Abstract
The molecular basis for breast cancer metastasis to the brain is largely unknown1,2. Brain relapse typically occurs years after the removal of a breast tumour2–4, suggesting that disseminated cancer cells must acquire specialized functions to overtake this organ. Here we show that breast cancer metastasis to the brain involves mediators of extravasation through non-fenestrated capillaries, complemented by specific enhancers of blood–brain barrier crossing and brain colonization. We isolated cells that preferentially infiltrate the brain from patients with advanced disease. Gene expression analysis of these cells and of clinical samples, coupled with functional analysis, identified the cyclooxygenase COX2 (also known as PTGS2), the epidermal growth factor receptor (EGFR) ligand HBEGF, and the α2,6-sialyltransferase ST6GALNAC5 as mediators of cancer cell passage through the blood–brain barrier. EGFR ligands and COX2 were previously linked to breast cancer infiltration of the lungs, but not the bones or liver5,6, suggesting a sharing of these mediators in cerebral and pulmonary metastases. In contrast, ST6GALNAC5 specifically mediates brain metastasis. Normally restricted to the brain7, the expression of ST6GALNAC5 in breast cancer cells enhances their adhesion to brain endothelial cells and their passage through the blood–brain barrier. This co-option of a brain sialyltransferase highlights the role of cell-surface glycosylation in organ-specific metastatic interactions.
Brain metastasis affects an estimated 10% of cancer patients with disseminated disease2,8,9. Even small lesions can cause neurological disability, and the median survival time of patients with brain metastasis is short. The two main sources of brain metastasis—adenocarcinomas of the lung or the breast—represent different models of the course of the disease. Metastasis from lung adenocarcinomas develops within months of diagnosis and affects several organs besides the brain10. This course suggests that aggressive pro-metastatic functions foster the colonization of several organs at once. In breast cancer, a long period of remission often precedes distant relapse3,4, suggesting that breast cancer cells initially lack the full competence for outgrowth in distant organs but develop this under the selective pressure of different organ microenvironments. Breast cancer metastasis frequently becomes prevalent in one organ long before it does in others, and brain metastasis tends to be a late event2. The barriers to metastasis are distinct in different organs. Capillary endothelia are backed by a basement membrane in the lung11 and also by tight junctions and astrocyte foot processes in the blood–brain barrier (BBB)2,8, whereas the capillaries in the bone marrow and the liver are fenestrated11,12. The composition of the parenchyma also varies extensively between these organs. The protracted progression of disseminated cancer cells in different environments may give rise to metastatic speciation, as suggested by the coexistence of malignant cells with different organ tropisms in fluids from patients with advanced disease5,13. Analysis of such malignant cell populations has revealed genes that selectively mediate breast cancer metastasis to bones13 or the lungs5. Here we adopted this approach to test the hypothesis that breast cancer infiltration of the brain requires general mediators of extravasation, complemented by specific enhancers of cell passage through the BBB.
We used oestrogen-receptor-negative (ER−) pleural malignant cells from a Memorial Sloan-Kettering Cancer Center (MSKCC) breast cancer patient (CN34 sample), and also from MDA-MB-231 cells (MDA231 for brevity)—an ER− breast cancer pleural cell line previously used for the isolation of bone and lung metastatic cells5,13 and brain metastatic cells14. CN34 and MDA231 cells were inoculated into the arterial circulation of immunodeficient female mice to isolate populations that target the brain (Fig. 1a–d). After tumour dissociation and expansion in culture, the resulting cell populations (brain metastatic derivative 1, BrM1) were subjected to a second round of in vivo selection, yielding BrM2 cell populations that showed a significant increase in brain metastatic activity (Fig. 1a, b). When grown as mammary tumours, CN34-BrM2 metastasized to brain in 42% (5 out of 12) of the mice, whereas parental CN34 mammary tumours yielded no brain metastases in ten mice. BrM2 cells showed no increase in bone or lung metastatic activity compared to the parental populations (Supplementary Table 1). MDA231 lung metastatic (LM2-4175) and bone metastatic (BoM-1833; refs 5, 13) derivatives were poorly metastatic to brain compared to BrM2 cells (Fig. 1b). The CN34-BrM2 and MDA231-BrM2 cell lines generated multifocal lesions in the cerebrum, the cerebellum and the brainstem (Fig. 1e, f and Supplementary Fig. 1a, b), and in the leptomeninges (Fig. 1g and Supplementary Fig. 1c, d). Larger nodules developed hemorrhagic cores and oedema (Fig. 1d). Astrogliosis occurred in the periphery of the tumours (Fig. 1h). All of these features are typical of brain metastasis in breast cancer patients2,15. Within 24 h of inoculation, BrM2 cells lodged in brain capillaries as single cells (Fig. 1i), suggesting that brain metastases resulted from an ability of these cells to breach the BBB.
Figure 1. Isolation and characterization of brain metastatic variants.
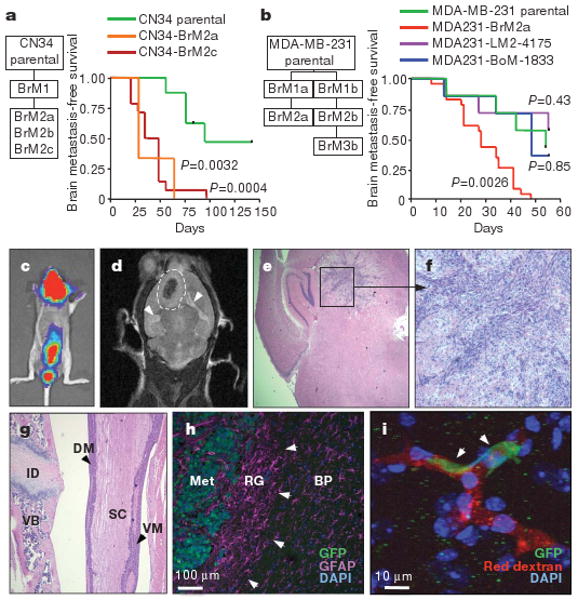
a, b, Flowcharts of the in-vivo-selected brain metastatic derivatives, and Kaplan–Meier survival curves for brain metastasis-free survival of representative CN34 (parental n = 8, BrM2c n =14, BrM2a n = 3) (a) and MDA231 (parental n = 7, LM n = 7, BoM n = 7, BrM2a n = 23) (b) cell line variants. A log-rank test was used to compare the survival curves of each cell line to the parental line. BoM, BrM and LM indicate bone, brain and lung metastatic derivative, respectively. c, Bioluminescence image of a mouse with brain and leptomeningeal metastasis by CN34-BrM2c cells. d, Magnetic resonance imaging (MRI) of a brain metastatic lesion (dashed line) showing a hemorrhagic core, and brain oedema (arrowheads). e, f, Representative haematoxylin and eosin (H&E)-stained sections of a mouse brain containing a CN34-BrM2c lesion (original magnification, ×2 (e) and ×10 (f)). g, H&E staining of a section showing MDA231-BrM2a cell colonization of the dorsal (DM) and ventral (VM) meninges. ID, intervertebral disc; SC, spinal cord; VB, vertebral body. Original magnification, ×5. h, MDA231-BrM2a brain metastatic lesion showing reactive glia (RG, arrowheads) around the metastatic lesion (Met). Tumour cells express green fluorescent protein (GFP), and glial cells are stained with the glial marker glial fibrillary protein (GFAP, purple). BP, brain parenchyma; DAPI, 4,6-diamidino-2-phenylindole. i, GPF+ MDA231-BrM2a cells (arrowheads) arrested in brain capillaries (red, rhodamine dextran) 24 h after intracardiac injection into mice. Nuclei were stained with DAPI (blue).
Comparative genome-wide expression analysis demonstrated 243 genes that were overexpressed or underexpressed in the brain metastatic populations of both cell lines, or were upregulated in one cell system and overexpressed in the other, or were downregulated in one system and underexpressed in the other (Supplementary Table 2). To prioritize these candidate genes, we screened for those whose expression in breast tumours was associated with brain relapse. Univariate analysis in a combined cohort of 368 clinically annotated breast tumours (MSK-82 and EMC-286 sets; Supplementary Table 3) showed 17 genes whose expression was correlated (P < 0.05) with brain relapse (Supplementary Table 4), and resembled the expression profile in the brain-metastatic-derived (BrM) cells (Supplementary Fig. 2a, b). A classifier trained with this brain metastasis gene set (BrMS) showed association with brain relapse in two independent breast tumour data sets (Fig. 2a and Supplementary Fig. 3a). The same procedures applied to randomly generated sets of 500 genes yielded no classifiers that performed in the data sets. The association of BrMS status with brain relapse remained significant within ER− tumours (Supplementary Table 5 and Supplementary Fig. 3b), and was stronger in patients who received no adjuvant therapy (P < 0.0001; Fig. 2b and Supplementary Table 6). BrMS+ tumours appeared in different molecular subtypes of breast cancer16 (Supplementary Fig. 4a–c).
Figure 2. COX2 and EGFR ligands as mediators of brain metastasis and BBB transmigration.
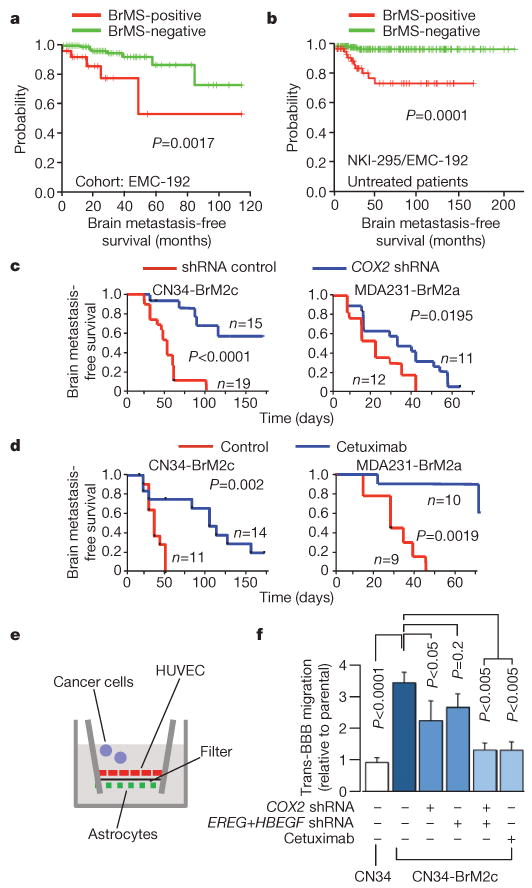
a, b, Kaplan–Meier curves for brain metastasis-free survival on the basis of BrMS status in an independent cohort of 192 breast tumours (a), and in a combined cohort of 262 breast tumours from patients who received no adjuvant therapy (b). c, Kaplan–Meier curves for brain metastasis-free survival of mice injected with the indicated cell lines expressing short hairpin RNA (shRNA) vector control or shRNA targeting COX2. d, Kaplan–Meier curves for brain metastasis-free survival of mice injected with the indicated cell lines and treated with cetuximab or vehicle control. e, Schematic of the in vitro BBB model assay system. HUVEC, human umbilical vein endothelial cells. f, In vitro BBB transmigration activity of the indicated cell lines and conditions. The number of transmigrated cells relative to the parental cell lines is plotted. Error bars, s.e.m.; n = 6–20. P values were determined by log rank test (a–d) and one-tailed unpaired t-test (f).
We do not interpret the results of the gene expression analysis as reflecting the only possible 17 genes associated with brain relapse. However, the expression of these 17 genes in breast tumours was not associated with relapse to bones, liver or lymph nodes (Supplementary Fig. 5a). Notably, six of these genes were shared with an 18-gene lung metastasis signature (LMS) that is associated with relapse to the lungs, but not to bones, liver or lymph nodes17. LMS+ status was weakly associated with relapse to the brain, and BrMS+ status with relapse to the lungs (Supplementary Fig. 5b–d). The shared genes include the prostaglandin-synthesizing enzyme cyclooxygenase-2 (COX2), which promotes extravasation in the lungs6; collagenase-1 (MMP1), which mediates invasion and extravasation6,18; angiopoietin-like 4 (ANGPTL4), which is induced by tumour-derived TGF-β and disrupts endothelial junctions19; latent TGF-β-binding protein (LTBP1), which controls TGF-β activation20; fascin-1 (FSCN1), which supports cancer cell migration21; and the putative metastasis suppressor RARRES3 (ref. 5). Furthermore, both gene sets include an EGFR ligand: heparin-binding EGF (HBEGF) in the BrMS, and epiregulin (EREG) in the LMS. EREG was highly expressed in CN34-BrM but not in MDA231-BrM cells.
These observations suggested a partial sharing of mediators of metastasis to the brain and lungs, a hypothesis that we tested by focusing on EGFR ligands and COX2. Prostaglandin production during inflammation increases BBB permeability22. HBEGF induces cancer cell motility and invasiveness23. The brain metastatic activity of BrM2 cells (Fig. 2c, d) was decreased by RNA interference (RNAi)-mediated knockdown of COX2 expression6 (Supplementary Fig. 6a, b), or by treatment of mice with cetuximab, which targets human EGFR24. To investigate cancer cell passage through the BBB, we used an in vitro model consisting of human primary endothelial cells and astrocytes (Fig. 2e and Supplementary Fig. 7a). This model generates a tight barrier that expresses brain endothelial markers and lacks permeability to albumin (Supplementary Fig. 7b–d)25. CN34-BrM2 and MDA231-BrM2 cells were three- to fourfold more active than their parental lines at migrating through this barrier. COX2 knockdown inhibited this transmigration in both BrM2 lines, as did the addition of cetuximab to CN34-BrM2 cells, or the combination of COX2, HBEGF and EREG knockdowns (Fig. 2f and Supplementary Fig. 7e).
The ability of COX2 and EGFR ligands to prime breast cancer cells for extravasation into the brain may explain the association of lung and brain relapse in breast cancer9,26. However, given the course of breast cancer, we postulated that brain relapse also depends on selective mediators of infiltration through the unique barriers of the brain. As candidates we selected genes whose expression was increased > threefold in all CN34-BrM2 and MDA231-BrM2 isolates, but not in bone metastatic13 or lung metastatic MDA231 derivatives5. After excluding histone genes, we arrived at a set of 26 candidates (Supplementary Table 7). This set largely consists of cell–cell interaction components. The α2,6-sialyltransferase ST6GALNAC5 stood out because its expression is normally restricted to the brain both in mice7 and in humans (Supplementary Fig. 8). ST6GALNAC5 messenger RNA levels were notably higher in brain metastatic derivatives than in parental cell lines in MDA231 samples (30-fold ± 1 higher; mean ± s.d.), CN34 cells (>100-fold), and in two other pleural-derived samples that were subjected to one cycle of selection for brain infiltration in mice (CN37-BrM1, 95-fold ± 23; CN41-BrM1, 72-fold ± 12).
Sambucus nigra agglutinin (SNA), a lectin that binds to α2,6-linked sialyl groups, specifically stained mammary tumours and brain lesions formed by BrM2 cells (Fig. 3a, b and Supplementary Fig. 9a, b). Fifty per cent (6 out of 12) of brain metastatic samples from breast cancer patients had areas with strong SNA staining, whereas 18% (2 out of 11) of lung metastatic samples had areas of low SNA staining, and the remaining 9 samples were negative (Fig. 3c, d). Furthermore, in a set of ER− breast cancer metastasis to various sites, ST6GALNAC5 mRNA levels roughly equalled that of the BrM2 cell lines in 23% (3 out of 12) of brain metastases, but not in metastases to other sites (Fig. 3e; P = 0.04, Fisher's exact test).
Figure 3. ST6GALNAC5 expression and activity in brain metastasis from breast cancer.
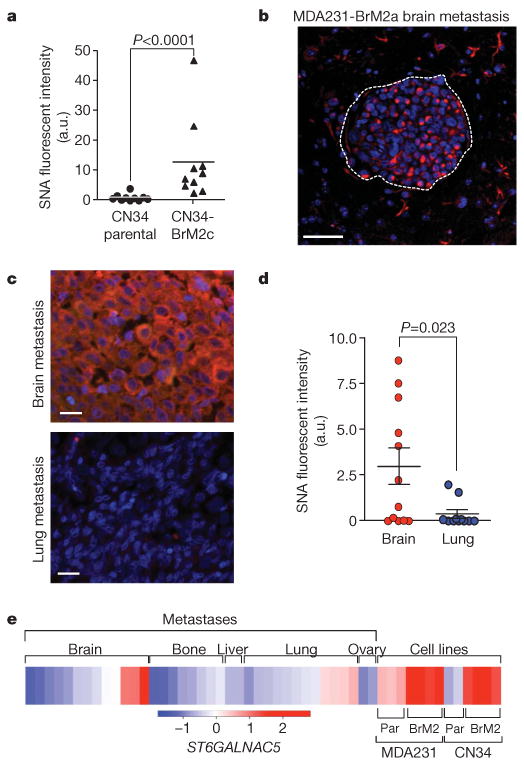
a, Quantification of SNA staining in mammary fat pad tumours formed by parental CN34 or CN34-BrM2c cells in mice. a.u., arbitrary units. b, SNA staining of a mouse brain metastasis after intracardiac inoculation of MDA231-BrM2a cells. Scale bar, 50 μm c, SNA staining of representative human brain and lung metastases samples from the same breast cancer patient. Scale bars, 20μm. d, Distribution of SNA staining intensity, quantified by Metamorph analysis, in 12 brain and 11 lung metastases resected from breast cancer patients. P values (a, d) were determined by Mann–Whitney one-tailed test. e, Heat map showing the relative ST6GALNAC5 expression levels in a panel of 13 brain, 8 bone, 3 liver, 12 lung and 2 ovary human metastases from breast cancer patients. Included for comparison are the parental (par) and brain metastatic derivatives from MDA231 and CN34 cells. Data are on the basis of Affymetrix probe intensity.
Sialyltransferases catalyse the addition of sialic acid to gangliosides and glycoproteins27, and cell-surface sialylation has been implicated in cell–cell interactions28. CN34-BrM2 cells were more adhesive to monolayers of human primary brain endothelial cells than to parental line or ST6GALNAC5-knockdown CN34-BrM2 cells (Supplementary Fig. 10a). Notably, the knockdown of ST6GALNAC5 decreased the BBB transmigration activity of CN34-BrM2 cells to ground level (Fig. 4a), and also decreased the brain metastatic activity of CN34-BrM2 cells (Fig. 4b). Brain metastasis was further decreased by combination with cetuximab treatment (Fig. 4b). ST6GALNAC5 knockdown did not inhibit the growth of CN34-BrM2c cells in culture or as mammary tumours, the basal lung-seeding ability of these cells, or the aggressive lung-colonizing ability of the MDA231 derivative line LM2-4175 (ref. 5) (Supplementary Fig. 10b–f). However, transduction of LM2-4175 cells with an ST6GALNAC5 expression vector increased the ability of these cells to transmigrate across a BBB (Fig. 4c) and to infiltrate the brain (Supplementary Fig. 11a). Infiltrated brains showed a prevalence of micrometastases in mice inoculated with LM2-ST6 and of larger lesions in mice inoculated with BrM2 cells (Fig. 4d and Supplementary Fig. 11b). ST6GALNAC5 therefore mediates infiltration into the brain, and further mediators may be required for the expansion of the resulting foci into macrometastases.
Figure 4. The brain-specific sialyltransferase ST6GALNAC5 as a BBB extravasation and brain metastasis gene.
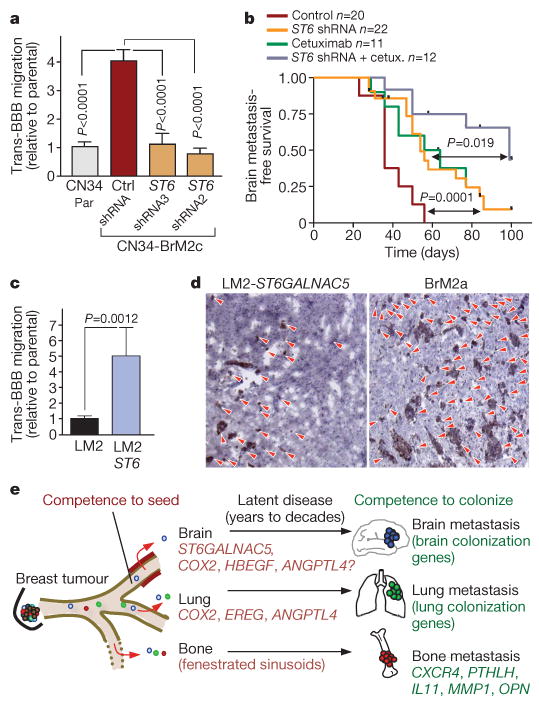
a, In vitro BBB transmigration activity of the indicated cell lines. Ctrl, control; par, parental; ST6, ST6GALNAC5. Error bars, s.e.m.; n = 9–27; P values were determined by Mann–Whitney one-tailed test. b, Kaplan–Meier curves for brain metastasis-free survival of mice injected with CN34-BrM2 cells expressing an shRNA targeting ST6GALNAC5 or an empty vector control, and then treated with cetuximab or vehicle. P values were determined using log-rank test. c, In vitro BBB transmigration activity of LM2 cells transduced with an empty vector or with ST6GALNAC5. Error bars, s.e.m.; n = 22–27; P values were determined by Mann–Whitney one-tailed test. d, Anti-GFP immunostaining of representative lesions from mice injected intracardially with the indicated cell lines. Red arrowheads show individual tumour foci; original magnification, ×10. e, Schematic model of organ-specific metastatic extravasation of breast cancer cells. Extravasation into the bone marrow is a relatively permissive process owing to the fenestrated endothelium lining the sinusoid capillaries. Extravasation into the pulmonary or brain parenchyma requires specific functions for breaching the non-fenestrated capillary walls of these organs. Shared mediators of extravasation include, among others, COX2 and EGFR ligands such as epiregulin and HBEGF. Passage through the BBB requires further mediators including, but not limited to, the brain-specific sialyltransferase ST6GALNAC5. Competence to colonize each organ requires additional mediators.
Breast cancer cells can disseminate to the lungs from early stages of tumour development29, indicating that cancer cells departing from breast tumours are competent for extravasation through lung micro-capillary walls. Our results indicate that the expression of COX2 and HBEGF in primary tumours enhances cancer cells for extravasation through the non-fenestrated capillaries of the brain and lungs, whereas ST6GALNAC5 expression is co-selected with, and acts as a specific mediator of, cancer cell infiltration through the blood–brain barrier (Fig. 4e). These findings draw attention to the role of cell-surface sialylation as a previously unrecognized participant in brain metastasis, and to the possibility of therapeutically disrupting these interactions. Our work also points to other candidate brain metastasis genes, including genes implicated in vascular permeability and leukocyte infiltration during brain inflammatory processes, and genes implicated in neurite extension and astrocyte cell processes. The role of these genes in brain metastasis and their interest as therapeutic targets is open to further analysis.
Methods Summary
CN34 tumour cells were isolated from the pleural effusion of a breast cancer patient treated at our institution, after written consent in accordance with Institutional Review Board (IRB) regulations. Brain metastatic populations from these cells and MDA-MB-231cells were obtained by consecutive rounds of in vivo selection in 6–7-week-old beige nude and athymic mice, respectively. All animal work was done in accordance with the MSKCC Institutional Animal Care and Use Committee. Methods for RNA extraction, labelling and hybridization for DNA microarray analysis have been described previously17. Bioinformatics analyses with detailed descriptions can be found in the Methods. Knockdown and overexpression of candidate genes, and cetuximab inhibitor studies were performed as previously described6. The in vitro BBB model was set up as previously described25, and modified to enable tumour cell counting. Sambucus nigra lectin staining was performed using standard histochemical techniques, and quantified using Metamorph software analysis. The Methods section provides further information, including malignant cell isolation from pleural fluids, tumour cell extraction and cell culture protocols, animal inoculation and bioluminescence imaging, generation of retroviral gene knockdown and overexpression vectors, transfections and infections, RNA and protein expression, in vitro BBB transmigration assay, endothelial cell adhesion assay, and metastatic tissue staining and quantification.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of our colleague W. Gerald. We thank E. Eugenin, E.Brogi, M.Drobnjac, K. LaPerle, M.Smid, A. Viale and K. Manova-Todorova for advice and support. We thank L. DeAngelis, A. Lassman, E. Holland, J. Posner and members of the Massagué laboratory for discussions. This work was supported by grants from the National Institutes of Health (U54 CA126518), the Kleberg Foundation and the Hearst Foundation, and the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO). J.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Information The clinical microarray data on the brain metastatic cell lines have been deposited in NCBI's Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) under the GEO series accession number GSE12237.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author Contributions P.D.B. and J.M. designed experiments, analysed data and wrote the manuscript. J.M. supervised the research. X.H.-F.Z. performed bioinformatics analyses. P.D.B. performed experiments. W.S. assisted with experiments. C.N. and R.R.G. isolated metastatic cells from clinical samples. D.X.N. helped with gliosis immunostaining and confocal microscopy. A.J.M. identified LMS clinical correlation with brain relapse. W.L.G., J.A.F. and M.V.d.V. obtained, classified and processed breast tumour samples. All authors discussed the results and commented on the manuscript.
References
- 1.Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weil RJ, et al. Breast cancer metastasis to the central nervous system. Am J Pathol. 2005;167:913–920. doi: 10.1016/S0002-9440(10)61180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karrison TG, Ferguson DJ, Meier P. Dormancy of mammary carcinoma after mastectomy. J Natl Cancer Inst. 1999;91:80–85. doi: 10.1093/jnci/91.1.80. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt-Kittler O, et al. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci USA. 2003;100:7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta GP, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 7.Okajima T, et al. Molecular cloning of brain-specific GD1α synthase (ST6GalNAc V) containing CAG/glutamine repeats. J Biol Chem. 1999;274:30557–30562. doi: 10.1074/jbc.274.43.30557. [DOI] [PubMed] [Google Scholar]
- 8.El Kamar FG, Posner JB. Brain metastases. Semin Neurol. 2004;24:347–362. doi: 10.1055/s-2004-861530. [DOI] [PubMed] [Google Scholar]
- 9.Lassman AB, DeAngelis LM. Brain metastases. Neurol Clin. 2003;21:1–23. doi: 10.1016/s0733-8619(02)00035-x. [DOI] [PubMed] [Google Scholar]
- 10.Feld R, Rubinstein LV, Weisenberger TH. Sites of recurrence in resected stage I non-small-cell lung cancer: a guide for future studies. J Clin Oncol. 1984;2:1352–1358. doi: 10.1200/JCO.1984.2.12.1352. [DOI] [PubMed] [Google Scholar]
- 11.Inoue S, Osmond DG. Basement membrane of mouse bone marrow sinusoids shows distinctive structure and proteoglycan composition: a high resolution ultrastructural study. Anat Rec. 2001;264:294–304. doi: 10.1002/ar.1166. [DOI] [PubMed] [Google Scholar]
- 12.Paku S, Dome B, Toth R, Timar J. Organ-specificity of the extravasation process: an ultrastructural study. Clin Exp Metastasis. 2000;18:481–492. doi: 10.1023/a:1011858925376. [DOI] [PubMed] [Google Scholar]
- 13.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 14.Yoneda T, et al. A bone-seeking clone exhibits different biological properties from the MDA-MB-231 parental human breast cancer cells and a brain-seeking clone in vivo and in vitro. J Bone Miner Res. 2001;16:1486–1495. doi: 10.1359/jbmr.2001.16.8.1486. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald DP, et al. Reactive glia are recruited by highly proliferative brain metastases of breast cancer and promote tumor cell colonization. Clin Exp Metastasis. 2008;25:799–810. doi: 10.1007/s10585-008-9193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan C, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 17.Minn AJ, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci USA. 2007;104:6740–6745. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nature Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 19.Padua D, et al. TGFβ primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saharinen J, Hyytiainen M, Taipale J, Keski-Oja J. Latent transforming growth factor-β binding proteins (LTBPs)–structural extracellular matrix proteins for targeting TGF-β action. Cytokine Growth Factor Rev. 1999;10:99–117. doi: 10.1016/s1359-6101(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 21.Adams JC. Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 22.de Vries HE, et al. The influence of cytokines on the integrity of the blood-brain barrier in vitro. J Neuroimmunol. 1996;64:37–43. doi: 10.1016/0165-5728(95)00148-4. [DOI] [PubMed] [Google Scholar]
- 23.Miyamoto S, et al. Heparin-binding epidermal growth factor-like growth factor as a novel targeting molecule for cancer therapy. Cancer Sci. 2006;97:341–347. doi: 10.1111/j.1349-7006.2006.00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein NI, et al. Biological efficacy of a chimeric antibody to the epidermal growth factor receptor in a human tumor xenograft model. Clin Cancer Res. 1995;1:1311–1318. [PubMed] [Google Scholar]
- 25.Eugenin EA, Berman JW. Chemokine-dependent mechanisms of leukocyte trafficking across a model of the blood-brain barrier. Methods. 2003;29:351–361. doi: 10.1016/s1046-2023(02)00359-6. [DOI] [PubMed] [Google Scholar]
- 26.Slimane K, et al. Risk factors for brain relapse in patients with metastatic breast cancer. Ann Oncol. 2004;15:1640–1644. doi: 10.1093/annonc/mdh432. [DOI] [PubMed] [Google Scholar]
- 27.Harduin-Lepers A, et al. The human sialyltransferase family. Biochimie. 2001;83:727–737. doi: 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- 28.Dall'Olio F, Chiricolo M. Sialyltransferases in cancer. Glycoconj J. 2001;18:841–850. doi: 10.1023/a:1022288022969. [DOI] [PubMed] [Google Scholar]
- 29.Husemann Y, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


