Abstract
The Mre11–Rad50–Nbs1 (MRN) complex functions in the repair of DNA double-strand breaks (DSBs) by homologous recombination (HR) at postreplicative stages of the cell cycle. During HR, the MRN complex functions directly in the repair of DNA DSBs and in the initiation of DSB responses through activation of the ataxia telangiectasia-mutated (ATM) serine-threonine kinase. Whether MRN functions in DNA damage responses before DNA replication in G0/G1 phase cells has been less clear. In developing G1-phase lymphocytes, DNA DSBs are generated by the Rag endonuclease and repaired during the assembly of antigen receptor genes by the process of V(D)J recombination. Mice and humans deficient in MRN function exhibit lymphoid phenotypes that are suggestive of defects in V(D)J recombination. We show that during V(D)J recombination, MRN deficiency leads to the aberrant joining of Rag DSBs and to the accumulation of unrepaired coding ends, thus establishing a functional role for MRN in the repair of Rag-mediated DNA DSBs. Moreover, these defects in V(D)J recombination are remarkably similar to those observed in ATM-deficient lymphocytes, suggesting that ATM and MRN function in the same DNA DSB response pathways during lymphocyte antigen receptor gene assembly.
DNA double-strand breaks (DSBs) are generated by genotoxic agents and as intermediates of several important physiological processes including antigen receptor gene assembly in developing lymphocytes. The cellular response to DNA DSBs relies on the sensing of these breaks and the subsequent initiation of effector pathways that enforce cell cycle checkpoints, promote DSB repair, and mediate cell death when DSBs are not efficiently repaired (1–4). DNA DSBs generated at stages of the cell cycle after DNA replication are repaired primarily by homologous recombination (HR), a process that uses the sister chromatid as a template for precise repair (4). DNA DSBs generated before DNA replication in G0/G1-phase cells are repaired primarily through nonhomologous end joining (NHEJ), which religates broken DNA ends in a manner that can be imprecise (4). Very little overlap exists in the protein machinery that carries out DSB repair by NHEJ and HR.
The Mre11, Rad50, and Nbs1 proteins form the Mre11–Rad50–Nbs1 (MRN) complex, which is thought to function primarily in DNA DSB responses in cells that have undergone DNA replication (5, 6). MRN is recruited to the sites of newly generated DNA DSBs where it functions to recruit and activate the ataxia telangiectasia-mutated (ATM) serine/threonine kinase, which is a major initiator of DNA damage responses (7–12). However, ATM may be activated in an MRN-independent fashion in response to some DSBs like those generated at stalled replication forks, suggesting that the requirement for MRN in initiating DSB responses may be context dependent (9). MRN also has several DNA damage response functions that are downstream of ATM activation. In this regard, analyses of Nbs1 mutants have implicated MRN in the regulation of cell cycle checkpoints and the activation of apoptotic pathways in response to DNA DSBs (13, 14). Mre11 has endonuclease and exonuclease activities that are important for DNA end processing, and Mre11 dimers may align and bridge two DNA ends during HR-mediated DSB repair (5, 6, 15). Rad50 also has DNA binding activities that may be involved in tethering sister chromatids during HR (16–20). In response to DSBs, ATM phosphorylates Mre11, Rad50, and Nbs1, which could potentially modulate their functions in DSB responses (21–24). Importantly, although Mre11, Nbs1, and Rad50 have distinct functions, these individual components are thought to function only in the context of the MRN holocomplex.
Whether the MRN complex functions in the response to and NHEJ-mediated repair of DSBs in G0/G1-phase cells has been much less clear. DNA end joining by NHEJ components purified from human cellular extracts was augmented by MRN in vitro; however, the repair activity of purified Xenopus laevis NHEJ components was not affected by the addition of MRN (25, 26). MRX, the yeast orthologue of MRN, functions during NHEJ in the budding yeast Saccharomyces cerevisiae but not in the fission yeast Schizosaccharomyces pombe (27–29). In mammalian cells, MRN is not recruited to the site of DSBs generated by the I-sceI endonuclease in G1-phase cells, and MRN does not appear to be required for the NHEJ-mediated repair of these DSBs (30, 31).
DNA DSBs are generated in all developing lymphocytes during the assembly of the second exon of antigen receptor genes from component V, J, and, in some cases, D gene segments (32). This occurs through the process of V(D)J recombination, which is initiated by the Rag-1 and Rag-2 proteins, which together form an endonuclease, hereafter referred to as Rag (33, 34). Rag introduces DSBs at the borders of two recombining gene segments and their associated Rag recognition sequences, which are termed recombination signals (RSs). The generation of these DSBs is restricted to cells at the G1 phase of the cell cycle as a result of the rapid degradation of Rag-2 upon entry into S phase (35). DNA cleavage by Rag leads to the formation of two hairpin-sealed coding ends and two blunt phosphorylated signal ends. These DNA ends are processed and joined by the NHEJ pathway of DNA DSB repair into a coding joint and signal joint, respectively (36, 37). The critical dependence of V(D)J recombination on NHEJ is indicated by the severe joining defects in NHEJ-deficient cell lines and the profound immunodeficiency observed in mice deficient for NHEJ factors required for the repair of Rag-mediated DNA DSBs (36).
Mice with homozygous-null mutations in the Mre11, Nbs1, or Rad50 genes exhibit early embryonic lethality; however, mice and humans with hypomorphic Mre11 or Nbs1 mutations are viable and exhibit mild immunodeficiency, suggesting that the MRN complex could function in the response to or repair of Rag-mediated DSBs generated during V(D)J recombination (9, 38–45). In developing lymphocytes, Rag DSBs activate ATM, which initiates a broad genetic program and functions in the repair of these breaks (46–53). In this regard, MRN could function in the activation of ATM in response to Rag-mediated DSBs and also downstream of ATM in the repair of these DSBs. Consistent with this notion, Nbs1 associates with Rag-mediated DSBs generated at T cell receptor loci in thymocytes and MRX function is required for the joining of signal ends generated by Rag cleavage in yeast (54, 55). However, analyses of V(D)J recombination of extrachromosomal substrates in mammalian nonlymphoid cells deficient in MRN have failed to reveal any significant defects in coding or signal joint formation (56–58). In this paper, we show that MRN-deficient lymphocytes exhibit defects in V(D)J recombination at endogenous antigen receptor loci and chromosomally introduced recombination substrates. These findings establish a function for MRN in the response to Rag-mediated DSBs generated in G1-phase lymphocytes.
RESULTS
Generation of Nbs1m/m and Mre11ATLD1/ATLD1 abl pre–B cells
We have developed an approach for analyzing the response to and repair of chromosomal Rag-mediated DSBs in v-abl–transformed pre–B cell lines, which are hereafter referred to as abl pre–B cells (48–50). Treatment of these cells with the v-abl kinase inhibitor STI571 leads to G1 cell cycle arrest, induction of Rag expression, and the generation of Rag DSBs at the endogenous IgL-κ locus and at chromosomally introduced retroviral recombination substrates (48–50, 59). In WT G1-phase abl pre–B cells these DSBs are rapidly repaired, whereas in cells deficient in specific DNA repair proteins these DSBs can persist unrepaired for substantial periods of time (48–50).
Mice with homozygous-null mutations at the Mre11, Nbs1, or Rad50 loci exhibit early embryonic lethality, preventing the generation of abl pre–B cells that are completely deficient of any of these proteins (9, 43–45). However, mice that express hypomorphic Nbs1 or Mre11 alleles are viable (9, 38–40). We generated several abl pre–B cell lines from mice homozygous for hypomorphic Mre11 (Mre11ATLD1) and Nbs1 (Nbs1m) alleles (38, 40). The Mre11ATLD1 allele has a gene-targeted point mutation that generates a premature stop codon, resulting in a C-terminal truncation of the Mre11 protein, which mimics a mutation that causes ataxia-telangiectasia–like disease (40). Exons 2 and 3 of the Nbs1 allele were replaced with a neomycin resistance gene generating the Nbs1m allele, which encodes a truncated Nbs1 protein (38).
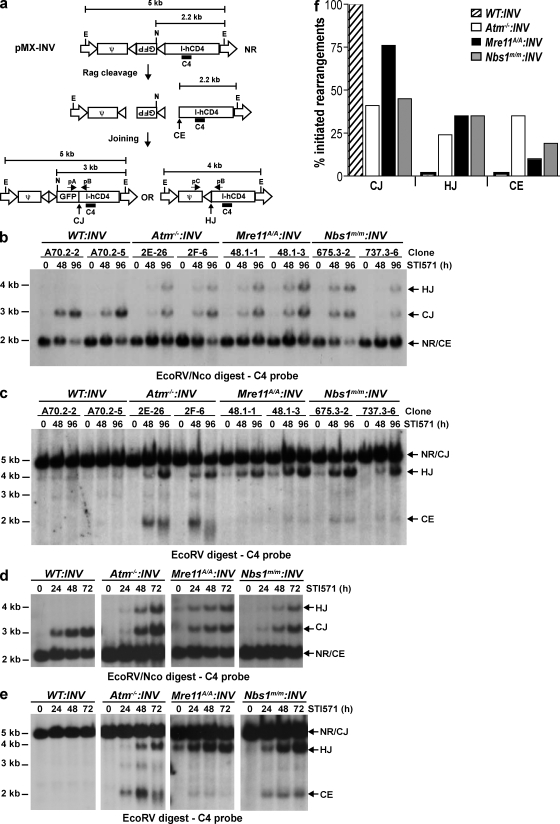
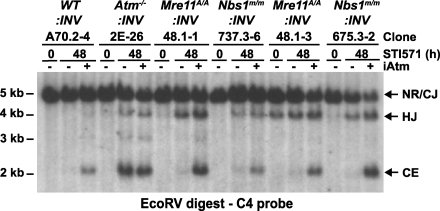
Independently derived WT, Atm−/−, Nbs1m/m, and Mre11ATLD1/ATLD1 (referred to as Mre11A/A in figures) abl pre–B cell lines were transduced with the pMX-INV retroviral recombination substrate (Fig. 1 a). The RSs in pMX-INV are oriented such that rearrangement occurs by inversion, with the coding and signal joints remaining in the chromosome (Fig. 1 a). Bulk populations of abl pre–B cells with pMX-INV integrants were purified by flow cytometric cell sorting for the expression of human CD4, which is encoded by the retrovirus (50). Abl pre–B cells with single pMX-INV integrants were isolated by limiting dilution. We analyze pMX-INV rearrangement both in clones with single pMX-INV integrants (Fig. 1, b and c; and Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20081326/DC1) and in original bulk populations (Fig. 1, d and e) of abl pre–B cells that have pMX-INV integrants at broadly heterogenous genomic locations (50).
Figure 1.
Rearrangement of pMX-INV in Nbs1m/m and Mre11ATLD1/ATLD1 abl pre–B cells. (a) Schematic of the pMX-INV retroviral recombination substrate, rearrangement intermediates, and products. The retroviral packaging signal (ψ), GFP complementary DNA, and IRES-human CD4 (I-hCD4) cassette are labeled. The viral LTRs (open arrows) and the RSs (open triangles) are shown. The nonrearranged (NR) pMX-INV and pMX-INV coding ends (CE), coding joint (CJ), and hybrid joint (HJ) are indicated. The relative position of the pA, pB, and pC oligonucleotides, the C4 probe (bar), and EcoRV (E) and NcoI (N) endonuclease restriction sites are shown, as are the expected sizes of hybridizing fragments. (b and c) Southern blot analysis of EcoRV–NcoI-digested (b) or EcoRV-digested (c) genomic DNA from WT, Atm−/−, Mre11ATLD1/ATLD1 (Mre11A/A), and Nbs1m/m abl pre–B cell clones containing the pMX-INV (INV) retroviral recombination substrate that had been treated with STI571 for the indicated time (hours). The abl pre–B cell clones analyzed each have single pMX-INV integrants and were derived from parental lines as indicated (parental line, clone number). Expected sizes for bands generated by nonrearranged pMX-INV (NR), coding joints (CJ), hybrid joints (HJ), and coding ends (CE) are indicated. (d and e) Southern blot analysis of EcoRV–NcoI-digested (d) or EcoRV-digested (e) genomic DNA from WT (line A70.2), Atm−/− (line 2F), Mre11ATLD1/ATLD1 (Mre11A/A; line 48.1), and Nbs1m/m (line 737.3) abl pre–B cells containing the pMX-INV (INV) retroviral recombination substrate that had been treated with STI571 for the indicated time (hours). (f) Quantification of rearrangement products from blots in d and e. Products are expressed as a percentage of the total fraction of pMX-INV substrates that had initiated V(D)J recombination (CJ + HJ + CE) to normalize for differences in cleavage efficiency between the cell lines. The data presented are representative of at least two experiments.
Atypical joining of Rag-mediated DSBs in MRN-deficient abl pre–B cells
Induction of Rag expression leads to robust pMX-INV rearrangement in Nbs1m/m:INV and Mre11ATLD1/ATLD1:INV abl pre–B cell clones (Fig. 1, b and c; and Fig. S1). Strikingly, pMX-INV hybrid joints form at high levels in both Nbs1m/m:INV and Mre11ATLD1/ATLD1:INV abl pre–B cell clones (Fig. 1, b and c; and Fig. S1). Hybrid joints are atypical nonproductive rearrangements formed by the ligation of a signal end generated at one Rag DSB to the coding end generated at the other (Fig. 1 a) (60, 61). pMX-INV hybrid joint formation in Nbs1m/m:INV and Mre11ATLD1/ATLD1:INV abl pre–B cell clones occurs at levels approximating those observed in Atm−/−:INV clones (Fig. 1, b and c; and Fig. S1). In contrast, Southern blot analyses did not reveal pMX-INV hybrid joint formation in WT able pre–B cell clones (WT:INV; Fig. 1, b and c; and Fig. S1). Similar results were observed when analyzing rearrangement in bulk populations of abl pre–B cells with pMX-INV integrants at numerous heterogenous chromosomal locations (Fig. 1, d–f; and Fig. S2). As was observed in Atm−/− abl pre–B cells, the increase in hybrid joint formation in Nbs1m/m and Mre11ATLD1/ATLD1 abl pre–B cells occurs during rearrangements by inversion but not during rearrangements by deletion (unpublished data) (50).
Coding joint formation in MRN-deficient abl pre–B cells
To assess the impact of MRN deficiency on coding joint formation, we initially assayed for the accumulation of unrepaired coding ends in Nbs1m/m:INV and Mre11ATLD1/ATLD1:INV abl pre–B cell clones (Fig. 1 c and Fig. S1 b). After Rag induction, pMX-INV coding ends were readily detected in all of the Nbs1m/m:INV and some of the Mre11ATLD1/ATLD1:INV abl pre–B cell clones analyzed (Fig. 1 c and Fig. S1 b). Unrepaired coding ends were also observed after Rag induction in Atm−/−:INV abl pre–B cell clones but not in WT:INV abl pre–B cell clones (Fig. 1 c and Fig. S1 b) (50). Analysis of rearrangement in bulk populations of abl pre–B cells was in general agreement with the clonal analyses (Fig. 1, e and f). In this regard, unrepaired pMX-INV coding ends were detected after Rag induction in Nbs1m/m:INV, Mre11ATLD1/ATLD1:INV, and Atm−/−:INV abl pre–B cells but not WT:INV abl pre–B cells (Fig. 1 e). Quantification revealed that Atm−/−:INV abl pre–B cells had the highest level of coding end accumulation, whereas Mre11ATLD1/ATLD1:INV cells had the lowest level of coding ends (Fig. 1 f). Sequence analyses of coding joints from WT:INV, Nbs1m/m:INV, Mre11ATLD1/ATLD1:INV, and Atm−/−:INV abl pre–B cells revealed that they were all diversified through N- and P-nucleotide additions and nucleotide loss (Fig. S3). Notably, several of the coding joints formed in the Atm−/−:INV abl pre–B cells exhibited excessive nucleotide loss (>50 bp), and one coding joint from the Nbs1m/m:INV abl pre–B cell had incorporated a 108-bp DNA fragment from the IgL-κ locus (Fig. S3). Together, these data demonstrate that coding joint formation is mildly perturbed in Nbs1m/m:INV and Mre11ATLD1/ATLD1:INV abl pre–B cells but that most of the coding joints formed in these cells do not exhibit marked alterations in sequence diversification.
Signal joint formation in MRN-deficient abl pre–B cells
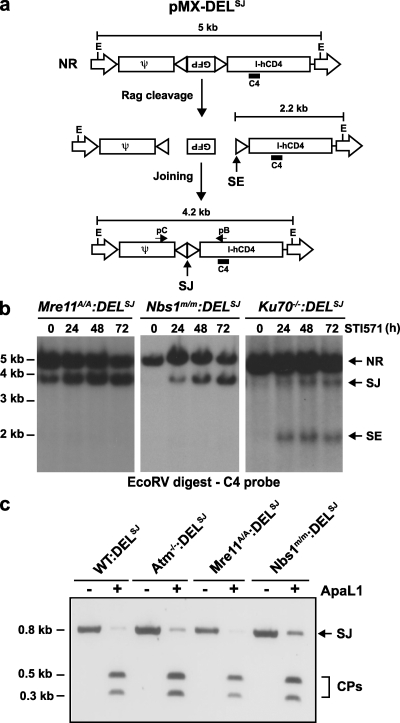
To assess the function of MRN during signal joint formation, we introduced the pMX-DELSJ retroviral recombination substrate into Nbs1m/m (Nbs1m/m:DELSJ) and Mre11ATLD1/ATLD1 (Mre11ATLD1/ATLD1:DELSJ) abl pre–B cells (Fig. 2 a). pMX-DELSJ is a derivative of pMX-INV in which one RS has been inverted so that rearrangement occurs by deletion with the signal joint remaining in the chromosome (Fig. 2 a) (50). As was previously observed in Atm−/−:DELSJ abl pre–B cells, induction of Rag expression in these cells leads to robust pMX-DELSJ signal joint formation with no detectable unrepaired signal ends (Fig. 2 b) (50). In contrast, unrepaired signal ends were readily detected after Rag induction in Ku70-deficient abl pre–B cells (Ku70−/−:DELSJ), as would be expected given the requirement for Ku70 in signal joint formation (Fig. 2 b). The majority of pMX-DELSJ signal joint PCR products amplified from Nbs1m/m:DELSJ, Mre11ATLD1/ATLD1:DELSJ, WT:DELSJ, and Atm−/−:DELSJ abl pre–B cells can be digested with ApaLI, which is indicative of precise formation (Fig. 2 c). Indeed, sequence analyses confirmed that most of the signal joints were generated by the precise joining of two signal ends; however, a small fraction of imprecise joints were present in all cells (Fig. 2 c and Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20081326/DC1). Together, these findings demonstrate that the compromised MRN function in the Nbs1m/m and Mre11ATLD1/ATLD1 abl pre–B cells does not lead to substantial quantitative or qualitative defects in signal joint formation.
Figure 2.
Signal joint formation in MRN-deficient abl pre–B cells. (a) Schematic of the pMX-DELSJ retroviral recombination substrate. All components of the retrovirus are as indicated in the Fig. 1 legend. The signal end (SE) and signal joint (SJ) are indicated. (b) Southern blot analysis of EcoRV-digested genomic DNA from Nbs1m/m (line 675.3), Mre11ATLD1/ATLD1 (line 48.1), and Ku70−/− (line 0.2) abl pre–B cells containing the pMX-DELSJ (DELSJ) retroviral recombination substrate that had been treated with STI571 for the indicated number of hours). (c) PCR analysis of pMX-DELSJ signal joints generated in WT:DELSJ, Atm−/−:DELSJ, Mre11ATLD1/ATLD1:DELSJ, and Nbs1m/m:DELSJ abl pre–B cells treated with STI571 for 72 h. PCR was performed using primers pB and pC (a), and PCR products were either not digested (−) or digested with ApaLI (+). Products were visualized by ethidium bromide staining, and undigested (SJ) and digested (CP) signal joint products are indicated. The data presented are representative of at least two experiments.
ATM activation in response to Rag DSBs in MRN-deficient abl pre–B cells
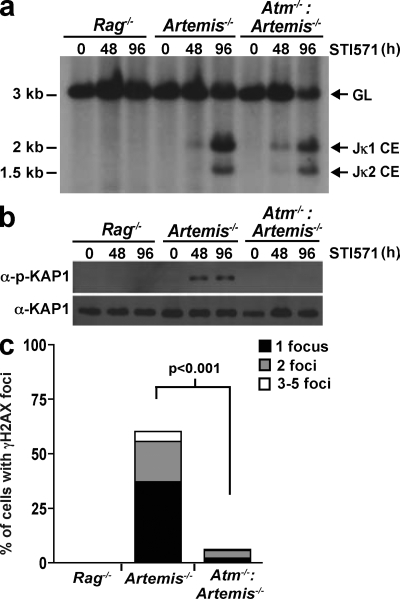
To determine whether ATM is activated in response to Rag DSBs in MRN-deficient abl pre–B cells, we generated several independently derived Mre11ATLD1/ATLD1 and Nbs1m/m abl pre–B cells that are also deficient in Artemis (Mre11ATLD1/ATLD1:Artemis−/− and Nbs1m/m:Artemis−/− abl pre–B cells, respectively). As we have previously demonstrated, induction of Rag in Artemis−/− abl pre–B cells leads to the accumulation of unrepaired coding ends at the endogenous IgL-κ locus as a result of the function of Artemis in opening hairpin-sealed DNA ends (Fig. 3 a) (48, 49, 62). These unrepaired coding ends activate ATM, as indicated by the phosphorylation of KAP-1 and H2AX and the nuclear translocation of NF-κB in response to these DSBs in Artemis−/−, but not Atm−/−:Artemis−/−, abl pre–B cells (Fig. 3 and Fig. 4 c) (48, 63).
Figure 3.
Atm activation by Rag DSBs. (a) Southern blot analysis showing Jκ1 and Jκ2 coding ends (CE) generated in Rag2−/−, Artemis−/−, and Artemis−/−:Atm−/− abl pre–B cells treated with STI571 for the indicated times (hours). SacI- and EcoRI-digested genomic DNA was hybridized to the JκIII probe. The bands corresponding to the IgL-κ locus in the germline configuration (GL) and Jκ1 and Jκ2 CEs are indicated. (b) Western blot analysis of phospho-KAP-1 (α-p-KAP-1) and KAP-1 (α-KAP-1) from STI571-treated cells shown in a. (c) Quantification of γ-H2AX nuclear foci in the cells from a after 24 h of STI571 treatment. Shown is the percentage of cells containing one, two, or three to five γ-H2AX foci. The total number of cells analyzed for each genotype was 500 and p-values were calculated using a two-tailed Fisher's Exact test. Note that γ-H2AX foci were not detected in any of the STI571-treated Rag−/− abl pre–B cells. The data presented are representative of at least two experiments.
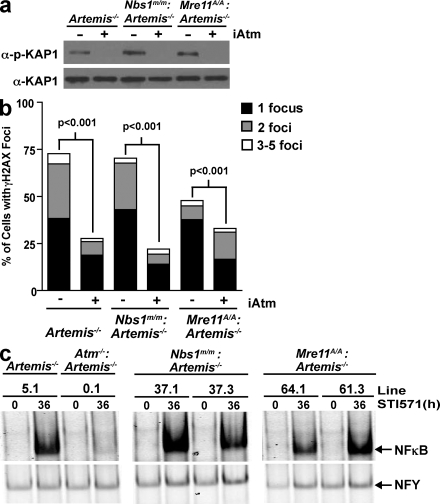
Figure 4.
Rag-DSB-mediated Atm activation in MRN-deficient cells. (a) Western blot analysis for phospho–KAP-1 (α–p–KAP-1) and KAP-1 (α–KAP-1) in Artemis−/− (line 5.1), Nbs1m/m:Artemis−/− (line 37.1), and Mre11ATLD1/ATLD1:Artemis−/− (line 64.1) abl pre–B cell lines treated with STI571 for 96 h and either DMSO (−) or the Atm inhibitor (iAtm) KU-55933 (+). (b) Percentage of Artemis−/− (line 5.3), Nbs1m/m:Artemis−/− (line 37.4), and Mre11ATLD1/ATLD1:Artemis−/− (line 61.2) abl pre–B cells with one, two, or three to five nuclear γ-H2AX foci after treatment with STI571 for 24 h and either DMSO (−) or the Atm inhibitor (iAtm) KU-55933 (+). The total number of cells analyzed for each genotype was 500 and p-values were calculated using a two-tailed Fisher's Exact test. (c) NF-κB EMSA of nuclear lysates from Artemis−/−, Atm−/−: Artemis−/−, and two independent Nbs1m/m:Artemis−/− and Mre11ATLD1/ATLD1:Artemis−/− abl pre–B cell lines treated with STI571 for 36 h. NFY EMSA is shown as a control. The data presented are representative of at least two experiments.
The induction of Rag DSBs in Mre11ATLD1/ATLD1:Artemis−/− and Nbs1m/m:Artemis−/− abl pre–B cell lines led to robust but variable levels of KAP-1 phosphorylation (Fig. 4 a and Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20081326/DC1). The variability in KAP-1 phosphorylation in these cells does not appear to be a result of differences in the levels of KAP-1, Nbs1, or Mre11 protein but may, in part, reflect differences in the levels of unrepaired coding ends generated in these cells after Rag induction (Fig. 4 a and Figs. S5–S7). Importantly, KAP-1 phosphorylation in Artemis−/−, Mre11ATLD1/ATLD1:Artemis−/−, and Nbs1m/m:Artemis−/− abl pre–B cells is nearly completely abrogated by the Atm inhibitor KU-55933, demonstrating that ATM is activated by Rag DSBs in all of these cells (Fig. 4 a and Fig. S5). Moreover, Rag DSBs promote an Atm-dependent phosphorylation of H2AX and nuclear translocation of NF-κB in Mre11ATLD1/ATLD1:Artemis−/− and Nbs1m/m:Artemis−/− abl pre–B cell lines (Fig. 4, b and c; Fig. S8; and not depicted). Finally, treatment of Mre11ATLD1/ATLD1:INV and Nbs1m/m:INV abl pre–B cells with KU-55933 results in a significant increase in the accumulation of unrepaired pMX-INV coding ends (Fig. 5). Collectively, our findings demonstrate that ATM is activated by Rag DSBs in Mre11ATLD1/ATLD1 and Nbs1m/m abl pre–B cells.
Figure 5.
Atm inhibition in MRN-deficient cells leads to increased coding end accumulation. Southern blot analysis of EcoRV-digested genomic DNA from WT, Atm−/−, and two Mre11ATLD1/ATLD1 (labeled as Mre11A/A) and Nbs1m/m abl pre–B clones containing pMX-INV. Cells were treated with STI571 for the indicated time (hours) and either DMSO (−) or the Atm inhibitor (iAtm) KU-55933 (+). Expected sizes for bands generated by nonrearranged substrate (NR), coding joints (CJ), hybrid joints (HJ), and coding ends (CE) are indicated. The data presented are representative of at least two experiments.
Defects in V(D)J recombination in developing MRN-deficient lymphocytes
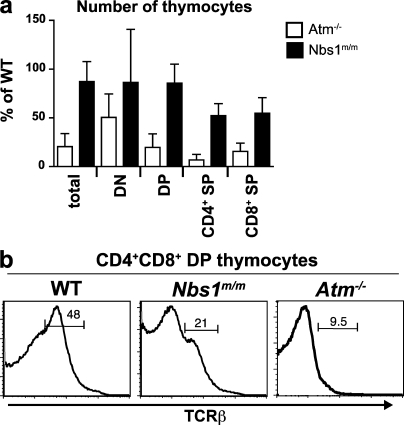
We have demonstrated that deficiencies in MRN function lead to defects in V(D)J recombination in Mre11ATLD1/ATLD1 and Nbs1m/m abl pre–B cells. To determine whether similar defects occur in developing MRN-deficient lymphocytes, we first analyzed thymocyte development in Nbs1m/m mice. Nbs1m/m mice have a mild reduction in total thymocyte number but a significant reduction in CD4+ and CD8+ (single positive) thymocytes (Fig. 6 a) (38). In addition, they have diminished numbers of preselection αβ-TCR–expressing (TCR-βint [TCR-β intermediate]) CD4+/CD8+ (double positive [DP]) thymocytes (Fig. 6 b). Defects in thymocyte development that are similar in nature but more severe are observed in Atm−/− mice (Fig. 6) (9, 46, 51, 52, 64). The reduction in TCR-βint DP thymocytes in Nbs1m/m mice could be a result of defects in the survival of these cells or in the developmental transition of thymocytes through this compartment. However, the reduction in TCR-βint DP thymocytes in Nbs1m/m mice could also be explained by defects in TCR-α chain gene assembly as have been observed previously in Atm−/− thymocytes (46, 51, 52).
Figure 6.
Thymocyte development in Nbs1m/m mice. (a) Number of thymocytes in Atm−/− (n = 16) and Nbs1m/m (n = 9) mice at the indicated stages of development, expressed as a percentage of the number of thymocytes found at each stage in WT Atm+/+ (n = 12) and Nbs1+/+ (n = 3) littermate controls. Calculations were done using mean numbers of thymocytes from mice of each genotype and propagated error. Error bars represent SEM. DN (double negative), CD4−CD8−; DP, CD4+CD8+; SP, single positive. (b) Representative flow cytometric analysis of TCR-β expression on CD4+CD8+ DP thymocytes from WT, Nbs1m/m, and Atm−/− mice. Numbers indicate percentage of DP cells that are TCR-βint. Results are representative of the analyses of at least three mice.
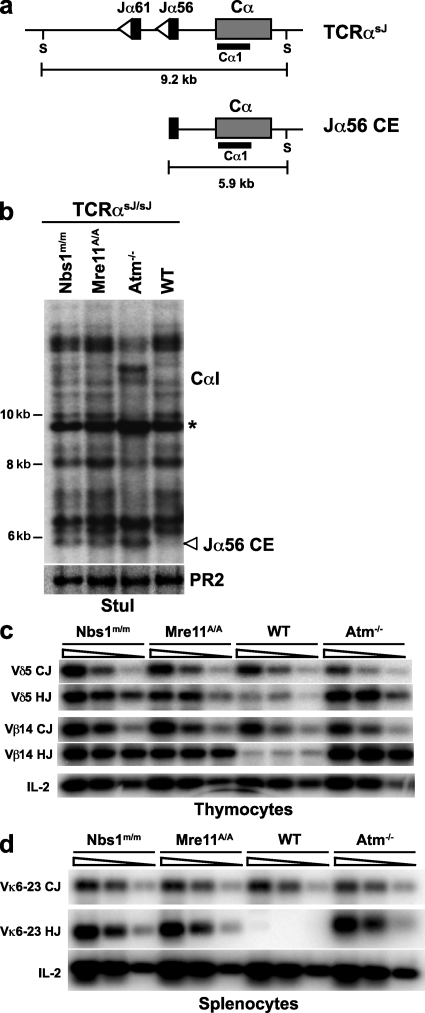
To directly determine whether MRN deficiency leads to defects in the repair of Rag-mediated DSBs in developing lymphocytes in vivo, we generated Mre11ATLD1/ATLD1 and Nbs1m/m mice homozygous for a modified TCR-α allele, TCR-αsJ (Mre11ATLD1/ATLD1:TCR-αsJ/sJ and Nbs1m/m:TCR-αsJ/sJ). The TCR-αsJ allele is identical to the TCR-α allele, except that the 61 Jα gene segments have been replaced by two closely linked Jα gene segments (Jα61 and Jα56) (65). As all Vα rearrangements must use one of these two Jα gene segments, accumulation of unrepaired Rag-mediated DSBs generated at these Jα gene segments can be readily detected by Southern blotting of genomic DNA from developing TCR-αsJ/sJ thymocytes (46).
Genomic DNA isolated from WT:TCR-αsJ/sJ, Atm−/−:TCRαsJ/sJ, Nbs1m/m:TCR-αsJ/sJ, and Mre11ATLD1/ATLD1:TCRαsJ/sJ thymocytes was digested with StuI and hybridized to the Cα1 probe, which is directed to the TCR-α constant region gene immediately downstream of the Jα56 gene segment on the TCR-αsJ/sJ allele (Fig. 7, a and b). These analyses revealed many nongermline-sized hybridizing fragments as a result of diverse Vα to Jα rearrangements in thymocytes from all of these mice (Fig. 7 b). In addition, the Atm−/−:TCR-αsJ/sJ, Nbs1m/m:TCR-αsJ/sJ, and Mre11ATLD1/ATLD1:TCRαsJ/sJ thymocytes each have a distinct 5.9-kb band, which was not present in WT:TCR-αsJ/sJ thymocytes, that we have previously demonstrated is generated by unrepaired Jα56 coding ends (Fig. 7 b) (46). The presence of unrepaired Jα56 coding ends in Nbs1m/m:TCR-αsJ/sJ and Mre11ATLD1/ATLD1:TCR-αsJ/sJ thymocytes was confirmed by ligation-mediated PCR (unpublished data). Notably, as was observed for pMX-INV coding ends in the abl pre–B cells, the level of unrepaired Jα56 coding ends was generally lower in the Nbs1m/m:TCR-αsJ/sJ and Mre11ATLD1/ATLD1:TCRαsJ/sJ thymocytes, as compared with the Atm−/−:TCR-αsJ/sJ thymocytes (Fig. 7 b). Unrepaired Jα56 signal ends do not accumulate at higher levels in Atm−/−:TCR-αsJ/sJ, Nbs1m/m:TCR-αsJ/sJ, and Mre11ATLD1/ATLD1:TCR-αsJ/sJ thymocytes as compared with WT:TCR-αsJ/sJ thymocytes (unpublished data) (46).
Figure 7.
V(D)J recombination defects in MRN-deficient lymphocytes in vivo. (a) Schematic of Rag cleavage at the Jα56 gene segment on the TCR-αsJ allele. The gene segments are shown as black rectangles, RSs are open triangles, and the Cα constant region is the gray rectangle. The StuI restriction sites (S) and the Cα1 probe (black bar) used for analysis are also shown. (b) Southern blot analysis of TCR-α rearrangement in WT, Atm−/−, Nbs1m/m, and Mre11ATLD1/ATLD1 thymocytes, all on the TCR-αsJ/sJ background. The expected sizes for germline TCR-αsJ allele (*) and Jα56 coding ends (Jα56 CE) are indicated. Genomic DNA was digested with StuI and probed with the Cα1 probe. Hybridization to a Rag-2 probe (PR2) is shown as a DNA loading control. Results are representative of three experiments. (c) PCR analysis of Vδ5DδJδ1 coding joints (Vδ5 CJ), Vδ5Dδ1 hybrid joints (Vδ5 HJ), Vβ14DβJβ2.7 coding joints (Vβ14 CJ), and Vβ14Dβ2 hybrid joints (Vβ14 HJ) in WT, Atm−/−, Nbs1m/m, and Mre11ATLD1/ATLD1 thymocytes. (d) PCR analysis of Vκ6-23 to Jκ1 coding joints (Vκ6-23 CJ) and hybrids joints (Vκ6-23 HJ) in WT, Atm−/−, Nbs1m/m, and Mre11ATLD1/ATLD1 splenocytes. The IL-2 gene PCR is shown as a DNA quantity control. The data presented are representative of analyses of at least two mice of each genotype.
Like MRN-deficient abl pre–B cells, Mre11ATLD1/ATLD1 and Nbs1m/m thymocytes also exhibited an increase in hybrid joint formation during rearrangements that occur by inversion (Fig. 7 c). In this regard, we find that hybrid joints involving the Vβ14 gene segment (TCR-β locus) and the Vδ5 gene segment (TCR-δ locus), which both rearrange by inversion, are increased in Mre11ATLD1/ATLD1 and Nbs1m/m thymocytes as compared with WT thymocytes (Fig. 7 c). Furthermore, the level of Vβ14 and Vδ5 hybrid joints is similar to that observed in Atm−/− thymocytes (Fig. 7 c). The increase in hybrid joint formation during rearrangements that occur by inversion was not restricted to MRN-deficient T cells as it is also observed during IgL-κ locus rearrangements in Mre11ATLD1/ATLD1 and Nbs1m/m B cells (Fig. 7 d). Together, these findings demonstrate that developing MRN-deficient lymphocytes exhibit defects in V(D)J recombination similar to those observed in Atm-deficient lymphocytes, which are characterized by an accumulation of unrepaired coding ends and an increase in hybrid joint formation during rearrangements by inversion.
DISCUSSION
In this paper, we have demonstrated that the MRN complex functions in the repair of chromosomal Rag-mediated DNA DSBs generated during V(D)J recombination in cultured pre–B cell lines and in developing lymphocytes. This is indicated by the accumulation of unrepaired coding ends and the increase in aberrant hybrid joint formation during V(D)J recombination in lymphocytes expressing hypomorphic Mre11 or Nbs1 proteins. As these defects are observed in pre–B cells arrested at the G1 phase of the cell cycle, our findings establish that MRN can function in the repair of DNA DSBs generated at prereplicative stages of the cell cycle.
The V(D)J recombination defects observed in MRN-deficient lymphocytes are remarkably similar to those observed in ATM-deficient lymphocytes, suggesting that MRN and ATM function in the same pathway during the repair of Rag-mediated DSBs (50). Although the defects in V(D)J recombination in ATM- and MRN-deficient cells are qualitatively similar, unrepaired coding ends accumulate at higher levels in Atm−/− abl pre–B cells as compared with Nbs1m/m or Mre11ATLD1/ATLD1 abl pre–B cells. This could reflect the activity of a repair pathway that depends on the activity of ATM but not MRN. Alternatively, the Mre11ATLD1 and Nbs1m hypomorphic proteins could have residual functions in the repair of Rag-mediated DSBs that may be modulated by ATM activation. The notion that the Mre11ATLD1 and Nbs1m hypomorphic proteins have residual MRN function is supported by the observation that Nbs1m/m or Mre11ATLD1/ATLD1 mice are viable, whereas mice null for Mre11 or Nbs1 exhibit early embryonic lethality (9, 38–40, 44, 45).
Unrepaired coding ends accumulate at greater levels in Nbs1m/m abl pre–B cells, as compared with Mre11ATLD1/ATLD1 abl pre–B cells. This could reflect distinct requirements for Mre11 and Nbs1 in the repair of Rag-mediated DSBs. However, because most of the known functions of Mre11, Nbs1, and Rad50 rely on their association in an MRN holocomplex, it is likely that the expression of the individual Mre11 or Nbs1 hypomorphs have differing effects on global MRN function. In this regard, it is conceivable that defects in coding joint formation in Nbs1m/m and Mre11ATLD1/ATLD1 abl pre–B cells are, in part, a result of diminished Rad50 function.
In contrast to our findings, previous studies of V(D)J recombination in fibroblasts from patients that express hypomorphic Nbs1 alleles and mouse embryonic fibroblasts that are deficient in Mre11 failed to reveal defects in coding joint formation (56, 58, 66). Importantly, these analyses assessed the repair of Rag-mediated DSBs generated on extrachromosomal plasmid recombination substrates in nonlymphoid cells, whereas we analyzed the repair of chromosomal Rag-mediated DSBs in developing lymphocytes and lymphocyte cell lines. Notably, analysis of V(D)J recombination on extrachromosomal substrates in ATM-deficient fibroblasts also did not reveal defects in coding joint formation, but these defects are clearly evident during chromosomal V(D)J recombination in ATM-deficient lymphocytes (46, 47, 50–52, 67). Thus, there may be differing requirements for the repair of Rag-mediated DSBs on extrachromosomal substrates and those generated in the chromosome. In this regard, ATM and MRN may function primarily in the repair of chromosomal Rag DSBs. Finally, these discrepancies may reflect differences in the requirement for MRN and ATM in the repair of Rag-mediated DSBs in lymphoid and nonlymphoid cells, as has recently been described for Cernunnos (also called XRcc4-like factor) (68).
How does MRN function during V(D)J recombination? ATM phosphorylates a large number of proteins, including all of the components of the MRN complex, which participate in diverse and broadly functional DNA damage response pathways (2, 21–24). MRN functions to activate ATM, and thus the primary function of MRN in V(D)J recombination may be to activate ATM in response to Rag-mediated DSBs (10–12). However, we find that ATM is activated in response to Rag DSBs generated in Mre11ATLD1/ATLD1 and Nbs1m/m abl pre–B cells, as indicated by the robust ATM-dependent phosphorylation of KAP-1 and H2AX and the nuclear translocation of NF-κB. Thus, the V(D)J recombination defects in MRN-deficient cells are not a result of a general inability of Rag DSBs to activate ATM in these cells. Importantly, although we find that ATM is activated by Rag-mediated DSBs in Mre11ATLD1/ATLD1 and Nbs1m/m abl pre–B cells, these cells could have isolated defects in the phosphorylation of other specific targets required for the repair of Rag-mediated DSBs.
In addition to activating ATM, MRN could have downstream functions in the repair of Rag-mediated DSBs. In this regard, ATM promotes coding joint formation by stabilizing coding ends in postcleavage complexes, and although ATM could perform this function directly, it likely regulates the activity of downstream proteins, such as MRN, that could perform these functions (50). Rad50 has N- and C-terminal Walker A and B nucleotide binding motifs, respectively, that form a DNA binding domain upon their intramolecular association (5, 6). This association leads to the formation of a central Rad50 hook domain, which can facilitate the tethering of two distinct MRN-bound DNA molecules through an intermolecular interaction between two Rad50 hook domains (16–20). The interaction between hook domains of Rad50 proteins bound to two coding ends could stabilize these DNA ends until they are joined. Moreover, it has recently been shown that Mre11 dimers provide an interface that can bridge and juxtapose two broken DNA ends (15). Thus, it is possible that the MRN complex functions to stabilize coding ends in postcleavage complexes through the activities of both Rad50 and Mre11. This stabilizing activity could be modulated by the phosphorylation of Rad50, Mre11, and/or Nbs1 by ATM in response to Rag-mediated DSBs (21–24).
MATERIALS AND METHODS
Mice.
Animals were housed in a specific pathogen-free animal facility at Washington University. Animal protocols were approved by the Washington University Institutional Animal Care and Use Committee. All mice were analyzed between 4 and 8 wk of age. The Nbs1m/m, Mre11ATLD1/ATLD1, Atm−/−, Artemis−/−, TCRαsJ/sJ, and Ku70−/− mice have been described previously (38, 40, 65, 69, 70). The Nbs1m/m mice were obtained from Y. Xu (University of California San Diego, La Jolla, CA). The Mre11ATLD1/ATLD1 mice were obtained from J. Petrini (Memorial Sloan Kettering Cancer Institute, New York, NY).
Flow cytometric analyses and sorting.
Flow cytometric analysis of thymocyte development was performed on a FACSCalibur (BD) using FITC-conjugated anti-CD8, PE-conjugated anti-CD4, and PE-conjugated anti–TCR-β chain (all obtained from BD). Cells containing pMX-INV or pMX-DELSJ were isolated by FACS (FACSVantage; BD) based on expression of the human CD4 (hCD4) and detected using PE-conjugated anti–hCD4 (BD).
Cell lines and culture conditions.
Nbs1m/m (lines 675.3 and 737.3), Mre11ATLD1/ATLD1 (line 48.1), WT (line A70.2), Atm−/− (lines Atm2E and Atm2F), Ku70−/− (line 0.2), Artemis−/− (lines 5.1, 0.1, 5.3, and 2.1), Atm−/−:Artemis−/− (line 0.1), Nbs1m/m:Artemis−/− (lines 37.1, 37.2, 37.3, and 37.4), and Mre11ATLD1/ATLD1:Artemis−/− (lines 61.2, 61.3, 64.1, and 64.2) v-abl–transformed pre–B cells were generated by culturing bone marrow of 3–5-wk-old mice with the pMSCV v-abl retrovirus as described previously (50). All cells were generated from mice harboring the Eμ-Bcl-2 transgene (71). All abl pre–B cells (106/ml) were transduced with retroviral recombination substrates by cocentrifugation at 1,800 rpm for 90 min. Clonal populations of cells with single pMX-INV integrants were isolated by limiting dilution of WT:INV (A70.2-2, -4, -5, and -6), Atm−/−:INV (Atm2E-26 and Atm2F-3, -6, and -11), Mre11ATLD1/ATLD1:INV (48.1–1, -3, -6, and -7), and Nbs1m/m:INV (675.3–2 and -9 and 737.3–4 and -6) abl pre–B cell lines. Cells were treated with 3 µM STI571 (Novartis) for the indicated times at 106 cells/ml. KU-55933 (Sigma-Aldrich) was used at 15 µM.
Southern blot and PCR analyses.
Southern blot analyses were performed on genomic DNA from cells harboring pMX-INV and pMX-DELSJ using the indicated restriction enzymes and the C4 probe, as previously described (50). Southern blot analyses of thymocytes from mice expressing the TCR-αsJ allele were performed using the StuI restriction enzyme and the CαI probe as reported previously (65). Southern blot analyses for IgL-κ locus Jκ coding ends was performed on SacI- and EcoRI-digested genomic DNA using the JκIII probe as previously described (48). pMX-INV coding joints were amplified using the pA and pB oligonucleotides and pMX-DELSJ signal joints were amplified used primers pB and pC oligonucleotides as previously described (50). Vδ5, Vβ14, and Vκ6-23 coding and hybrid joints and the IL-2 gene were amplified by PCR as previously described (50).
Immunofluorescent detection of γ-H2AX foci.
Nuclear γ-H2AX foci were detected using standard protocols with minor modifications (54, 72, 73). In brief, cells were cytospun onto poly-l-lysine–coated slides (Sigma-Aldrich), fixed in 4% paraformaldehyde for 10 min, washed in PBS, permeabilized with 0.15% Triton X-100 in PBS, and blocked in PBS with 2% bovine serum albumin. Cells were incubated in anti γ-H2AX antibody (Millipore) at a 1–2-µg/ml concentration for 3 h at 37°C in a moist chamber, washed with PBS, and further incubated with anti–mouse FITC conjugate (Vector Laboratories) for 45 min. After washing with PBS, slides were mounted in Vectashield mounting medium with DAPI (Vector Laboratories). Foci were observed and imaged as described previously on a microscope (Axioplan 2; Carl Zeiss, Inc.) using ISIS imaging software (MetaSystems) (72).
Western blot and EMSA analyses.
Western blots were done on whole cell lysates using antibodies to mouse KAP-1 (GeneTex, Inc.), phosphorylated KAP-1 (Bethyl Laboratories, Inc.), Mre11 (Novus Biologicals), and Nbs1 (Cell Signaling Technology). The secondary reagents were horseradish peroxidase–conjugated goat anti–mouse IgG (Invitrogen) or donkey anti–rabbit IgG (GE Healthcare). NF-κB EMSA were run as described previously and were analyzed with an Infrared Scanner (Odyssey; LI COR Biosciences) (48).
Online supplemental material.
Fig. S1 shows Southern blot analyses of pMX-INV rearrangement on additional abl pre–B cell clones. Fig. S2 shows longer exposure of the WT:INV and Atm-/-:INV Southern blot analyses. Fig. S3 shows sequences of pMX-INV coding joints from WT:INV, Atm-/-:INV, Mre11A/A:INV, and Nbs1m/m:INV abl pre–B cells. Fig. S4 shows sequences of pMX-DELSJ signal joints from WT:DELSJ, Atm-/-:DELSJ, Mre11A/A:DELSJ, and Nbs1m/m:DELSJ abl pre–B cells. Fig. S5 shows analysis of KAP-1 phosphorylation carried out as described in Fig. 4 a on additional Artemis-/-, Artemis-/-:Nbs1m/m, and Artemis-/-:Mre11A/A abl pre–B cell lines. Fig. S6 shows Nbs1 and Mre11 western blot analyses in the different Artemis-/-, Artemis-/-:Nbs1m/m, and Artemis-/-:Mre11A/A abl pre–B cells analyzed. Fig. S7 shows Southern blot analyses of IgL-κ locus Jκ coding ends after Rag induction in the different Artemis-/-, Artemis-/-:Nbs1m/m, and Artemis-/-:Mre11A/A abl pre–B cell lines. Fig. S8 shows EMSA analyses of NF-κB translocation to the nucleus in the additional Artemis-/-, Artemis-/-:Nbs1m/m, and Artemis-/-:Mre11A/A abl pre–B cells not analyzed in Fig. 4 c. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081326/DC1.
Acknowledgments
We thank Dr. Yang Xu for providing the Nbs1m/m mice and Dr. John Petrini for providing the Mre11ATLD1/ATLD1 mice.
This work is supported by the National Institutes of Health grants AI074953 and AI47829 (B.P. Sleckman) and CA129537 and CA123232 (T.K. Pandita). C.-Y. Huang and A.L. Bredemeyer were supported by a postdoctoral training grant from the National Institutes of Health. J.J. Bednarski is supported by a National Institutes of Health Ruth L. Kirschstein National Research Service Award (5 T32 HD007499) from the National Institute of Child Health and Human Development.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: ATM, ataxia telangiectasia mutated; DP, double positive; DSB, double-strand break; HR, homologous recombination; MRN, Mre11–Rad50–Nbs1; NHEJ, nonhomologous end joining; RS, recombination signal.
References
- Zhou B.B., Elledge S.J. 2000. The DNA damage response: putting checkpoints in perspective.Nature. 408:433–439 [DOI] [PubMed] [Google Scholar]
- Shiloh Y. 2003. ATM and related protein kinases: safeguarding genome integrity.Nat. Rev. Cancer. 3:155–168 [DOI] [PubMed] [Google Scholar]
- Rouse J., Jackson S.P. 2002. Interfaces between the detection, signaling, and repair of DNA damage.Science. 297:547–551 [DOI] [PubMed] [Google Scholar]
- Wyman C., Kanaar R. 2006. DNA double-strand break repair: all's well that ends well.Annu. Rev. Genet. 40:363–383 [DOI] [PubMed] [Google Scholar]
- Stracker T.H., Theunissen J.W., Morales M., Petrini J.H. 2004. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together.DNA Repair (Amst.). 3:845–854 [DOI] [PubMed] [Google Scholar]
- D’Amours D., Jackson S.P. 2002. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling.Nat. Rev. Mol. Cell Biol. 3:317–327 [DOI] [PubMed] [Google Scholar]
- Maser R.S., Monsen K.J., Nelms B.E., Petrini J.H. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks.Mol. Cell. Biol. 17:6087–6096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms B.E., Maser R.S., MacKay J.F., Lagally M.G., Petrini J.H. 1998. In situ visualization of DNA double-strand break repair in human fibroblasts.Science. 280:590–592 [DOI] [PubMed] [Google Scholar]
- Difilippantonio S., Celeste A., Fernandez-Capetillo O., Chen H.T., Reina San Martin B., Van Laethem F., Yang Y.P., Petukhova G.V., Eckhaus M., Feigenbaum L., et al. 2005. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models.Nat. Cell Biol. 7:675–685 [DOI] [PubMed] [Google Scholar]
- Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. 2003. Requirement of the MRN complex for ATM activation by DNA damage.EMBO J. 22:5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Paull T.T. 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex.Science. 304:93–96 [DOI] [PubMed] [Google Scholar]
- Lee J.H., Paull T.T. 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex.Science. 308:551–554 [DOI] [PubMed] [Google Scholar]
- Stracker T.H., Morales M., Couto S.S., Hussein H., Petrini J.H. 2007. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex.Nature. 447:218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S., Celeste A., Kruhlak M.J., Lee Y., Difilippantonio M.J., Feigenbaum L., Jackson S.P., McKinnon P.J., Nussenzweig A. 2007. Distinct domains in Nbs1 regulate irradiation-induced checkpoints and apoptosis.J. Exp. Med. 204:1003–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.S., Moncalian G., Williams J.S., Yamada Y., Limbo O., Shin D.S., Groocock L.M., Cahill D., Hitomi C., Guenther G., et al. 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.Cell. 135:97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.E., Trujillo K.M., Sung P., Erickson H.P. 2001. Structure of the Rad50 x Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy.J. Biol. Chem. 276:37027–37033 [DOI] [PubMed] [Google Scholar]
- de Jager M., van Noort J., van Gent D.C., Dekker C., Kanaar R., Wyman C. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends.Mol. Cell. 8:1129–1135 [DOI] [PubMed] [Google Scholar]
- Wiltzius J.J., Hohl M., Fleming J.C., Petrini J.H. 2005. The Rad50 hook domain is a critical determinant of Mre11 complex functions.Nat. Struct. Mol. Biol. 12:403–407 [DOI] [PubMed] [Google Scholar]
- Hopfner K.P., Craig L., Moncalian G., Zinkel R.A., Usui T., Owen B.A., Karcher A., Henderson B., Bodmer J.L., McMurray C.T., et al. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair.Nature. 418:562–566 [DOI] [PubMed] [Google Scholar]
- Moreno-Herrero F., de Jager M., Dekker N.H., Kanaar R., Wyman C., Dekker C. 2005. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA.Nature. 437:440–443 [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., III, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage.Science. 316:1160–1166 [DOI] [PubMed] [Google Scholar]
- Gatei M., Young D., Cerosaletti K.M., Desai-Mehta A., Spring K., Kozlov S., Lavin M.F., Gatti R.A., Concannon P., Khanna K. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure.Nat. Genet. 25:115–119 [DOI] [PubMed] [Google Scholar]
- Wu X., Ranganathan V., Weisman D.S., Heine W.F., Ciccone D.N., O’Neill T.B., Crick K.E., Pierce K.A., Lane W.S., Rathbun G., et al. 2000. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response.Nature. 405:477–482 [DOI] [PubMed] [Google Scholar]
- Lim D.S., Kim S.T., Xu B., Maser R.S., Lin J., Petrini J.H., Kastan M.B. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway.Nature. 404:613–617 [DOI] [PubMed] [Google Scholar]
- Huang J., Dynan W.S. 2002. Reconstitution of the mammalian DNA double-strand break end-joining reaction reveals a requirement for an Mre11/Rad50/NBS1-containing fraction.Nucleic Acids Res. 30:667–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M., Gautier J. 2005. Repair of double-strand breaks by nonhomologous end joining in the absence of Mre11.J. Cell Biol. 171:765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K., Haber J.E. 1996. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae.Mol. Cell. Biol. 16:2164–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S.J., Jackson S.P. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing.EMBO J. 17:1819–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolis K.G., Nimmo E.R., Hartsuiker E., Carr A.M., Jeggo P.A., Allshire R.C. 2001. Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe.EMBO J. 20:210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue A., Lafrance M., Gauthier M.C., McDonald D., Hendzel M., West S.C., Jasin M., Masson J.Y. 2006. Interplay between human DNA repair proteins at a unique double-strand break in vivo.EMBO J. 25:222–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.G., Saidi A., Frappart P.O., Min W., Barrucand C., Dumon-Jones V., Michelon J., Herceg Z., Wang Z.Q. 2006. Conditional deletion of Nbs1 in murine cells reveals its role in branching repair pathways of DNA double-strand breaks.EMBO J. 25:5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. 1983. Somatic generation of antibody diversity.Nature. 302:575–581 [DOI] [PubMed] [Google Scholar]
- Kim D.R., Park S.J., Oettinger M.A. 2000. V(D)J recombination: site-specific cleavage and repair.Mol. Cells. 10:367–374 [PubMed] [Google Scholar]
- Fugmann S.D., Lee A.I., Shockett P.E., Villey I.J., Schatz D.G. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition.Annu. Rev. Immunol. 18:495–527 [DOI] [PubMed] [Google Scholar]
- Desiderio S., Lin W.C., Li Z. 1996. The cell cycle and V(D)J recombination.Curr. Top. Microbiol. Immunol. 217:45–59 [DOI] [PubMed] [Google Scholar]
- Rooney S., Chaudhuri J., Alt F.W. 2004. The role of the non-homologous end-joining pathway in lymphocyte development.Immunol. Rev. 200:115–131 [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Swat W., Alt F.W. 2002. The mechanism and regulation of chromosomal V(D)J recombination.Cell. 109(Suppl):S45–S55 [DOI] [PubMed] [Google Scholar]
- Kang J., Bronson R.T., Xu Y. 2002. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair.EMBO J. 21:1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.R., Mirzoeva O.K., Morgan W.F., Lin J., Dunnick W., Petrini J.H. 2002. A murine model of Nijmegen breakage syndrome.Curr. Biol. 12:648–653 [DOI] [PubMed] [Google Scholar]
- Theunissen J.W., Kaplan M.I., Hunt P.A., Williams B.R., Ferguson D.O., Alt F.W., Petrini J.H. 2003. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice.Mol. Cell. 12:1511–1523 [DOI] [PubMed] [Google Scholar]
- Digweed M., Sperling K. 2004. Nijmegen breakage syndrome: clinical manifestation of defective response to DNA double-strand breaks.DNA Repair (Amst.). 3:1207–1217 [DOI] [PubMed] [Google Scholar]
- Taylor A.M., Groom A., Byrd P.J. 2004. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis.DNA Repair (Amst.). 3:1219–1225 [DOI] [PubMed] [Google Scholar]
- Luo G., Yao M.S., Bender C.F., Mills M., Bladl A.R., Bradley A., Petrini J.H. 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation.Proc. Natl. Acad. Sci. USA. 96:7376–7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Weaver D.T. 1997. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells.Nucleic Acids Res. 25:2985–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Petersen S., Tessarollo L., Nussenzweig A. 2001. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice.Curr. Biol. 11:105–109 [DOI] [PubMed] [Google Scholar]
- Huang C.Y., Sharma G.G., Walker L.M., Bassing C.H., Pandita T.K., Sleckman B.P. 2007. Defects in coding joint formation in vivo in developing ATM-deficient B and T lymphocytes.J. Exp. Med. 204:1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E., Jankovic M., Difilippantonio S., Daniel J.A., Chen H.T., Celeste A., Pellegrini M., McBride K., Wangsa D., Bredemeyer A.L., et al. 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes.Cell. 130:63–75 [DOI] [PubMed] [Google Scholar]
- Bredemeyer A.L., Helmink B.A., Innes C.L., Calderon B., McGinnis L.M., Mahowald G.K., Gapud E.J., Walker L.M., Collins J.B., Weaver B.K., et al. 2008. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes.Nature. 456:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer A.L., Huang C.Y., Walker L.M., Bassing C.H., Sleckman B.P. 2008. Aberrant V(D)J recombination in ataxia telangiectasia mutated-deficient lymphocytes is dependent on nonhomologous DNA end joining.J. Immunol. 181:2620–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer A.L., Sharma G.G., Huang C.Y., Helmink B.A., Walker L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H., Sleckman B.P. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination.Nature. 442:466–470 [DOI] [PubMed] [Google Scholar]
- Vacchio M.S., Olaru A., Livak F., Hodes R.J. 2007. ATM deficiency impairs thymocyte maturation because of defective resolution of T cell receptor {alpha} locus coding end breaks.Proc. Natl. Acad. Sci. USA. 104:6323–6328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei I.R., Gladdy R.A., Nutter L.M., Canty A., Guidos C.J., Danska J.S. 2007. ATM deficiency disrupts Tcra locus integrity and the maturation of CD4+CD8+ thymocytes.Blood. 109:1887–1896 [DOI] [PubMed] [Google Scholar]
- Perkins E.J., Nair A., Cowley D.O., Van Dyke T., Chang Y., Ramsden D.A. 2002. Sensing of intermediates in V(D)J recombination by ATM.Genes Dev. 16:159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.T., Bhandoola A., Difilippantonio M.J., Zhu J., Brown M.J., Tai X., Rogakou E.P., Brotz T.M., Bonner W.M., Ried T., Nussenzweig A. 2000. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX.Science. 290:1962–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy A.E., Valencia-Burton M.A., Haber J.E., Oettinger M.A. 2005. The MRE11-RAD50-XRS2 complex, in addition to other non-homologous end-joining factors, is required for V(D)J joining in yeast.J. Biol. Chem. 280:20247–20252 [DOI] [PubMed] [Google Scholar]
- Harfst E., Cooper S., Neubauer S., Distel L., Grawunder U. 2000. Normal V(D)J recombination in cells from patients with Nijmegen breakage syndrome.Mol. Immunol. 37:915–929 [DOI] [PubMed] [Google Scholar]
- Donahue S.L., Tabah A.A., Schmitz K., Aaron A., Campbell C. 2007. Defective signal joint recombination in fanconi anemia fibroblasts reveals a role for Rad50 in V(D)J recombination.J. Mol. Biol. 370:449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J.M., Chang S., Ferguson D.O. 2008. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation.Cell. 135:85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo S.A., Schlissel M.S. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines.Nat. Immunol. 4:31–37 [DOI] [PubMed] [Google Scholar]
- Morzycka-Wroblewska E., Lee F.E., Desiderio S.V. 1988. Unusual immunoglobulin gene rearrangement leads to replacement of recombinational signal sequences.Science. 242:261–263 [DOI] [PubMed] [Google Scholar]
- Lewis S.M., Hesse J.E., Mizuuchi K., Gellert M. 1988. Novel strand exchanges in V(D)J recombination.Cell. 55:1099–1107 [DOI] [PubMed] [Google Scholar]
- Ma Y., Pannicke U., Schwarz K., Lieber M.R. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination.Cell. 108:781–794 [DOI] [PubMed] [Google Scholar]
- Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. 2006. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway.Nat. Cell Biol. 8:870–876 [DOI] [PubMed] [Google Scholar]
- Borghesani P.R., Alt F.W., Bottaro A., Davidson L., Aksoy S., Rathbun G.A., Roberts T.M., Swat W., Segal R.A., Gu Y. 2000. Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice.Proc. Natl. Acad. Sci. USA. 97:3336–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.Y., Sleckman B.P., Kanagawa O. 2005. Revision of T cell receptor {alpha} chain genes is required for normal T lymphocyte development.Proc. Natl. Acad. Sci. USA. 102:14356–14361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo T.C., Xia D., Hassouneh S., Yang X.O., Sabath D.E., Sperling K., Gatti R.A., Concannon P., Willerford D.M. 2000. V(D)J rearrangement in Nijmegen breakage syndrome.Mol. Immunol. 37:1131–1139 [DOI] [PubMed] [Google Scholar]
- Hsieh C.L., Arlett C.F., Lieber M.R. 1993. V(D)J recombination in ataxia telangiectasia, Bloom’s syndrome, and a DNA ligase I-associated immunodeficiency disorder.J. Biol. Chem. 268:20105–20109 [PubMed] [Google Scholar]
- Li G., Alt F.W., Cheng H.L., Brush J.W., Goff P.H., Murphy M.M., Franco S., Zhang Y., Zha S. 2008. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination.Mol. Cell. 31:631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., Wynshaw-Boris A. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia.Cell. 86:159–171 [DOI] [PubMed] [Google Scholar]
- Rooney S., Sekiguchi J., Zhu C., Cheng H.L., Manis J., Whitlow S., DeVido J., Foy D., Chaudhuri J., Lombard D., Alt F.W. 2002. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice.Mol. Cell. 10:1379–1390 [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris A.W., Cory S. 1991. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship.Cell. 67:889–899 [DOI] [PubMed] [Google Scholar]
- Pandita R.K., Sharma G.G., Laszlo A., Hopkins K.M., Davey S., Chakhparonian M., Gupta A., Wellinger R.J., Zhang J., Powell S.N., et al. 2006. Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair.Mol. Cell. Biol. 26:1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt C.R., Pandita R.K., Laszlo A., Higashikubo R., Agarwal M., Kitamura T., Gupta A., Rief N., Horikoshi N., Baskaran R., et al. 2007. Hyperthermia activates a subset of ataxia-telangiectasia mutated effectors independent of DNA strand breaks and heat shock protein 70 status.Cancer Res. 67:3010–3017 [DOI] [PubMed] [Google Scholar]