Summary
Because neurotropic viruses naturally traverse neural pathways, they are extremely valuable for elucidating neural circuits. Naturally occurring herpes and rabies viruses have been used for transneuronal circuit tracing for decades. Depending on the type of virus and strain, virus can travel preferentially in the anterograde or the retrograde direction. More recently, genetic modifications have allowed for many improvements. These include: reduced pathogenicity; addition of marker genes; control of synaptic spread; pseudotyping for infection of selected cells; addition of ancillary genetic elements for combining circuit tracing with manipulation of activity or functional assays. These modifications, along with the likelihood of future developments, suggest that neurotropic viruses will be increasingly important and effective tools for future studies of neural circuits.
I. Introduction
A major goal of systems neuroscience is to understand how neural circuits generate perception and behavior. To this end, considerable effort has been devoted to revealing brain circuits. Such studies are most useful when they also allow links to be made to function, either by correlating activity with circuitry or by manipulating circuit components and monitoring changes in function or behavior. Such efforts are difficult, however, due to the extreme complexity of the brain. For example, conventional anterograde and retrograde neuronal tracers have allowed the collection of extensive information about connectivity between visual cortical areas in the macaque monkey [1]. These data reveal what areas are directly connected, the locations of the cells that provide the connections and the laminar zones of termination of afferent axons. This information has been used to guide studies in which likely functional interactions between areas are inferred and tested with lesions or electrical stimulation [2] [3]. But conventional tracers do not have sufficient resolution to reveal connectivity or guide functional studies at finer levels of complexity. For example, they do not reveal whether axons that terminate in a particular location do, or do not, make synaptic contacts onto particular cell types or onto cells that in turn connect to other cells. Instead, methods are required that can reveal multisynaptic pathways and can also identify connections to and from particular cell types. Ideally, such methods can also be integrated with functional studies.
There are many approaches that have been successfully exploited to reveal connectivity at high resolution [4]. Each of these has advantages and limitations. The purposes of this review are: to highlight the advantages and limitations of neurotropic viruses; to provide sufficient basic information about the properties and behavior of neurotropic viruses to allow an understanding of the sources of these advantages and limitations; and to review recent progress in the exploitation of this understanding to allow the development of novel tracing systems that are based on neurotropic viruses. The advantages and limitations of other methods will not be directly addressed.
Neurotropic viruses have two main advantages that have been exploited in studies of neural circuits. These are the ability to traverse multisynaptic pathways and the ability of viral replication to amplify signals at each step in the process [5–7]. Furthermore, depending on the species and strain of the virus, viral spread can be highly specific to synaptically connected partners. Limitations also depend on the species and strain of virus and include a lack of synaptic specificity of spread, inability to target infection to particular cell types, dependence of the speed of spread on both strength and numbers of connections, and induction of cell death. More recently, the fact that viruses are genetic machines has been exploited to eliminate limitations of naturally occurring neurotropic viruses. Genetic modifications allow control over which cells are initially infected, control over the extent of viral spread, and control of the direction of spread. It is also possible to express useful genes directly from the viral genome.
II. Basic properties of neurotropic viruses
In order to understand the advantages and limitations of neurotropic viruses for tracing neural circuits, it is informative to first understand their basic properties and the similarities and differences between viral species and strains. Although there are numerous neurotropic viruses, only two major classes have typically been used to trace neural circuits experimentally, and these will be the focus of this review. These are herpes viruses and rabies virus. (It is important to note that one particular strain of herpes virus has been named pseudorabies virus [PRV]. This particular strain will be referred to as PRV throughout this review to distinguish it from other herpes viruses and to make clear that it is not a rabies virus; it is a herpes virus. “Herpes” will be used to refer generically to all herpes viruses, including PRV.) The primary feature that makes both herpes and rabies viruses useful for tracing neural circuits is their natural ability to spread between neurons. Their natural life cycles begin with infection at the periphery and continue with spread to neurons and subsequent transneuronal spread within the nervous system. Experimentally, virus is often injected directly into the central nervous system, where it can directly infect neurons and then spread transneuronally.
Beyond their ability to infect neurons and spread in the nervous system, herpes and rabies are fundamentally different viruses. Herpes viruses belong to the family of double-stranded DNA viruses, while rabies belongs to the family of negative-sense, single-stranded RNA viruses [8]. From a practical standpoint, they also differ in other respects that will be discussed more fully below. These practical differences generally follow from the basic structural and genetic differences between viral species. It is therefore informative to begin with consideration of these differences.
Although rabies and herpes are fundamentally different, they do share one very important structural trait; they are both enveloped viruses [8]. This means that nascent viral particles consisting of proteins and genetic material (more later) bud out of host cells, and are enveloped in host cell membrane. This host cell membrane is the viral envelope. Along with the host cell membrane, viral particles also take with them envelope proteins (also called glycoprotein) that are coded in the viral genome, produced by the host cell, and embedded in the enveloped membrane.
Envelope proteins play a crucial role in mediating subsequent infection of additional neurons, as infection is mediated by an interaction between envelope proteins and cell surface receptors. Herpes and rabies differ from each other in the envelope proteins coded in their genome. Rabies has only a single gene coding for an envelope protein, called RG (rabies glycoprotein) [9]. Thus, rabies can only infect neurons via receptors for RG, which appear to be restricted to presynaptic nerve terminals [10]. This greatly limits the avenues by which rabies can infect neurons, probably contributing importantly to its apparent synaptic specificity (see further below). In contrast, herpes viruses have multiple genes coding for envelope proteins [11].
The fundamental difference in the genetic makeup of herpes (dsDNA) and rabies (negative-sense ssRNA), has the practical implication that standard DNA manipulation techniques can be used to modify the herpes genome (e.g. [12]), while more indirect methods are required to modify the rabies genome (e.g. [13,14]). Furthermore, the herpes genome can be manipulated so that it is subject to recombination with bacterial recombinases (e.g. cre-recombinase) during infection of cells in the host animal [15]. This is not possible with rabies.
Herpes and rabies also differ markedly in the size of their genomes. Rabies has a genome of only about 12Kb that codes for only 5 genes [9], while herpes viruses have genomes of about 150Kb that code for more than 70 genes [11]. The relative simplicity of rabies limits its repertoire both in nature and in the experimental setting, but is advantageous to the neuroscientist because it makes it more straightforward to understand and control how the virus works.
Because rabies is an RNA virus, it is able to complete its life cycle within the cytoplasm of the host cell [8]. The viral particle carries it own polymerase, which, following infection, is able to immediately begin transcription of mRNA from the viral genome. The host cell’s machinery then translates this mRNA to allow further production of the proteins coded in the viral genome, including more polymerase. The practical result is fast and highly efficient amplification of the rabies genome, mRNAs, protein, and nascent viral particles which bud out of the host cell and spread to other neurons. It is reported that a single particle of the CVS strain of rabies virus injected into the brain is sufficient to spread and eventually kill an animal [16]. It would follow that the infection of a single neuron must be sufficient to allow spread to other neurons (see further below).
In contrast to rabies, herpes has a complex life cycle, with the function of many genes still not understood [17]. For example, some of the herpes genes code for transcription factors, which influence gene expression both from the viral genome and from the host cell’s genome. Thus, the response to viral infection is not fixed and is dependent on the cell type and species infected. From a practical standpoint, it is also relevant that the herpes life cycle involves both retrograde transport of viral particles from the periphery and, following a latent phase, subsequent reactivation and anterograde transport of nascent viral particles back to the periphery [8]. The relatively convoluted processes controlling direction of spread are at least partly understood at the cellular and molecular level [7,18–20], providing the potential for selection or design of viruses in which this is controlled.
III. Selectivity and reliability of viral spread
One of the most important characteristics to consider for any transneuronal tracer, including neurotropic viruses, is whether spread is restricted to neurons that are connected by synaptic contacts. Because the transfer of information between neurons is primarily dependent on synapses (and gap junctions), the most relevant circuit diagram is based exclusively on connectivity, not proximity. This issue is not fully resolved but has been most carefully explored for the PRV Bartha strain of herpes virus (“PRV”) and for rabies virus [6,7,21,22].
Both PRV and rabies virus can be used to selectively label cells via transneuronal spread that is apparently restricted to cells that are synaptically connected [6,7,21,22]. These properties are likely to hold across rabies strains. However, herpes viruses used for circuit tracing are more diverse and might vary in the specificity of transneuronal spread. Furthermore, it is possible that selectivity may vary depending on the circuit studied or other unknown factors. One should therefore be cautious in the interpretation of any studies using these tools.
The factors that influence the mode of viral spread are not completely understood, however one likely contributor is lysis of infected cells. Herpes viruses typically have a lytic phase in which nascent viral particles may be released from the host cell and can infect nearby cells. It is thought that preferential uptake of PRV Bartha by glia may protect neighboring neurons from non-specific spread [22]. The specificity of herpes spread may therefore differ depending on the relationships between neurons and glia in various circuits. The extent and timing of lysis also depends on the viral strain [22]. The improved utility of the Bartha strain of PRV is apparently due largely to its slower spread and reduced cytopathic effects [7,22].
The available evidence suggests that the spread of rabies virus may be exclusively transsynaptic (restricted to synapses) [6,21]. The mechanisms responsible for this restriction are not fully understood. One possible contributor is that receptors for the rabies glycoprotein may be restricted to presynaptic nerve terminals. Although various, more or less prevalent, presynaptic elements have been identified as rabies receptors, including the P75 neurotrophin receptor and NCAM, there may also be additional, as yet unidentified, receptors [10]. Additional factors may also contribute to synapse specificity. For example, ensheathment of nerve terminals by glia may limit infection by rabies particles that bud from extrasynaptic sites. Another formal possibility is that budding of viral particles from the host cell is limited to postsynaptic sites, however, the ability to produce rabies in cells which do not have synapses, argues against this.
IV. Direction of viral spread and other differences between viruses and strains
The virulence and direction of transneuronal spread of neurotropic viruses depends both on the type of virus and the strain. Rabies virus spreads exclusively in the retrograde direction, from postsynaptic to presynaptic cells, regardless of the strain [6,23]. Different strains of rabies virus do differ, however, in their rate of spread and their safety when used in a laboratory setting. Because the rate of spread of rabies virus can vary dramatically depending not only on strain, but also on passage history, it is important for any investigator to first directly test the virus in a well established system to determine the rate of spread, before using it in the experimental system of interest. For example, when tracing circuitry in the primate motor system, the CVS strain of rabies spread across one synaptic step approximately every 24 hours, while the N2C strain spread far more rapidly, across one synaptic step every 10–12 hours [24,25]. (It is important to note the rate of spread is also dependent on the strength of connections under study [21].)
Rabies strains also differ in their safety in a laboratory setting. Most strains of rabies, such as CVS, are derived initially from a small number of strains isolated from the wild and propagated in culture. The strains isolated from the wild are, in general, deadly to both humans and other animals. As a result, extreme caution is required for their use, and investigators working with them need to be vaccinated. Such strains are not, however, the only rabies viruses that are useful for circuit tracing. Notably, the SAD-B19 strain of rabies virus has been used extensively to vaccinate animals in the wild. Vaccinated animals develop an immune response to the virus before it can spread to the central nervous system; counterintuitively, this heightened immune response appears to result from more rapid replication of SAD-B19 than strains that cause disease and death [26]. More slowly replicating strains are apparently able to spread into the CNS before initiating an effective immune response. Furthermore, the SAD-B19 strain does not appear to spread from vaccinated animals to other animals in close proximity [27]. Nevertheless, when injected directly into the nervous system, SAD-B19 rabies spreads between neurons in a manner that is similar to other rabies viruses. This makes SAD-B19 rabies both effective for experimental investigation and relatively safe. Nevertheless, appropriate caution should still be exercised in the use of this virus [23].
Herpes viruses are a large family of viruses [8]. The most commonly used of these for circuit tracing are strains of either Herpes Simplex Virus type 1 (HSV-1) which is endemic in humans, or PRV (pseudorabies virus), which does not infect humans or non-human primates. Most naturally occurring herpes viruses can spread both anterogradely and retrogradely. However, the McIntyre-B strain of HSV-1 spreads exclusively in the retrograde direction, while the H129 strain appears to spread preferentially in the anterograde direction [28–30]. There is evidence, however, that there might not be complete elimination of retrograde spread of the H129 strain [18,29].
Because PRV does not infect primates (including humans), it is much safer than other herpes viruses and often preferred for experimental studies [7,22]. Although PRV is far less likely to cause problems for human health, it remains necessary to avoid exposure of healthy animals to infected animals, because PRV spreads rapidly between animals and is lethal in host species, including laboratory animals such as rats and mice. This difference obviously limits the utility of PRV to non-primate species.
The Bartha strain of PRV is an attenuated strain, which has proven to be particularly useful for circuit tracing [7,22]. This increased utility results first, from the restriction of transneuronal spread to the retrograde direction. As a result, experiments are more straightforward to interpret than they would be for a virus spreading bidirectionally. Another trait that appears to improve utility is a reduction in toxicity. Cells appear to remain healthy longer following infection and animals survive longer. This allows tracing across a greater number of synaptic steps than if animals died more quickly and might also limit non-synaptic spread (see above).
V. Tracing connections between specific cell types and controlling the extent of spread
Due to the complexity of the nervous system, it is extremely valuable to be able to trace connections to or from particular cell types. To some degree, neurotropic viruses make this possible simply by virtue of the fact that they can spread across multiple synaptic steps. For example, if a virus that spreads retrogradely is injected into area A, the initial infection will be restricted to cells in area A and to cell types in other areas, say area B, that project axons to area A. As the virus propagates, it might be possible to infer that subsequent spread within area B or to areas that provide input to area B, results specifically from connections to the area B cell type which projects to area A. (See, for example [31,32].) Nevertheless, this situation is far from ideal and there are many caveats that require caution in making such interpretations. Most notable among these are that most brain areas are connected with multiple structures, making it increasingly difficult to determine the exact route of spread as virus traverses increasing numbers of synaptic steps [32]. And even when the number of synaptic steps is small, the rate of viral spread depends on the strength of connections [21]. As Ugolini notes [21]: “The speed of transfer of rabies virus to these cell groups was strongly correlated with the strength of their input… with rabies, as with HSV-1 [33]…different groups of second-order neurons were labelled at different times. This generated an overlap … between the delayed labelling of the last groups of second-order neurons and the onset of labelling of higher order neurons…”.
The utility of neurotropic viruses could therefore be greatly improved by controlling what cell types are initially infected or are permissive for viral replication, and by controlling the number of synaptic steps that are crossed. Effective genetic modifications for achieving these goals have been applied to both PRV Bartha and to the SAD-B19 strain of rabies virus.
To trace connections to specific cell types, De Falco et al. [15] developed and used a modified form of PRV Bartha, called Ba2001, which is unable to replicate unless its genome is recombined by cre-recombinase. This strategy utilizes the fact that viral thymidine kinase (TK) is required for viral replication and also takes advantage of the fact that there are mouse lines which express cre-recombinase only in specific cell types. Combining these tools allows tracing of the inputs to cre-expressing cells. In particular, the thymidine kinase (TK) gene was deleted from the PRV genome and replaced with a “floxed stop” sequence followed by coding sequences for both TK and green fluorescent protein (GFP). Thus, in Ba2001, cre-recombinase will excise the floxed stop and allow TK plus GFP expression only in Cre-expressing cells. When Ba2001 is injected into a brain area, it infects all cell types. But only the cell types expressing cre, recombine the viral genome, allowing the subsequent GFP expression to mark the recombined virus and TK expression to allow replication. Recombined viral particles can then spread transneuronally, and the infected neurons will also be marked by GFP and replicate recombined virus for further retrograde spread. The spread of the GFP-expressing virus can therefore be used to trace direct and indirect inputs to a specific cell type [15,34].
To trace connections to specific cell types with rabies virus, it is not possible to use a strategy based on cre-recombination. This is because the RNA genome of rabies cannot be recombined with cre and there is no DNA phase in the viral life cycle. Therefore, Wickersham et al. [35] developed a strategy in which the initial viral infection is restricted to particular cells. This was done by deleting the RG gene from the SAD-B19 rabies genome, replacing it with GFP, and then producing viral particles with a different envelope protein in their envelope (pseudotyping). The RG-deleted virus was pseudotyped with an avian virus envelope protein, called EnvA; this new virus is called EnvA-SADdG-GFP. When this virus is injected into the brain of a normal animal, it does not infect any neurons because the mammalian brain has no receptors for EnvA. But by misexpressing the EnvA receptor, TVA, in particular cells, it was possible to selectively infect those cells.
Because RG is required for formation of new viral particles and transsynaptic spread [36,37], and SAD-dG-GFP has no coding sequence for RG, RG expression is also required to allow viral spread from the targeted neurons. When neurons expressed TVA and not RG, infection with EnvA-SADdG-GFP (and GFP expression from the rabies genome) was restricted to TVA cells and did not spread from those cells [35]. When neurons expressed both TVA and RG, the initial infection was restricted to TVA cells, and RG expression in those cells allowed transsynaptic spread to, and GFP labeling of additional neurons that were directly presynaptic to the TVA cells. Continued spread beyond the directly presynaptic neurons did not occur, because the presynaptic cells do not express RG and there is no RG coding sequence carried in the viral genome. Thus, this system has the additional advantage that rabies spread is monosynaptically-restricted, eliminating any confound between strength of connections and numbers of synapses crossed; this eliminates any ambiguity about numbers of synaptic steps that have been crossed. This method can be used in combination with, for example, cre- or Tta-expressing mouse lines to obtain cell type specific expression of TVA and RG (cf. [4]). Subsequently, the targeted cell type can be specifically infected with EnvA-SADdG-GFP and direct inputs labeled by transsynaptic spread of the virus and GFP expression.
In view of the fact that neighboring neurons of the same type often connect with different presynaptic neurons (e.g. [38,39]), it is highly desirable to have methods that can identify the connections to single neurons. Experimental evidence indicates that this is feasible with the EnvA-SADdG-GFP rabies virus described above [35]. One needs to express TVA and RG in a single neuron and then infect that neuron selectively with EnvA-SADdG-GFP. Over the course of several days, the GFP expressing virus spreads to and labels neurons that are directly presynaptic to the single parent neuron. This method is now being combined with single cell electroporation [40] or other strategies for obtaining sparse cell infection, in order to trace connections to single neurons in vivo.
In contrast to EnvA-SADdG-GFP, the Ba2001 system does allow continued transneuronal spread across multiple synaptic steps [15,34]. Although this can be advantageous, it has the disadvantage of possible ambiguity in interpretation of results [21,33] (see above). An alternative that has been tested in the author’s lab, but not published, is to use the PRV virus Ba2000, which was described and used as a control by De Falco et al. [15]. Unlike Ba2001, Ba2000 has the TK gene deleted from its genome and cannot express TK under any circumstances. But Ba2000 does express GFP following cre-recombination, as do any viral progeny that are produced from the recombined genome. To use this virus for monosynaptically-restricted spread from cre-expressing neurons, it is necessary to simply provide a means for TK to be expressed selectively in the cre-expressing neurons. Under these circumstances, GFP-expressing virus is replicated only in cre-expressing cells and it can then spread retrogradely to neurons providing direct input to those cells. These directly presynaptic neurons can express GFP from the viral genome, thus marking them, but the virus cannot replicate and spread any further.
VI. Expression of additional gene products
A final advantage of genetically modified herpes and rabies viruses is that essentially any gene of interest can be inserted into the genome, so that it is expressed in selected cells with known connectivity. The most straightforward gene products are those to mark cells, such as fluorescent proteins. Since these come in many colors [41], it is possible to selectively mark various neuronal populations that have been labeled with different viruses. But these strategies can be extended to the full range of genetic tools that are available for monitoring or manipulating neurons [4], leaving one apparently limited only by creativity and perseverance. For example, it should be possible to express light sensitive ion channels or genetic activity indicators in the cells that converge onto a single neuron, so that their activity can be manipulated or monitored optically.
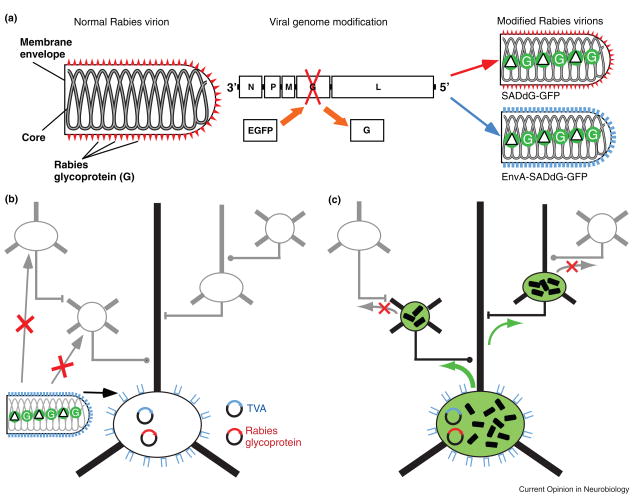
Pseudoptyped Rabies Virus for Tracing Connections of Targeted Neurons.
A. Normal and modified rabies virions. The normal rabies virion (left) includes an RNA core within the viral envelope. The envelope is coated with the rabies glycoprotein (RG). It is possible to produce modified rabies virions in which the RG gene has been deleted from the genome and replaced with coding sequence for EGFP (middle). When these modified virions are grown in culture they can be coated with RG, conferring the normal infectious properties of rabies virus (right, top), or pseudotyped with an envelope protein from another virus (e.g. EnvA), conferring the infectious properties of that virus (right, bottom). Although these modified virions are capable of infecting cells and replicating to produce large quantities of EGFP, they are not able to spread out of those cells without the help of another DNA expression vector that provides RG. This is because RG is absolutely essential for viral spread. B. Selective Infection. EnvA Pseudotyped rabies virus can be used to selectively infect neurons which have been targeted for expression of the EnvA receptor, TVA. Because there are not endogenous receptors for EnvA in the mammalian brain, other cells are not infected. Because, the RG gene has been deleted from the rabies genome, complementation is required to allow spread of the virus from infected cells. This can be accomplished by targeted expression of RG in the same cells which express TVA. C. Monosynaptically-restricted spread. Following infection and RG complementation in the initially infected neurons, the rabies virus is able to spread retrogradely to directly presynaptic neurons. However, because these presynaptic neurons lack RG expression, the virus cannot spread beyond these cells. After Wickersham et al. [35].
Acknowledgments
Work in the author’s laboratory is supported by the National Institutes of Health. The author is grateful to current and former members of his lab, and particularly Ian Wickersham, for numerous discussions and insights.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- 2.Newsome WT, Wurtz RH, Dursteler MR, Mikami A. Deficits in visual motion processing following ibotenic acid lesions of the middle temporal visual area of the macaque monkey. J Neurosci. 1985;5:825–840. doi: 10.1523/JNEUROSCI.05-03-00825.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salzman CD, Britten KH, Newsome WT. Cortical microstimulation influences perceptual judgements of motion direction. Nature. 1990;346:174–177. doi: 10.1038/346174a0. [DOI] [PubMed] [Google Scholar]
- 4.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuypers HG, Ugolini G. Viruses as transneuronal tracers. Trends Neurosci. 1990;13:71–75. doi: 10.1016/0166-2236(90)90071-h. [DOI] [PubMed] [Google Scholar]
- 6.Ugolini G. Use of rabies virus as a transneuronal tracer of neuronal connections: implications for the understanding of rabies pathogenesis. Dev Biol (Basel) 2008;131:493–506. [PubMed] [Google Scholar]
- 7.Enquist LW. Exploiting circuit-specific spread of pseudorabies virus in the central nervous system: insights to pathogenesis and circuit tracers. J Infect Dis. 2002;186 (Suppl 2):S209–214. doi: 10.1086/344278. [DOI] [PubMed] [Google Scholar]
- 8.Knipe DM, Howley PM, editors. Fields Virology. 5. Philadelphia: Lippincott Williams and Wilkins; 2007. [Google Scholar]
- 9.Conzelmann KK, Cox JH, Schneider LG, Thiel HJ. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. [DOI] [PubMed] [Google Scholar]
- 10.Lafon M. Rabies virus receptors. J Neurovirol. 2005;11:82–87. doi: 10.1080/13550280590900427. [DOI] [PubMed] [Google Scholar]
- 11.McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, Frame MC, McNab D, Perry LJ, Scott JE, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69 (Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 12.Sauer B, Whealy M, Robbins A, Enquist L. Site-specific insertion of DNA into a pseudorabies virus vector. Proc Natl Acad Sci U S A. 1987;84:9108–9112. doi: 10.1073/pnas.84.24.9108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnell MJ, Mebatsion T, Conzelmann KK. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mebatsion T, Schnell MJ, Cox JH, Finke S, Conzelmann KK. Highly stable expression of a foreign gene from rabies virus vectors. Proc Natl Acad Sci U S A. 1996;93:7310–7314. doi: 10.1073/pnas.93.14.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15**.DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, Enquist L, Friedman JM. Virus-Assisted Mapping of Neural Inputs to a Feeding Center in the Hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. This manuscript demonstrates the production and utility of a genetically modified pseduorabies virus, with conditional replication, for tracing the inputs to specific neuron types. [DOI] [PubMed] [Google Scholar]
- 16.Coulon P, Rollin P, Aubert M, Flamand A. Molecular basis of rabies virus virulence. I. Selection of avirulent mutants of the CVS strain with anti-G monoclonal antibodies. J Gen Virol. 1982;61 (Pt l):97–100. doi: 10.1099/0022-1317-61-1-97. [DOI] [PubMed] [Google Scholar]
- 17.Lilley CE, Branston RH, Coffin RS. Herpes simplex virus vectors for the nervous system. Curr Gene Ther. 2001;1:339–358. doi: 10.2174/1566523013348346. [DOI] [PubMed] [Google Scholar]
- 18.Garner JA, LaVail JH. Differential anterograde transport of HSV type 1 viral strains in the murine optic pathway. J Neurovirol. 1999;5:140–150. doi: 10.3109/13550289909021996. [DOI] [PubMed] [Google Scholar]
- 19.Enquist LW, Tomishima MJ, Gross S, Smith GA. Directional spread of an alpha-herpesvirus in the nervous system. Vet Microbiol. 2002;86:5–16. doi: 10.1016/s0378-1135(01)00486-2. [DOI] [PubMed] [Google Scholar]
- 20.LaVail JH, Tauscher AN, Sucher A, Harrabi O, Brandimarti R. Viral regulation of the long distance axonal transport of herpes simplex virus nucleocapsid. Neuroscience. 2007;146:974–985. doi: 10.1016/j.neuroscience.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Ugolini G. Specificity of rabies virus as a transneuronal tracer of motor networks: transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. J Comp Neurol. 1995;356:457–480. doi: 10.1002/cne.903560312. This manuscript describes rigorous tests of the synapse specificity of the transneuronal spread of rabies and compares this with the spread of herpes virus. [DOI] [PubMed] [Google Scholar]
- 22.Ekstrand MI, Enquist LW, Pomeranz LE. The alpha-herpesviruses: molecular pathfinders in nervous system circuits. Trends Mol Med. 2008;14:134–140. doi: 10.1016/j.molmed.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly RM, Strick PL. Rabies as a transneuronal tracer of circuits in the central nervous system. J Neurosci Methods. 2000;103:63–71. doi: 10.1016/s0165-0270(00)00296-x. [DOI] [PubMed] [Google Scholar]
- 24.Kelly RM, Strick PL. Cerebellar loops with motor cortex and prefrontal cortex of a nonhuman primate. J Neurosci. 2003;23:8432–8444. doi: 10.1523/JNEUROSCI.23-23-08432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoshi E, Tremblay L, Feger J, Carras PL, Strick PL. The cerebellum communicates with the basal ganglia. Nat Neurosci. 2005;8:1491–1493. doi: 10.1038/nn1544. [DOI] [PubMed] [Google Scholar]
- 26.Jackson AC, Rasalingam P, Weli SC. Comparative pathogenesis of recombinant rabies vaccine strain SAD-L16 and SAD-D29 with replacement of Arg333 in the glycoprotein after peripheral inoculation of neonatal mice: less neurovirulent strain is a stronger inducer of neuronal apoptosis. Acta Neuropathol. 2006;111:372–378. doi: 10.1007/s00401-005-0006-z. [DOI] [PubMed] [Google Scholar]
- 27.Vos A, Neubert A, Aylan O, Schuster P, Pommerening E, Muller T, Chivatsi DC. An update on safety studies of SAD B19 rabies virus vaccine in target and non-target species. Epidemiol Infect. 1999;123:165–175. doi: 10.1017/s0950268899002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zemanick MC, Strick PL, Dix RD. Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc Natl Acad Sci U S A. 1991;88:8048–8051. doi: 10.1073/pnas.88.18.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaVail JH, Topp KS, Giblin PA, Garner JA. Factors that contribute to the transneuronal spread of herpes simplex virus. J Neurosci Res. 1997;49:485–496. [PubMed] [Google Scholar]
- 30.Song CK, Schwartz GJ, Bartness TJ. Anterograde Transneuronal Viral Tract Tracing Reveals Central Sensory Circuits From White Adipose Tissue. Am J Physiol Regul Integr Comp Physiol. 2008 doi: 10.1152/ajpregu.90786.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nassi JJ, Lyon DC, Callaway EM. The parvocellular LGN provides a robust disynaptic input to the visual motion area MT. Neuron. 2006;50:319–327. doi: 10.1016/j.neuron.2006.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nassi JJ, Callaway EM. Multiple circuits relaying primate parallel visual pathways to the middle temporal area. J Neurosci. 2006;26:12789–12798. doi: 10.1523/JNEUROSCI.4044-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ugolini G, Kuypers HG, Simmons A. Retrograde transneuronal transfer of herpes simplex virus type 1 (HSV 1) from motoneurones. Brain Res. 1987;422:242–256. doi: 10.1016/0006-8993(87)90931-0. [DOI] [PubMed] [Google Scholar]
- 34*.Campbell RE, Herbison AE. Definition of brainstem afferents to gonadotropin-releasing hormone neurons in the mouse using conditional viral tract tracing. Endocrinology. 2007;148:5884–5890. doi: 10.1210/en.2007-0854. This manuscripts describes use of the cre-dependent pseudorabies virus developed by De Falco et al. [15] to identify inputs to a specific class of neurons in the rostral preoptic area. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. This paper demonstrates the production and utility of glycoprotein deleted rabies virus, pseudotyped with EnvA, to allow selective infection of specific cell types. Glycoprotein expression in the same targeted neurons allows complementation and monsynaptically restricted spread to label the direct inputs to a single neuron or genetically targeted population. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Etessami R, Conzelmann KK, Fadai-Ghotbi B, Natelson B, Tsiang H, Ceccaldi PE. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J Gen Virol. 2000;81:2147–2153. doi: 10.1099/0022-1317-81-9-2147. [DOI] [PubMed] [Google Scholar]
- 37*.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007;4:47–49. doi: 10.1038/NMETH999. The authors demonstrate the utility of a glycoprotein deleted, GFP expressing rabies virus as a retrograde tracer which reveals detailed morphology. The results also demonstrate that rabies glycoprotein is required for transneuronal viral spread. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshimura Y, Callaway EM. Fine-scale specificity of cortical networks depends on inhibitory cell type and connectivity. Nat Neurosci. 2005;8:1552–1559. doi: 10.1038/nn1565. [DOI] [PubMed] [Google Scholar]
- 39.Yoshimura Y, Dantzker JL, Callaway EM. Excitatory cortical neurons form fine-scale functional networks. Nature. 2005;433:868–873. doi: 10.1038/nature03252. [DOI] [PubMed] [Google Scholar]
- 40.Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Targeted patch-clamp recordings and single-cell electroporation of unlabeled neurons in vivo. Nat Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci U S A. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]