promoter
E-box E2, in contrast to E0 and E4, appears to play a major role as a repressor. Mutation of E2 within the Dll3 promoter resulted in two types of ectopic expression of the reporter gene that we term temporal and tissue ectopic expression (Fig. 4, Dll3-mE2). Temporal ectopic expression appears in the VZ, indicating that expression initiates in cells more immature than in those seen with the wild-type promoter (Fig. 4, Dll3-mE2, arrowhead). In contrast, tissue ectopic expression appears in mesenchymal tissue surrounding the neural tube (Fig. 4, Dll3-mE2, arrow). E2 thus appears to serve an important function in Dll3 regulation by restricting its expression to neural progenitors of the appropriate stage.
The presence of an N-box, the consensus binding site for Hairy/En(S)/HES factors, in the promoter provided another candidate repressor pathway to examine since these factors typically suppress neurogenesis (Kageyama et al., 1997; Sasai et al., 1992). Mutation of the N-box resulted in temporal ectopic expression (Fig. 4), consistent with the presence of Hes1 and Hes5 in the neural tube VZ at this time (Ohtsuka et al., 1999). Notably, the tissue ectopic expression seen when E2 was mutated was not detected with the N-box mutation demonstrating that multiple mechanisms restrict activity of the Dll3 promoter.
The ectopic expression seen when the activator E-boxes E0 and E4 were the only E-boxes present (Fig. 3, Dll3-mET+E0,4) strongly mimics the individual E2 mutant (Fig. 4, Dll3-mE2), implicating E2 function in attenuating E0/E4 activity. We tested this hypothesis by constructing a transgene containing E2 plus E0 and E4 (Fig. 4, Dll3-mE3,5,6). The presence of E2 dramatically repressed the ectopic expression seen with the E0/E4 only mutant. The overwhelming loss of expression in Dll3-mE3,5,6 suggests that in the context of the wild-type promoter, the repressive activity of E2 must be modulated not only by the activator E-boxes E0 and E4 but also by a combination of the other E-boxes (E3, E5, and E6). In summary, of the seven E-boxes tested, a dramatic affect on enhancer activity was detected for three; E0 and E4 have enhancer activity, and E2 has repressor activity.
Dll3 promoter E-boxes are differentially bound by bHLH factors in vitro
Although ChIP analysis established that Ascl1 is bound to the Dll3 promoter in vivo, it is unable to spatially resolve interactions with specific E-boxes or to provide insight into the specific complexes that are involved. To determine the ability of Ascl1 and Neurog2 to interact with specific E-boxes, we used EMSA with in vitro translated Ascl1, Neurog2, and Tcfe2a-E12 (E12) proteins, as well as nuclear extracts from E10.5 neural tube. A summary of the data obtained with in vitro translated protein lysates is presented in Fig. 5A. There was surprising variability in the binding of bHLH heterodimer complexes to each E-box. An example of a typical experiment showing the classical behavior of an E-box/ClassII bHLH interaction, using the E5 E-box probe, is shown in Fig. 5B. E5 can be bound efficiently by E12 homodimer (lane 2) and Ascl1/E12 heterodimer (lane 4), much less efficiently with Neurog2/E12 heterodimer (lane 7), and not at all by Ascl1/Ascl1 homodimer (lane 3). Using this assay, we demonstrate that each E-box has distinct properties with respect to the bHLH/E-box complexes that can form in vitro.
Ascl1/E12 (Fig. 5A, black bars) and to a lesser extent Neurog2/E12 (Fig. 5A, dark gray bars) bound five of the seven E-boxes with varying efficiencies. E-box E2 stood out as a strong Ascl1/E12 binding site. In contrast, E0 and E4, the major enhancer E-boxes, were not efficiently bound by Ascl1/E12 or Neurog2/E12. These findings were surprising since Ascl1/E12 and Neurog2/E12 are known activators of transcription (Gradwohl et al., 1996; Johnson et al., 1992).
In an attempt to identify an E-box binding bHLH transcription activator that might act through the enhancer E-boxes E0 and E4, the bHLH factor Nhlh1 (previously Nscl1, Hen1) was tested. We tested Nhlh1 since it is expressed in the neural tube just lateral to the VZ as cells become post-mitotic (Begley et al., 1992), an expression pattern similar to Dll3wt-GFP. Nhlh1 bound efficiently as a heterodimer with E12 specifically to E4, but not the other E-boxes (Fig. 5A). Thus, Nhlh1 is one candidate that might upregulate Dll3 through this E-box sequence.
Evidence for novel DNA binding complexes of Ascl1 including homodimers and multi-factor complexes with Neurog2
The requirement of E-boxes E0, E2 and E4 for wild-type activity of the Dll3 promoter was demonstrated in transgenic mice (Fig. 4). The efficient binding of Ascl1 and Neurog2 heterodimers to the E-box with an apparent repressor activity (E2) but not to E-box E0 and E4 with enhancer activity (Fig. 5A) presents an apparent contradiction since Ascl1 and Neurog2 are transcriptional activators (Gradwohl et al., 1996; Johnson et al., 1992). To gain further insight into the complexes that can form on these E-box sequences, we used EMSA with proteins from E10.5 neural tube nuclear extracts and specific antibodies to Ascl1, Neurog2, and Tcfe2a-E12 (Fig. 6). This analysis revealed novel Ascl1 and Neurog2 DNA binding complexes can form at least in vitro, particularly to E2, the repressor E-box, and E4, an activator E-box.
Figure 6. EMSA using nuclear extracts reveal the formation of Ascl1/Neurog2 DNA binding complexes.
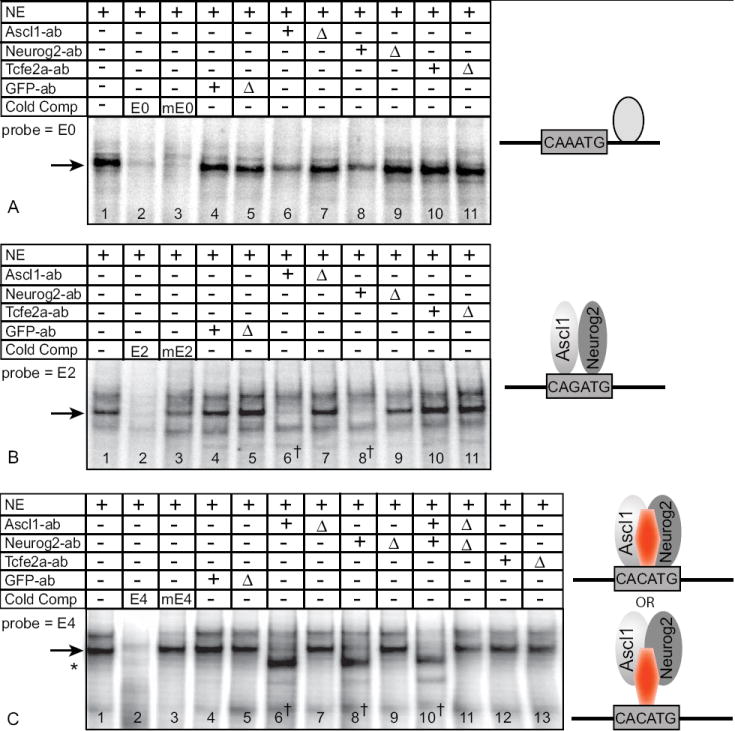
Nuclear extracts (NE) from E10.5 mouse neural tube contain DNA binding activities (lane 1) using oligonucleotides probes from E0 (A), E2 (B), and E4 (C). Except for E0, complex formation requires an intact E-box shown using cold competitor oligonucleotides (lanes 2,3). Extracts were preincubated with untreated or heat inactivated (Δ) antibodies (ab) specific to Ascl1, Neurog2, Tcfe2a-E12, or control GFP (lanes 4-13). Arrows indicate the position of the complexes containing Ascl1 and Neurog2. The † indicates the lanes where complexes are lost with addition of specific antibodies to Ascl1 and Neurog2 (B-lanes 6, 8; C-lanes 6, 8, 10). Asterisk in (C) indicates a new band revealed by depleting Ascl1 and Neurog2 from the extract.
Models shown on the right depict proposed complexes that bind each E-box.
Protein complexes with E-box E0
EMSA with nuclear extracts revealed a protein-DNA complex formed on E0, but it did not require an intact E-box since competition with a cold E-box mutant oligonucleotide efficiently competed for binding (Fig. 6, lanes 1-3). Furthermore, the complex was only slightly blocked with pretreatment of the nuclear extracts with antibodies to Ascl1 or Neurog2, and not at all with antibodies to Tcfe2a-E12 (Fig. 6A, lanes 4-11). These results are consistent with the EMSA with in vitro translated proteins where no bHLH was found to bind E0. Thus, although E0 is required for Dll3 promoter activity, the proteins involved in this activity were not identified (Fig. 5A).
Protein complexes with E-box E2
E-box E2 has strong negative activity that keeps the Dll3 promoter restricted to the neural tube and keeps it from turning on prematurely. Using in vitro translated proteins, E2 can be bound efficiently by E12/E12, Ascl1/E12, and Neurog2/E12 (Fig. 5A). Surprisingly, E2, but none of the other E-boxes tested, was also efficiently bound by an Ascl1/Ascl1 homodimer, a complex whose existence has not been previously reported (Fig. 5C, lanes 2-5).
EMSA performed with E10.5 nuclear extracts also revealed E-box dependent complexes binding E2, but the complex identified includes both Ascl1 and Neurog2 (Fig. 6B, lanes 1-3). Pretreating the extracts with antibodies specific to Ascl1 or to Neurog2 completely blocked the formation of the same band demonstrating the existence of a novel Ascl1/Neurog2 E-box binding complex (Fig. 6B, lanes 6 and 8). To verify that we were detecting a specific interaction of the antisera to Ascl1 and Neurog2 in the complex, we heat inactivated antisera prior to use (Δ), and we tested an unrelated anti-GFP antiserum. In both cases, there was no attenuation of the protein-DNA complex (Fig. 6B, lanes 4-9). In addition, cross detection of Ascl1 by Neurog2 antisera was not seen using in vitro transcribed and translated protein (Fig. 5C, lanes 5-6). Antibodies to Tcfe2a-E12 had little if any effect on the formation of the complexes (Fig. 6B, lanes 10-11). The ability of an Ascl1/Neurog2 heterodimer to bind E2 E-box DNA was confirmed using in vitro translated proteins (Fig. 5C, lanes 8-12). Thus, E2 can be bound by multiple Ascl1 containing complexes, including the classical Ascl1/E12 heterodimer as well as an Ascl1/Ascl1 homodimer and Ascl1/Neurog2 heterodimer.
Protein complexes with E-box E4
E-box E4 has strong enhancer activity in the Dll3 promoter (Fig. 4, Dll3-mE4). Using in vitro translated proteins, only the heterodimer Nhlh1/E12 bound E4 efficiently (Fig. 5A). EMSA with nuclear extracts, however, revealed a novel E-box binding transcription factor complex that again includes Ascl1 and Neurog2, and also suggest it requires at least one additional unidentified factor (Fig. 6C). The presence of Ascl1 and Neurog2 in the protein-E4 complex was demonstrated by the complete disruption of the complex specifically with antibodies to both Ascl1 and Neurog2, but not to Tcfe2a-E12 or to a control GFP (Fig. 6C, lanes 4-13). Surprisingly, blocking the Ascl1 and Neurog2 interaction with DNA by addition of specific antisera revealed a new protein-DNA complex with faster mobility than the wild-type complex (Fig. 6C, lanes 6,8,10 asterisk). This new complex also requires an intact E-box (data not shown). The Ascl1/Neurog2 independent complex revealed could normally be a component of a higher order complex with these bHLH factors, or it could represent a binding activity only revealed after Ascl1 and Neurog2 are removed from the extract. However, consistent with the interpretation that an additional factor is required in the Ascl1/Neurog2 complex with E4, in vitro translated proteins alone can not form an Ascl1/Neurog2 heterodimer with E4 (Fig. 5A). Thus, a novel multimeric complex containing Ascl1, Neurog2, and possibly another unidentified factor, or modification of the heterodimer, may play a role in Dll3 promoter activity.
Ascl1 homodimer and Ascl1/Neurog2 heterodimers function as transcriptional activators
Two novel Ascl1 DNA binding complexes were identified via in vitro EMSA analysis: Ascl1/Ascl1 homodimer and Ascl1/Neurog2 heterodimer. Both complexes can bind E2, the E-box that contains repressor activity. To test a model whereby one or both of these two novel heterodimeric complexes function to repress Dll3 expression, a cell culture based luciferase assay was utilized. Ascl1, Neurog2, and Tcfe2a-E12 were expressed in HEK293 cells with luciferase reporters containing hexamers of either wild-type E2 or mutant mE2. The results are shown as the fold induction of luciferase activity from the wild-type E2 reporter relative to that from the mutant mE2 (Fig. 7). Singly Ascl1, Neurog2, and E12 are all activators, with Neurog2 being by far the strongest. Co-expressing Ascl1 with E12 dramatically increases the transcriptional activation activity through the E2 sequence, consistent with the known function of the Ascl1/E12 heterodimer as an activator complex. To bias the formation of specific Ascl1 complexes, expression constructs were designed to tether Ascl1 with a peptide to either Ascl1 itself to favor the homodimer, to Neurog2 to favor the Ascl1/Neurog2 heterodimer, or to E12 to favor the Ascl1/E12 heterodimer. The Ascl1 tethered homodimer (Ascl1tAscl1) and the Ascl1 tethered to E12 (Ascl1tE12) were both strong activators in this assay. Ascl1 tethered to Neurog2 (Ascl1tNeurog2) also activated transcription but to a much lesser extent. Taken together, in these reporter assays, all Ascl1 complexes appear to act as activators, not repressors, but with varying efficiencies. Thus, the repressor activity of E2 can not easily be explained by binding of the novel Ascl1 complexes, suggesting other factors bind E2 to repress ectopic expression of Dll3.
Figure 7. Ascl1/Ascl1 homodimers and Ascl1/Neurog2 heterodimers function as transcriptional activators.
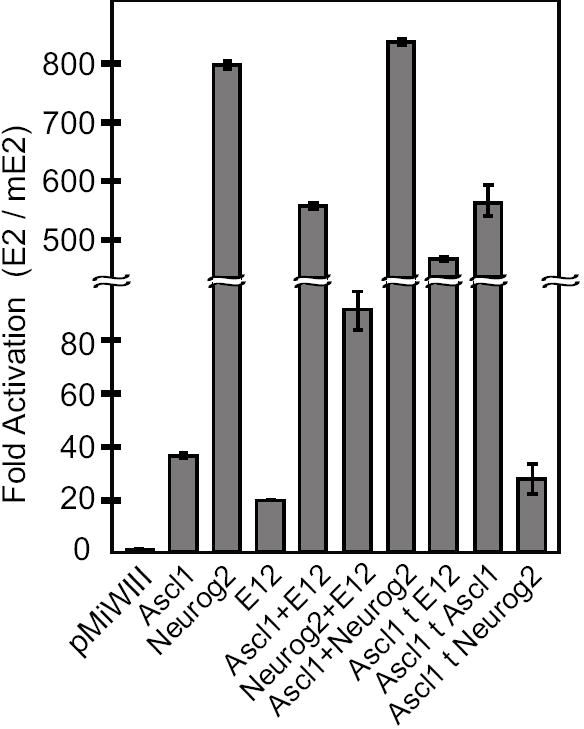
The activity of Firefly luciferase reporters with E-box E2 or mutant E2 were assayed in HEK293 cells expressing various bHLH factors. Firefly luciferase activity for each reporter was normalized to control Renilla luciferase activity, and then represented as the fold activation through the E2 elements versus the mutant E2. pMiWIII is the empty expression vector. Tethered constructs are indicated by ‘t’. All bHLH factors activated expression of the E2 reporter constructs but to varying extents. Mean values are shown for n=6 transfections.
Discussion
This study identifies an evolutionarily conserved promoter responsible for Dll3 expression and demonstrates the regulation through this promoter in the dorsal neural tube by the bHLH transcription factors Ascl1 and Neurog2. The Dll3 promoter contains multiple E-boxes, two of which are required and sufficient for activity of the promoter, and one that behaves as a repressor. Additional E-boxes appear to function redundantly since these sites had to be mutated in combination rather than individually to disrupt Dll3 promoter activity.
Dll3 expression is a reflection of integrating specific binding properties of multiple E-box binding complexes with distinct temporal expression characteristics of each factor in these complexes. bHLH transcription factors represent one class of E-box binding proteins whose importance were investigated in this study. The bHLH factors Ascl1, Neurog2, and Nhlh1 accumulate in the dorsal neural tube with distinct temporal characteristics. Ascl1 is present in neural precursor cells prior to Neurog2 and Nhlh1 (Begley et al., 1992; Brown et al., 1992; Helms et al., 2005) (Fig. 2). In the Ascl1 mutant, Dll3 expression is lost in the VZ in the dorsal domain where Ascl1 is normally expressed. This is compared to the loss of Dll3 only in the lateral, more differentiated cells in the Neurog2 mutant. Thus, integration of the activities of different bHLH complexes on the Dll3 promoter results in its dynamic expression pattern and suggests that the different bHLH factors act in a temporal cascade, possibly through similar E-box sites.
One unanswered question in this study is how E-box E2 functions to repress ectopic activity of the Dll3 promoter. E-box sites with repressor function have been described previously (Genetta et al., 1994; Weintraub et al., 1994). For example, the μE5 E-box within the IgH enhancer acts as a repressor directed at MyoD in muscle presumably to maintain specificity of expression in B-cells. This activity requires sequences within and adjacent to the E-box (Weintraub et al., 1994). In addition, the zinc finger factor Zeb1 is an E-box binding repressor that must be displaced for activation through the E-box (Genetta et al., 1994). In the Dll3 promoter, E2 could be acting in a similar way to repress activity of the promoter, a repression that can be overcome by specific activator bHLH complexes.
The identification of a single evolutionary conserved sequence that contains multiple E-box sites suggests a strategy for regulation of Dll3 expression distinct from that shown for the related factor Dll1. In the case of Dll1, two distinct and separable enhancers were identified (Castro et al., 2006). Each enhancer contains E-box sequences, but each enhancer is bound specifically by Ascl1 or Neurog2. When tested in transgenic mice, one enhancer directs expression of a reporter in the Ascl1 pattern while the other directs expression in a Neurog2 pattern. In contrast, the Dll3 promoter with its multiple E-box sites directs expression of a reporter reflecting an additive pattern from multiple bHLH factors.
Ascl1 is a component of several novel complexes that can bind the Dll3 promoter
We show that Ascl1 is present in at least four different DNA binding complexes that are capable of interacting with key regulatory E-boxes present in the Dll3 promoter. In addition to the classical Ascl1/E12 heterodimer, we show binding activities for an Ascl1/Ascl1 homodimer, an Ascl1/Neurog2 heterodimer, and a complex containing some unknown factor or modification in combination with Ascl1/Neurog2 heterodimers. It is important to note that the existence of these different DNA binding complexes can be demonstrated in vitro, but their existence in vivo, and specifics of their in vivo contribution to Dll3 expression has not been shown.
The primary active form of a class II bHLH factor such as Ascl1 is thought to be as a heterodimer with an E-protein, such as Tcfe2a-E12 (see review Massari and Murre, 2000). Here we provide evidence that an Ascl1/Ascl1 homodimer can exist as well. The homodimer binds DNA with a similar apparent affinity as the Ascl1/E12 heterodimer but appears more selective in its sequence requirements in that of the seven E-boxes tested, only E2 is a substrate for homodimer binding. The Ascl1 homodimer, however, is not the complex repressing through E2 as we detected no obvious difference in the ability of the homodimer and heterodimer to activate transcription.
Other novel DNA binding complexes were identified that contain Ascl1/Neurog2 heterodimers. Although a protein-protein interaction between Ascl1 and Neurog2 was previously reported, no DNA binding activity could be attributed to the complex (Gradwohl et al., 1996). Furthermore, there are no reports of a heterodimer of two class II bHLH factors forming to bind DNA. Here we show that Ascl1/Neurog2 heterodimers from nuclear extracts can specifically bind E-boxes E2 and E4. Interestingly, only E2 functions as a substrate for in vitro translated Ascl1/Neurog2 heterodimers. This suggests that there are additional factors present in the nuclear extract from E10.5 neural tube that stabilize the interaction with E4, factors that are not required for binding of the heterodimer to E2. The requirement for an additional factor for stable binding of bHLH factors has been shown for homodimer formation of Myod and Tcfe2a-E12 (Anand et al., 1997). In this case, the S5a subunit of the 26S proteasome complex is required for stable homodimer binding to the muscle creatine kinase enhancer E-box. Such a molecule may provide a similar function for the Ascl1/Neurog2 heterodimer, allowing it to bind E4. Nevertheless, with or without additional factors, the Ascl1/Neurog2 heterodimer clearly has sequence specific DNA binding capabilities.
Ascl1 target DNA sequences
A major hurdle in fully understanding the function of any transcription factor is the ability to identify its downstream targets. This is true for neural bHLH factors as well, particularly since the core recognition sequence of these factors is the degenerate E-box, CANNTG (Bertrand et al., 2002; Massari and Murre, 2000). Recent advances are revealing additional sequence recognition constraints as well as identifying compound sites that include transcription factor binding sites neighboring the E-box (Castro et al., 2006; Powell et al., 2004; Singson et al., 1994). In particular, Ascl1 has been shown to directly regulate Dll1 synergistically with Brn factors (Pou domain containing factors) in the telencephalon (Castro et al., 2006). Ascl1/E-protein heterodimers plus Brn-family factors regulate Dll1 by binding DNA through a compound site containing an E-box and the Brn consensus site, thereby activating transcription. Using this extended consensus DNA sequence, additional targets were identified that included E-box E3 of Dll3 (Castro et al., 2006). However, mutation of this E-box site alone did not dramatically disrupt enhancer activity as assayed in transgenic mice (Fig. 4, Dll3-mE3). It is possible that the two-nucleotide mutation tested here was not sufficient to completely disrupt complex formation at this site in vivo, particularly if the complex contains Ascl1 plus a Brn factor with multiple protein-DNA contact surfaces as suggested to occur in the telencephalon. Alternatively, there may be differences in the co-factors used to activate Dll3 expression in the telencephalon versus the neural tube.
Other direct targets of Ascl1 have been identified and include transcription factors Dl×1/2 and Hes6 (Poitras et al., 2007), a secreted factor PK2 (Zhang et al., 2007), and proprotein convertase Pace4 (Yoshida et al., 2001). Each of these targets contains a preferred Ascl1/E-protein heterodimer E-box site: RCAGSTGK. Recently, a bioinformatics approach was used to identify candidate co-factors for Ascl1 and Neurog2 during telencephalon development (Gohlke et al., 2008). This analysis was restricted to targets with the preferred E-box site as well. The existence of multiple Ascl1 transcription factor complexes that have different sequence preferences, and possibly different affinities and transcription activities will complicate models of Ascl1 and Neurog2 function. The combination of bHLH containing complexes at any given time as a cell differentiates will have distinct but overlapping target gene specificity, and together with the genomic organization of the E-boxes present, will determine the temporal and spatial characteristics of downstream target expression.
Supplementary Material
A stable transgenic mouse line, Dll3-GFP, was generated and embryos were characterized at multiple stages. (A-C) GFP expression in transgenic mouse embryos at E9.5 (A), E11.5 (B), and E13.5 (C) generated using the 640 bp Dll3 promoter to drive GFP. GFP at E9.5 is mostly in somites (A’), E11.5 expression is in neural tube and limbs, and E13.5 expression is seen in the forebrain (C’), the dorsal spinal cord (C”), the retina (C’”), and the hypothalamus (C””). (D-I) mRNA in situ hybridization of E11.5 Dll3-GFP transgenic embryos comparing the endogenous Dll3 mRNA (D-F) to the transgene GFP mRNA (G-I) in the telencephalon, limb, and olfactory epithelium (OE) where Dll3 is expressed at this stage. LV; lateral ventricle.
Transient transgenic embryos with a wild-type or E-box mutant Dll3 promoter driving GFP expression at E11.5 are shown in whole mount or as one half of a cross section through the neural tube. The blue circles represent each E-box, and the X indicates a mutation of the site. The relative expression in the neural tube is indicated by +, and expression in limb or in ectopic locations is indicated by Y (expression seen) or N (no expression seen). Temporal ectopic expression is early expression in the ventricular zone (arrowheads). The number of expressing embryos analyzed for each transgene is indicated (# Expressing). The images shown were obtained using identical exposure time for the whole mount embryos and identical imaging parameters on the confocal for the cross sections. Each embryo is representative of those obtained for each transgene. These data can be directly compared to data in Fig. 3.
Acknowledgments
We appreciate the critical comments and discussions on this manuscript by Drs. Helen Lai and G. Swift. We gratefully acknowledge the outstanding service of the UTSW Transgenic Facility in generating all transgenic mice in this study, and the outstanding technical assistance of Ms Preeti Parab and Judy Dumas. This study was supported by NIH NS032817 to JEJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anand G, Yin X, Shahidi AK, Grove L, Prochownik EV. Novel regulation of the helix-loop-helix protein Id1 by S5a, a subunit of the 26 S proteasome. J Biol Chem. 1997;272:19140–51. doi: 10.1074/jbc.272.31.19140. [DOI] [PubMed] [Google Scholar]
- Begley CG, Lipkowitz S, Gobel V, Mahon KA, Bertness V, Green AR, Gough NM, Kirsch IR. Molecular characterization of NSCL, a gene encoding a helix-loop-helix protein expressed in the developing nervous system. Proc Natl Acad Sci USA. 1992;89:38–42. doi: 10.1073/pnas.89.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres T, Masui T, Swift GH, Shi L, Henke RM, MacDonald RJ. PTF1 is an organ-specific and Notch-independent bHLH complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue RBP-L. Mol Cell Biol. 2006;26:117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neuroscience. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet J-L, Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Brinster RL, Chen HY, Trumbauer ME, Yagle MK, Palmiter RD. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci U S A. 1985;82:4438–42. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Espinosa R, Le Beau MM, Siciliano MJ, Baer R. HEN1 and HEN2: a subgroup of basic helix-loop-helix genes that are coexpressed in a human neuroblastoma. Proc Natl Acad Sci. 1992;89:8492–8496. doi: 10.1073/pnas.89.18.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, Matter JM, Guillemot F. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–44. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Henrique D, Harrison SM, Beddington RS. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124:3065–76. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20:483–94. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- Geffers I, Serth K, Chapman G, Jaekel R, Schuster-Gossler K, Cordes R, Sparrow DB, Kremmer E, Dunwoodie SL, Klein T, Gossler A. Divergent functions and distinct localization of the Notch ligands DLL1 and DLL3 in vivo. J Cell Biol. 2007;178:465–76. doi: 10.1083/jcb.200702009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke JM, Armant O, Parham FM, Smith MV, Zimmer C, Castro DS, Nguyen L, Parker JS, Gradwohl G, Portier CJ, Guillemot F. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol. 2008;6:15. doi: 10.1186/1741-7007-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Fode C, Guillemot F. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev Biol. 1996;180:227–241. doi: 10.1006/dbio.1996.0297. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–76. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Helms AW, Battiste J, Henke RM, Nakada Y, Simplicio N, Guillemot F, Johnson JE. Sequential roles for Mash1 and Ngn2 in the generation of dorsal spinal cord interneurons. Development. 2005;132:2709–19. doi: 10.1242/dev.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Birren SJ, Anderson DJ. Two rat homologues of Drosophila achaete-scute specifically expressed in neuronal precursors. Nature. 1990;346:858–861. doi: 10.1038/346858a0. [DOI] [PubMed] [Google Scholar]
- Johnson JE, Birren SJ, Saito T, Anderson DJ. DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle-specific enhancer. Proc Natl Acad Sci U S A. 1992;89:3596–600. doi: 10.1073/pnas.89.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Ishibashi M, Takebayashi K, Tomita K. bHLH transcription factors and mammalian neuronal differentiation. Int J Biochem Cell Biol. 1997;29:1389–1399. doi: 10.1016/s1357-2725(97)89968-2. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T. The Notch-Hes pathway in mammalian neural development. Cell Res. 1999;9:179–188. doi: 10.1038/sj.cr.7290016. [DOI] [PubMed] [Google Scholar]
- Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, Boulter J, Sun YE, Kintner C, Weinmaster G. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol. 2005;170:983–92. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendahl U. A growing family of Notch ligands. Bioessays. 1998;20:103–7. doi: 10.1002/(SICI)1521-1878(199802)20:2<103::AID-BIES1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cellular Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;120:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga E, Araki I, Nakamura H. Role of Pax3/7 in the tectum regionalization. Development. 2001;128:4069–77. doi: 10.1242/dev.128.20.4069. [DOI] [PubMed] [Google Scholar]
- Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Odenwald WF, Rasband W, Kuzin A, Brody T. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc Natl Acad Sci U S A. 2005;102:14700–5. doi: 10.1073/pnas.0506915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO Journal. 1999;18:2196–207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitras L, Ghanem N, Hatch G, Ekker M. The proneural determinant MASH1 regulates forebrain Dlx1/2 expression through the I12b intergenic enhancer. Development. 2007;134:1755–65. doi: 10.1242/dev.02845. [DOI] [PubMed] [Google Scholar]
- Powell LM, Zur Lage PI, Prentice DR, Senthinathan B, Jarman AP. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol Cell Biol. 2004;24:9517–26. doi: 10.1128/MCB.24.21.9517-9526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- Shutter JR, Scully S, Fan W, Richards WG, Kitajewski J, Deblandre GA, Kintner CR, Stark KL. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;14:1313–8. [PMC free article] [PubMed] [Google Scholar]
- Singson A, Leviten MW, Bang AG, Hua XH, Posakony JW. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev. 1994;8:2058–71. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- Sommer L, Ma Q, Anderson DJ. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogenity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- Suemori H, Kadodawa Y, Goto K, Araki I, Kondoh H, Nakatsuji N. A mouse embryonic stem cell line showing pluripotency of differentiation in early embryos and ubiquitous beta-galactosidase expression. Cell Differ Dev. 1990;29:181–6. doi: 10.1016/0922-3371(90)90120-l. [DOI] [PubMed] [Google Scholar]
- Timmer J, Johnson J, Niswander L. The use of in ovo electroporation for the rapid analysis of neural-specific murine enhancers. Genesis. 2001;29:123–132. doi: 10.1002/gene.1015. [DOI] [PubMed] [Google Scholar]
- Weintraub H, Genetta T, Kadesch T. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev. 1994;8:2203–11. doi: 10.1101/gad.8.18.2203. [DOI] [PubMed] [Google Scholar]
- Yoshida I, Koide S, Hasegawa SI, Nakagawara A, Tsuji A, Matsuda Y. Proprotein convertase PACE4 is down-regulated by the basic helix-loop-helix transcription factor hASH-1 and MASH-1. Biochem J. 2001;360:683–9. doi: 10.1042/0264-6021:3600683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Ng KL, Li JD, He F, Anderson DJ, Sun YE, Zhou QY. Prokineticin 2 is a target gene of proneural basic helix-loop-helix factors for olfactory bulb neurogenesis. J Biol Chem. 2007;282:6917–21. doi: 10.1074/jbc.C600290200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A stable transgenic mouse line, Dll3-GFP, was generated and embryos were characterized at multiple stages. (A-C) GFP expression in transgenic mouse embryos at E9.5 (A), E11.5 (B), and E13.5 (C) generated using the 640 bp Dll3 promoter to drive GFP. GFP at E9.5 is mostly in somites (A’), E11.5 expression is in neural tube and limbs, and E13.5 expression is seen in the forebrain (C’), the dorsal spinal cord (C”), the retina (C’”), and the hypothalamus (C””). (D-I) mRNA in situ hybridization of E11.5 Dll3-GFP transgenic embryos comparing the endogenous Dll3 mRNA (D-F) to the transgene GFP mRNA (G-I) in the telencephalon, limb, and olfactory epithelium (OE) where Dll3 is expressed at this stage. LV; lateral ventricle.
Transient transgenic embryos with a wild-type or E-box mutant Dll3 promoter driving GFP expression at E11.5 are shown in whole mount or as one half of a cross section through the neural tube. The blue circles represent each E-box, and the X indicates a mutation of the site. The relative expression in the neural tube is indicated by +, and expression in limb or in ectopic locations is indicated by Y (expression seen) or N (no expression seen). Temporal ectopic expression is early expression in the ventricular zone (arrowheads). The number of expressing embryos analyzed for each transgene is indicated (# Expressing). The images shown were obtained using identical exposure time for the whole mount embryos and identical imaging parameters on the confocal for the cross sections. Each embryo is representative of those obtained for each transgene. These data can be directly compared to data in Fig. 3.


