Abstract
1. Multidrug resistance-associated proteins 2-4 (MRP2-4) are membrane efflux transporters that are critical for the hepatic clearance of pharmaceuticals and endogenous chemicals. Little is known about the constitutive regulation of MRP2-4 mRNA in normal human liver.
2. The purpose of this study was to identify transcription factors whose expression significantly correlates with MRP2-4 mRNA in human liver specimens.
3. Ninety adult human livers were profiled for mRNA expression of MRP2-4 as well as AhR, CAR, PXR, PPARα and γ, LXRα, FXR, GR, RXRα, HNF1α and 4α, and Nrf2 transcription factors. Using linear regression and stepwise selection of partial R2 values, CAR, HNF1α, and PPARα mRNA exhibited the greatest correlation with MRP2, 3, and 4, respectively.
4. Interindividual variation in the expression of the identified transcription factors may account for the variability in constitutive mRNA levels of MRP2-4. The multivariate approach presented in this study should aid in predicting signaling pathways that participate either directly or indirectly in regulating hepatic drug disposition.
Keywords: ABCC, Transporters, CAR, PPARalpha, HNF-1alpha
Introduction
The safety and efficacy of pharmacotherapy regimens are influenced by interindividual variability in pharmacokinetics and pharmacodynamics. This variance is governed by many factors including gender, age, ethnicity, concomitant medications, environmental exposures, underlying disease states, and polymorphisms. However, there are likely additional determinants that dictate the constitutive expression of genes involved in hepatic drug clearance that have yet to be elucidated. One potential determinant may be the expression levels of transcription factors that regulate metabolism and transport genes.
The hepatic clearance of drugs involves a series of events including uptake of chemicals into the liver by passive diffusion or carrier-mediated processes. Subsequently, drugs are metabolized by Phase-I and -II enzymes to produce water-soluble forms that are readily excreted by ATP-dependent membrane transporters. The primary hepatic efflux transporters are the multidrug resistance-associated proteins (MRP, human; Mrp, rodent) that belong to the ATP-binding cassette, subfamily c gene family. MRP3 and 4 localize to the sinusoidal membrane and excrete substrates from the hepatocyte to the blood. In contrast, canalicular MRP2 transports chemicals into bile. MRP2-4 isoforms exhibit differences in substrate preference. For example, Mrp2/MRP2 prefers to excrete glutathione conjugates whereas Mrp3/MRP3 predominantly transports glucuronidated chemicals (Manautou et al. 2005; Zelcer et al. 2005; Lagas et al. 2006; Vlaming et al. 2006; Zamek-Gliszczynski et al. 2006; van de Wetering et al. 2007). Sulfated xenobiotics are good substrates for Mrp4/MRP4-mediated transport (Mennone et al. 2006; Zamek-Gliszczynski et al. 2006). Comparison of nucleotide sequences using the NIH BLAST2 algorithm revealed 81-89% similarity between mouse and human MRP2-4 genes, indicating potential overlap in substrate specificity between these two species (http://www.ncbi/nlm.nih.gov/blast/bl2seq/). The recent development of knockout mice will enable further analysis of the in vivo roles of Mrp2-4 in hepatic drug clearance.
Both in vivo rodent studies and in vitro research with primary human hepatocytes demonstrate the basal and inducible regulation of transporters by ligand-activated transcription factors (Wang and LeCluyse 2003; Klaassen and Slitt 2005). Transcription factors such as aryl hydrocarbon receptor (AhR), constitutive androstane receptor (CAR), pregnane X receptor (PXR), and peroxisome proliferator activated-receptor (PPAR) have documented roles in regulating Mrp expression and are expressed in liver (Klaassen and Slitt 2005). Human PXR has also been termed the steroid and xenobiotic receptor. The glucocorticoid receptor (GR) is an additional member of the nuclear hormone receptor family, although its ability to regulate Mrp transporters is unknown. A number of bile acid, sterol, and oxysterol sensors are also expressed in liver. Liver X receptor (LXR) and the farnesoid X receptor (FXR) may be relevant to the current study because MRP3 and MRP4 can transport bile acids into plasma and likely represent alternate bile acid transport pathways during cholestasis (Slitt et al. 2007). With the exception of GR, the receptors listed above heterodimerize with retinoid X receptor alpha (RXRα) and bind to responsive elements in the upstream regions of target genes and activate target gene transcription.
The liver also expresses other transcription factors that may control expression of nuclear hormone receptors and/or transporters. These include the hepatocyte nuclear factors (HNF) 1α and 4α (Cereghini 1996; Schrem et al. 2002). It was recently shown that HNF4α may function as a master regulator dictating expression of metabolism and transporter genes as well as nuclear hormone receptors (Odom et al. 2004; Wortham et al. 2007). Finally, the transcription factor nuclear factor E2-related factor 2 (Nrf2) regulates mouse Mrp2-4 expression during periods of glutathione depletion, chemical induction, and acetaminophen-induced hepatotoxicity (Maher et al. 2005; 2007; Aleksunes et al. 2008). Nrf2 is activated in liver by electrophiles and/or oxidative stress to coordinately up-regulate cytoprotective genes. Collectively, these transcription factors participate in regulating Mrp expression in rodent models and in vitro hepatocyte systems via chemical induction and/or hepatotoxicant exposure.
There is limited information regarding the constitutive regulation of MRP mRNA expression in human liver. A recent report by Wortham et al. used 20 human livers and observed a relationship between MRP2 and HNF4α mRNA expression (Wortham et al. 2007). The purpose of the current study is to identify the influence of variation in mRNA expression of transcription factors on constitutive levels of hepatic MRP2-4 mRNA in 90 adult human liver specimens. The multivariate statistical analyses employed in the present study identify transcription factors whose mRNA expression exhibits the highest association with MRP expression. Additional analysis examines potential interrelationships between receptors and transcription factors that demonstrate the greatest correlation with MRP mRNA expression.
Materials and Methods
Liver samples
Ninety human liver lysates, donor ages 17 years to 79, were purchased from Xenotech LLC (Lenexa, KS) in TRIzol® Reagent. Demographics of the donors included in this study are listed in Supplementary Table 1. Liver specimens were isolated from 52 males and 38 females. Of the 90 specimens, 69 were Caucasian, 9 were Hispanic, 9 were African American and 3 were of Asian descent. This study received institutional review board exemption status by the University of Kansas Medical Center Human Subjects Committee because the specimens were obtained commercially and not identified to the authors of the study.
RNA extraction
Total RNA was isolated from each sample using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. RNA was quantified by UV spectrophotometry at 260/280nm and diluted to 1μg/μl in diethyl pyrocarbamate-treated water. RNA integrity was assessed by formaldehyde-agarose gel electrophoresis, and integrity was confirmed by visualization of 18S and 28S rRNA bands.
QuantiGene® multiplex suspension array
Human liver mRNA expression was determined by the Quantigene multiplex suspension assay (Panomics Inc., Fremont, CA). Individual bead-based oligonucleotide probe sets specific for each human gene examined were developed by Panomics Inc. using previously published NCBI gene accession numbers (Supplementary Table 2). GAPDH mRNA expression was used as an internal control for each sample. Samples were analyzed by Panomics, Inc using a Bio-Plex System Array reader with Luminex 100 xMAP technology, and data was acquired using Bio-Plex Data Manager Software (Bio-Rad, Hercules, CA). Assays were performed according to the manufacturer's protocol (Panomics, Inc.). Briefly, 3 μg of total RNA was incubated overnight at 53°C with X-MAP beads containing oligonucleotide capture probes, label extenders, and blockers. The next day, beads and bound target RNA were washed and subsequently incubated with amplifier at 46°C for 1 hr. Next, samples were washed and incubated with label (biotin) and incubated at 46°C for 1 hr. Samples were washed and incubated with streptavidin-conjugated R-phycoerythrin, which binds biotinylated probes, and incubated at room temperature for 30 min. Streptavidin-conjugated R-phycoerythrin fluorescence was then detected for each analyte within each sample. All data were standardized to the internal control GAPDH. Data is expressed as the ratio of relative light units of target gene mRNA normalized to GAPDH mRNA.
Statistical analysis
GraphPad Prism (v4) (GraphPad Software, Inc, San Diego, CA) and JMP (v.7.0) (SAS Institute, Inc., Cary, NC) were used for creating graphs and conducting exploratory data analysis, respectively. Statistical tests were performed using SAS (v.9.1) (SAS Institute, Inc., Cary, NC). The following procedural statements were used in SAS: PROC CORR (linear correlations and Pearson partial correlations), PROC REG (stepwise selection of coefficients of partial determination), PROC MEANS (mean, standard error, and coefficient of variation), PROC GLM (analysis of variance with Bonferroni post-hoc multiple comparisons), and PROC TTEST (two level comparisons, such as gender). Linear correlations were used to determine the strength of association between MRP2-4 mRNA and each transcription factor. This analysis was followed by stepwise regression to estimate partial R2 values for each transcription factor in order to determine which transcription factors, among all factors tested, best explained the variability of each MRP gene. Interrelationships between the top two transcription factor genes identified by the stepwise regression were analyzed by first-order partial correlation coefficient analysis, which tests the strength of the association between one transcription factor and MRP, while controlling for variability in the second factor. Linear regressions and first-order partial correlations were deemed significant at p<0.005. All other comparisons were considered statistically significant at p<0.05.
Results
Distribution of MRP2-4 mRNA expression in adult human livers
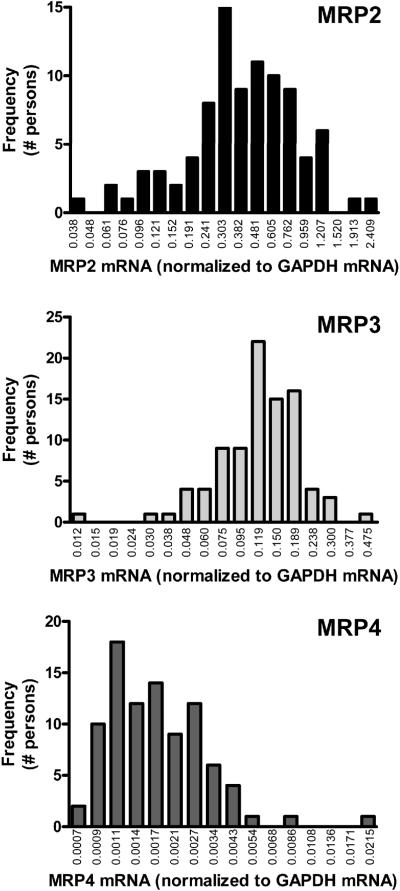
The frequency distributions for MRP2-4 mRNA expression in adult human liver are unimodal (Figure 1). There were no significant differences observed in MRP expression with respect to donor gender or ethnicity (data not shown). MRP2 and 4 mRNA expression were not different by age class (17-39, 40-59, or 60+ years old); however, MRP3 mRNA expression was 1.5-fold higher in the 60+ year old cohort compared to the 17-39 year old group (data not shown).
Figure 1. Frequency distribution of MRP2-4 mRNA in adult human livers.
mRNA levels of MRP2-4 transporters are presented as frequency distributions on a logarithmic scale normalized to GAPDH mRNA.
Correlation coefficient (linear regression)
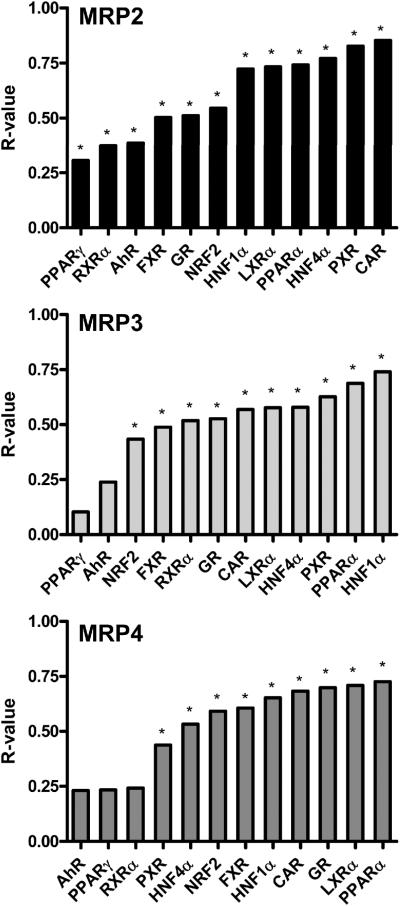
Regression analysis was performed to test the strength of linear relationships between each MRP gene and transcription factor combinations. Zero-order linear regressions are routinely used as an initial, and sometimes final, step towards elucidating potential correlations between two variables. Correlation coefficients (R-values) are shown in Figure 2. Notably, a substantial number of statistically significant correlations (p < 0.005) in mRNA levels between each MRP gene and the 12 transcription factors were observed. MRP2 mRNA correlated with all 12 transcription factors. The most notable correlations for MRP2 were CAR, PXR, HNF4α, PPARα, and LXRα. MRP3 mRNA correlated with 10 transcription factors. HNF1α, PPARα, PXR, HNF4α, LXRα, and CAR exhibited the highest correlation with MRP3 mRNA. MRP4 mRNA correlated with 9 transcription factors including PPARα, LXRα, GR, CAR, and HNF1α.
Figure 2. Correlation of MRP2-4 and transcription factor mRNA expression.
Comparisons between each MRP gene and a panel of transcription factors were determined by linear regression and presented as R-values. Statistical significance (*) was set at p < 0.005.
The highest R-value for correlation with MRP2 was CAR (0.853), with MRP3 was HNF1α (0.740), and with MRP4 was PPARα (0.725). R-values for the remaining transcription factors were variable. Despite the conservative limit for determining statistical significance among the regressions, the linear regression procedure did not identify a single transcription factor that best correlated with MRP mRNA expression.
Coefficient of partial determination (stepwise selection of partial R2)
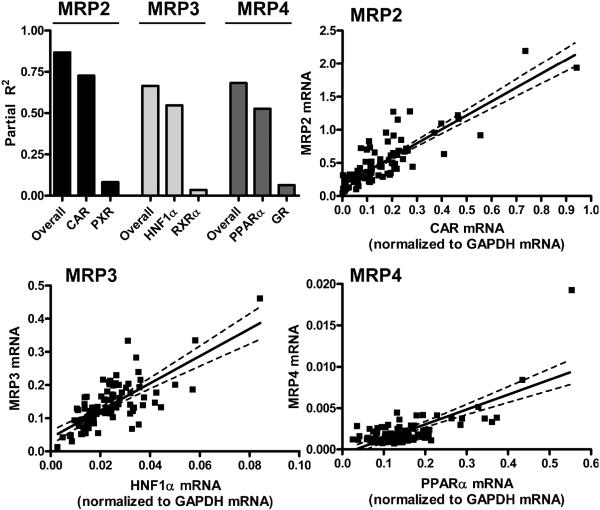
In order to rank candidate regulatory genes for each transporter, stepwise regression was used to compute partial R2 values. The stepwise procedure is a robust method of approximating a single linear function that includes multiple, statistically significant, independent variables. When the stepwise procedure is complete, the overall model R2 value is presented as the sum of the individual partial R2 values which are representative of each of the transcription factors included in the statistical model. The MRP2 stepwise regression model yielded the highest R2 of 0.87, meaning that 87% of the variability in MRP2 was explained by transcription factor mRNA levels. The MRP3 and MRP4 stepwise regression models exhibited overall model R2 values of 0.66 and 0.68, respectively.
The stepwise ranking procedure identifies the single independent variable, in this case the transcription factor, whose mRNA expression is most predictive of MRP expression relative to all other transcription factors within the multivariate linear model. With respect to each individual MRP gene, there was one transcription factor that comparatively best explained MRP expression. To distinguish between the top two transcription factors identified in each MRP stepwise regression, the pair of transcription factors were plotted in comparison to the overall model R2 (Figure 3). CAR was selected as the principal transcription factor (partial R2 = 0.73) in the MRP2 multivariate model, whereas HNF1α (partial R2 = 0.55) and PPARα (partial R2 = 0.53) were identified as the factors that most accounted for MRP3 and MRP4 expression, respectively. PXR, RXRα, and GR were selected second in the stepwise models for MRP2, 3, and 4, respectively. Linear regressions for MRP2-4 and the most correlative transcription factors (CAR, HNF1α and PPARα, respectively) in 90 adult human livers are shown in Figure 3.
Figure 3. MRP:transcription factor interactions.
Correlative transcription factors were ranked using stepwise selection and the factors with the highest partial R2 values are presented. Partial R2 values for each transcription factor are plotted in comparison to the overall model R2 on a scale of 0-1.0. Linear regressions for MRP2-CAR, MRP3-HNF1α, and MRP4-PPARα mRNA are shown. Data are presented as mean mRNA expression (normalized to GAPDH mRNA). Confidence intervals (95%) are depicted as dashed lines.
Upon further examination, there was no relationship between the MRP residuals and correlative transcription factor mRNA expression (data not shown). The residuals are the difference between actual MRP mRNA expression and predicted expression from the linear regression. The residual plots confirmed that the variability of MRP mRNA expression is random about the linear regression model and the residuals are not dependent on correlative transcription factor mRNA expression.
Partial correlation coefficient
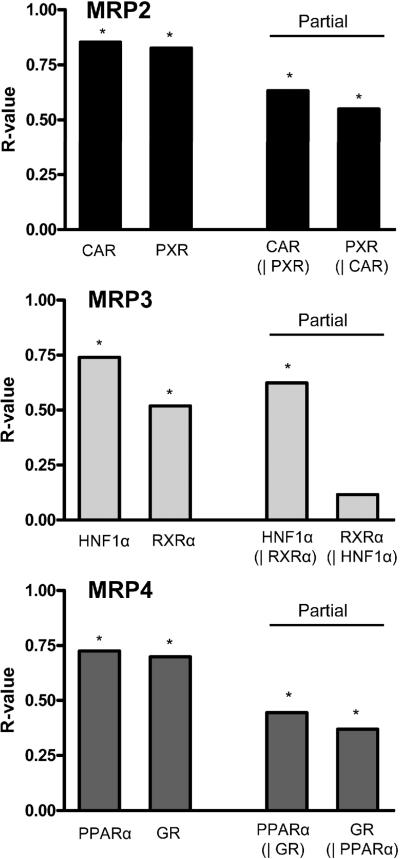
First order partial correlation coefficient analysis has been used in previous studies to identify master gene regulators of various cytochrome P450 enzymes and transporters in human liver (Wortham et al. 2007). For the purpose of this study, partial correlation coefficients were calculated for the two most correlative transcription factors for each MRP gene, as identified in the stepwise selection analysis (Figure 3). Partial correlation coefficients were determined for each receptor-MRP association by statistically controlling for the variability of the second receptor. Thus, the presence of a cooperative relationship and/or co-regulation between the transcription factors was tested. For example, by holding PXR constant, the influence of PXR on the relationship between CAR and MRP2 could be determined. It is important to further explore the inter-relationships between the transcription factors as MRP expression may be influenced indirectly via receptor-receptor interactions.
Correlation coefficients (linear regressions) and partial correlation coefficients for the two transcription factors with the highest correlation to each MRP gene are presented in Figure 4. CAR and PXR correlations to MRP2 are both reduced in the respective partial correlation analyses; however, the associations did not lose statistical significance (p < 0.005). The same trend was observed for MRP4, where each transcription factor-transporter correlation was reduced upon holding the variability of one receptor constant, however statistical significance was unchanged. The significant correlation between MRP3 and RXRα was lost upon controlling for the variability in HNF1α mRNA expression (R-value = 0.116, p = 0.280). This finding suggests that RXRα does not directly account for MRP3 variability, and that the association between RXRα and MRP3 revealed during the stepwise selection is more likely a result of the high correlation between HNF1α and RXRα (R-value = 0.618, p < 0.0001).
Figure 4. Transcription factor interrelationships and correlation to MRP2-4 mRNA.
Correlation coefficients (R-values) obtained from linear regression analysis in Figure 1 are presented for MRP2 (CAR and PXR), MRP3 (HNF1α and RXRα), and MRP4 (PPARα and GR) mRNA. Partial correlation coefficients (R-values) were calculated for each MRP-transcription factor association, controlling for variability in the second transcription factor. The symbol | denotes the partial variable. Statistical significance (*) was set at p < 0.005.
Discussion
This manuscript investigated for the first time the influence of variation in mRNA expression of transcription factors with basal levels of hepatic MRP2-4 in 90 adult human liver specimens. Linear regression identified significant correlations between the mRNA expression of 9 to 12 transcription factors for each MRP gene. Using a stepwise selection of partial R2 values, we were able to identify two transcription factors whose expression best correlated with the mRNA levels of each MRP. As exhibited in Figure 3, the combined expression of CAR and PXR predicted 81% of the variability in MRP2 levels in human livers. Similarly, HNF1α and RXRα expression accounted for 58% of the variability in MRP3 mRNA. Lastly, 59% of the variance in MRP4 expression was explained by PPARα and GR levels. In each of these cases, one transcription factor accounted for a significantly higher percentage of the variability in the corresponding MRP mRNA expression. Subsequent analysis was undertaken to identify potential relationships between each pair of transcription factors and the cooperative ability of these two transcription factors to influence MRP levels. CAR and PXR independently correlated with MRP2. Similarly, PPARα and GR independently correlated with MRP4. Interestingly, the correlation of the heterodimer partner RXRα and MRP3 was dependent on HNF1α expression. This interrelationship has not been previously reported and may reflect coordinated regulation of all three genes by an unknown upstream transcription factor.
In comparison to linear regression determination, the partial R2 analysis provided a clear distinction between the strength of relationships for CAR-MRP2 and PXR-MRP2. The transcription factors are directly compared to each other by combining multiple genes within a single MRP stepwise regression model. Zero-order linear regression yielded comparable correlation coefficients (R-values) for CAR-MRP2 (0.853) and PXR-MRP2 (0.826) (Figure 2), whereas the MRP2 partial R2 values for CAR (0.73) and PXR (0.08) were quite different (Figure 3). The inclusion of multiple, independent variables is not permitted in the zero-order linear regression procedure. By relying solely upon linear regression analysis, we would not have been able to differentiate the strength of relationships between CAR/PXR and MRP2. Although the highest correlation coefficients corresponded with the highest partial R2 value, the partial R2 analysis proved to be a much more robust procedure when it came to identifying and ranking the transcription factors that best explained the variability of MRP mRNA expression. Our use of first-order partial correlations is supported, in part, in a report by de la Fuente et al in which the false discovery rate (independent of statistical threshold, i.e., p = 0.01, 0.001 or 0.0001) using zero-order Pearson correlation analysis (linear regression) was approximately 40-50%. In their study, the false discovery rate was reduced to less than 15% when first-order partial correlations were used (de la Fuente et al. 2004). Because there are many regulatory factors to be considered in human drug metabolism (transcription factors, enzymes, transporters), a multivariate approach using first-order partial correlations is a useful tool to differentiate and rank gene-gene relationships.
Biological data support the associations observed between mRNA levels of MRP and nuclear receptor genes. For example, the rat Mrp2 gene contains a responsive element that is activated by co-transfection with the rat CAR gene (Kast et al. 2002). Treatment of mice with a daily dose of the CAR ligand TCPOBOP for 4 days induces hepatic mRNA expression for the CAR target, Cyp2b10, as well as Mrp2 (Maher et al. 2005). TCPOBOP induction of Mrp2 is absent in CAR-null mice (Huang et al. 2003). Little is known about the relationship between Mrp3 and HNF1α in rodent liver. However, HNF1α-null mice exhibit no basal differences in hepatic Mrp3 expression, suggesting that HNF1α may act through indirect means to regulate MRP3, or alternatively that constitutive rodent and human MRP3 are regulated via distinct pathways (Maher et al. 2006). As our data in Figure 4 suggest, the high correlation of MRP3 and HNF1α likely results from the strong interaction between HNF1α and RXRα. Lastly, there is evidence in mice that PPARα can regulate expression of Mrp4. The mouse Mrp4 gene contains multiple DR-1 elements in its upstream promoter region (Moffit et al. 2006). Treatment of mice with peroxisome proliferators, such as clofibrate and perfluorodecanoic acid, induces expression of Mrp4 mRNA and protein (Maher et al. 2005; Moffit et al. 2006). Using knockout mice, up-regulation of Mrp4 mRNA after clofibrate was shown to be dependent upon PPARα expression. Our analysis is particularly important because it not only confirms, but also extends prior research in rodent and in vitro models by comprehensively reviewing gene expression pathways in a relatively large database of human livers. The transcription factors identified as highly correlative to each MRP gene using stepwise selection may be biologically relevant.
There are some limitations of extrapolating rodent to human transcription factors and should be taken into consideration. Although the rodent and human findings are similar, it should be noted that the majority of the rodent data is generated using xenobiotic induction. Little is known about the dietary, environmental, and/or pharmaceutical exposure of humans from which the liver specimens were obtained. In addition, transcription factor function between human and rodent isoforms can be different. For example, transcriptional activation by rodent and human PXR show preferential responses to different sets of xenobiotics (rodent: pregnane 16α-carbonitrile, human: rifampicin) (Jones et al. 2000). There are also clear differences in rodent and human PPARα signaling. Whereas PPARα activation in mice causes peroxisome proliferation, increased liver weight, and liver carcinogenesis, human PPARα activation is not associated with these sequelae (Gonzalez and Shah 2008).
It is worth noting that the stepwise regression models did not select Nrf2 or AhR as a top transcription factor for any MRP gene. Although Nrf2 and AhR were included in the final stepwise models for MRP2 and MRP4, respectively, the partial R2 values were too low for further analysis using partial correlation coefficients. Possible AhR and Nrf2 co-regulation of transporter genes, such as Mrps, is an interesting research topic that has recently been reviewed (Kohle and Bock 2007). However, reports of Nrf2 or AhR mediated gene induction are commonly based on studies where rodents or cells are administered prototypical inducers such as oltipraz or 2,3,7,8-tetrachlorodibenzo-p-dioxin, respectively.
Recent work from our laboratory and others has identified a critical role for Nrf2 in regulating the inducible expression of mouse Mrp2-4 genes. This conclusion was based upon in vitro chromatin immunoprecipitation of Nrf2 bound to antioxidant response elements in the upstream promoter regions of these genes, along with in vivo experiments demonstrating the up-regulation of Mrp2-4 mRNA and protein in livers of wild-type mice treated with the Nrf2 activator, oltipraz. Importantly, inducible expression was lost in Nrf2-null mice (Maher et al. 2007). Likewise, increased levels of Mrp3 and Mrp4 mRNA and protein were observed in wild-type mice treated with a hepatotoxic dose of acetaminophen, with no change in Nrf2-null mice (Aleksunes et al. 2008). Given the data generated in this manuscript along with rodent studies it can be hypothesized that Nrf2 is more important in regulating the inducible expression of some MRP/Mrp genes rather than constitutive levels as studied in this human liver study. An alternative explanation for this discordance may be differences in regulation between species.
Co-regulation of nuclear receptor and MRP mRNA levels by upstream transcription factors is an alternate interpretation of these data. Recent work suggests that drug metabolic and excretion pathways are coordinately regulated. Therefore, it is possible that the relationships identified between MRP2-CAR, MRP3-HNF1α, and MRP4-PPARα are an extension of this coordination. Additional experimentation is necessary to dissect the direct and indirect signaling pathways in human hepatocytes and identify potential upstream regulatory mechanisms for the nuclear receptor-MRP pairs.
There are a number of advantages with the methods employed in the current study. Use of the stepwise selection of partial R2 values is a novel method that was able to discriminate amongst numerous transcription factors that appeared to predict MRP mRNA levels based upon linear regression. We conducted a second multivariate analysis including two likely unrelated genes (steroid receptor coactivator-1, delta-aminolevulinate synthase 1) in addition to the various transcription factors. Inclusion of these variables in the multivariate analysis did not alter the stepwise selection of CAR, HNF1α, and PPARα, for MRP2, 3, and 4, respectively (unpublished observations). This suggests that the approach presented in this manuscript can discriminate variables when unlikely correlates are included. In addition, this unbiased study included all 90 samples with diversity in gender, age, and ethnicity. Although basic demographic information was provided for the donors, undiagnosed comorbid conditions as well as xenobiotic induction are potential confounders to the data interpretation that cannot be controlled.
Spliced transcripts and allelic variants have been previously identified for CAR, PPARα, and HNF1α (Ringeisen et al. 1993; Auerbach et al. 2003; Flavell et al. 2005). The presence of these variants in the current panel of 90 liver samples was not investigated and could explain some variability in expression of MRP2-4. Polymorphisms in a particular MRP gene may have also impacted our analysis (Lee et al. 2004; Meier et al. 2006; Fukushima-Uesaka et al. 2007). For example, two polymorphisms in MRP2 (V1188E, C1515Y) are associated with increased expression of MRP2 in human liver (Meier et al. 2006). The presence of these variants in this human liver panel is one possible explanation for specimens that did not show correlation between CAR and MRP2 mRNA levels.
It is the intent of this project to identify correlations between levels of transcription factors and MRP genes with the long-term goal to better predict an individual's response to drug therapy (both efficacy and side effects). Future studies need to determine whether the identified transcription factor:MRP relationships occur via direct gene activation because a causal relationship cannot be definitively made from the current analysis. Along these lines, the protein expression and functional activity of MRP2-4 in panels of liver specimens need to be quantified in order to correlate nuclear receptor mRNA expression with transport activity. The methods used in the current study may serve as a novel approach for additional investigations into the regulation of biotransformation enzymes as well as other hepatobiliary transporters in normal adult human liver.
Table 1.
Sample demographics (Xenotech, Lenexa, KS)
| Gender | |
|---|---|
| Male | 52 |
| Female | 38 |
| Ethnic Descent | |
|---|---|
| Caucasian | 69 |
| African American | 9 |
| Hispanic | 9 |
| Asian | 3 |
| Age category (years) | |
|---|---|
| 17-39 | 19 |
| 40-59 | 46 |
| 60+ | 25 |
Table 2.
Gene accession numbers
| Gene | Species | Accession Number | Symbol |
|---|---|---|---|
| MRP2 | Human | NM_000392 | ABCC2 |
| MRP3 | Human | NM_003786 | ABCC3 |
| MRP4 | Human | NM_005845 | ABCC4 |
| AhR | Human | NM_001621 | ARR |
| CAR | Human | NM_005122 | NR1I3 |
| FXR | Human | NM_005123 | NR1H4 |
| GR | Human | NM_000176 | NR3C1 |
| HNF1α | Human | NM_000545 | TCF1 |
| HNF4α | Human | NM_000457 | HNF4A |
| LXRα | Human | NM_005693 | NR1H3 |
| Nrf2 | Human | NM_006164 | NFE2L2 |
| PPARα | Human | NM_005036 | PPARA |
| PPARγ | Human | NM_138712 | PPARG |
| RXRα | Human | NM_002957 | RXRA |
| PXR | Human | NM_003889 | NR1I2 |
| GAPDH | Human | NM 002046 | GAPD |
Acknowledgments
The authors thank Dr. David Buckley for isolation of human RNA and gene analysis, Dr. Jon Maher for analysis of nucleotide similarity between mouse and human MRP2-4 genes, and Dr. Matthew Mayo for assistance with statistical analysis. This work was supported by NIH grants ES-007079, ES-009716, and DK-080774.
Non-Standard Abbreviations
- AhR
aryl hydrocarbon receptor
- CAR
constitutive androstane receptor
- FXR
farnesoid X receptor
- GR
glucocorticoid receptor
- HNF
hepatocyte nuclear factors
- LXR
liver X receptor
- MRP
multidrug resistance-associated proteins
- Nrf2
nuclear factor E2-related factor 2
- PPAR
peroxisome proliferator activated receptor
- PXR
pregnane X receptor
- RXRα
retinoid X receptor alpha
Footnotes
Declaration of Interest The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Aleksunes LM, Slitt AL, Maher JM, Augustine LM, Goedken MJ, Chan JY, Cherrington NJ, Klaassen CD, Manautou JE. Induction of Mrp3 and Mrp4 transporters during acetaminophen hepatotoxicity is dependent on Nrf2. Toxicol Appl Pharmacol. 2008;226:74–83. doi: 10.1016/j.taap.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach SS, Ramsden R, Stoner MA, Verlinde C, Hassett C, Omiecinski CJ. Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Research. 2003;31:3194–3207. doi: 10.1093/nar/gkg419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;10:267–282. [PubMed] [Google Scholar]
- de la Fuente A, Bing N, Hoeschele I, Mendes P. Discovery of meaningful associations in genomic data using partial correlation coefficients. Bioinformatics. 2004;20:3565–3574. doi: 10.1093/bioinformatics/bth445. [DOI] [PubMed] [Google Scholar]
- Flavell DM, Ireland H, Stephens JW, Hawe E, Acharya J, Mather H, Hurel SJ, Humphries SE. Peroxisome proliferator-activated receptor alpha gene variation influences age of onset and progression of type 2 diabetes. Diabetes. 2005;54:582–586. doi: 10.2337/diabetes.54.2.582. [DOI] [PubMed] [Google Scholar]
- Fukushima-Uesaka H, Saito Y, Maekawa K, Hasegawa R, Suzuki K, Yanagawa T, Kajio H, Kuzuya N, Noda M, Yasuda K, Tohkin M, Sawada J. Genetic variations of the ABC transporter gene ABCC3 in a Japanese population. Drug Metab Pharmacokinet. 2007;22:129–135. doi: 10.2133/dmpk.22.129. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Shah YM. PPARalpha: mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 2008;246:2–8. doi: 10.1016/j.tox.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhang J, Chua SS, Qatanani M, Han Y, Granata R, Moore DD. Induction of bilirubin clearance by the constitutive androstane receptor (CAR) Proc Natl Acad Sci U S A. 2003;100:4156–4161. doi: 10.1073/pnas.0630614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SA, Moore LB, Shenk JL, Wisely GB, Hamilton GA, McKee DD, Tomkinson NC, LeCluyse EL, Lambert MH, Willson TM, Kliewer SA, Moore JT. The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol Endocrinol. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, Tontonoz P, Kliewer S, Willson TM, Edwards PA. Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J Biol Chem. 2002;277:2908–2915. doi: 10.1074/jbc.M109326200. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Slitt AL. Regulation of hepatic transporters by xenobiotic receptors. Curr Drug Metab. 2005;6:309–328. doi: 10.2174/1389200054633826. [DOI] [PubMed] [Google Scholar]
- Kohle C, Bock KW. Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem Pharmacol. 2007;73:1853–1862. doi: 10.1016/j.bcp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Lagas JS, Vlaming ML, van Tellingen O, Wagenaar E, Jansen RS, Rosing H, Beijnen JH, Schinkel AH. Multidrug resistance protein 2 is an important determinant of paclitaxel pharmacokinetics. Clin Cancer Res. 2006;12:6125–6132. doi: 10.1158/1078-0432.CCR-06-1352. [DOI] [PubMed] [Google Scholar]
- Lee YM, Cui Y, Konig J, Risch A, Jager B, Drings P, Bartsch H, Keppler D, Nies AT. Identification and functional characterization of the natural variant MRP3-Arg1297His of human multidrug resistance protein 3 (MRP3/ABCC3) Pharmacogenetics. 2004;14:213–223. doi: 10.1097/00008571-200404000-00001. [DOI] [PubMed] [Google Scholar]
- Maher JM, Cheng X, Slitt AL, Dieter MZ, Klaassen CD. Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab Dispos. 2005;33:956–962. doi: 10.1124/dmd.105.003798. [DOI] [PubMed] [Google Scholar]
- Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, Dalton TP, Yamamoto M, Klaassen CD. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology. 2007;46:1597–1610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- Maher JM, Slitt AL, Callaghan TN, Cheng X, Cheung C, Gonzalez FJ, Klaassen CD. Alterations in transporter expression in liver, kidney, and duodenum after targeted disruption of the transcription factor HNF1alpha. Biochem Pharmacol. 2006;72:512–522. doi: 10.1016/j.bcp.2006.03.016. [DOI] [PubMed] [Google Scholar]
- Manautou JE, de Waart DR, Kunne C, Zelcer N, Goedken M, Borst P, Elferink RO. Altered disposition of acetaminophen in mice with a disruption of the Mrp3 gene. Hepatology. 2005;42:1091–1098. doi: 10.1002/hep.20898. [DOI] [PubMed] [Google Scholar]
- Meier Y, Pauli-Magnus C, Zanger UM, Klein K, Schaeffeler E, Nussler AK, Nussler N, Eichelbaum M, Meier PJ, Stieger B. Interindividual variability of canalicular ATP-binding-cassette (ABC)-transporter expression in human liver. Hepatology. 2006;44:62–74. doi: 10.1002/hep.21214. [DOI] [PubMed] [Google Scholar]
- Mennone A, Soroka CJ, Cai SY, Harry K, Adachi M, Hagey L, Schuetz JD, Boyer JL. Mrp4-/- mice have an impaired cytoprotective response in obstructive cholestasis. Hepatology. 2006;43:1013–1021. doi: 10.1002/hep.21158. [DOI] [PubMed] [Google Scholar]
- Moffit JS, Aleksunes LM, Maher JM, Scheffer GL, Klaassen CD, Manautou JE. Induction of hepatic transporters multidrug resistance-associated proteins (Mrp) 3 and 4 by clofibrate is regulated by peroxisome proliferator-activated receptor alpha. J Pharmacol Exp Ther. 2006;317:537–545. doi: 10.1124/jpet.105.093765. [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringeisen F, Rey-Campos J, Yaniv M. The transactivation potential of variant hepatocyte nuclear factor 1 is modified by alternative splicing. J Biol Chem. 1993;268:25706–25711. [PubMed] [Google Scholar]
- Schrem H, Klempnauer J, Borlak J. Liver-enriched transcription factors in liver function and development. Part I: the hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol Rev. 2002;54:129–158. doi: 10.1124/pr.54.1.129. [DOI] [PubMed] [Google Scholar]
- Slitt AL, Allen K, Morrone J, Aleksunes LM, Chen C, Maher JM, Manautou JE, Cherrington NJ, Klaassen CD. Regulation of transporter expression in mouse liver, kidney, and intestine during extrahepatic cholestasis. Biochim Biophys Acta. 2007;1768:637–647. doi: 10.1016/j.bbamem.2006.10.008. [DOI] [PubMed] [Google Scholar]
- van de Wetering K, Zelcer N, Kuil A, Feddema W, Hillebrand M, Vlaming ML, Schinkel AH, Beijnen JH, Borst P. Multidrug resistance proteins 2 and 3 provide alternative routes for hepatic excretion of morphine-glucuronides. Mol Pharmacol. 2007;72:387–394. doi: 10.1124/mol.107.035592. [DOI] [PubMed] [Google Scholar]
- Vlaming ML, Mohrmann K, Wagenaar E, de Waart DR, Elferink RP, Lagas JS, van Tellingen O, Vainchtein LD, Rosing H, Beijnen JH, Schellens JH, Schinkel AH. Carcinogen and anticancer drug transport by Mrp2 in vivo: studies using Mrp2 (Abcc2) knockout mice. J Pharmacol Exp Ther. 2006;318:319–327. doi: 10.1124/jpet.106.101774. [DOI] [PubMed] [Google Scholar]
- Wang H, LeCluyse EL. Role of orphan nuclear receptors in the regulation of drug-metabolising enzymes. Clin Pharmacokinet. 2003;42:1331–1357. doi: 10.2165/00003088-200342150-00003. [DOI] [PubMed] [Google Scholar]
- Wortham M, Czerwinski M, He L, Parkinson A, Wan YJ. Expression of constitutive androstane receptor, hepatic nuclear factor 4{alpha}, and P450 oxidoreductase genes determines interindividual variability in basal expression and activity of a broad scope of xenobiotic metabolism genes in the human liver. Drug Metab Dispos. 2007;35:1700–1710. doi: 10.1124/dmd.107.016436. [DOI] [PubMed] [Google Scholar]
- Zamek-Gliszczynski MJ, Nezasa K, Tian X, Bridges AS, Lee K, Belinsky MG, Kruh GD, Brouwer KL. Evaluation of the role of multidrug resistance-associated protein (Mrp) 3 and Mrp4 in hepatic basolateral excretion of sulfate and glucuronide metabolites of acetaminophen, 4-methylumbelliferone, and harmol in Abcc3-/- and Abcc4-/- mice. J Pharmacol Exp Ther. 2006;319:1485–1491. doi: 10.1124/jpet.106.110106. [DOI] [PubMed] [Google Scholar]
- Zelcer N, van de Wetering K, Hillebrand M, Sarton E, Kuil A, Wielinga PR, Tephly T, Dahan A, Beijnen JH, Borst P. Mice lacking multidrug resistance protein 3 show altered morphine pharmacokinetics and morphine-6-glucuronide antinociception. Proc Natl Acad Sci U S A. 2005;102:7274–7279. doi: 10.1073/pnas.0502530102. [DOI] [PMC free article] [PubMed] [Google Scholar]