Abstract
Introduction
Gene profiling may improve prognostic accuracy in patients with early breast cancer, but this technology is not widely available. We used commercial assays for qRT-PCR to assess the performance of the gene profiles included in the 70-Gene Signature, the Recurrence Score and the Two-Gene Ratio.
Methods
153 patients with early breast cancer and a minimum follow-up of 5 years were included. All tumours were positive for hormonal receptors and 38% had positive lymph nodes; 64% of patients received adjuvant chemotherapy. RNA was extracted from formalin-fixed paraffin-embedded (FFPE) specimens using a specific kit. qRT-PCR amplifications were performed with TaqMan Gene Expression Assays products. We applied the three gene-expression-based models to our patient cohort to compare the predictions derived from these gene sets.
Results
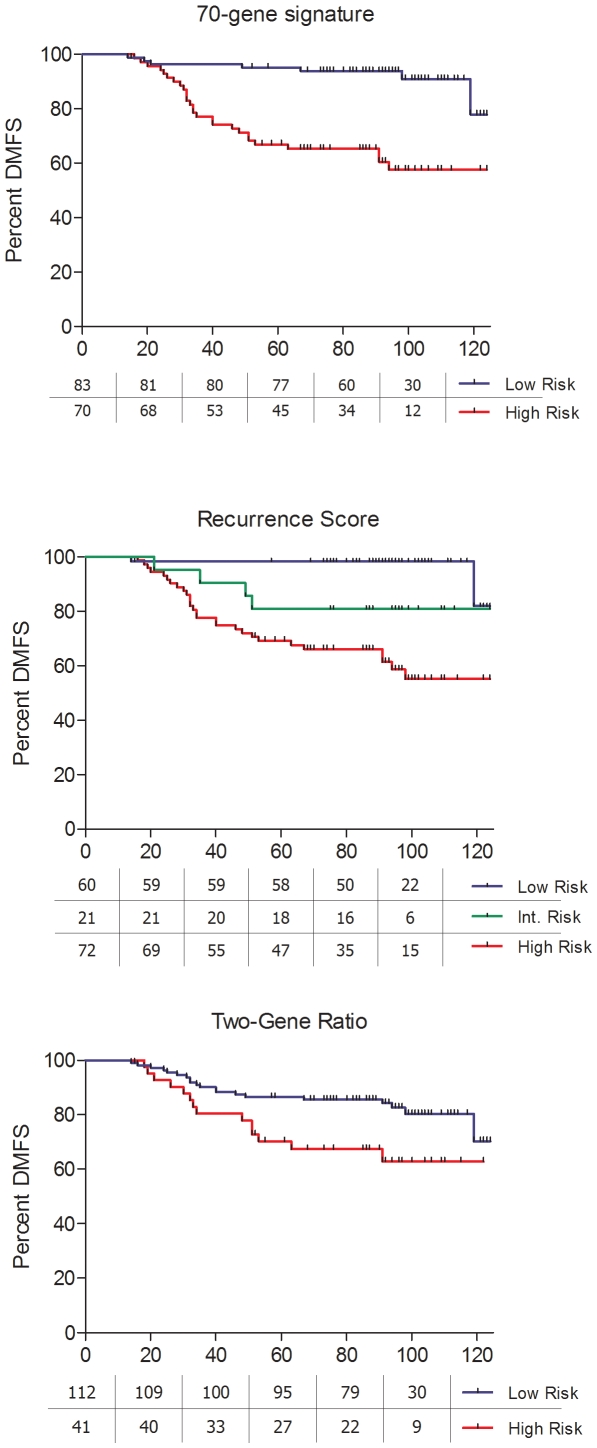
After a median follow-up of 91 months, 22% of patients relapsed. The distant metastasis-free survival (DMFS) at 5 years was calculated for each profile. For the 70-Gene Signature, DMFS was 95% -good prognosis- versus 66% -poor prognosis. In the case of the Recurrence Score, DMFS was 98%, 81% and 69% for low, intermediate and high-risk groups, respectively. Finally, for the Two-Gene Ratio, DMFS was 86% versus 70%. The 70-Gene Signature and the Recurrence Score were highly informative in identifying patients with distant metastasis, even in multivariate analysis.
Conclusion
Commercially available assays for qRT-PCR can be used to assess the prognostic utility of previously published gene expression profiles in FFPE material from patients with early breast cancer. Our results, with the use of a different platform and with different material, confirm the robustness of the 70-Gene Signature and represent an independent test for the Recurrence Score, using different primer/probe sets.
Introduction
A key area in the management of women with early breast cancer is the selection of adjuvant therapy, which depends on the use of prognostic and predictive factors. Chemotherapy is usually recommended in the presence of adverse factors, such as positive lymph nodes, size>1 cm, histological grade>1 or negative hormonal receptors. These factors are included in guidelines or specific software to help decision making [1], [2], but using these criteria leads to unnecessary treatment in many women, either because they would not relapse in the absence of adjuvant chemotherapy or because they would suffer a relapse anyway [3].
Gene expression profiles may improve prognostic and predictive information in breast cancer patients. Two of these profiles, the 70-Gene Signature (MammaPrint™) and the Recurrence Score (OncoType DX™) are being evaluated in phase III studies, with randomization based on the results of the assays [4], [5]. The 70-Gene Signature is suitable for patients with either ER-positive or ER-negative tumours, whereas the Recurrence Score has to be used in ER-positive tumours. Another group has reported a Two-Gene Ratio (HOXB13/IL17BR) predicting disease-free survival in patients with early-stage, ER-positive breast cancer who received adjuvant tamoxifen [6]. An RT-PCR based method to assess this ratio from paraffin-embedded tissue samples is now commercially available (Theros H/I; bioTheranostics).
Some centres already use gene profiles to identify patients with very low risk of relapse who may not need adjuvant chemotherapy. The major restraints for the widespread application of such tests are some reservations regarding their cost/effectiveness ratio and the lack of availability, because samples must be sent to central laboratories for processing. The need for fresh-frozen (FF) material in some cases adds to these restraints, as the process of collecting, processing and storing FF samples for large scale studies is difficult. However, the use of gene profiles would be facilitated if centralized processing was not required and formalin-fixed paraffin-embedded (FFPE) samples could be used. FFPE samples are stable at room temperature, easily storable and, most importantly, they constitute a widely available archive of clinical samples linked to clinical information.
In the present study, we assessed the performance of the above referred gene expression profiles by using commercially available assays for qRT-PCR in FFPE samples.
Results
One hundred fifty-three patients diagnosed between February-95 and March-03 were included. Table 1 shows their clinical features. Median age was 58 years and median follow-up was 91 months for the whole group. Sixty-six patients (43%) had a mastectomy, whereas the remaining underwent a conservative surgery followed by adjuvant radiation. All patients received adjuvant tamoxifen for five years and 97 (63%) underwent adjuvant chemotherapy. Thirty-four patients (22%) had a distant relapse, of which 17 died and 7 were lost for follow-up after the relapse. Among 119 patients without distant relapse, four had a local/regional recurrence successfully treated with surgery.
Table 1. Clinical characteristics of the patients.
| Number of patients (percentage) | |
| Age | Median 58, range: 29–82 |
| T | |
| 1 | 77 (50.3%) |
| 2 | 76 (49.7%) |
| N | |
| 0 | 96 (62.7%) |
| 1 | 57 (37.3%) |
| Stage | |
| I | 61 (39.9%) |
| IIa | 51 (33.3%) |
| IIb | 41 (26.8%) |
| Hormone receptors | |
| ER+/PgR− | 30 (19.6%) |
| ER+/PgR+ | 110 (71.9%) |
| ER+/PgR unknown | 12 (7.9%) |
| ER−/PgR+ | 1 (0.7%) |
| Grade | |
| 1 | 29 (18.9%) |
| 2 | 64 (41.8%) |
| 3 | 59 (38.6%) |
| x | 1 (0.7%) |
| Chemotherapy | |
| No chemotherapy | 56 (36.6%) |
| CMF | 42 (27.4%) |
| Anthracycline-based | 55 (35.9%) |
All tumours were positive for oestrogen and/or progesterone receptors. All patients had received adjuvant tamoxifen.
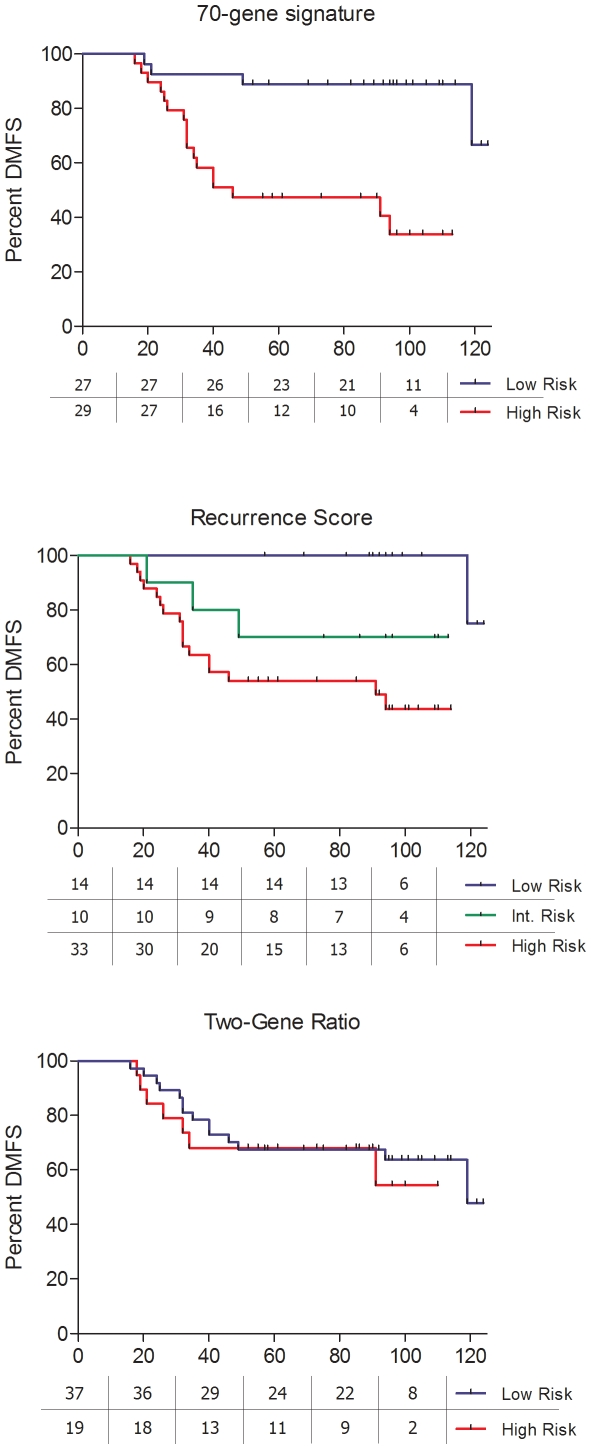
qRT-PCR reactions worked correctly for all genes included in the Recurrence Score, both genes of the Two-Gene Ratio, and all 60 we could include from the 70-Gene Signature. Raw data appear in supplementary Table S1. In the univariate analysis, lymph-node status, tumour grade, size and the three gene profiles were significant predictors of DMFS (lymph-node status p = 0.001; tumour grade p<0.001; size p = 0.002; 70-Gene Signature p<0.001; Recurrence Score<0.001; Two-gene Ratio p = 0.023). DMFS at five years for every profile was as follows: for the 70-Gene Signature, 95% good prognosis versus 66% poor prognosis; for the Recurrence Score profile, 98% low-risk versus 81% intermediate risk versus 69% high-risk; and in the case of the Two-Gene Ratio, 86% favourable versus 70% unfavourable (figure 1). We restricted the univariate analysis to patients who did not receive adjuvant chemotherapy and the three profiles still found significant differences (figure 2).
Figure 1. Kaplan-Meier plots for distant metastasis-free survival for the whole group.
Figure 2. Survival analysis in patients who did not receive chemotherapy.
Cramer's V statistic was performed in the profile-to-profile comparison and, for this purpose, we combined the low and intermediate Recurrence Score categories. The concordance of the Two-Gene Ratio with either the 70-Gene Signature or the Recurrence Score was 0.3 in both cases. Concordance was 0.6 between the 70-Gene Signature and the Recurrence Score, indicating a strong correlation. Most tumours classified as having a low risk of recurrence by one the three models were classified as such by the other two, although the low-risk group defined by the Two-Gene Ratio was poorly predicted by the 70-Gene Signature and the Recurrence Score. When comparing high risk tumours, again there was a good correlation between the 70-Gene Signature and the Recurrence Score, but rather poor for the Two-Gene Ratio. Table 2 shows the intersection between profiles.
Table 2. Concordance among the three profiles.
| Good prognosis by… | Coincidence cases | Coincidence cases |
| 70-Gene Signature | Recurrence Score | Two-Gene Ratio |
| n = 83; 54% | n = 66; 79% | n = 71; 85% |
| Recurrence Score | 70-Gene Signature | Two-Gene Ratio |
| n = 81; 53% | n = 66; 81% | n = 68; 84% |
| Two-Gene Ratio | 70-Gene Signature | Recurrence Score |
| n = 112; 73% | n = 71; 63% | n = 68; 61% |
| Poor prognosis by… | Coincidence cases | Coincidence cases |
| 70-Gene Signature | Recurrence Score | Two-Gene Ratio |
| n = 70; 46% | n = 55; 78% | n = 29; 41% |
| Recurrence Score | 70-Gene Signature | Two-Gene Ratio |
| n = 72; 47% | n = 55: 76% | n = 28; 39% |
| Two-Gene Ratio | 70-Gene Signature | Recurrence Score |
| n = 41; 27% | n = 29; 71% | n = 28; 68% |
To assess the discrimination capability of each prognostic profile at 5 years, Harrell's bias corrected concordance index was calculated. The values were: Recurrence Score = 0.73>70-Gene Signature = 0.70>Two-Gene Ratio = 0.59.
We also performed a multivariate Cox proportional hazards analysis to evaluate the prognostic value of each gene-expression based model individually. The analysis also included tumour size, nodal status and tumour grade. The 70-Gene Signature and the Recurrence Score were significant predictors of DMFS (Table S2, supporting information), indicating that those gene expression profiles added important prognostic information beyond that provided by clinical factors. The Two-Gene Ratio was not a significant predictor in the multivariate analysis. Node status remained the only clinical factor with significant value in all cases. Histological grade was significant for the cases of Recurrence Score (p = 0.043) and Two-Gene Ratio (p = 0.002).
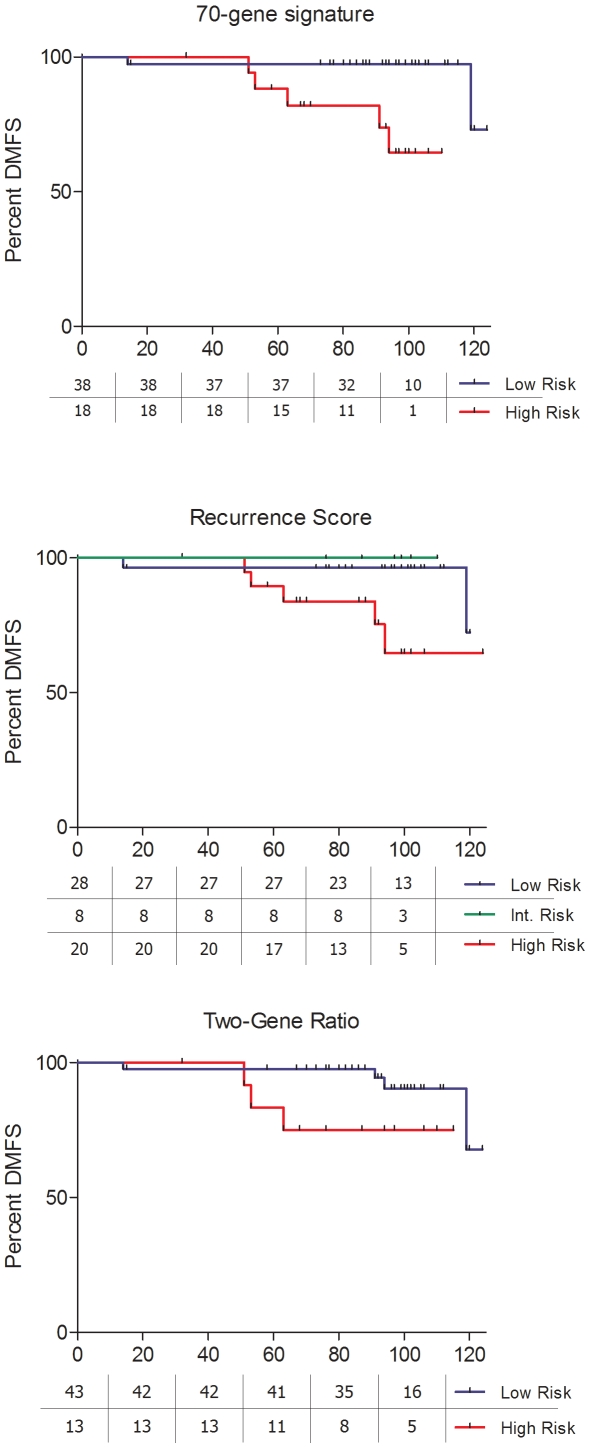
Likewise, the 70-Gene Signature and the Recurrence Score detected striking differences for the subgroup of patients with positive lymph nodes, whereas differences were not significant in the case of the Two-Gene Ratio (figure 3).
Figure 3. Survival analysis in patients with positive lymph nodes.
Adjuvant! Online estimated that chemotherapy would reduce the rate of relapse by less than 5% in 37% of patients. A threshold of 5% was selected because it is used by many clinicians to discourage the use of chemotherapy. By contrast, the three gene profiles allocated more patients in low-risk groups: 46% the 70-Gene Signature, 53% the Recurrence Score (low + intermediate risk) and 73% the Two-Gene Ratio. The multivariate analysis including Adjuvant! Online showed that both the 70-Gene Signature and the Recurrence Score remained significant (p<0.005), while the Two-Gene Ratio was not (p = 0.078) (Table 3). In our population, the Nottingham Prognosis Index did not show significant value to define prognosis in the univariate analysis (p = 0.061), so that it was not included in the multivariate analysis.
Table 3. Multivariate analysis comparing Adjuvant! Online with gene expression classifiers.
| Adjuvant! Online | |||
| HR | CI 95% | p value | |
| Adjuvant! Online | 10.814 | 2.589–45.173 | 0.001 |
| 70-Gene Signature | 4.218 | 1.800–9.883 | 0.001 |
| Adjuvant! Online | 7.458 | 1.764–31.533 | 0.006 |
| Recurrence Score | 0.003 | ||
| (Interm. risk vs. Low risk) | 4.132 | 0.745–22.906 | 0.105 |
| (High risk vs. Low risk) | 10.098 | 2.343–43.518 | 0.002 |
| Adjuvant! Online | 6.451 | 1.529–27.226 | 0.011 |
| Two-Gene Ratio | 1.864 | 0.932–3.727 | 0.078 |
| Adjuvant! Online | 10.051 | 2.400–42.101 | 0.002 |
The gain in predictive accuracy from adding each of the analyzed signatures to the Nottingham Prognostic Index and Adjuvant! Online clinical systems is presented in Table 4. The individual performance for DMFS of Nottingham Prognostic Index and Adjuvant! Online was comparable (0.239 and 0.236, respectively). The performance of the 70-Gene Signature (0.234) was in line with that of both clinical staging systems, whereas the Recurrence Score showed the best performance (0.225). The addition of the 70-Gene Signature and the Recurrence Score to any of the clinical staging systems decreased their predictive inaccuracy. The gain by adding the 70-Gene Signature in explained variation was 9.2%, and over 10% for the Recurrence Score in this dataset. The Two-Gene Ratio did not improve the clinical staging systems explained variation.
Table 4. Explained variation and predictive inaccuracy for DMFS.
| Predicitve inaccuracy | Explained variation (%) | |
| No Predictor | 0.275 | |
| NPI | 0.239±0.006 | 13.3±1.0 |
| Adjuvant! | 0.236±0.002 | 13.9±0.9 |
| 70-Gene S. | 0.234±0.003 | 14.8±1.0 |
| RS | 0.225±0.003 | 18.1±0.9 |
| Two-Gene Ratio | 0.264±0.002 | 3.4±0.9 |
| NPI & 70-Gene S. | 0.213±0.004 | 22.4±1.3 |
| Adjuvant! & 70-Gene S. | 0.211±0.003 | 23.1±1.2 |
| NPI & RS | 0.210±0.005 | 23.7±1.5 |
| Adjuvant! & RS | 0.204±0.004 | 25.8±1.4 |
| NPI & Two-Gene Ratio | 0.237±0.006 | 13.8±1.5 |
| Adjuvant! & Two-Gene Ratio | 0.232±0.004 | 15.5±1.5 |
| Gain by adding 70-Gene S. to NPI | 0.025 | 9.2 |
| Gain by adding 70-Gene S. to Adjuvant! | 0.025 | 9.2 |
| Gain by adding RS to NPI | 0.028 | 10.4 |
| Gain by adding RS to Adjuvant! | 0.033 | 11.8 |
| Gain by adding Two-Gene Ratio to NPI | 0.001 | 0.5 |
| Gain by adding Two-Gene Ratio to Adjuvant! | 0.004 | 1.5 |
NPI: Nottingham Prognostic Index, RS: Recurrence Score, 70-Gene S.: 70-Gene Signature. Data presented as the mean±standard error.
Discussion
Gene profiles help to determine prognosis in patients with early breast cancer and some of them are being used to make clinical decisions. If gene profiles become part of the standard pathological workup in early breast cancer in the future, general availability will be required and the use of FFPE material would be very useful. Several studies have demonstrated the feasibility of using FFPE tissues to perform gene expression profiling by qRT-PCR. Although RNA degradation leads to a loss of amplifiable templates, optimized normalization strategies could effectively compensate for this bias [4], [7], [8].
We compared the performance of three gene profiles -70-Gene Signature, Recurrence Score and Two-Gene Ratio- in patients with early breast cancer. The material was FFPE tissue and the technique qRT-PCR in all cases, in an attempt to simplify technical procedures. The 70-Gene Signature was initially described in young women who had not received adjuvant therapy, but further research has confirmed its validity in other groups [5], [9], [10], [11], and its applicability in patients with either positive or negative hormonal receptors [12]. On the other hand, the Recurrence Score is aimed at tumours expressing hormonal receptors and provides prognostic and predictive information in patients treated with tamoxifen [4], [13], [14]. The Two-Gene Ratio was developed and validated in early-stage hormonal receptor positive patients that had received adjuvant tamoxifen therapy [6], [15]. So the populations where each of these profiles was initially described vary according to their clinical, pathological and therapeutic features, thus hindering any direct comparison. In our study, the three profiles identified groups of patients with significant differences in DMFS, although only the 70-Gene Signature and the Recurrence Score offered significant value in the multivariate analysis, showing a high correlation between them. The other profile -Two-Gene Ratio- has recently been improved by incorporating five genes related to tumour grade [16]. This five-gene set was comparable to a more complex 97-gene genomic grade index in multiple data sets [17].
Clinical comparisons have not been performed among profiles, although one study by Fan et al. evaluated five gene sets in a data set of patients [18]. This study demonstrated significant agreement among four out of the five profiles, namely, intrinsic subtypes, 70-Gene Signature, Recurrence Score and Wound-Signature. The conclusion was that they probably track a common set of biologic phenotypes. In agreement with the study by Fan et al, we find a high concordance between the 70-Gene Signature and the Recurrence Score and a lack of reproducibility of the Two-Gene Ratio. However, there are important differences between both studies. Firstly, we hereby present an independent set of patients who received more aggressive therapy than those included in the other study; this could have improved outcome in our patients. Secondly, all of our samples derived from FFPE tissue. Thirdly, the technique we used was qRT-PCR for the three profiles, whereas Fan et al used the original assay for the 70-Gene Signature and translated the PCR values of the Recurrence Score into a microarray dataset. In other words, we used a different platform for the 70-Gene Signature and different, commercial available probes for the Recurrence Score. To our knowledge, our study is the first fully independent application of the Recurrence Score algorithm, because other investigators sent samples to a central laboratory for processing [19].
Fan et al. showed that microarray data would be applicable to assess the Recurrence Score and the Two-Gene Ratio (originally developed with PCR), so the opposite assumption may be also correct. On the other hand, we previously demonstrated that qRT-PCR can be used to assess the 70-Gene Signature profile [20]. Likewise, the present study proves that the 70-Gene Signature can also be determined from FFPE material. This work solves a technical difference that gave certain advantage to the Recurrence Score over the 70-Gene Signature in their attempt to burst in the clinic, i.e., the use of FFPE material. Our results with the use of a different platform and with different material confirm the robustness of the 70-Gene Signature and the Recurrence Score. In other words, the value of these profiles cannot be attributed to the use of a specific platform, but to the algorithms and the genes themselves. Our results are in agreement with those of other investigators seeing that previously reported prognostic signatures, despite differences in gene lists, carry similar prognostic information [21], [22].
In our series, gene profiles outperformed clinical data to identify patients with low risk of relapse. The use of Adjuvant! Online would have led to recommend chemotherapy in 63% of patients whereas few patients lied in high-risk groups determined by gene expression profiles. A recent study with hormone receptor-positive breast cancer and zero to three positive axillary nodes also demonstrated that the Recurrence Score is a more accurate predictor of relapse than standard features included in Adjuvant! Online [23].This is important considering that our series consisted of patients with small tumours and none or up to three positive lymph nodes, i.e., good prognosis tumours. Differences could have been bigger in a non-selected population including tumours with unfavourable clinical features, as shown in other studies [13], [24]. Although subgroup analysis must be viewed with caution, we also found that in patients with positive lymph nodes, a favourable 70-Gene Signature or a low-risk Recurrence Score was still associated with an excellent prognosis. Similar results were seen in the group not receiving chemotherapy.
Additionally, we showed that the Recurrence Score and the 70-Gene Signature improved the predictive accuracy of commonly used clinical systems, whereas the Two-Gene Ratio did not. This is relevant because experts in the field agree that these new techniques should provide more accurate information than classical factors to be incorporated into clinical practice. Moreover, these gene expression classifiers should not be regarded as a tool to replace standard pathological and clinical criteria, but should instead be integrated with clinical parameters [25].
Phase III trials have been initiated to determine whether two of them –the 70-Gene Signature and the Recurrence Score- may reduce the need for adjuvant chemotherapy in patients with low risk of relapse. Of course, this does not support the indiscriminate use of this new technology in the clinic, at least by present standards. But if phase III trials demonstrate the validity of either the 70-Gene Signature or the Recurrence Score, a substantial number of patients could be treated without adjuvant chemotherapy in the future. Patients with N0 disease would benefit first from this strategy, as many of them (particularly young women) are offered chemotherapy if there is an adverse factor, such as high grade or size >2 cm. There is also a possibility that some patients with positive lymph nodes could do so in the future. The field is evolving very rapidly and new contributions will be needed to improve the accuracy of the information provided by currently available profiles.
In summary, we verified the performance of some gene profiles, particularly the 70-Gene Signature and the Recurrence Score by using qRT-PCR in FFPE samples with commercially available assays. Our study opens the possibility to simplify the procedures to perform high-throughput techniques in the general population of patients with breast carcinoma. However, before this technology is taken to the clinic, ongoing phase III trials should validate the utility of prognostic gene profiles.
Materials and Methods
Ethics statement
At the time of initial diagnosis, all patients had provided consent in the sense that their tumour samples could be used for investigational purposes. Institutional approval from our ethical committee was obtained for the conduct of the study (Comité Ético de Investigación Clínica). Data were analyzed anonymously. Patients provided written consent so that their samples and clinical data could be used for investigational purposes.
Patients and clinical data
The study population consisted of women with early breast cancer. Inclusion criteria were: invasive ductal carcinoma, stage I or II (TNM classification, 2002), positive for oestrogen and/or progesterone receptors and appropriate therapy. Appropriate therapy should include either mastectomy or tumorectomy plus adjuvant radiotherapy, adjuvant hormonal therapy for 5 years in all patients, and adjuvant anthracycline-based chemotherapy in N+ or in N0 patients with poor prognostic features. A minimum follow-up of 5 years was also required. The following data were recorded and tabulated: age at diagnosis, size of primary tumour, lymph node stage, number of positive nodes, grade of differentiation, hormonal receptors, adjuvant therapy (either radiation, hormones or chemotherapy), date of relapse or last follow-up, site of relapse, cause of death.
RNA Isolation and cDNA Synthesis
An experienced pathologist evaluated H&E preparations to select samples with at least 70% of tumour cells. Fifteen sections 5 µm each from every FFPE sample were deparaffinized with xylene and washed with ethanol in decreasing concentrations (100%, 90% and 70%). RNA was then extracted with the Master Pure™ Kit (Epicentre).
Isolated total RNA was quantified and qualitatively assessed using spectrophotometer OD260 measurements and agarose gel electrophoresis. We normalized to total RNA input; therefore, first-strand cDNA was synthesized from 1 µg of total RNA, using random primers, according to the High Capacity cDNA Reverse Transcription Kit protocol (Applied Biosystems). The complete reaction mixes were incubated at 25°C for 10 min and 37°C for 120 min.
Quantitative RT-PCR
qRT-PCR amplifications were performed with TaqMan Gene Expression Assays products in an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems). The reactions were carried out using the TaqMan Low Density Arrays (TLDAs, Applied Biosystems) containing 50 µL TaqMan Universal PCR Master Mix (Applied Biosystems) and 50 µL of a cDNA template corresponding to 100 ng total RNA per channel of the microfluidic card.
Gene selection
We configured a TLDA series to analyze those genes included in the 70-Gene Signature [5], the Recurrence Score [4] and the Two-Gene Ratio [6] with TaqMan Gene Expression Assays available. Table S1 (supporting information) shows the assays we used to study the genes included in each profile as well as the housekeeping genes.
Although RNA degradation in FFPE samples leads to a loss of amplifiable templates, optimized normalization strategies can effectively compensate for this bias [7], [8]. We have generated a normalization model called NorMean (unpublished data). This model ranks housekeeping genes according to their capacity to control for several levels of experimental variability in qRT-PCR. Using this ranking, we calculated different normalization factors by stepwise inclusion of control genes and geometric averaging of their expression levels. The optimal number of control genes for normalization was determined by comparing the percentage of significantly correlated genes between FF and FFPE materials using each normalization factor.
Calculations for gene expression profiles
Average cycling threshold (Ct) values, defined as the point at which the fluorescence rises above the background fluorescence [26], were obtained using the SDS 2.2 software (Applied Biosystems). The maximum Ct value was set at 40. These Ct values were recorded in Microsoft Excel 2003 for subsequent calculations. The methods originally described for the 70-Gene Signature [10], the Recurrence Score [4] and the Two-Gene Ratio [6], [15] were used to analyze the performance of these profiles.
70-Gene Signature
Sixty out of the 70 genes were included [10], as the remaining 10 were not available for TLDAs by the time the experiment was performed. One previous study demonstrated that a reduction of the signature had little impact on the performance of the classifier [12]. Relative expression level of each target gene was expressed as ΔCt = Ctref−Cttarget. Normalization was performed using the geometric mean of the best housekeeping gene set (IPO8, POLR2A, UBC and SDHA). As the 70-gene signature was defined using microarrays, normalized gene Cts values were z-score transformed. The mean good-prognosis profile was calculated averaging the gene expression values of each gene for the patients without recurrence. Pearson's correlation coefficient between this mean good-prognosis profile and each patient's gene expression profile was calculated. A value threshold = 0 was used, as previously described [12]. Although the validity of calculating correlation coefficients in this way is questionable [27], we decided to use it to resemble as much as possible the methodology described by van't Veer et al [5] and others [12].
Recurrence Score
Briefly, expression of each gene was normalized relative to the expression of the reference genes (ACTB, GAPDH, GUS, RPLP0 and TFRC). Reference-normalized expression measurements were calculated as described by Paik et al., so that one unit increase in reference-normalized expression measurements reflects approximately a 2-fold increase in RNA. We substituted GSTM1 by GSTM3 because no GSTM1 probe was available for the TLDAs, as described by the authors in the patent information (Baker J et al, http://www.freepatentsonline.com/20050048542.html)
The RS algorithm was then used to generate an unscaled Recurrence Score for each patient. Values were scaled and patients were assigned to the low, intermediate or high-risk group using the RS cut-offs previously described [4].
Two-Gene Ratio
Relative expression levels of HOXB13 and IL17BR were expressed as ΔCt = Ctref−Cttarget. Cts for the four reference genes (ACTB, HMBS, SDHA and UBC) were averaged to obtain Ctref. These values were z-transformed for each gene and the Two-Gene Ratio was calculated taking the difference and using a cut off point of 1.0, as described [15].
Statistical Analysis
The prognostic value of each gene-expression–based model was evaluated by log-rank test. We also applied multivariate Cox proportional-hazards analysis to each profile individually in a model including tumour grade (2 vs. 1 and 3 vs. 1), size (>2 cm vs. ≤2 cm) and nodal status (one to three positive nodes vs. no positive nodes). We also applied multivariate Cox analyses to each profile in a model including all clinical parameters that resulted significant in the univariate analysis. Distant metastasis-free survival (DMFS) was the end point, similarly to other studies of gene profiles in breast cancer.
Two-way contingency-table analyses and the calculation of Cramer's V statistic were also performed to measure the strength of the association between the different profiles [18]. As both the 70-Gene Signature and the Two-Gene Ratio establish two groups with high and low risk, this analysis required that the low and intermediate groups of the Recurrence Score were combined.
To assess model accuracy (discrimination) at five years, Harrell's bias corrected concordance index was calculated. Models were refit 500 times with the bootstrap resampling technique. The concordance index is the percentage of patient pairs in which the predicted and observed outcomes are in agreement; i.e., the probability that for two patients chosen at random, the patient who had the event first had a higher probability of having the event according to the model. c = 0.50 represents agreement by chance; c = 1.0 represents perfect discrimination [28]. Concordance is essentially the Wilcoxon-Mann-Whitney statistic for comparing predictions in the two outcome groups, and it is identical to the area under a receiver operating characteristic curve (ROC) [29].
The gain in predictive accuracy of each classifier, as compared with common clinical staging systems, was investigated using the method of Schemper and Henderson, implemented in the R package software as previously described [21], [25], [30], [31].
All statistical analyses were performed with the SPSS v9.1 software package, GraphPad Prism v5.00 and “R” v 2.2 with the Design software package v2.0-12. All P values were two-sided, and P<0.05 was considered statistically significant.
Supporting Information
Assays and raw data.
(0.34 MB XLS)
Multivariate analysis with individual clinical factors
(0.05 MB DOC)
Acknowledgments
We thank Ricardo Ramos for his technical expertise in qRT-PCR analyses and Rocío López and Esther Díaz for their assistance in the manipulation of the tumour samples.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Grant support: This work was supported by FIS Grant PI050668 (Instituto de Salud Carlos III, Ministerio de Sanidad y Consumo, Spain) (EE, ISN, JAFV), by Fundación SEOM grant 2006 (EE), Red Temática de Investigación Cooperativa en Cáncer (RTICC, RD06-0020-1022) and by unrestricted grants from Astra-Zeneca, Bristol-Myers Squibb, Cephalon-Pharma, Glaxo SmithKline, Roche Farma, Sanofi-Aventis and Schering-Plough. JAFV is supported by Grant award FIS CP05/00248 from Fondo de Investigación Sanitaria (Instituto de Salud Carlos III, Ministerio de Sanidad y Consumo, Spain). AGP is the recipient of a fellowship by Ministerio de Educación y Ciencia, Spain. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Goldhirsch A, Wood WC, Gelber RD, Coates AS, Thurlimann B, et al. Progress and promise: highlights of the international expert consensus on the primary therapy of early breast cancer 2007. Ann Oncol. 2007;18:1133–1144. doi: 10.1093/annonc/mdm271. [DOI] [PubMed] [Google Scholar]
- 2.Olivotto IA, Bajdik CD, Ravdin PM, Speers CH, Coldman AJ, et al. Population-based validation of the prognostic model ADJUVANT! for early breast cancer. J Clin Oncol. 2005;23:2716–2725. doi: 10.1200/JCO.2005.06.178. [DOI] [PubMed] [Google Scholar]
- 3.Bergh J, Holmquist M. Who should not receive adjuvant chemotherapy? International databases. J Natl Cancer Inst Monogr. 2001:103–108. doi: 10.1093/oxfordjournals.jncimonographs.a003445. [DOI] [PubMed] [Google Scholar]
- 4.Paik S, Shak S, Tang G, Kim C, Baker J, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 5.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 6.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 7.Antonov J, Goldstein DR, Oberli A, Baltzer A, Pirotta M, et al. Reliable gene expression measurements from degraded RNA by quantitative real-time PCR depend on short amplicons and a proper normalization. Lab Invest. 2005;85:1040–1050. doi: 10.1038/labinvest.3700303. [DOI] [PubMed] [Google Scholar]
- 8.Cronin M, Pho M, Dutta D, Stephans JC, Shak S, et al. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004;164:35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buyse M, Loi S, van't Veer L, Viale G, Delorenzi M, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 10.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 11.Mook S, Schmidt MK, Viale G, Pruneri G, Eekhout I, et al. The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1–3 positive lymph nodes in an independent validation study. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-0130-2. [DOI] [PubMed] [Google Scholar]
- 12.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albain K, Barlow W, Shak S, Hortobagyi G, R L, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal, node-positive, ER-positive breast cancer. 2007 doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paik S, Tang G, Shak S, Kim C, Baker J, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 15.Ma XJ, Hilsenbeck SG, Wang W, Ding L, Sgroi DC, et al. The HOXB13:IL17BR expression index is a prognostic factor in early-stage breast cancer. J Clin Oncol. 2006;24:4611–4619. doi: 10.1200/JCO.2006.06.6944. [DOI] [PubMed] [Google Scholar]
- 16.Ma XJ, Salunga R, Dahiya S, Wang W, Carney E, et al. A Five-Gene Molecular Grade Index and HOXB13:IL17BR Are Complementary Prognostic Factors in Early Stage Breast Cancer. Clin Cancer Res. 2008;14:2601–2608. doi: 10.1158/1078-0432.CCR-07-5026. [DOI] [PubMed] [Google Scholar]
- 17.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 18.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 19.Habel LA, Shak S, Jacobs MK, Capra A, Alexander C, et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 2006;8:R25. doi: 10.1186/bcr1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espinosa E, Vara JA, Redondo A, Sanchez JJ, Hardisson D, et al. Breast cancer prognosis determined by gene expression profiling: a quantitative reverse transcriptase polymerase chain reaction study. J Clin Oncol. 2005;23:7278–7285. doi: 10.1200/JCO.2005.01.4746. [DOI] [PubMed] [Google Scholar]
- 21.Reyal F, van Vliet MH, Armstrong NJ, Horlings HM, de Visser KE, et al. A comprehensive analysis of prognostic signatures reveals the high predictive capacity of the proliferation, immune response and RNA splicing modules in breast cancer. Breast Cancer Res. 2008;10:R93. doi: 10.1186/bcr2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein LJ, Gray R, Badve S, Childs BH, Yoshizawa C, et al. Prognostic utility of the 21-gene assay in hormone receptor-positive operable breast cancer compared with classical clinicopathologic features. J Clin Oncol. 2008;26:4063–4071. doi: 10.1200/JCO.2007.14.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mook S, Schmidt MK, Viale G, Pruneri G, Eekhout I, et al. Breast cancer patients with 1–3 positive lymph nodes and a low risk 70-gene profile have an excellent survival. Proc of 2007 SABCS. 2007:abstract 50. [Google Scholar]
- 25.Dunkler D, Michiels S, Schemper M. Gene expression profiling: does it add predictive accuracy to clinical characteristics in cancer prognosis? Eur J Cancer. 2007;43:745–751. doi: 10.1016/j.ejca.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 26.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koscielny S. Critical review of microarray-based prognostic tests and trials in breast cancer. Curr Opin Obstet Gynecol. 2008;20:47–50. doi: 10.1097/GCO.0b013e3282f39d9e. [DOI] [PubMed] [Google Scholar]
- 28.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Pencina MJ, D'Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23:2109–2123. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 30.Lusa L, Miceli R, Mariani L. Estimation of predictive accuracy in survival analysis using R and S-PLUS. Comput Methods Programs Biomed. 2007;87:132–137. doi: 10.1016/j.cmpb.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Schemper M, Henderson R. Predictive accuracy and explained variation in Cox regression. Biometrics. 2000;56:249–255. doi: 10.1111/j.0006-341x.2000.00249.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Assays and raw data.
(0.34 MB XLS)
Multivariate analysis with individual clinical factors
(0.05 MB DOC)