Abstract
Centromeric chromatin – spindle microtubule interactions mediated by kinetochores drive chromosome segregation. We have developed a two-color fluorescence light microscopy method that measures average label separation, Delta, at < 5 nm accuracy — to elucidate the protein architecture of human metaphase kinetochores. Delta analysis, when correlated with tension states of spindle-attached sister kinetochore pairs, provided information on mechanical properties of protein linkages within kinetochores. Treatment with taxol—which suppresses microtubule dynamics, eliminates tension at kinetochores, and activates the spindle checkpoint—resulted in specific large-scale changes in kinetochore architecture. Cumulatively, Delta analysis revealed compliant linkages close to the centromeric chromatin, suggests a model for how the KMN (KNL1/Mis12 complex/Ndc80 complex) network provides microtubule attachment and generates pulling forces from depolymerization, and reveals architectural changes induced by taxol treatment. The methods described here should also be applicable to other intermediate-scale biological machines in cells.
Keywords: centromere, chromosome, mitosis, cell division, tubulin, spindle, taxol, fluorescence microscopy
Introduction
Kinetochores are protein assemblies at the periphery of centromeric chromatin that are required for segregating chromosomes in all eukaryotes (Maiato et al., 2004). Robust spindle microtubule (MT) plus-end attachment is “end-on” and MTs bound to kinetochores are known as kinetochore microtubules (kMTs)(Rieder, 1982). The number of MTs bound per kinetochore can be as few as one for budding yeast and up to 20 in humans (Maiato et al., 2004; Rieder, 1982). By electron microscopy, human kinetochores show a multilayered disk structure (Dong et al., 2007; Maiato et al., 2004). The periphery of centromeric chromatin has higher density and is called the inner plate, followed by a low contrast gap of 20–30 nm, an outer plate 40–50 nm thick, and a peripheral fibrous corona extending 100–200 nm away from the outer plate. Protofilaments at plus ends of kMTs curve inside-out by various degrees near the inner surface of the outer plate and the kMTs rarely penetrate centromeric chromatin (Dong et al., 2007; Maiato et al., 2006; McIntosh et al., 2008; VandenBeldt et al., 2006). Multiple sites for end-on kMT attachment are formed by weak lateral interactions between individual sites at the centromere periphery (Brinkley et al., 1992; Cimini et al., 2004). These observations suggest that protein linkages along the inner-outer kinetochore axis connect centromeric chromatin to spindle MTs.
Information about relative inside-out locations of different proteins at kinetochores has been obtained from epistasis assays in conjunction with protein-protein interaction data (Cheeseman and Desai, 2008; Cheeseman et al., 2008; Foltz et al., 2006; Liu et al., 2006; Maiato et al., 2004; Musacchio and Salmon, 2007). The inner domain of the kinetochore is assembled from proteins constitutively present at centromeres during the cell cycle. First is CENP-A, a variant of histone H3, that replaces H3 in nucleosomes of chromatin at the base of the kinetochore (Blower et al., 2002). Following CENP-A are 14 proteins known as the constitutive centromere-associated network (CCAN) (Cheeseman and Desai, 2008; Foltz et al., 2006). Unlike the constitutively localized proteins, outer kinetochore proteins are assembled at kinetochores beginning at prophase and leave kinetochores at the end of mitosis. Important are three highly conserved protein complexes (the KMN Network) which assemble stably within the outer kinetochore to produce core attachment sites for kMTs (Cheeseman et al., 2006; Cheeseman and Desai, 2008; DeLuca et al., 2006; McAinsh et al., 2006; Tanaka and Desai, 2008): the Mis12 complex of four proteins (hMis12, hDsn1Q9H410, hNnf1PMF1, hNsl1DC31), hKnl1/Blinkin (Cheeseman et al., 2008; Kiyomitsu et al., 2007), and the 4-subunit Ndc80 complex (comprised of Hec1/hNdc80, Nuf2, Spc24 and Spc25). Other outer domain components transiently associate with kinetochores after entry into mitosis (Liu et al., 2006; Musacchio and Salmon, 2007) and include spindle assembly checkpoint (SAC) components, MT motor proteins, the large coiled-coil protein CENP-F, and MT-binding proteins that regulate plus-end assembly dynamics (Musacchio and Salmon, 2007).
Here we describe kinetochore architecture in human metaphase cells using a fluorescence microscopy method that we developed that measures the separation, Delta, between proteins or protein domains labeled with two different fluorophores. The accuracy of the Delta calculation was also verified using a technique developed for measuring separation distances within single molecules (SHREC, Churchman et al., 2005). We found that our assay is capable of measuring at <5 nm accuracy the average separations of fluorescent protein markers along the inner-outer axis of sister kinetochores. At metaphase, sister kinetochore pairs exhibit directional instability that produces oscillations in the stretch of centromeric chromatin between sisters (Maiato et al., 2004). Correlating Delta measurements with centromere stretch provided information on stiffness of protein linkages within kinetochores. We also identify major changes in kinetochore architecture induced by the MT-stabilizing drug taxol (Waters et al., 1998) which activates the SAC and eliminates centromeric tension between sister kinetochores without loss of bound kMTs (Maiato et al., 2004). Finally, we employed a modified two-color co-localization assay to locate the ends of kMTs relative to the MT-binding Ndc80 complex. Cumulatively, our assays provide a molecular resolution view of a macromolecular machine with a central role in cell division and reveal the mechanical nature of specific protein linkages at kinetochores as well as large scale changes in kinetochore architecture induced by suppression of MT dynamics.
RESULTS
Delta Measurements of the Ndc80 complex at Human Metaphase Kinetochores
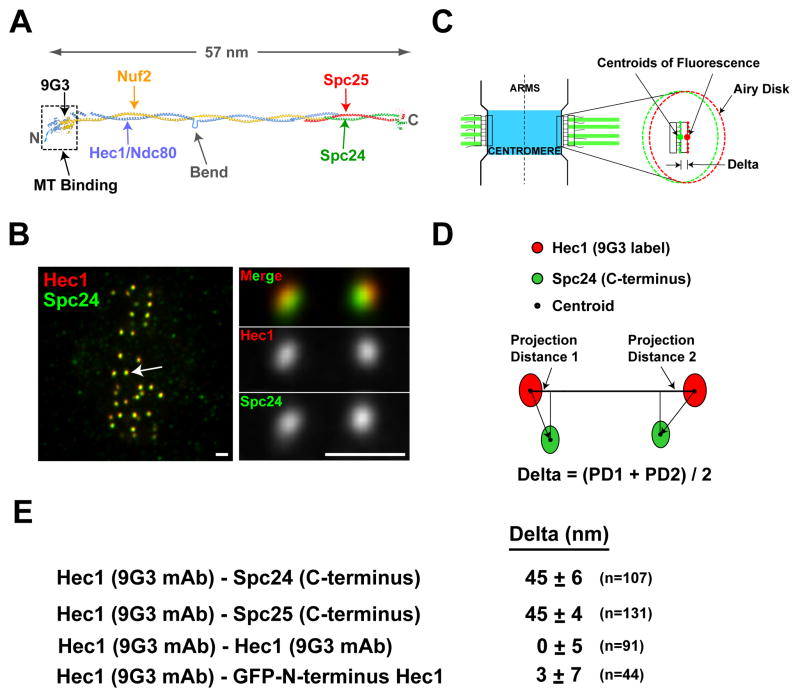
We initially developed the Delta assay to analyze the 4-subunit Ndc80 complex, which is the major MT-binding component of the outer kinetochore. Electron microscopy and atomic structure studies indicate that the Ndc80 complex is 57 nm long and has a dumbbell-shape (Fig. 1A, Ciferri et al., 2008; Wei et al., 2007). MT-binding activity resides in the Hec1 and Nuf2 N-terminal regions at one end of the complex (Cheeseman et al., 2006; Ciferri et al., 2008; Wei et al., 2007). The 9G3 monoclonal antibody binds aa 200–215, roughly 2.5 nm inside the Hec1 N-terminus (DeLuca et al., 2006; Ciferri et al., 2008) and localizes by immuno-EM to the kinetochore outer plate (DeLuca et al., 2005). Globular C-terminal regions of Spc24 and Spc25 are at the opposite end of a long alpha-helical coiled-coil rod (Fig. 1A). Previous work suggests that the 9G3 label is external to the Spc24 label along the inner-outer kinetochore axis (DeLuca et al., 2006), which is consistent with binding orientation of the complex relative to MT polarity (Wilson-Kubalek et al., 2008).
Figure 1. Delta Measurements for the Ndc80 Complex.
(A) Molecular structure of the tetrameric Ndc80 complex. (B) Images of metaphase HeLa cells fixed and stained for Hec1 9G3 (red) and anti-Spc24 (green). Scale bar = 1.1 μm. (C) Schematic of a pair of sister kinetochores at metaphase with one kinetochore expanded to show Airy disk images of 9G3 and anti-Spc24 labels and Delta, the distance between their centroids along the inner-outer kinetochore axis. (D) Method for calculation of Delta that eliminates, locally, errors from lateral chromatic aberration in microscope optics. (E) Average Delta measurements for different label pairs along the Ndc80 complex.
We analyzed metaphase HeLa cells where kinetochore inner-outer linkages to kMTs are mostly parallel to the axis between sister kinetochores (Fig. 1B,C). Two-color immunofluorescence was performed using the 9G3 antibody to Hec1 and polyclonal antibodies to the C-terminal heads of Spc24 or Spc25 (Supp. Fig. 1–2). We acquired high-resolution, low noise, 3-D image stacks of cells where the metaphase spindle was parallel to the coverslip surface—ensuring that several sister kinetochore pairs were in focus within the same set of optical sections (Fig. 1B). The image in each color is a convolution of the label distribution within the kinetochore and the objective point spread function (Fig. 1C, Maddox et al., 2003). As kinetochore proteins are distributed across the width of kinetochores (Fig. 1C), the “Airy Disk” image is elongated into an elliptical cross-section whose major axis defines the orientation of the kinetochore face and whose centroid defines the average position of the label along the inner-outer kinetochore axis (Fig. 1C; Suppl. Figs. 3,4). Sister pairs in or close to the same image plane were selected for analysis. A 3-D Gaussian fitting algorithm was used to obtain the 4 centroid coordinates needed to calculate the separation of the two color labels, Delta (Fig. 1D,E). This calculation scheme automatically corrected for the considerable chromatic aberration—the green label was on average shifted down and to the left of red by ~30 nm, but exact shifts varied by position within the cell. Delta measurement simulations showed that staggering of attachment site linkages by up to 150 nm along the interkinetochore (K-K) axis (Supp Fig. 3D) had little effect on Delta accuracy. However, tilt of the kinetochore face that inclines attachment linkage to the K-K axis (Supp Fig. 3D) was a source of measurement error (1% in untreated cells) that was corrected to obtain the final average value of Delta (Supp. Methods).
The average Delta value (Fig. 1E) measured between the 9G3 label near the Hec1 head and the Spc24/Spc25 C-terminus was 45 +/− 6 nm and 45 +/− 4 nm respectively (SD; n=107 sister kinetochore pairs for each combination). We also obtained the same value after correcting for the majority of lateral chromatic aberration between the 9G3 and Spc24 images (Supp. Methods and Supp. Fig 5). To assess the efficacy of the correction scheme and the accuracy of the K-SHREC method using antibody labeling we labeled Hec1 with 9G3 and used equal amounts of red and green secondary antibodies. The average Delta measured in this case was 0 +/− 5 nm (SD; n=91 sister kinetochore pairs). In a second test for accuracy, cells expressing GFP-Hec1 were labeled with anti-GFP and 9G3 – in this case, the average Delta was 3 +/− 7 nm, a value consistent with structure (Fig. 1A, Ciferri et al., 2008). Average Delta values typically had 95% confidence intervals of +/− 1–2 nm (Supp. Table 1) and two averages that differed by 3 nm or more were significantly different (paired t-test with p value <.02) because of the high signal-to-noise ratio (>~30) of the kinetochore fluorescence (Supp. Fig. 4) and the averaging of many (>100) individual Delta values. A major assumption of our Delta assay is that both sister kinetochores have the same protein architecture and stiffness. We verified this assumption by direct measurements of label separation within individual kinetochores of sister pairs for the Ndc80 complex and several other proteins (Supp. Methods and Supp. Fig 5). These and other tests (Supp. Methods) establish the Delta assay as a technique for analyzing kinetochore architecture with an accuracy of <5 nm. The 45 nm Delta value measured for Spc24/25 and Hec1 indicates that the Ndc80 complex adopts an elongated shape along the inner-outer kinetochore axis.
Correlation of Delta Measurements to Centromere Stretch Indicates that the Ndc80 Complex is Non-Compliant
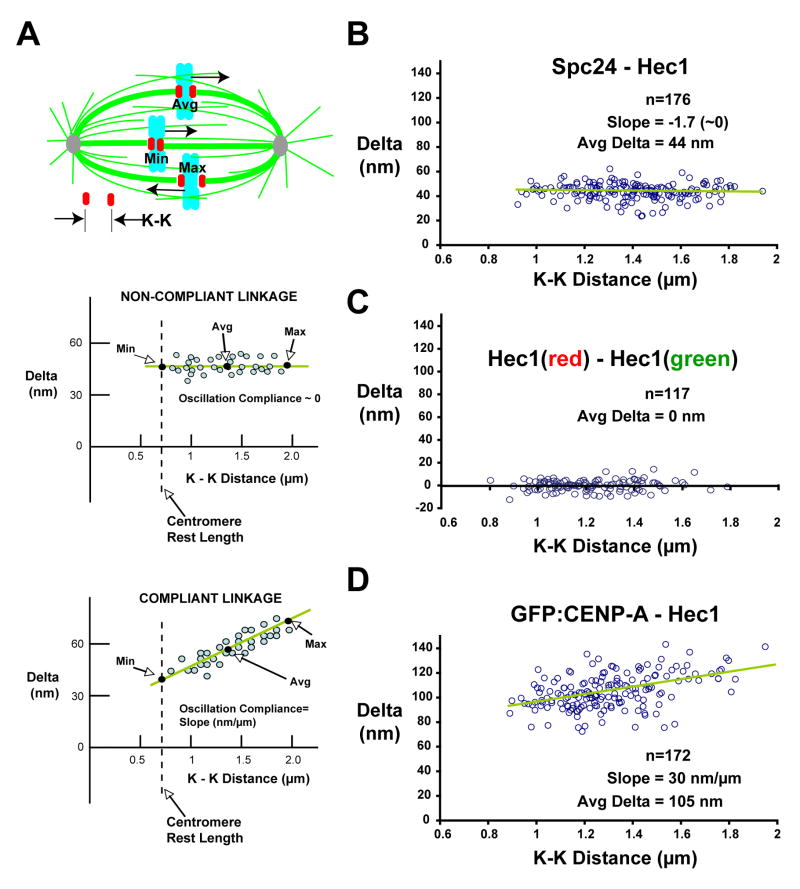
Sister kinetochores of metaphase bioriented chromosomes exhibit directional instability – oscillations characterized by abrupt switches between persistent phases of poleward and anti-poleward movement (Inoue and Salmon, 1995; Maiato et al., 2005; Skibbens et al., 1993). This directional instability produces oscillations in the stretch of centromeric chromatin between sister kinetochores that are asynchronous between different chromosomes. Consequently, a fixed image of a metaphase cell has within it sister kinetochore pairs in different mechanical states (Fig. 2A). It is straightforward to assess the mechanical state of each sister pair by measuring the K-K distance using the 9G3 label, which varies during metaphase oscillations between a minimum of ~0.8 to a maximum of ~2 μm in HeLa cells; the rest length in nocodazole-treated cells is ~0.7 μm. Thus, correlating Delta values with the inter-kinetochore K-K distance provides direct information on mechanical properties of specific protein linkages. The slope of the least-squares line through a plot of Delta versus K-K distance represents stiffness during oscillation of a particular protein linkage at metaphase, which we refer to as “Oscillation Compliance” (Fig. 2A).
Figure 2. Correlation of Delta Values with Centromere Stretch: the Ndc80 Complex Is Not Compliant and Maintains a Constant Shape.
(A) Schematic of sister kinetochore pairs in different mechanical states (top); Predictions of Delta measurements for non-compliant (middle) and compliant (bottom) protein linkages. (B) Delta values of separations between Hec1 9G3 and antibody to Spc24 C-terminus across the entire range of K-K distances. (C) Delta values for 9G3 anti-Hec1 labeling with a mixture of red/green fluorescent secondary antibodies are individually plotted versus K-K distance. For (B) and (C), measurements are insensitive to K-K separation. (D) Delta values of separations between Hec1 9G3 and GFP-CENP-A across the entire range of K-K distances show oscillation compliance. The whole dataset of Delta values uncorrected for tilt are shown; average Deltas are within 1–2 nm of corrected values (Supp. Table 1).
When Delta values measured for 9G3 and Spc24 antibody labels were plotted as a function of K-K distance, the slope was near zero, indicating that tension changes during sister kinetochore oscillations do not affect the conformation of the Ndc80 complex (Fig. 2B). The slope was also near zero for cells labeled with 9G3 and equal amounts of red and green secondary antibody (Fig. 2C), indicating that Delta measurements were insensitive to K-K separation. We conclude that the Ndc80 complex is a stiff mechanical entity within kinetochores with one end attached to the plus ends of kMTs and the other end located toward the inner kinetochore.
Human Metaphase Kinetochore Architecture from Delta Analysis of 16 Kinetochore Proteins
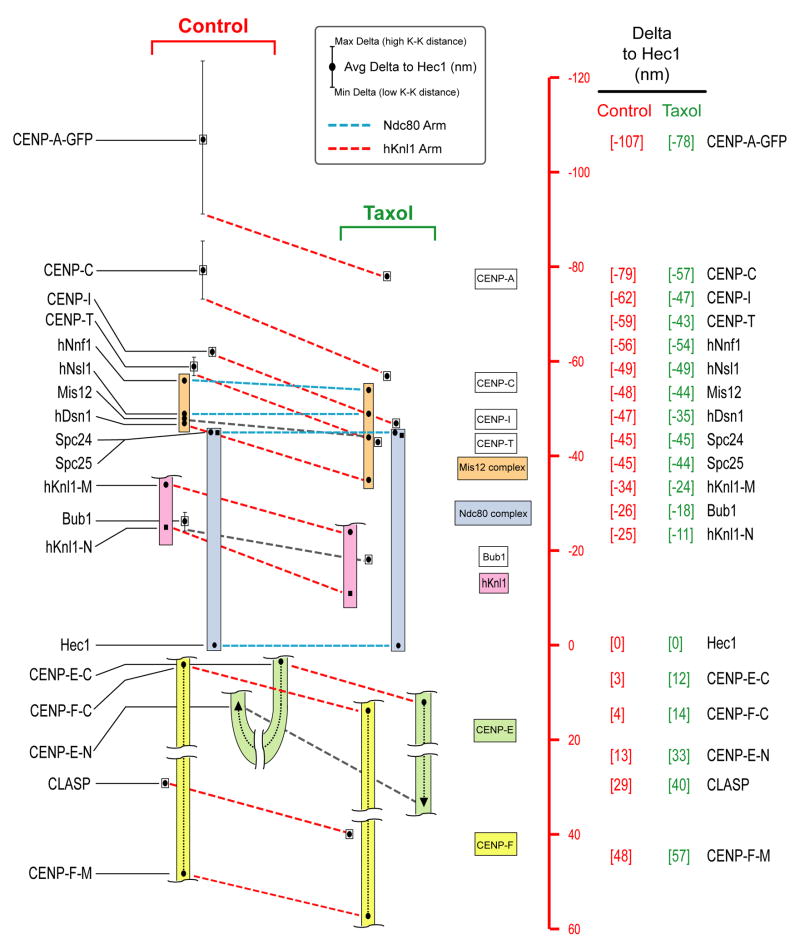
We next extended the analysis of position and mechanical properties to 19 epitopes in 16 proteins representing all of the distinct groups comprising core kinetochore structure (Supp. Table 1). For three large proteins (hKnl1, CENP-E and CENP-F), we analyzed two different regions using independent antibodies (Supp. Table 2 and Supp. Fig. 6). The entire dataset is summarized in Figure 3; the positions along the kinetochore inner-outer axis for all analyzed epitopes are plotted relative to Hec1 9G3. Positive values are outside (i.e., towards the spindle side) of the position of the 9G3 centroid and negative values are inside (i.e., towards the centromeric chromatin). All average Delta values in this position map have been corrected for inclination-tilt (Supp. Figs. 3,7; Supp. Table 1). The data for control cells is on the left of Figure 3 with dots indicating average values. The mechanical properties of each Delta value are summarized by vertical lines through the average dots which indicate minimum to maximum variation with K-K separation. The average position data for taxol-treated cells is on the right and insights derived from this perturbation are discussed in the next results section. With this overview of the entire dataset, we describe specific aspects of the measurements for each protein/protein complex.
Figure 3. Summary of Delta Measurements in Control and Taxol-Treated HeLa Cells.
Summary of Delta measurements for 19 epitopes in 16 kinetochore proteins in control cells (left) and taxol-treated cells (right). Scale (red) on the far-right is set equal to zero at the position of the Hec1 9G3 centroid; positive values are outward (towards the spindle MTs) while negative values are inward (towards the centromeric chromatin). Color-coded boxes indicate complexes. Colored dotted lines indicate proposed “arms” of the structural kinetochore. Black dots indicate average Delta values corrected for tilt. Vertical lines indicate minimum and maximum Delta values measured during oscillations in centromere stretch – for most linkages that do not show significant compliance, the vertical lines do not extend beyond the symbol used to indicate the average.
I. Constitutive Centromere-Associated Proteins
CENP-A and CENP-C
The centromeric histone H3 variant CENP-A and closely associated conserved CENP-C protein are present at centromeres throughout the cell cycle and provide a foundation for kinetochore assembly. We labeled CENP-A in two ways that gave similar results - in the first, CENP-A-GFP was stably expressed in HeLa cells (Gerlich et al., 2003) and GFP was detected with antibodies (Fig. 3). In the second, a primary antibody to CENP-A was used. As this antibody required a different fixation condition (cold methanol) than the other epitopes, we focused on data acquired with the GFP fusion and the standard aldehyde-based fixation procedure. In control cells, average Delta of CENP-A-GFP relative to the Hec1 head was 107 nm and there was a pronounced upward slope of Delta with centromere stretch (increased K-K distance) (Figs. 2D, 3). The average Delta for CENP-C, which has direct DNA binding activity (Politi et al., 2002) and lacks extended coiled-coils, was 79 nm and the oscillation compliance was ~40% of that exhibited by CENP-A (Fig. 3). From the entire set of proteins analyzed (Fig. 3), these were the only two proteins that exhibited significant oscillation compliance relative to the Ndc80 complex.
CENP-I and CENP-T
In addition to CENP-A and CENP-C, 13 additional CCAN proteins (CENP-H, -I and –K to –U) localize to centromeres throughout the cell cycle and play important roles in chromosome segregation (Foltz et al., 2006; Hori et al., 2008; Okada et al., 2006). We analyzed CENP-I and CENP-T, two representatives of different subclasses of the CCAN (Okada et al., 2006). In control cells, the average positions of CENP-I and CENP-T were about 17 and 14 nm inside the Spc24/Spc25 end of the Ndc80 complex, respectively (Fig. 3). In contrast to CENP-A and CENP-C, CENP-I and CENP-T did not exhibit significant oscillation compliance (Fig. 3, Fig. 4C, Supp. Fig 8A). This result suggests that components of the CCAN (excluding CENP-C) assemble in the 34 nm gap between the centroid of CENP-C and the Spc24/Spc25 end of the Ndc80 complex and they exhibit a stiff linkage to the Ndc80 complex during oscillations.
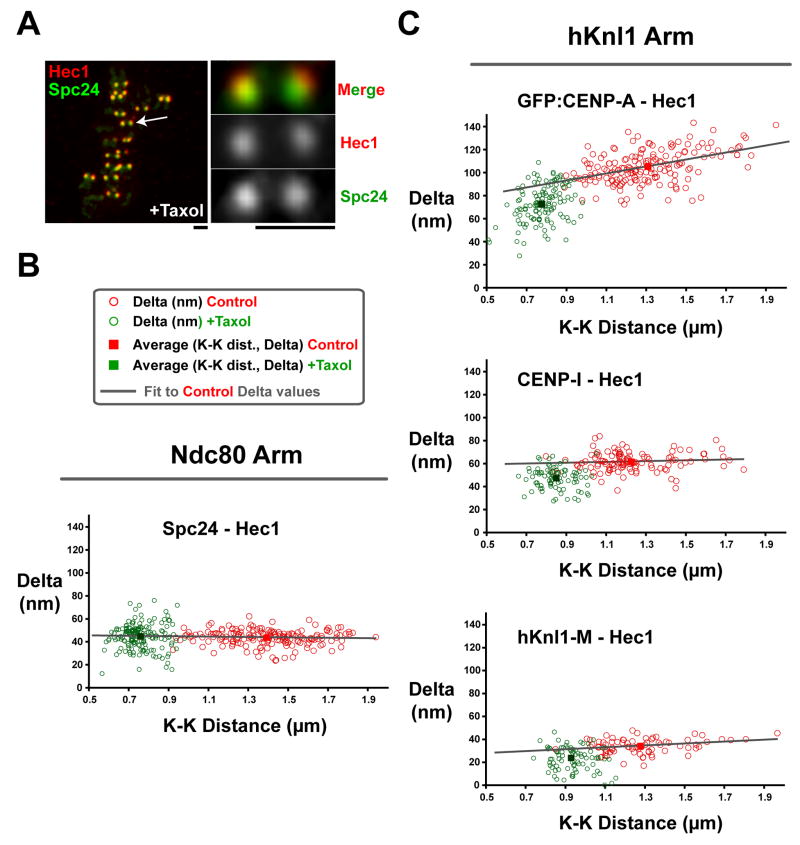
Figure 4. Changes in Kinetochore Architecture Associated with Taxol Treatment.
(A) Images of a metaphase HeLa cell treated with 10 μm taxol and then fixed and stained for Hec1 9G3 (red) and anti-Spc24 (green). Scale = 1.1 μm. (B) Graph of measured Delta value vs. K-K distance for the Hec1 and Spc24 labels in untreated (red) and taxol-treated (green) cells. (C) Graph of measured Delta value vs. K-K distance for the Hec1 label vs. GFP-CENP-A (top) and antibodies to CENP-I (middle), and hKnl1 (bottom) in untreated (red) and taxol-treated (green) cells. The whole dataset of Delta values uncorrected for tilt are shown; the average Deltas are within 1–2 nm of corrected values (Supp. Table 1).
II. The KMN Network and the Spindle Checkpoint Kinase Bub1
The KNL1/Mis12 Complex/Ndc80 Complex (KMN) proteins play a central role in kinetochore architecture and in MT binding. As described above, the two ends of the Ndc80 complex are ~45 nm apart and this complex appeared stiff with no oscillation compliance (Fig. 2B). The 4 subunits of the Mis12 complex extend from the Spc24/25 end of the Ndc80 complex to about 11 nm inside it, next to the centroid of CENP-T (Fig. 3) and also do not exhibit oscillation compliance. hKnl1 (also known as Blinkin/CASC5/AF15Q14) is a large (2342 aa) protein recruited to kinetochores by the Mis12 complex (Cheeseman et al., 2008) and is suggested to bind to the hDsn1 subunit of the Mis12 complex at its C-terminal end (Cheeseman et al., 2008; Kiyomitsu et al., 2007). hKnl1 has a predicted coiled-coil domain of 300 amino acids near its C-terminus (Cheeseman et al., 2006). We measured the position of the central region of hKnl1 using an antibody to aa 1220–1440 and the position of the N-terminus using a monoclonal antibody to the first 43 aa (Supp. Fig. 6). These epitopes were found to be on average 34 nm and 25 nm inside of the Hec1 head (Fig. 3) and neither exhibited significant oscillation compliance.
hKnl1 is part of the kinetochore binding site for SAC kinase Bub1 (Kiyomitsu et al., 2007). A Bub1 antibody raised to a region near its kinetochore-targeting domain (Supp. Fig. 6) was 26 nm internal to the Hec1 head and just inside of the N-terminal epitope in hKnl1 (Fig. 3). The distance between hDsn1, a likely marker for the C-terminal region of hKnl1 (Cheeseman et al., 2008; Kiyomitsu et al., 2007) and the N-terminus of hKnl1 was a constant 21–23 nm across the entire range of K-K distances indicating lack of oscillation compliance. This result indicates that hKnl1 is stiff like the Ndc80 complex and that its long axis is oriented primarily along the kinetochore inner-outer axis.
III. Corona Proteins
The Motor CENP-E
CENP-E is a plus-end directed dimeric motor protein, with a very long (225 nm) coiled-coil stalk and a globular tail domain (Kim et al., 2008). CENP-E has been localized by immunogold EM to the outer plate and fibrous corona (Cooke et al., 1997) and contributes to kMT attachment and kinetochore movements toward the spindle equator (Kapoor et al., 2006; Kim et al., 2008). The C-terminal kinetochore-binding domain interacts with CENP-F and BubR1 (Chan et al., 1998) and CENP-E depends on Bub1 for targeting to kinetochores (Johnson et al., 2004; Liu et al., 2006). We found the centroid of an antibody to aa 1571–1859, about 100 nm from its C-terminal kinetochore-targeting domain (Supp. Fig. 6) was about 3 nm outside of the Hec1 head (Fig. 3). Surprisingly, the centroid of an antibody to aa 663–973, about 50 nm from the motor domain of CENP-E (Supp. Fig. 6), was only ~13 nm outside the Hec1 head. Neither epitope on CENP-E exhibited any oscillation compliance (Supp. Fig. 8A, Fig. 3). This result indicates that the 225-nm long CENP-E molecule is bent with both its tail and motor domains located near the Hec1/Nuf2 heads.
The Coiled-Coil Protein CENP-F
CENP-F is a major component of the fibrous corona as determined by immunogold EM (Rattner et al., 1993). It is large (300kD, 3210aa) and possesses extensive central alpha helical coiled-coil domains (Supp. Fig. 6). CENP-F localizes to kinetochores via its C-terminus and requires hKnl1 and Bub1 (Cheeseman et al., 2008; Johnson et al., 2004). An antibody to the C-terminal 561 aa localized ~4 nm outside the Hec1 head, and a different antibody to a region in the middle of the molecule (Supp. Fig. 6, Fig. 3) was ~48 nm outside the Hec1 head, indicating that CENP-F is primarily oriented perpendicular to the outer plate.
The MT Polymerization-Promoting Protein CLASP
CLASP is a MT-binding protein that promotes polymerization, suppresses depolymerization, and concentrates at the growing tips of MTs in the cytoplasm of interphase cells (Maiato et al., 2005). CLASP is concentrated within the very periphery of the kinetochore during mitosis. We find that the centroid of CLASP is 29 nm outside of the Hec1 head in control metaphase cells (Fig. 3). This position is within the C-terminal half of CENP-F or the loop of CENP-E extending into the corona, but not near the location expected for the plus ends of the majority of kMTs, which is near the inner surface of the kinetochore outer plate (Dong et al., 2007; VandenBeldt et al., 2006).
Large Changes in Kinetochore Protein Architecture Are Observed Following Taxol Treatment
The anti-cancer agent taxol suppresses polymerization dynamics of MTs. Taxol treatment (10 μM) at metaphase eliminates tension at kinetochores and activates the spindle checkpoint (Waters et al., 1998). The mean inter-kinetochore distance in taxol-treated metaphase cells is about 0.75 μm (Fig. 4A), only slightly less than the minimum 0.8 μm value that transiently occurs during oscillations (Fig. 2). This similarity suggests that Delta measurements in taxol-treated cells should resemble values observed at lowest K-K distance for linkages with oscillation compliance (CENP-A and CENP-C) and similar to average values for stiff non-compliant linkages. Consistent with this, the dimensions of the stiff Ndc80 complex remained constant in taxol (Figs. 3, 4B). However, somewhat surprisingly, there were several significant changes induced in kinetochore architecture in taxol-treated cells. Centroids of CENP-A, CENP-C, CENP-I, and CENP-T all moved ~16 nm closer to the position of the Spc24/25 end of the Ndc80 complex (Figs. 3, 4C). This movement did not change the relative separation distance between CENP-I and CENP-T centroids that do not exhibit oscillation compliance. In addition, separation between CENP-I/CENP-C centroids and CENP-I/CENP-A centroids was the same as that observed at minimal centromere stretch (K-K distance) during normal control oscillations. Thus, the entire inner kinetochore is ~16 nm closer to the Spc24/25 end of the Ndc80 complex in taxol-treated cells.
The Mis12 complex also exhibited a striking change in taxol-treated cells (Fig. 3). In control cells, the distance along the inner-outer axis from the hNsl1 subunit at the interior end of the Mis12 complex and the hDsn1 subunit at the exterior end was constant during oscillations at ~9 nm; in taxol this length became 19 nm. Two components at one end of this elongated complex, hNnf1, and hNsl1/Mis14 did not significantly change position relative to Spc24 after taxol treatment; Mis12 showed modest (4 nm) outward movement. In contrast, a striking effect was observed for hDsn1, which shifted ~12 nm outward in taxol-treated cells.
hDsn1 likely directly binds to the C-terminal region of hKnl1 (Kiyomitsu et al., 2007); consistent with this, hKnl1 also exhibited outward movement similar to hDsn1. The centroid of the Bub1 epitope also moved outward, but less than expected for its putative binding site near the N-terminus of hKnl1; this lack of movement may indicate changes in Bub1 binding sites following taxol treatment.
The C-terminal regions of large corona proteins CENP-E and CENP-F also moved ~10 nm outward relative to Hec1/Nuf2 heads after taxol treatment (Fig. 3). The most striking change observed was with CENP-E: the epitope close to the motor domain moved from near the Hec1 head (and its own C-terminus) in controls to 33 nm beyond the Hec1 head. This result suggests that CENP-E changes from being bent in control metaphase cells to an extended conformation in taxol-treated cells.
A major pattern emerging from comparison of the taxol-treated and control cell Delta measurements suggests relative movement of two protein sets whose constituents behave as if they are coupled – we refer to each of these protein sets as “arms” in order to connote a multi-part mechanical entity. The Ndc80 arm (whose average Delta values are linked using blue dotted lines between control and taxol treatment in Fig. 3) is comprised of the Ndc80 complex and hNnf1, hNsl1 and part of Mis12—none of these show significant taxol-specific changes in separation. The hKnl1 arm (whose average Delta values are linked using red dotted lines between control and taxol treatment in Fig. 3) is comprised of CENP-A, CENP-C, CENP-I, CENP-T, hDsn1 and part of the hMis12 subunits, hKnl1, Bub1, CENP-F, and the C-terminal end of CENP-E – all of these move ~10–16 nm outward relative to the Hec-1 head (for compliant CENP-A and CENP-C linkages, this movement is relative to the Delta value measured at minimal K-K stretch, i.e. lowest tension state in control cells—see point where lines linking the control and taxol data sets are drawn in Fig. 3). As distances between the inner kinetochore proteins (e.g. CENP-I/CENP-T) and proteins in the Ndc80 arm decrease in taxol-treated cells, we conclude that the Ndc80 arm moves in towards the centromeric chromatin in taxol-treated cells while the hKnl1 arm does not. This shift, which is not observed at minimal K-K distance in control cells, suggests a mechanism in taxol-treated cells leading to uncoupling and separation of these normally coupled protein arms.
In summary, taxol treatment did not recapitulate kinetochore architecture at lowest K-K distance but instead revealed large-scale changes in different regions of the kinetochore. These changes may arise from persistent absence of mechanical tension (as opposed to transient absence in oscillating kinetochores), activation of the SAC, or both.
Location of the Plus Ends of Kinetochore MTs Relative to the Ndc80 Complex
To determine the position of bound kMTs we analyzed metaphase PtK2 cells cooled to 6°C (Supp. Methods). Under these conditions, only kMTs persist, a full complement of kMTs penetrating the outer plate is present (Rieder, 1981), and kMT fibers mainly orient perpendicular to kinetochores, which makes them remain in focus several μm beyond kinetochores (Fig. 5A,B). Such images could not be obtained in metaphase HeLa cells because of the high degree of spindle and kinetochore fiber curvature. In cold-treated PtK2 cells, centromeres were unstretched with average K-K distance similar to that in nocodazole – thus the measurements of kMT position only apply to this low-tension state.
Figure 5. A Two-Color Method for Locating the Plus Ends of kMTs Relative to the Hec1 Head in Cooled PtK2 Cells.
(A) Fluorescent image of PtK2 cells stably expressing GFP-α-tubulin cooled to 6°C, fixed, and stained with the Hec1 9G3 antibody and a red fluorescent secondary. The image shows a kinetochore fiber and its kinetochore in the same focal plane. At right is a magnified image of the boxed region, showing how linescans were drawn down centers of the fibers through the centroids of Hec1 9G3 fluorescence. (B) Sample linescan of (A) showing GFP-α-tubulin intensity (green) and Hec1 9G3 (red) fluorescent intensity along the linescan. (C) Plot of all normalized linescans (n=92), with Hec1 9G3 centroid set to zero for each on the x-axis and the error function (purple) that best fits the data set. Blue lines mark x and y positions of the 50% amplitude of the error function.
Preliminary experiments indicated that antibodies to tubulin do not penetrate kinetochores as fluorescence ended in front of the Hec1 9G3 label (data not shown). Therefore, we directly imaged fluorescence of GFP-α-tubulin incorporated into kMTs (Rusan et al., 2001) and obtained intensity profiles along bundles of green fluorescent kMTs through the centroid of the red Hec1 9G3 label (Fig 5B,C). Different linescans (n= 92) were plotted on the same axes by setting the position of the 9G3 centroid to zero and normalizing intensity values (Supp. Methods). The position of the 50% intensity point was determined by fitting an error function to the cumulative data (Fig. 5C). The average position of the kMT end determined with this method was 62.3 +/− 15 nm (95% confidence interval) inside of the 9G3 label centroid (Fig. 5C). The variance is large because fibers oriented at different angles within the field of view had different amounts of lateral chromatic aberration, which was averaged out by an equal number of fibers facing in all directions (Supp. Fig. 9). These results indicate that, on average at 6°C in PtK2 cells, the plus ends of kMTs extend 10–15 nm inside the Spc24/Spc25 ends of the Ndc80 complexes. In taxol-treated cells, this position would be at the periphery of the inner centromere (Fig. 3).
DISCUSSION
The Chromatin-Proximal Region of the Kinetochore has Distinct Structural and Mechanical Domains
At minimal centromere stretch, separation between centroids of CENP-I/T and CENP-C is ~11 nm and between CENP-I/T and CENP-A is ~30 nm (Fig. 3). As biochemical studies support close associations between CENP-I/T and CENP-A nucleosomes (Foltz et al., 2006; Obuse et al., 2004; Okada et al., 2006), these values suggest that only a small fraction of chromatin-bound CENP-A and CENP-C is exposed at the peripheral surface of the centromere on a metaphase chromosome in a position to bind components of the CCAN. The depth of CENP-C and CENP-A chromatin from the base of the kinetochore increases with centromere stretch (from ~22 and ~60 nm, respectively, at minimal stretch to 46 and 128 nm, respectively, at maximal stretch; Figs. 2,3) assuming a uniform distribution of each within chromatin (Supp. Fig. 10A). These values are small compared to the 420 nm diameter of a green fluorescent Airy disk and would not be resolvable by conventional fluorescence microscopy. However, they are consistent with past reports that CENP-C concentrates near the base of the kinetochore (Saitoh et al., 1992), while CENP-A extends further inside the centromere (Blower et al., 2002; Amor et al., 2004) and that only 10% of the CENP-A is sufficient to build a functional kinetochore (Liu et al., 2006).
In contrast to the compliance of chromatin at the base of the kinetochore containing CENP-A and CENP-C, protein linkages between CENP-I and CENP-T and between these CCAN subunits and the outer kinetochore were stiff in control cells (Fig. 3). Thus, for oscillating metaphase chromosomes, the CCAN is assembled at the very periphery of the CENP-A/C-containing centromeric chromatin (Supp. Fig. 10A) and is tightly linked to the outer kinetochore.
The Ndc80 Complex is Likely Bent Along its Length and Connected to the Inner Kinetochore by a Flexible Linkage
During metaphase in both untreated and taxol-treated human cells the Delta between markers at the two ends of the 57 nm-long Ndc80 complex was a constant 45 nm. A previous study of isolated Drosophila chromosomes used a linescan method and reported ~22 nm separation between the ends of the Ndc80 complex (Schittenhelm et al., 2007). We have measured a Delta of ~18 nm between the Spc24-C terminus and 9G3 just inside the Hec1 head in nocodazole-treated HeLa cells (data not shown), whereas this distance from the structure is 54.5 nm. The lower numbers in nocodazole may indicate flexibility in orientation of the Ndc80 complex and bending of the rod domain at a kink site (Fig. 1A, Ciferri et al., 2008; Wang et al., 2008) in the absence of attached kMTs. However, they may also result from measurement errors induced by severe tilt and/or curvature of the kinetochore face relative to the inner-outer kinetochore axis—such curvature has been observed following extended mitotic arrest in the absence of MTs (DeLuca et al., 2005).
A 45 nm average Delta value is predicted if the 57 nm-long Ndc80 complex extends straight from the surface of the bound kMT at an angle θ = ~34°, similar to the angle that the rod domain of the Ndc80/Nuf2 dimer exhibits when bound in vitro to purified MTs (Cheeseman et al., 2006; Wilson-Kubalek et al., 2008). However, this inclined straight conformation puts the C-terminal ends of Spc24/Spc25 ~32 nm radially outside the surface of a kMT. A pulling force, F, at heads bound to the MT lattice generates at the Spc24/Spc25 end both a pulling force, F, along the inner-outer kinetochore axis and a radial inward force (equal to Fsin(θ)/cos(θ)). For θ = 34°, the radial force is ~67% of F. If unopposed, the radial force would move Spc24/Spc25 ends up close to the surface of the kMT, resulting in an average Delta value similar to the label separation of 54.5 nm along the length of the Ndc80 complex. However, we did not observe this even under maximal centromere stretch suggesting that this radial force is opposed by a mechanism anchoring the Spc24/25 end of the Ndc80 complex or that the complex does not adopt a straight conformation. Anchorage by lateral linkages between adjacent kMT-attachment sites (a “load sharing mechanism”) that are of similar strength to the inner-outer linkages is unlikely because the kinetochore is weak laterally. During merotelic attachments, when a single kinetochore is pulled towards opposite poles by kMTs, lateral stretch of kinetochore proteins and peripheral centromeric chromatin often occurs for >1 μm (Cimini et al., 2004; Cimini et al., 2001; Supp. Fig. 10B).
An alternative explanation for the 45 nm separation of the labels at the two ends is that the Ndc80 complex is bent (Fig. 6A). There are a number of reasons to favor this idea. There is a conserved break in the coiled-coil rod domain about 16 nm inside the Nuf2/Hec1 heads (Fig. 1A; Ciferri et al., 2008). Recent EM analyses of purified Ndc80 complexes in vitro (Wang et al., 2008) indicate that flexible bending occurs at this site within the rod domain that connects the two globular ends of the Ndc80 complex (Fig. 1A). The existence of a flexible bend does not, however, explain constancy of Ndc80 complex dimensions across the entire range of centromere stretch and in taxol - a fixed angle bend would have to exist even under tension in order to account for this constancy and 45 nm separation of the two end labels. This consideration assumes that the majority of Ndc80 complexes are bound to kMTs, which is compatible with the requirement of this complex for the SAC and inactivation of the SAC at metaphase (Musacchio and Salmon, 2007).
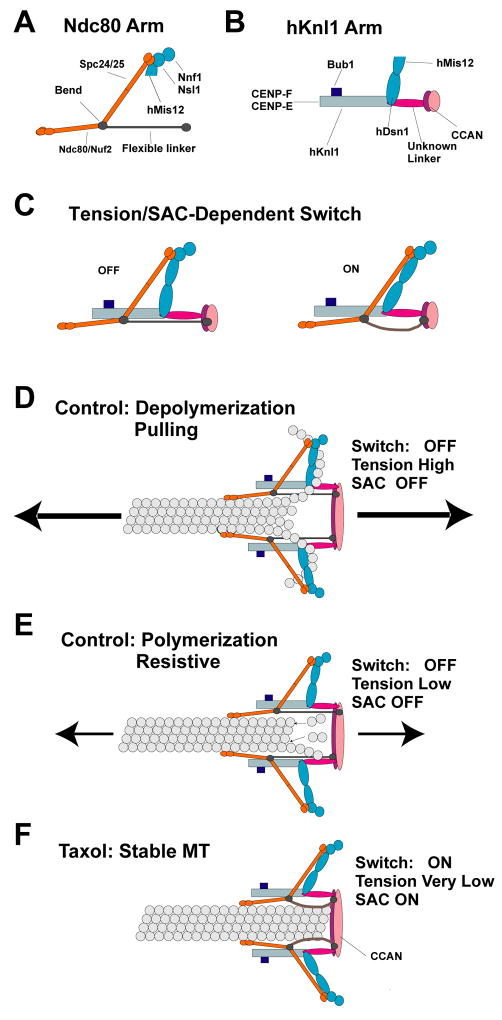
Figure 6. Centromere and Kinetochore Protein Architecture, Mechanics, Tension Sensing and Force Generation.
Schematics of the Ndc80 arm (A) and the hKnl1 arm (B) revealed by comparison of control and taxol-treated metaphase cells. (C) A tension/SAC activation-dependent intra-kinetochore switch produced by a 15 nm translocation of the Ndc80 arm relative to the hKnl1 arm. This translocation is proposed to occur by rotation of Mis12 and hDsn1 subunits coupled to relaxation of a flexible filament-like linkage between the Ndc80 Arm and the inner kinetochore. (D–F) Models of the protein architecture of the KMN network of proteins within a kinetochore MT attachment site for depolymerizing ends (D), polymerizing ends (E), and taxol-stabilized ends (F).
A proposal we favor to account for the 45 nm distance is that there is a protein complex bound at the bend site that stabilizes the bend and links the Ndc80 complex to the inner kinetochore (Fig. 6A). Upon binding of the Ndc80 heads to the kMT lattice, the coil-coiled region between the head and the bend/linker attachment site extends and transmits a pulling force in an outer-inner direction to the bend site and along the hypothetical linker protein to the inner kinetochore – in this case there is no inward radial force as the direction of force transmission is along the inner-outer axis. In this configuration, the Spc24/Spc25 end of the complex bends outward from the kMT lattice by ~20 nm to produce the ~45 nm inner-outer separation between the two end labels. This model produces little to no radial force at either end of the molecule and binding of the linker may force a constant bend angle at the otherwise flexible break in the coiled-coil. It is possible that the CENP-H subunit of the CCAN, which is primarily coiled-coil and has been shown to interact with the Ndc80 complex (Cheeseman et al., 2008; Hori et al., 2008; Okada et al., 2006), may constitute such a linker. Nevertheless, as discussed below, such a linker must be flexible.
Movement of the Ndc80 Arm Relative to the Inner Kinetochore and the hKnl1 Arm: A Low Tension/Checkpoint-Activated Switch
We propose that the Ndc80 arm includes a flexible filament-like linkage between the bend in the Ndc80 complex and the inner kinetochore (Fig. 6A, C–F). Such a linker would explain why the Ndc80 arm is able to move 15 nm towards the inner kinetochore in taxol (the filament buckles either due to persistent low tension and/or due to SAC activation), but remains constant in position relative to the inner kinetochore at high tension (the extended filament is stiff). A flexible linker (not shown for clarity in Fig. 6) may also exist between hNsl1 of the Mis12 complex and the inner kinetochore as suggested by recent EM images (McIntosh et al., 2008). In contrast, the hKnl1 arm connects separately to the inner kinetochore and this connection is stiff and does not change in taxol-treated cells (Fig. 6F).
Since the four subunits of the Mis12 complex are linked together (Kline et al., 2006), the best way to merge the control and taxol configurations is to assume that in controls the Mis12 complex extends mostly in a lateral direction (Figs. 3, 6C–F). In taxol, when the Ndc80 arm moves relative to the hKnl1 arm, the Mis12 complex rotates such that the hDsn1 subunit makes the largest translation (Fig. 6C, F). Movement of the Ndc80 complex toward the inner centromere can be explained by outward movement of the hKnl1 arm along stable kMTs until further movement is blocked by the kMT end (Figs. 3, 6F). This outward movement may be driven by the minus motor activity of cytoplasmic dynein linked to Knl1 by other proteins (Stehman et al., 2007; Vergnolle and Taylor, 2007; Kiyomitsu et al., 2007; Cheeseman et al. 2008).
The low-tension/SAC switch within the kinetochore may be part of the tension-sensing mechanism that controls the stability of kMT attachment and/or SAC signaling at kinetochores - the Ndc80 complex is required for both of these essential functions (Musacchio and Salmon, 2007). Another process the switch may regulate is the conformation of the CENP-E motor - in controls, the 225 nm-long CENP-E molecule is bent with markers for its tail and motor domains located near the Hec1 heads, but in taxol the marker proximal to the motor domain of CENP-E moved more than 33 nm beyond the Hec1 heads into the coronal region (Fig. 3). This dramatic conformational change is likely to involve kinase activity (Espeut et al., 2008; Kim et al., 2008), which in turn may be modulated by the switch that we describe here.
The Ndc80 Arm Could Contribute in Two Ways to MT Attachment and Pulling Force Generation
Multiple Ndc80 arms may contribute to a “Hill-like” mechanism, where the dynamic binding of their Hec1 and Nuf2 heads to a kMT helps hold the attachment site near either a growing or shortening plus end (Asbury and Davis, 2008; Hill, 1985; Fig. 6D-E). In budding yeast, there are 8 Ndc80 complexes per kMT (Joglekar et al., 2006) which supports this possibility - the number and distribution along the inner-outer axis in human cells are not yet established. The Ndc80 arm within HeLa kinetochores could also act as a force transducer. In budding yeast, recent analysis of kinetochores at metaphase indicates that the Ndc80 complex is extended its full length along the axis of kMTs (Ajit Joglekar, personal communication). A DAM/DASH ring has received much attention as a force transducer in budding yeast for the rearward peeling of tubulin protofilaments (Asbury and Davis, 2008; Efremov et al., 2007; Tanaka and Desai, 2008). However, in mammalian kinetochores no such rings or corresponding protein subunits have been identified and in fission yeast, the DAM/DASH proteins are non-essential (McIntosh, 2005; McIntosh et al., 2008). Within HeLa kinetochores, the bent configuration of the Ndc80 complex and its lateral linkage by an elongated Mis12 complex to the hKnl1 homolog (Fig. 6E) could act in place of a ring for transmitting pulling forces generated by curling protofilaments to the inner kinetochore (Fig. 6D).
Experimental Procedures
The basic procedures used for Delta measurements of the separation of fluorescent labels on kinetochore proteins in HeLa cells at metaphase and the plus ends of kMTs within PtK2 cells at 6°C are described in the Results section along with the rationale behind the development of the Delta assay. More details about the different measurement techniques, including the use of the SHREC method (Churchman et al., 2005) for two-color label co-localization within individual kinetochores, are given in Supplementary Methods.
Supplementary Material
Acknowledgments
We are very grateful to a number of scientists who provided well characterized reagents including Dr. Steve Taylor, Dr. Mitsuhiro Yanagida, Dr. Tomomi Kiyomitsu, Dr. Aaron Straight, and Dr. Jan Ellenberg. We also thank Dr. Kerry Bloom, Dr. Andrea Musacchio, and members of the Salmon and Desai labs for their critiques of early versions of this manuscript. Funded by NIHGMS 24364 (EDS); GM06627 (BFM); GM074215 (AD). APJ holds a Career Award at the Scientific Interface from the Burroughs-Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amor DJ, Kalitsis P, Sumer H, Choo KH. Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol. 2004;14:359–368. doi: 10.1016/j.tcb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Asbury CL, Davis TN. Insights into the kinetochore. Structure. 2008;16:834–836. doi: 10.1016/j.str.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley BR, Ouspenski I, Zinkowski RP. Structure and molecular organization of the centromere-kinetochore complex. Trends Cell Biol. 1992;2:15–21. doi: 10.1016/0962-8924(92)90139-e. [DOI] [PubMed] [Google Scholar]
- Chan GK, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K Complex Coordinately Direct Kinetochore Assembly in Vertebrates. Mol Biol Cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. PNAS. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Schaar B, Yen TJ, Earnshaw WC. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Dong Y, Vanden Beldt KJ, Meng X, Khodjakov A, McEwen BF. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov A, Grishchuk EL, McIntosh JR, Ataullakhanov FI. In search of an optimal ring to couple microtubule depolymerization to processive chromosome motions. PNAS. 2007;104:19017–19022. doi: 10.1073/pnas.0709524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J, Gaussen A, Bieling P, Morin V, Prieto S, Fesquet D, Surrey T, Abrieu A. Phosphorylation relieves autoinhibition of the kinetochore motor Cenp-E. Mol Cell. 2008;29:637–643. doi: 10.1016/j.molcel.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. PNAS. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Okada M, Maenaka K, Fukagawa T. CENP-O Class Proteins Form a Stable Complex and Are Required for Proper Kinetochore Function. Mol Biol Cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Salmon ED. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol Biol Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Moree B, Canman JC, Salmon ED. Spinning disk confocal microscope system for rapid high-resolution, multimode, fluorescence speckle microscopy and green fluorescent protein imaging in living cells. Methods Enzymol. 2003;360:597–617. doi: 10.1016/s0076-6879(03)60130-8. [DOI] [PubMed] [Google Scholar]
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- Maiato H, Hergert PJ, Moutinho-Pereira S, Dong Y, Vandenbeldt KJ, Rieder CL, McEwen BF. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma. 2006;115:469–480. doi: 10.1007/s00412-006-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh AD, Meraldi P, Draviam VM, Toso A, Sorger PK. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. Embo J. 2006;25:4033–4049. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR. Rings around kinetochore microtubules in yeast. Nat Struct Mol Biol. 2005;12:210–212. doi: 10.1038/nsmb0305-210. [DOI] [PubMed] [Google Scholar]
- McIntosh JR, Grishchuk EL, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Politi V, Perini G, Trazzi S, Pliss A, Raska I, Earnshaw WC, Della Valle G. CENP-C binds the alpha-satellite DNA in vivo at specific centromere domains. J Cell Sci. 2002;115:2317–2327. doi: 10.1242/jcs.115.11.2317. [DOI] [PubMed] [Google Scholar]
- Rattner JB, Rao A, Fritzler MJ, Valencia DW, Yen TJ. CENP-F is a .ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil Cytoskeleton. 1993;26:214–226. doi: 10.1002/cm.970260305. [DOI] [PubMed] [Google Scholar]
- Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1981;84:145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- Rieder CL. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- Rusan NM, Fagerstrom CJ, Yvon AM, Wadsworth P. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol Biol Cell. 2001;12:971–980. doi: 10.1091/mbc.12.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Tomkiel J, Cooke CA, Ratrie H, 3rd, Maurer M, Rothfield NF, Earnshaw WC. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Schittenhelm RB, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, Lehner CF. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 2007;116:385–402. doi: 10.1007/s00412-007-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehman SA, Chen Y, McKenney RJ, Vallee RB. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J Cell Biol. 2007;178:583–594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Desai A. Kinetochore-microtubule interactions: the means to the end. Curr Opin Cell Biol. 2008;20:53–63. doi: 10.1016/j.ceb.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBeldt KJ, Barnard RM, Hergert PJ, Meng X, Maiato H, McEwen BF. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr Biol. 2006;16:1217–1223. doi: 10.1016/j.cub.2006.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle MA, Taylor SS. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr Biol. 2007;17:1173–1179. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and Flexibility of the Yeast Ndc80 Kinetochore Complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and Structure of the Ndc80 Complex on the Microtubule Lattice. J Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.