Abstract
HECT (homologous to the E6AP C terminus) ubiquitin ligases have diverse functions in eukaryotic cells. In screens for proteins that bind to the HECT ubiquitin ligase WWP1, we identified Spartin, which is also known as SPG20. This protein is truncated in a neurological disease, Troyer syndrome. In this study, we show that SPG20 associates with the surface of lipid droplets (LDs) and can regulate their size and number. SPG20 binds to another LD protein, TIP47, and both proteins compete with an additional LD protein, adipophilin/adipocyte differentiation-related protein, for occupancy of LDs. The mutant SPG20 present in Troyer syndrome does not possess these activities. Depletion of SPG20 using RNA interference increases the number and size of LDs when cells are fed with oleic acid. Binding of WWP1 to SPG20 and the consequent ubiquitin transfer remove SPG20 from LDs and reduce the levels of coexpressed SPG20. These experiments suggest functions for ubiquitin ligases and SPG20 in the regulation of LD turnover and potential pathological mechanisms in Troyer syndrome.
Introduction
HECT (homologous to the E6AP C terminus) ubiquitin ligases share a common protein organization, encoding an N-terminal C2 domain involved in membrane binding, several centrally located WW domains that bind to PPxY motifs, and a C-terminal HECT domain possessing the ubiquitin binding and ligase activities. HECT ubiquitin ligases have diverse functions in eukaryotic cells (Ingham et al., 2004), largely through their ability to modify target proteins with mono- or polyubiquitin, thereby affecting protein localization, interactions, and stability. For example, HECT ubiquitin ligases modulate the levels or localization of proteins involved in the control of the actin cytoskeleton (RhoA; Zhang et al., 2004), surface receptors such as CXCR4 (Marchese et al., 2003), transcription factors in TGF-β signaling (Zhang et al., 2001), and factors that modulate Notch signaling (Bray, 2006) as well as several tumor suppressors, including Smad4 (Moren et al., 2005) and p53 (Laine and Ronai, 2007). HECT ubiquitin ligases are also exploited by several enveloped viruses to initiate the recruitment of the ESCRT (endosomal sorting complex required for transport) machinery that mediates viral budding (Martin-Serrano et al., 2005; Chung et al., 2008; Usami et al., 2008).
In two independent screens for proteins that bind to HECT ubiquitin ligases, we identified Spartin as a WWP1-binding protein. Spartin, also know as SPG20, is mutated in a rare neuronal disease, termed Troyer syndrome (Patel et al., 2002). This syndrome is a complicated hereditary spastic paraparesis, which is characterized by distal muscle wasting, mental retardation, and dysarthria in addition to the lower extremity paraparesis and spasticity that characterize “pure” hereditary spastic paraparesis. SPG20 is ubiquitously expressed and encodes a microtubule interaction and trafficking (MIT) domain at its N terminus, which is also found in the related protein SPG4 (Spastin; Ciccarelli et al., 2003) as well as in the class E vacuolar protein sorting ATPase, VPS4 (Scott et al., 2005; Obita et al., 2007; Stuchell-Brereton et al., 2007), both of which bind to components of the ESCRT machinery (Obita et al., 2007; Stuchell-Brereton et al., 2007; Hurley and Yang, 2008). Additionally, SPG20 encodes a domain of unknown function toward its C terminus, which is defined by homology to proteins that are up-regulated during plant senescence (Ciccarelli et al., 2003). There is some confusion as to the function of SPG20 in cells, as some investigators have reported that it binds to EPS15 and plays a role in endocytosis (Bakowska et al., 2005, 2007), whereas others have suggested that it localizes to mitochondria (Lu et al., 2006). Additional experiments have suggested that SPG20 has a complex pattern of subcellular distribution, including transient localization with the TGN, as well as synaptic vesicles and discrete undefined puncta within terminally differentiated neuroblastoma cells (Robay et al., 2006).
In this study, we show that rather than associating with endosomal compartments and ESCRT proteins, SPG20 associates with the surface of lipid droplets (LDs). LDs are dynamic organelles that are the primary site for the storage of neutral lipids (Welte, 2007). They are believed to form within membranes of the ER, are transported along microtubules, and can be mobilized for lipolysis by adipose triglyceride lipase in response to changes in metabolic conditions (Brasaemle, 2007). LD surfaces display several specific proteins, including proteins of the perilipin (PAT) family such as adipophilin/adipocyte differentiation-related protein (ADRP) as well as TIP47 (tail-interacting protein of 47 kD), OXPAT/myocardial LD protein, and S3-12. These proteins may dynamically exchange between cytoplasmic- and LD-associated fractions (Londos et al., 2005; Listenberger et al., 2007).
We also find that SPG20 binds to the LD-localized protein TIP47. Interestingly, a mutant SPG20 present in Troyer syndrome fails to localize to LDs and does not bind TIP47. Moreover, ADRP and SPG20 appear to compete for LD surfaces and mutually exclude each other from LDs. Importantly, depletion of SPG20 from cells using RNA interference causes increases in both the number and size of LD when cells are fed with oleic acid (OA). Finally, SPG20 is ubiquitinated, does not accumulate on the surfaces of LDs, and is degraded under conditions of HECT ubiquitin ligase (WWP1) overexpression, suggesting that the protein composition of LDs, and by inference lipid storage, may be regulated by HECT ubiquitin ligases. Overall, these experiments suggest potential molecular mechanisms for the pathology of Troyer syndrome and a role for ubiquitin ligases and SPG20 in the regulation of LD turnover.
Results
Identification of Spartin/SPG20 as a HECT ubiquitin ligase–binding protein
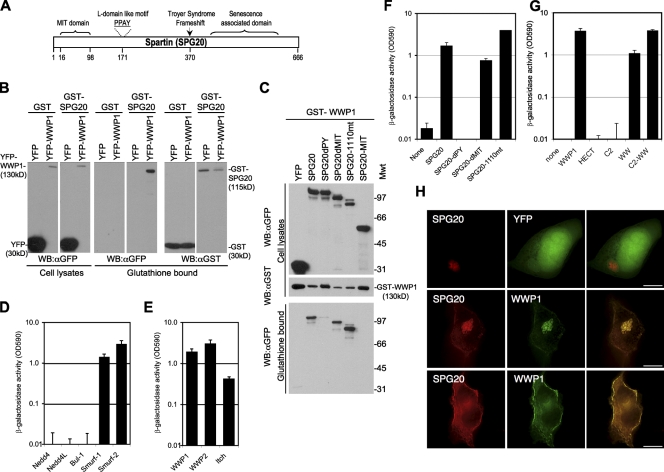
In screens to identify proteins that interact with HECT ubiquitin ligases, we used yeast two-hybrid and tandem affinity purification (TAP) approaches using full-length WWP1 as bait. Both screens identified several cellular proteins (unpublished data), but only one, namely SPG20, was identified by both approaches. SPG20 consists of an N-terminal MIT domain linked to a poorly characterized domain, which also contains a PPxY motif (Fig. 1 A). In the case of the yeast two-hybrid screen, an N-terminally truncated SPG20 (a deletion of residues 1–129, i.e., lacking the MIT domain; Fig. 1 A) was identified.
Figure 1.
SPG20 interacts with the WW domains of WWP1 via its PPxY motif. (A) Schematic representation of the Spartin/SPG20 protein showing the MIT domain (aa 16–98), PPAY sequence (aa 171–174), and Troyer syndrome frameshift mutant (deletion of nt 1,110 at aa 370 to truncate the 666-aa protein at aa 398). (B) Coprecipitation of WWP1 and SPG20. Unfused GST or GST-SPG20 was coexpressed with YFP or YFP-WWP1 in 293T cells, and cell lysates as well as the glutathione-bound fraction were analyzed by Western blotting with α-GST and α-GFP antibodies. (C) GST-WWP1 coprecipitation experiments with YFP fused to wild-type SPG20, SPG20 lacking the PPxY domain (SPG20dPY), SPG20 lacking the MIT domain (SPG20dMIT), the Troyer syndrome mutant SPG20 (SPG20-1110mt), or the isolated SPG20 MIT domain (SPG20-MIT). Mwt, molecular weight; WB, Western blot. (D–G) Yeast two-hybrid analysis of interactions between SPG20 and HECT ubiquitin ligases. β-Galactosidase activity measured in yeast transformed with Gal4 DNA-binding domain fusion proteins and VP16 activation domain fusion proteins expressed in optical density units (OD590). (D) Yeast two-hybrid analysis interactions between a Gal4-SPG20 and WW domains of Nedd4, Nedd4-L, Bul-1, Smurf1, and Smurf2. (E) Yeast two-hybrid analysis interactions between a Gal4-SPG20 and full-length WWP1, WWP2, and Itch. (F) Yeast two-hybrid analysis of Gal4-WWP1 binding to the various SPG20 mutants described in C. (G) Yeast two-hybrid analysis of Gal4-SPG20 binding to the various domains of WWP1: the N-terminal C2 domain, the central (PPxY binding) WW domains, and the C-terminal catalytic HECT domain. (H) Images of 293T cells transiently overexpressing CFP-SPG20 and either unfused YFP (top) or YFP-WWP1 (middle and bottom). Two different optical sections from a stack of deconvolved images are shown in the middle and bottom panels. Error bars represent the standard deviation of the mean. Bars, 10 µm.
Coprecipitation experiments using full-length SPG20 fused to GST expressed in 293T cells showed that GST-SPG20 could coprecipitate YFP-WWP1 (Fig. 1 B). In reciprocal experiments, GST-WWP1 coprecipitated YFP-SPG20 (Fig. 1 C). The coprecipitation of SPG20 and WWP1 was dependent on the SPG20 PPxY motif, but deletion of either of the MIT domain or residues C terminal to position 1,110 (mimicking the SPG20 mutation in Troyer syndrome) had no effect on WWP1 binding (Fig. 1 C). These results were confirmed using yeast two-hybrid assays (Fig. 1 F). Further yeast two-hybrid assays revealed that the central WW domains of WWP1 were necessary and sufficient for interaction with SPG20 (Fig. 1 G). Thus, SPG20–WWP1 interaction appeared to be mediated exclusively by the PPxY motif in SPG20 and the WW domains in WWP1. Yeast two-hybrid analyses revealed that SPG20 could bind to the WW domains from several HECT ubiquitin ligases, specifically WWP1, WWP2, Itch, Smurf1, and Smurf2, but did not bind to WW domains of other HECT ubiquitin ligases, namely Nedd4, Nedd4-L, and Bul-1 (Fig. 1, D and E).
Colocalization of SPG20 and WWP1
Overexpression of CFP-SPG20 in 293T cells resulted in the formation of large, apparently spherical, clustered vesicular structures within the cytoplasm (Fig. 1 H). Upon coexpression of CFP-SPG20 with YFP-WWP1, the localization of both proteins changed, and extensive colocalization was observed (Fig. 1 H). Ordinarily, YFP-WWP1 localizes primarily at the plasma membrane (Martin-Serrano et al., 2005). However, when coexpressed with CFP-SPG20, a portion of WWP1 relocalized to the CFP-SPG20–labeled vesicular structures. Moreover, a portion of CFP-SPG20 was relocalized to the plasma membrane when coexpressed with WWP1 (Fig. 1 H). Colocalization experiments using overexpressed WWP1 fragments (YFPWW-HECT, YFPC2-WW, YFPHECT, and YFPWW) revealed that fragments that included the WW domain colocalized extensively with CFP-SPG20, and YFPC2-WW overexpression caused CFP-SPG20 to relocalize to the plasma membrane (Fig. S1). Conversely, upon coexpression of CFP-SPG20 with YFPHECT, there was no colocalization or any change in the distribution of either protein (Fig. S1, bottom). Thus, the WW domain of WWP1 meditates WWP1 recruitment to SPG20-containing structures, which is consistent with the finding that the WW domains of WWP1 bind to SPG20.
Use of virus release-based assays to demonstrate HECT ubiquitin ligase recruitment by Spartin/SPG20
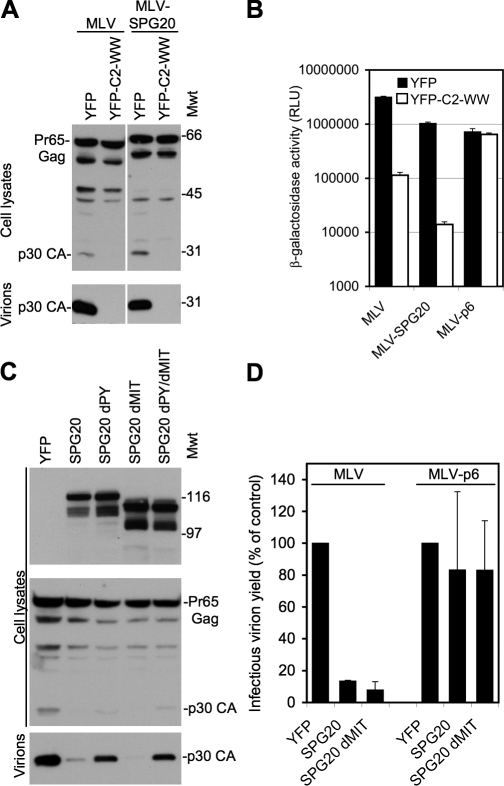
Several enveloped viruses, including murine leukemia virus (MLV), engage cellular machinery required for virus particle release using PPxY-based “late-domain” peptide motifs that recruit ubiquitin ligases. If the SPG20 PPxY motif indeed bound to HECT ubiquitin ligases in a cellular environment, we reasoned that it should substitute for a motif within the MLV Gag protein that is known to recruit HECT ubiquitin ligases and thereby induce viral budding. Therefore, we substituted the MLV late domain with a motif corresponding to 14 aa residues centered around the PPxY motif in SPG20. Notably, this construct, termed MLV-SPG20, efficiently generated infectious MLV virions, at least 200-fold more efficiently than the background level generated by an MLV lacking the PPPY motif (Fig. 2, A and B). Moreover, MLV-SPG20 release was highly sensitive to the inhibition by a C-terminally truncated version of WWP1 lacking the HECT domain (Fig. 2, A and B), which is a potent inhibitor of wild-type MLV budding (Martin-Serrano et al., 2005). Importantly, an isogenic control virus, MLV-p6, that contains a PTAP late domain and does not require HECT ubiquitin ligases for budding (Fig. 2 B) was not inhibited by the truncated WWP1 protein. Thus, transplantation of the SPG20 PPxY motif into a heterologous context conferred the ability to recruit HECT ubiquitin ligases.
Figure 2.
Virus particle release assays to demonstrate interactions between SPG20 and HECT ubiquitin ligases within cells. (A) Release of virions from cells transfected with MLV constructs encoding either the unmodified PPPY domain or one in which the PPPY motif was replaced with a region encompassing the SPG20-PPxY sequence (MLV-SPG20). Virions were generated in the presence or absence of the WWP1dHECT protein, and blots were probed with an α-MLV capsid antibody. Mwt, molecular weight. (B) Measurement of infectious virions generated by MLV, MLV-SPG20, or an MLV encoding the HIV-p6–derived PTAP motif (MLV-p6). Virions containing the pMSCV/Tat vector were generated in the presence or absence of the WWP1dHECT protein and were quantitated by measuring β-galactosidase activity after inoculation of TZM cells. RLU, relative light unit. (C) Western blot analysis of cell lysates and released MLV virion particles from 293T cells cotransfected with an unmodified MLV provirus along with YFP alone or YFP fused to SPG20, SPG20dPY, SPG20dMIT, or SPG20dPYdMIT. Blots were probed with α-GFP (top) or α-MLV capsid (bottom) antibodies. (D) Infectious virion release from cells transfected with MLV or MLV-p6 in the presence or absence of SPG20. Infectious virions were measured using TZM indicator cells as in B and are expressed as a percentage of control uninhibited virion release. Error bars represent the standard deviation of the mean.
The MLV Gag protein can efficiently recruit WWP1 to nascent viruslike particles at the plasma membrane in a PPxY-dependent manner. We reasoned that if the interaction of SPG20 and WWP1 was authentic, SPG20 might compete with MLV Gag that normally binds to WWP1, block WWP1 recruitment, and inhibit virus particle release. Coexpressed CFP-tagged MLV Gag and YFP-WWP1 extensively colocalized in puncta at the plasma membrane (Fig. S2 A). Notably, simultaneous overexpression of SPG20, but not SPG20 possessing a mutated PPPY domain (SPG20dPY), blocked YFP-WWP1 recruitment to MLV Gag-CFP puncta (Fig. S2, B and C). Additionally, overexpression of either full-length SPG20 or the MIT domain–deleted variant substantially reduced the release of MLV virions (Fig. 2, C and D) but had no effect on MLV-p6 (Fig. 2 D). This inhibition of MLV release was largely dependent on the SPG20 PPxY motif, as expression of SPG20dPY had only marginal effects (Fig. 2 C).
Intact SPG20 but not the Troyer syndrome variant is an LD-associated protein
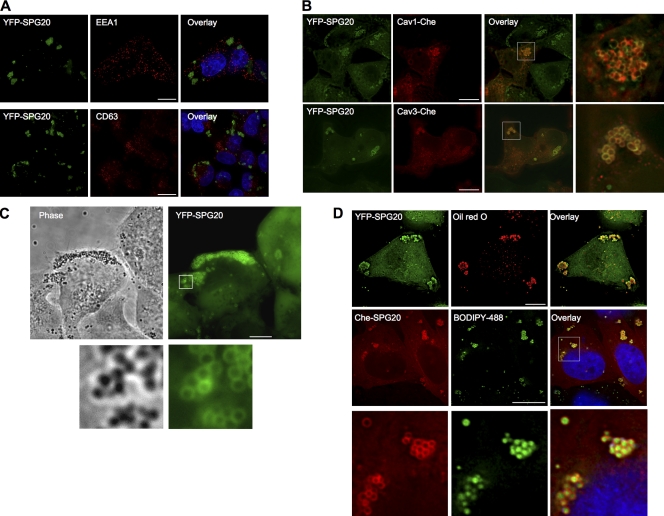
We generated cell lines constitutively expressing SPG20 with both N- and C-terminal fusions of YFP, Cherry fluorescent protein (Che), or the HA antigen tag. In each cell line, the tagged SPG20 protein localized in spherical, apparently vesicular structures. Because SPG20 was reported to bind EPS15 and because proteins encoding MIT domains often bind to cellular factors involved in endosomal trafficking, we determined whether stably expressed YFP-SPG20 colocalized to endosome markers. These included endogenous markers for early and late endosomes (EEA1 and CD63) and late endosome to Golgi transport vesicles (mannose-6-phosphate receptor [M6PR]) as well as transfected markers for recycling endosomes (CFP-Rab11) and markers of secretory granules (Che-Vamp2 and pCDNA-prolactin). We found no colocalization with these markers (Fig. 3 A and not depicted). However, some of the SPG20-positive vesicles were positive for transiently expressed Che-tagged Cav1 (Fig. 3 B). Additionally, although we did not detect any YFP-SPG20 signal in Cav3 puncta at the plasma membrane, we did observe substantial colocalization of YFP-SPG20 with Che-Cav3 on spherical intracellular structures (Fig. 3 B).
Figure 3.
SPG20 localizes to LDs. (A) Immunofluorescent localization of endosome markers (EEA1 and CD63; red) in a cell line (HeLa/YFP-SPG20) stably expressing a YFP-SPG20 fusion protein (green; images represent projections of deconvolved image stacks). (B) Localization of Che-tagged caveolin proteins (Cav1-Che and Cav3-Che; red) after transient expression in HeLa/YFP-SPG20 cells. Expanded views of the boxed areas in the overlay images are shown to the right. (C) Phase-contrast (left) and fluorescent (right) images of HeLa/YFP-SPG20 cells. An expanded view of the boxed area is shown below. (D) Staining of HeLa cells with LD markers. HeLa/YFP-SGP20 cells were stained with the LD marker Oil red O (top), whereas HeLa/Che-SPG20 cells were stained using the LD marker BODIPY 493/503 (middle). An expanded view of the boxed area in the BODIPY 493/503–stained HeLa/Che-SPG20 is shown in the bottom panels. Bars, 10 µm.
We also noticed that YFP-SPG20 colocalized perfectly with spheres that were easily visible using phase-contrast microscopy (Fig. 3 C). The unique appearance of these vesicles as well as the colocalization with caveolins (Martin and Parton, 2005) suggested that YFP-SPG20–labeled spheres might represent LDs. Upon staining with the LD marker Oil red 0, we found that YFP-SPG20 perfectly encircled Oil red 0–positive spheres (Fig. 3 D, top). Similarly, examination of a Che-SPG20–expressing HeLa cell line revealed that Che-SPG20 also localized as spheres. These spheres could be stained with boron-dipyrromethene (BODIPY) 493/503, another fluorescent marker of LDs, with the Che-SPG20 signal surrounding the BODIPY 493/503–stained LDs (Fig. 3 D, middle and bottom).
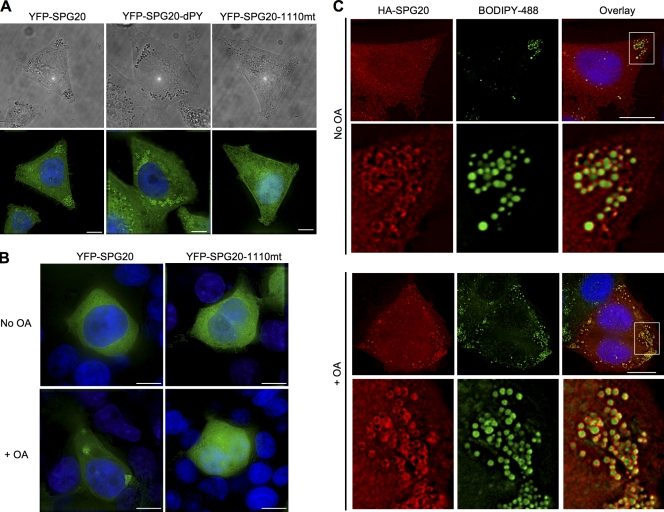
Next, we compared the localization of the wild-type YFP-SPG20 protein with two YFP-SPG20 mutants. In particular, we used the YFP-SPG20dPY mutant that no longer binds to WWP1 and an SPG20 mutant found in individuals with Troyer syndrome (YFP-SPG20-1110mt), in which a single nucleotide deletion at position 1,110 of the 2,001-nt SPG20 mRNA causes a frame shift after aa 369 and expression of a truncated 398-aa protein (wild-type SPG20 is 666 aa). The PPxY motif, and by inference the ability of SPG20 to bind to WWP1, was not required for LD localization (Fig. 4 A). However, the Troyer syndrome SPG20 protein was diffusely distributed and did not localize to LDs (Fig. 4 B).
Figure 4.
The C-terminal region of SPG20 that is absent in the Troyer syndrome variant is required for LD targeting. (A) HeLa cell lines stably expressing YFP-SPG20, YFP-SPG20dPY, or YFP-SPG20-1110mt were visualized using phase contrast (top) and fluorescence (bottom). LDs, which are present in all cells, can be seen as dark spheres and are more prominent in the presence of YFP-SPG20 and YFP-SPG20dPY. (B) 293T cells stably expressing YFP-SPG20 (left) or YFP-SGP20-1110mt (right) under normal culture conditions (top) or after incubation for 18 h with OA (bottom). Note that in 293T cells, expression of the YFP fusion proteins is lower than in HeLa cells, and YFP-SPG20 appears diffuse under normal culture conditions (top left). (C) Localization of HA-tagged SPG20. HeLa cells stably expressing HA-SPG20 were cultured in the absence (top) or presence (bottom) of OA and stained with antibody to the HA epitope tag (red) and with BODIPY 493/53 (green). An expanded view of LDs present in the area of the images bounded by the white boxes is shown below each image set. Bars, 10 µm.
Stable expression of YFP-SPG20 in 293T cells by transduction with a retroviral vector resulted in lower levels of SPG20 than in HeLa/YFP-SPG20 cells, as indicated by Western blotting (unpublished data). Under these conditions, YFP-SPG20 was not localized to LDs and instead was diffusely distributed in the cytoplasm (Fig. 4 B). The addition of the fatty acid OA has been shown to stimulate LD formation in a variety of nonadipose cell types (Xu et al., 2005; Smirnova et al., 2006; Bostrom et al., 2007; Fei et al., 2008), and, strikingly, a significant proportion of the diffuse YFP-SPG20 was redistributed to LDs upon addition of OA to the culture medium of 293T/YFP-SPG20 cells (Fig. 4 B). However, stably expressed YFP-SPG20-1110mt did not respond to OA in this way, confirming that the recruitment of SPG20 to LDs was dependent on the C-terminal portion of the protein that is missing in Troyer syndrome (Fig. 4 B).
To ensure that the LD localization of YFP-SPG20 and Che-SPG20 was not an artifact of fluorescent protein tagging, we generated a HeLa cell line stably expressing HA-tagged SPG20 (HA-SPG20). The levels of HA-SPG20 in this cell line were lower than YFP-SPG20 and Che-SPG20 stably expressed in HeLa cells (Fig. S3). Indeed, HA-SPG20 was expressed at approximately equivalent levels to the endogenous HeLa cell SPG20 (Fig. S3 A). In these cells, a diffuse cytoplasmic staining was evident with some degree of HA-SPG20 localization on LDs (Fig. 4 C, top). However, notably, when the HeLa/HA-SPG20 cells were cultured in the presence of OA, localization of HA-SPG20 to BODIPY-positive LDs became prominent (Fig. 4 C, bottom) without effects on the HA-SPG20 expression levels (Fig. S3 B). Interestingly, OA reproducibly caused the appearance of low levels of higher molecular weight forms of HA-SPG20/SPG20, perhaps suggesting that SPG20 ubiquitination had been induced.
SPG20 interacts with and colocalizes with the LD protein TIP47
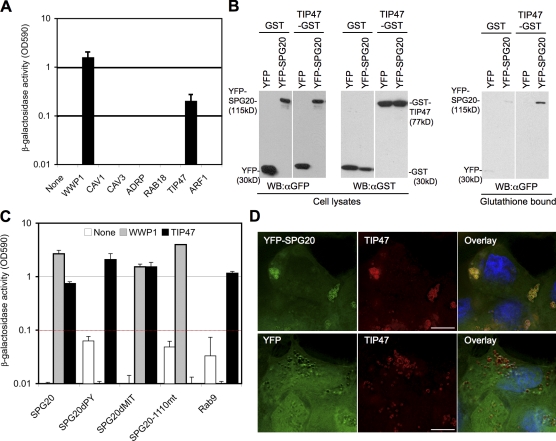
Next, we used yeast two-hybrid experiments to test for interactions between SPG20 and a selection of proteins known to be associated with LDs, including ADRP, TIP47, Rab18, Arf-1, Cav1, and Cav3. Notably, SPG20 bound to TIP47 but not to the other proteins tested (Fig. 5 A). Confirmatory assays in which 293T cells were cotransfected with plasmids expressing TIP47-GST and YFP-SPG20 (Fig. 5 B) revealed that the two proteins could be coprecipitated. Interestingly, the Troyer syndrome SPG20 variant (SPG20-1110mt) did not bind TIP47 (Fig. 5 C), whereas SPG20 proteins lacking either the MIT domain or the PPxY motif bound to TIP47 as efficiently as intact SPG20.
Figure 5.
SPG20 interacts with TIP47. (A) Directed yeast two-hybrid analysis of interactions between Gal4-SPG20 and candidate LD-associated proteins (Cav1, Cav3, ADRP, Rab18, TIP47, and Arf-1) fused to the VP16 activation domain. WWP1 is included as a positive control. (B) GST coprecipitation of YFP-SPG20 and TIP47-GST. Unfractionated 293T cell lysates and proteins bound to glutathione-agarose were detected by Western blotting with α-GFP and α-GST antibodies. WB, Western blot. (C) Yeast two-hybrid analysis of interactions between SPG20 and TIP47. The DNA-binding constructs containing full-length SPG20, SPG20dPY, SPG20dMIT, and SPG20-1110mt were tested against the VP16 activation domain fused to WWP1 or TIP47. Gal4-Rab9 was included in the experiment as a control for TIP47 binding. (D) Colocalization of SPG20 and TIP47. Endogenous TIP47 is stained with an α-TIP47 monoclonal antibody in HeLa cells stably expressing either YFP-SPG20 (top) or unfused YFP (bottom). Error bars represent the standard deviation of the mean. Bars, 10 µm.
A transiently expressed Che-tagged TIP47 (TIP47-Che) exhibited a mixture of diffuse and LD-localized fluorescence (confirmed by labeling with BODIPY 493/503; Fig. S4 and not depicted). However, coexpression of TIP47-Che with YFP-SPG20 in 293T cells resulted in substantial colocalization of the two overexpressed proteins on LDs that coalesced together (Fig. S4). Moreover, endogenous TIP47 detected by immunofluorescent staining nearly perfectly colocalized with YFP-SPG20 on LDs in HeLa/YFP-SPG20 cells (Fig. 5 D). Notably, the distribution of TIP47 in these cells was distinct from that in HeLa cells expressing YFP alone (Fig. 5 D), as SPG20/TIP47-positive LDs appeared to group together.
SPG20 and TIP47 mark LD surfaces distinct from those occupied by ADRP
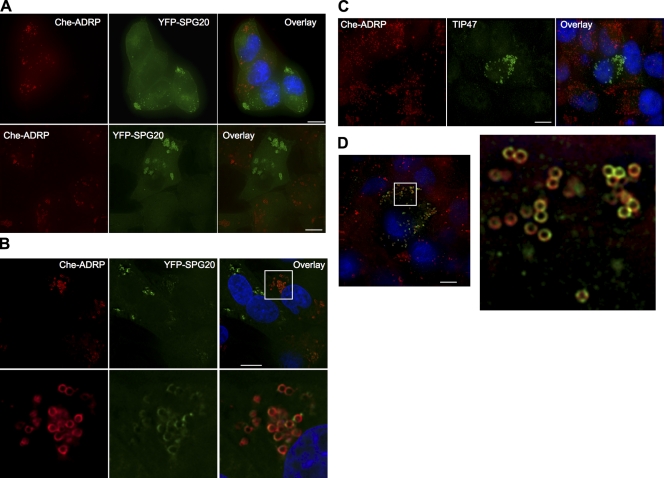
To determine whether SPG20 colocalized with another marker of LDs, we transduced HeLa YFP-SPG20 cells with a retroviral vector expressing Che-tagged ADRP (Che-ADRP). Notably, YFP-SPG20 was displaced from LDs in cells expressing Che-ADRP (Fig. 6 A, top). In a reciprocal experiment, a HeLa cell line stably expressing Che-ADRP was transduced with a YFP-SPG20–expressing retroviral vector, and a reduced Che-ADRP association with LDs was observed in cells expressing high levels of YFP-SPG20 (Fig. 6 A, bottom). Again, the retroviral vector expressing only YFP had no effect on Che-ADRP distribution (unpublished data). In cells expressing lower levels of YFP-SPG20, some of the LDs were labeled with both YFP-SPG20 and Che-ADRP (Fig. 6 B). Transduction with retroviral vectors expressing either YFP-SPG20dPY or YFP-SPG20dMIT resulted in the displacement of ADRP from LDs, similar to wild-type YFP-SPG20, but the YFP-SPG20-1110mt that is deficient in LD localization did not affect Che-ADRP localization (unpublished data).
Figure 6.
Reciprocal exclusion of SPG20, TIP47, and ADRP from LDs. (A) Expression of ADRP displaces SPG20 from LDs and vice versa. HeLa/YFP-SPG20 cells were transduced with a retroviral vector expressing Che-ADRP and visualized several days after infection. The images shown represent a projection of a deconvolved vertical stack of images, and a field containing both Che-ADRP–positive and Che-ADRP–negative cells is shown (top). The reciprocal experiment, in which a HeLa Che-ADRP cell line was transduced with a YFP-SPG20–expressing retroviral vector, is shown in the bottom panels. (B) Coresidence of Che-ADRP and YFP-SPG20 in cells expressing lower levels of YFP-SPG20. An expanded view of the boxed area is shown below. (C and D) HeLa/Che-ADRP cells stained with antibodies against endogenous TIP47 (green). Note that in C, TIP47 localization is only clearly visible in cells expressing no or low levels of Che-ADRP. In D, an expanded area of a cell (boxed area) expressing moderate levels of ADRP is shown, revealing coexistence of TIP47 and ADRP on individual LDs but apparently occupying distinct areas of the LD surface. Bars, 10 µm.
Additionally, immunofluorescent analysis of HeLa cells stably expressing Che-ADRP revealed that the LDs in the majority of Che-ADRP–expressing cells did not display TIP47 on their surfaces (Fig. 6 C), which is in contrast to the untransduced HeLa cells in which TIP47 was clearly present on the LDs in the majority of cells. Similar results have recently been reported by others, in which ADRP was shown to reduce the association of several proteins with LD surfaces (Listenberger et al., 2007). In a small subset of cells expressing detectable levels of TIP47 on LDs, apparently reduced levels of Che-ADRP were present (Fig. 6 D). In these cells, in which both Che-ADRP and TIP47 were detectable, they tended to localize to different areas on the surface of individual LDs (Fig. 6 D).
WWP1 can induce SPG20 ubiquitination and degradation and prevents SPG20 accumulation on LDs
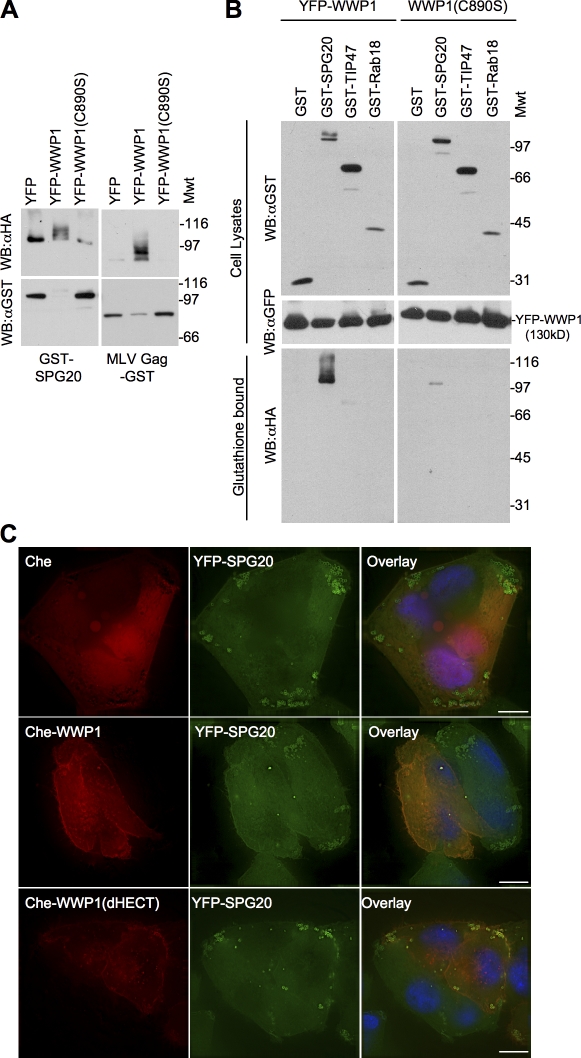
The HECT ubiquitin ligase WWP1 and SPG20 can clearly bind to each other and colocalize (Figs. 1 and 2); therefore, we determined whether there was a functional consequence of this interaction. To accomplish this, we coexpressed a GST-tagged version of SPG20 (or MLV Gag as a control) with WWP1 and HA-tagged ubiquitin (HA-ubiquitin). Western blot analysis of glutathione-agarose–precipitated proteins revealed that a small fraction of SPG20 was monoubiquitinated in the absence of overexpressed WWP1. Notably, SPG20 ubiquitination was stimulated by overexpression of WWP1 and inhibited by a catalytically inactive WWP1 point mutant (C890S; Fig. 7 A). The predominant ubiquitinated SPG20 species observed was monoubiquitinated, as indicated by the ∼10-kD larger form of SPG20 that was sometimes detected in whole cell lysates (Fig. 7, A and B) and the dominant anti-HA reactive species in glutathione-agarose precipitates. However, additional ubiquitinated species were detected in the glutathione-agarose–precipitated GST-SPG20 fraction (Fig. 7, A and B), and the total amount of cellular SPG20 protein was often reduced upon overexpression of WWP1 as compared with a control protein (YFP) or WWP1 (C890S), which is presumably the result of proteosome-mediated degradation. Other LD resident proteins (expressed as GST fusion proteins), including TIP47, were either not ubiquitinated or only exhibited traces of ubiquitination upon coexpression with WWP1 (Fig. 7 B).
Figure 7.
WWP1 can ubiquitinate SPG20 and prevent its accumulation on LDs. (A) Ubiquitin transfer experiment performed in 293T cells cotransfected with GST-SPG20 or MLV Gag–GST along with HA-ubiquitin and unfused YFP, YFP-WWP1, or catalytically inactive YFP-WWP1 (C890S). Glutathione-bound proteins were analyzed by Western blotting with α-HA and α-GST antibodies. Mwt, molecular weight. WB, Western blot. (B) Similar to A except that cell lysates and glutathione-bound fractions were analyzed by Western blotting. 293T cells transfected with GST-SPG20, GST-TIP47, or GST-Rab18 along with YFP-WWP1 or YFP-WWP1 (C890S) as well as HA-ubiquitin. GST-bound proteins were probed with antibodies against the HA tag (bottom) to detect specific ubiquitin transfer. Cellular lysates were probed with antibodies against GST (top) and YFP (middle) to detect the corresponding fusion proteins. (C) Overexpression of WWP1 depletes SPG20 from LDs. HeLa YFP-SPG20 cells were transduced with retroviruses expressing unfused Che (top), Che-WWP1 (middle), or Che-WWP1dHECT (bottom). Bars, 10 µm.
Microscopic examination of HeLa/YFP-SPG20 cells coexpressing Che-fused WWP1 (Che-WWP1) revealed that overexpression of Che-WWP1 inhibited the accumulation of YFP-SPG20 protein on LDs (or alternatively induced the removal of YFP-SPG20 from LDs; Fig. 7 C, middle), whereas unfused Che had no effect (Fig. 7 C, top). This could have been caused by the specific removal of the LD-associated YFP-SPG20 fraction or, alternatively, caused by the reduction in the overall YFP-SPG20 levels and consequent reduction in LD association. Che-WWP1 lacking the catalytic HECT domain (Che-WWP1dHECT) did not prevent YFP-SPG20 accumulation on LDs and actually appeared to enhance the YFP-SPG20 signal on LDs, where Che-WWP1dHECT was colocalized (Fig. 7 C, bottom).
Overexpression or underexpression of SPG20 induces LD accumulation
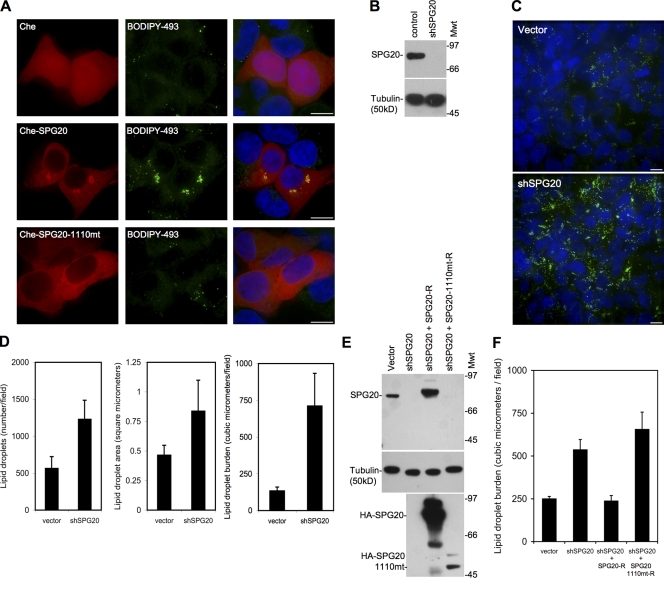
To determine whether SPG20 plays a role in LD formation or turnover, we overexpressed Che-SPG20 in 293T cells and subsequently incubated these cells with BODIPY 493/503 to label LDs. The presence of high levels of SPG20 (∼10-fold higher than in the HeLa/YFP-SPG20 cells; unpublished data) caused the accumulation and coalescence of LDs into perinuclear clusters where Che-SPG20 was also concentrated (Fig. 8 A). Overexpression of HA-SPG20 also induced perinuclear clusters of HA-SPG20–coated LDs (Fig. S5). This change was not observed upon overexpression of the Che-SPG20-1110mt or the unfused Che protein (Fig. 8 A).
Figure 8.
Over- or underexpression of SPG20 perturbs LDs. (A) 293T cells transfected with Che (top), Che-SPG20 (middle), or Che-SPG20-1110mt (bottom) and stained with the LD marker BODIPY 493/503. (B) SPG20-depleted cells generated by transduction of 293T cells with lentiviral vectors expressing an shRNA targeting SPG20 (shSPG0). Western blot analysis of cell lysates probed with an antibody against SPG20 (top) and against tubulin (bottom) as a loading control. Mwt, molecular weight. (C) Representative fields of control vector–containing (top) and shSPG20-containing (bottom) 293T cells cultured in the presence of 500 µM OA for 48 h and stained with BODIPY 493/503 to detect LDs (green). (D) The total number and size of LDs were quantified in 10 randomly selected fields using the softWoRx software. (E and F) Similar experiments as shown in B–D elaborated by the inclusion of shSPG20 cells reconstituted with an shRNA-resistant form of wild-type HA-tagged SPG20 (SPG20-R) or the Troyer syndrome variant (SPG20-1110mt-R). (E) Western blots probed with antibodies to SPG20 (top), tubulin (loading control; middle), and HA-tagged proteins (bottom). (F) LD burden in SPG20-depleted/reconstituted cells determined as in D. Error bars represent the standard deviation of the mean. Bars, 10 µm.
To determine whether endogenous SPG20 affected LD accumulation, we generated 293T cell populations expressing a short hairpin RNA (shRNA) directed against the SPG20 coding sequence. Western blot analysis revealed that SPG20 protein was efficiently depleted using this approach (Fig. 8 B). Incubation of SPG20-depleted 293T cells with the fluorescent LD marker BODIPY 493/503 did not reveal any effect on LDs under basal conditions (unpublished data). However, when cells were cultured with OA for 2 d, SPG20 depletion had a marked effect on LD accumulation. Specifically, SPG20-depleted cells contained a clearly increased amount of BODIPY 493/503–stained LDs as compared with control vector–transduced cells (Fig. 8 C). We quantitated this difference by measuring the total number and size of LDs in randomly selected fields in the cell monolayer (Fig. 8 D). Both parameters were increased in SPG20-depleted cells as compared with control vector–transduced cells, and when both LD size and numbers were accounted for and assuming that LDs are spherical, we estimated that SPG20-depleted cells bore an approximately three- to fivefold greater total burden of LD volume as compared with control vector–transduced cells. As a control, we transduced the SPG20-depleted cells with retroviral vectors expressing an shRNA-resistant form of either wild-type SPG20 (HA-SPG20-R) or a mutant form (HA-SPG20-1110mt-R) analogous to that found in Troyer syndrome and performed similar experiments. Expression of HA-SPG20-R could relieve the LD accumulation that was observed in the OA-fed, SPG20-depleted cells, whereas HA-SPG20-1110mt-R did not (Fig. 8 E). Note that the HA-SPG20-1110mt-R protein was expressed at substantially lower levels in transduced cells than was the HA-SPG20-R protein, as has been previously reported for the endogenous Troyer syndrome form of SPG20 (Bakowska et al., 2008).
Discussion
The cellular function of SPG20 and the underlying basis for neurodegeneration observed in Troyer syndrome patients harboring a mutation in SPG20 is unknown. Previous work has suggested that SPG20 is involved in endocytic trafficking through interaction with EPS15 (Bakowska et al., 2005, 2007); however, we were not able to observe any effect on endocytic traffic upon siRNA-mediated knockdown of SPG20. Additionally, we were not able to detect any interaction of the SPG20-MIT domain with proteins that are involved in endocytic trafficking and are known to possess MIT interaction motifs (Row et al., 2007; Stuchell-Brereton et al., 2007; Hurley and Yang, 2008) such as the charged multivesicular body proteins that were previously shown to bind to the MIT domains of VPS4 (Martin-Serrano et al., 2003; von Schwedler et al., 2003) and the SPG20-related protein SPG4 (Spastin; Reid et al., 2005).
Instead, we uncovered an alternative function for SPG20. Notably, we found a striking localization of SPG20 on LDs in several cell lines, including HeLa, 293T, and SH-SY5Y neuroblastoma cells. At low expression levels, approximating those of the endogenous levels found in HeLa cells, SPG20 fusion proteins were found to be diffusely distributed in the cytoplasm with some degree of accumulation on LDs. However, SPG20 localization to LDs was stimulated by OA feeding without affecting SPG20 expression levels in a manner quite similar to that observed with PAT proteins such as TIP47 (Wolins et al., 2005; Brasaemle, 2007). Overexpressed YFP, Che, or HA-tagged SPG20 proteins clearly localized on LDs and, in fact, induced clustering of LDs.
Other studies have described diverse patterns of localization for SPG20, including within mitochondria (Lu et al., 2006), early and late endosomes (Bakowska et al., 2007), the nucleus, and the TGN as well as synaptotagmin-positive synaptic vesicles (Robay et al., 2006). However, in some of these previous studies, the morphological pattern of SPG20 localization depicted is reminiscent of the LD staining described in this study. Indeed, other proteins that had been previously thought to reside in the mitochondria or endosomes have since been reclassified as LD-localized proteins (Wolins et al., 2001; Puri et al., 2008). Moreover, in contrast to the previous studies, we did not detect an SPG20 colocalization with endosomal markers (EEA1 or CD63), swollen late endosomes induced by expression of dominant-negative VPS4, or with mitochondria marked by either a resident fluorescent protein (DsRed-Mito) or MitoTracker (unpublished data). However, we did detect LDs labeled with YFP-SPG20 in the vicinity of both VPS4-induced endosomes and mitochondria. Interestingly, two studies have found that LDs exist in close proximity to mitochondria within adipocytes (Cohen et al., 2004; Brasaemle, 2007), and it is quite likely that the localization of SPG20 to mitochondria or endosomal compartments that has been reported was actually SPG20 on LDs. This may have been difficult to resolve using the microscopic techniques used in those studies (Lu et al., 2006; Robay et al., 2006; Bakowska et al., 2007). Importantly, we found that the SPG20-1110mt protein failed to associate with LDs. The portion of SPG20 that is disrupted in the SPG20-1110mt protein contains a poorly defined plant-related senescence domain (Ciccarelli et al., 2003), and, therefore, it is plausible that the senescence domain is an LD-targeting domain. Indeed, targeting of SPG20 to LDs may stabilize SPG20 levels in vivo because a recent study has shown that cells isolated from Troyer syndrome patients contain undetectable levels of SPG20-1110mt mutant protein (Bakowska et al., 2008).
Although several recent studies have identified numerous proteins associated with LDs using either proteomic or functional genomic screens (Beller et al., 2006, 2008; Cermelli et al., 2006; Sato et al., 2006; Bartz et al., 2007; Greenberg and Obin, 2008; Guo et al., 2008), SPG20 has yet to be identified as LD associated. This is in keeping with the diffuse pattern of localization that we found upon low levels of SPG20 expression. Indeed, SPG20 only became highly enriched on LDs when overexpressed or when cells were fed with OA. Nonetheless, consistent with the notion that SPG20 can genuinely localize to LDs, we also found that SPG20 interacted with TIP47. Importantly, this interaction required the C-terminal region of SPG20 that is lost in the Troyer syndrome mutant protein and is also required for LD association. Moreover, we found that overexpression of a YFP-tagged version of SPG20 caused a dramatic accumulation of endogenous TIP47 to clustered LDs that were also SPG20 positive. Notably, previous work has shown that TIP47 and ADRP mark distinct populations of LDs that appear to be at different stages of maturation. Specifically, shortly after OA treatment of adipocytes, TIP47 has been found to be associated with small lipid accumulations at the cell periphery that subsequently develop into mature LDs, and TIP47 is simultaneously replaced by ADARP (Wolins et al., 2005; Ohsaki et al., 2006; Listenberger et al., 2007). In addition, another protein associated with the LD, Rab18, has been shown to displace ADRP from LDs upon overexpression (Ozeki et al., 2005). In this study, we found a similar relationship between SPG20 and ADRP, as the presence of SPG20 and ADRP on LDs appeared to be mutually exclusive. Cells expressing high levels of SPG20 had quite low or undetectable levels of ADRP on LDs and vice versa. Thus, SPG20 behaved similarly to TIP47 and bound to TIP47. Therefore, TIP47/SPG20 may compete with ADRP for binding sites on the LD surface. Curiously, a similar phenotype was observed upon overexpression of the hepatitis C virus core protein such that hepatitis C virus core proteins were found to coat the LD and to exclude ADRP as well as cause LD aggregation in a rather similar manner to SPG20 (Boulant et al., 2008). Notably, both overexpression and underexpression of SPG20 induced the accumulation of LDs in cultured cells. The reasons for this are not clear, but we speculate that perhaps SPG20 functions as part of a protein complex, whose stoichiometry (and function) could be disrupted by either overexpression or underexpression of SPG20. However, SPG20 overexpression appeared to induce tightly clustered LDs, whereas SPG20 depletion induced LDs that appeared to be dispersed throughout the cell. The precise mechanisms underlying these effects remain to be determined.
Using a variety of assays, we found that SPG20 binds to HECT ubiquitin ligases via a PPxY domain in SPG20 and the WW domains in the HECT ligases. We also found that binding of WWP1 to SPG20 results in ubiquitin transfer to SPG20. Previous experiments have shown that SPG20 is monoubiquitinated (Bakowska et al., 2007), and, concordantly, the major species that we observed resulting from WWP1 overexpression was monoubiquitinated SPG20. In addition, we found that WWP1 overexpression reduced the levels of coexpressed SPG20 in cells. Although we have not defined a direct role for WWP1 in LD morphogenesis, it is likely that HECT ubiquitin ligases control SPG20 localization and/or protein levels via ubiquitination and, therefore, could influence LD protein composition and turnover.
The symptoms of SPG20 mutation in Troyer syndrome include lower limb spasticity and are consistent with the degeneration of long axons. However, no defects in general lipid metabolism such as obesity or fat deposition have been reported. This is perhaps surprising given our findings with SPG20, although inspection of subcellular architecture, in particular that of LDs, in Troyer syndrome patients has not been undertaken as far as we are aware. It may be that axons may simply be more sensitive than other cell types to perturbations in LD turnover. There are interesting precedents linking genes involved in lipid turnover with diseases that have a similar presentation to Troyer syndrome. For example, a motor neuron disease can be caused by mutations of the gene encoding the neuropathy target esterase (NTE) or inhibition of NTE by organophosphorus compounds (Rainier et al., 2008). NTE is partially localized to the cytoplasmic face of the ER and appears to be important for the proper balance of membrane lipids and the maintenance of axon integrity (Glynn, 1999; Zaccheo et al., 2004). NTE is a phospholipase and is a member of the superfamily of patatin domain lipases that also includes the closely related lysophospholipase NTE-related esterase that is found on LDs and adipose triglyceride lipase, which can regulate LD size in nonadipocyte cells (Smirnova et al., 2006; Kienesberger et al., 2008). Additionally, mutation of another human protein, Seipin (BSCL2), has also been shown to be associated with motor neuron disorders, including wasting of distal limb muscles and lower limb spasticity (Agarwal and Garg, 2004; Ito and Suzuki, 2007). Interestingly, yeast lacking a functional homologue of Seipin (FLD1) contain increased levels of neutral lipids and possess irregularly clustered/fused and enlarged LDs, a defect that can be rescued by introduction of human or mouse Seipin (Szymanski et al., 2007; Fei et al., 2008). Thus, misregulation of lipid and/or LD morphogenesis/metabolism can apparently lead to axonal damage. Collectively, these studies and our study suggest a potentially important requirement for proper LD function in the health of motor neurons.
Overall, these data identify a likely function for SPG20 and expand the range of cellular activities that are potentially regulated by ubiquitination and, in particular, by the HECT ubiquitin ligases. Further work will be required to determine precisely how SPG20 mutation leads to disease, but data from this and other studies indicate that improper regulation of LDs can have profound effects on neuronal health.
Materials and methods
Cells and transfections
293T and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and gentamycin. Derivatives expressing YFP, Che, HA-tagged SPG20 or ADRP proteins, or TAP-YFP/WWP1 were generated by transduction with MLV-based LNCX2 or LPCX viral vectors and selected with either 1.0 mg/ml G418 or 500 µg/ml puromycin, respectively. Stable SPG20 knockdown cells were created by transducing 293T cells with an HIV-based retroviral vector, pLENTI-shSPG20. This vector was derived from pLENTI-3′-U6-EC-EP7 (provided by D. Boden, Aaron Diamond AIDS Research Center, New York, NY) by inserting an oligonucleotide that generated an shRNA sequence, 5′-GTAACTGCCTACAATATTAACAA-3′ (nt 1,795–1,817), targeting a 3′ region of the SPG20 mRNA. The HA-tagged shRNA-resistant forms of SPG20 and the SPG20 mutant (HA-SPG20-R and HA-SPG20-1110mt-R, respectively) were introduced into the SPG20 knockdown 293T cells by transduction with LHCX-based vectors and selected with 50 µg/ml hygromycin. Transient transfection experiments were performed using polyethylenimine as the transfection reagent, as previously described (Neil et al., 2006).
Plasmids and mutagenesis
Plasmids based on pCR3.1/YFP that express WWP1, other HECT ubiquitin ligases, and derivatives of them have previously been described (Martin-Serrano et al., 2005). The coding sequences of SPG20, TIP47, ADRP, Rab11, Rab18, and Cav1 and 3 were amplified by PCR from human placenta–derived cDNA and inserted into the pCR3.1/YFP/CFP/Che, LNCX2/YFP, or LNCX2/Che plasmids for transient or stable expression of fluorescent fusion proteins. Mutant forms of YFP-SPG20 were created by recombinant PCR-based techniques and include YFP-SPG20dPY (PPAY to PAAA at aa position 170–174), YFP-SPG20dMIT (deletion of aa 1–129), and YFP-SPG20-1110mt (deletion of A at nt position 1,110). The shRNA-resistant form of SPG20 contains the sequence 5′-GTcACgGCaTAtAAcATcAAtAA-3′ from position 1,795–1,817 of the cDNA sequence (silent mutations are in lower case).
Virus release assays
MLV particle release assays were conducted using wild-type MLV or a variant, termed MLV-p6, in which a fragment of HIV-1 was inserted in place of the PPPY motif (Yuan et al., 2000). Additionally, an MLV construct containing a fragment of SPG20, termed MLV-SPG20, was created by inserting a 14-aa stretch of residues centered around the SPG20 PPPY sequence (PAEAPPAYTPQAAE) in place of the MLV PPPY motif. These mutations were introduced into a full-length MLV proviral plasmid and as well as into an MLV GagPol expression vector. 293T cells were plated in 12-well dishes and transfected with 700 ng MLV, MLV-p6, MLV-SPG20 proviral, or GagPol expression vector along with 300 ng pCR3.1/YFP or pCR3.1/YFP-SPG20 and derivates. Included in the transfection was 350 ng of a transfer vector that was either pMSCV-Tat or pMSCV-GFP as well as 150 ng of a plasmid expressing the vesicular stomatitis glycoprotein envelope protein. The medium was replaced 24 h after transfection, and cells and extracellular virions were harvested at 45 h after transfection. Supernatants were filtered (0.2 µm), and virions were harvested by centrifugation through a 20% sucrose cushion at 14,000 rpm in a microfuge (5417R; Eppendorf) at 4°C for 90 min. Cells, along with the viral pellet, were lysed in protein sample buffer and analyzed by SDS-PAGE gels and Western blotting. When using the pMSCV-Tat vector, infectious virions in culture supernatants were measured using TZM cells as targets (TZM cells contain an integrated HIV long terminal repeat driving the expression of LacZ). At 48 h after infection, cells were lysed, and β-galactosidase activity was measured. Infectious virions generated using the pMSCV-GFP vector were measured using HeLa cells as targets. 48 h after infection, cells were trypsinized, fixed, and analyzed by FACS to enumerate infected (GFP positive) cells.
Yeast two-hybrid assays
Full-length WWP1 was cloned into the pGBKT7 Gal4 DNA-binding domain vector and transformed into the yeast strain Y190 to be used as bait against a pACT2-based human testis cDNA library (Clontech Laboratories, Inc.) with a complexity of ∼2 × 106 independent clones. The screen was performed as previously described (Eastman et al., 2005). Directed yeast two-hybrid experiments were also conducted in the Y190 strain transformed with the pGBKT7 DNA-binding domain and pVP16 activation domain vectors as previously described (Martin-Serrano et al., 2001).
TAP
TAP of WWP1-associated proteins was performed using modifications of published procedures (Rigaut et al., 1999; Puig et al., 2001). Specifically, three confluent 10-cm dishes of 293T cells constitutively expressing either TAP-YFP or TAP-YFP-WWP1 were harvested, washed three times in PBS, lysed in buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40, and 10% glycerol) for 30 min on ice, cleared of nuclei by centrifugation at 1,000 rpm for 10 min, and incubated with an IgG–Sepharose bead slurry buffer for 20 min under rotation at 4°C. The column containing the beads was then washed three times with buffer without glycerol and washed once with tobacco etch virus (TEV) buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, and 1 mM DTT) before incubation with 25 U TEV protease (Invitrogen) in TEV buffer under rotation at 4°C overnight. The TEV eluate was then combined with 3 ml of calmodulin-binding buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40, and 10 mM β-mercaptoethanol) along with 3 µl of 1 M CaCl2 and incubated with a calmodulin bead slurry for 1 h under rotation at 4°C. The column was then washed three times with calmodulin-binding buffer and eluted with 10 mM Tris, pH 8.0, 150 mM NaCl, 1 mM magnesium acetate, 1 mM imidazole, 25 mM EGTA, 0.1% NP-40, and 10 mM β-mercaptoethanol by rotation for 30 min at 4°C. The eluate was combined with SDS sample buffer and run on 4–12% Tris-glycine SDS-PAGE gradient gels (Invitrogen) and stained (CodeBlue). Protein species that were present in the TAP-YFP-WWP1 sample but not the in the TAP-YFP samples were excised and subjected to mass spectrophotometry analysis (The Rockefeller University Core Facility, New York, NY).
GST coprecipitation and ubiquitin transfer assays
The procedure for the GST coprecipitation assays was essentially the same as for the ubiquitin transfer experiments, with the exception of the input DNA. Specifically, 293T cells within 12-well dishes were transfected with 500 ng of a pCAGGS-based plasmid expressing a GST fusion protein along with 750 ng of a pCR3.1/YFP-based plasmid expressing a YFP fusion protein. For ubiquitin transfer assays, 750 ng pCR3.1/YFP, pCR3.1/YFP-WWP1, or pCR3.1/YFP-WWP1 C890S and 500 ng plasmid HA-ubiquitin were transfected along with the plasmid expressing a GST fusion protein. At 45 h after transfection, cells were lysed in 1 ml of GST lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, and 5% glycerol supplemented with 5 mM N-ethylmaleimide to inhibit deubiquitination) for 10 min followed by centrifugation at 14,000 rpm. The postcentrifugation supernatant was incubated with glutathione-Sepharose for 3 h at 4°C and washed three times in GST wash buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, and 01% Triton X-100), and proteins were eluted with protein sample buffer, separated on SDS-PAGE gels, and analyzed by Western blotting.
Western blotting
Cellular lysates and viral pellets were separated on 10 or 12% SDS-PAGE gels, transferred to nitrocellulose, probed with monoclonal antibodies raised against GFP (Roche), GST (Santa Cruz Biotechnology, Inc.), HA (Covance), and MLV Gag p30 (R187; Rat Hybridoma no. CRL-1912; American Type Culture Collection) and a polyclonal antisera recognizing SPG20 provided by C. Blackstone (National Institutes of Health, Bethesda, MD) and J. Bakowska (Loyola University, Chicago, IL) followed by peroxidase-conjugated secondary antibodies (Millipore), and developed using chemiluminescence (Thermo Fisher Scientific).
Microscopy
Microscopy-based experiments were performed as previously described (Eastman et al., 2005; Martin-Serrano et al., 2005). In brief, HeLa or 293T cells were plated on 35-mM dishes containing glass coverslips (MatTek) and were subsequently transfected via Lipofectamine 2000 (HeLa cells; Invitrogen) or polyethylenimine (293T cells). Alternatively, HeLa cells stably expressing fluorescent SPG20 or ADRP proteins were used. 24 h after transfection, cells were fixed with 4% paraformaldehyde for 20 min and rinsed with 10 mM glycine in PBS. Cells were imaged in PBS using a DeltaVision system (Applied Precision, LLC) that included a microscope (IX70; Olympus) equipped with U Apochromat/340 40× NA 1.35 and U Plan-Apochromat 100× NA 1.35 objectives and a camera (CoolMax HQ; Photometrics). Image acquisition and processing by iterative deconvolution were performed using softWoRx software (Applied Precision, LLC). In experiments that included immunofluorescent analysis of endogenous proteins or ectopically expressed tagged proteins, cells were permeabilized after fixation with 1% Triton X-100 in PBS and incubated with antibodies against EEA1 (BD), CD63 (H5C6; Developmental Studies Hybridoma Bank), TIP47 (ProteinTech Group, Inc.), or the HA epitope tag (Covance). Secondary antibodies that were used in immunofluorescence experiments were labeled with Alexa Fluor 488 or 594.
LD visualization, induction, and quantitation assays
To visualize LDs, cell monolayers in MatTek dishes were rinsed with PBS and incubated with BODIPY 493/503 (Invitrogen) to label LDs (Smirnova et al., 2006; Turró et al., 2006). Alternatively, in some experiments, LDs were visualized by staining fixed cells with Oil red O (0.6% in 60% isopropanol). To induce LD formation, HeLa or 293T cells were cultured with 300–500 µM OA (Sigma-Aldrich) in 0.3% BSA (OA/BSA) for 12–48 h. For quantitation of LD burden in cells, we used BODIPY 493/503–stained cells and the softWoRx program to count the total number of LDs present in a projected image (a composite stack of 50 deconvolved optical sections acquired at 40× magnification) and to measure the diameter of LDs present in all optical sections of the same deconvolved image. Thereafter, we calculated the mean number and diameter of LDs present in images acquired from 10 randomly chosen fields of cells. Typically, each field contained ∼50–70 cells.
Online supplemental material
Fig. S1 shows localization of CFP-SPG20 in 293T cells coexpressing YFP fusion proteins containing the various domains of WWP1. Fig. S2 shows disruption of WWP1 to sites of MLV viral-budding Gag by SPG20 overexpression. Fig. S3 shows YFP-SPG20, Che-SPG20, HA-SPG20, and endogenous SPG20 expression levels in transduced HeLa cells. Fig. S4 shows colocalization of transiently coexpressed TIP47-Che and YFP-SPG20. Fig. S5 shows clustered LDs induced by overexpression of HA-SPG20. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200808041/DC1.
Acknowledgments
We thank Craig Blackstone, Joanna Bakowska, Daniel Boden, and Michael Rindler for providing reagents and members of the Bieniasz laboratory for helpful discussions.
This work was supported by a grant from the National Institutes of Health (R01AI52774 to P.D. Biensiasz) and a Ruth L. Kirschstein National Research Service Award (F32 AI065094 to S.W. Eastman).
Footnotes
Abbreviations used in this paper: ADRP, adipophilin/adipocyte differentiation-related protein; BODIPY, boron-dipyrromethene; ESCRT, endosomal sorting complex required for transport; HECT, homologous to the E6AP C terminus; LD, lipid droplet; MIT, microtubule interaction and trafficking; MLV, murine leukemia virus; NTE, neuropathy target esterase; OA, oleic acid; shRNA, short hairpin RNA; TAP, tandem affinity purification; TEV, tobacco etch virus.
References
- Agarwal A.K., Garg A. 2004. Seipin: a mysterious protein.Trends Mol. Med. 10:440–444 [DOI] [PubMed] [Google Scholar]
- Bakowska J.C., Jenkins R., Pendleton J., Blackstone C. 2005. The Troyer syndrome (SPG20) protein spartin interacts with Eps15.Biochem. Biophys. Res. Commun. 334:1042–1048 [DOI] [PubMed] [Google Scholar]
- Bakowska J.C., Jupille H., Fatheddin P., Puertollano R., Blackstone C. 2007. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking.Mol. Biol. Cell. 18:1683–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowska J.C., Wang H., Xin B., Sumner C.J., Blackstone C. 2008. Lack of spartin protein in Troyer syndrome: a loss-of-function disease mechanism? Arch. Neurol. 65:520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz R., Zehmer J.K., Zhu M., Chen Y., Serrero G., Zhao Y., Liu P. 2007. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation.J. Proteome Res. 6:3256–3265 [DOI] [PubMed] [Google Scholar]
- Beller M., Riedel D., Jansch L., Dieterich G., Wehland J., Jackle H., Kuhnlein R.P. 2006. Characterization of the Drosophila lipid droplet subproteome.Mol. Cell. Proteomics. 5:1082–1094 [DOI] [PubMed] [Google Scholar]
- Beller M., Sztalryd C., Southall N., Bell M., Jäckle H., Auld D.S., Oliver B. 2008. COPI complex is a regulator of lipid homeostasis.PLoS Biol. 6:e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom P., Andersson L., Rutberg M., Perman J., Lidberg U., Johansson B.R., Fernandez-Rodriguez J., Ericson J., Nilsson T., Boren J., Olofsson S.O. 2007. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity.Nat. Cell Biol. 9:1286–1293 [DOI] [PubMed] [Google Scholar]
- Boulant S., Douglas M.W., Moody L., Budkowska A., Targett-Adams P., McLauchlan J. 2008. Hepatitis C virus core protein induces lipid droplet redistribution in a microtubule- and dynein-dependent manner.Traffic. 9:1268–1282 [DOI] [PubMed] [Google Scholar]
- Brasaemle D.L. 2007. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis.J. Lipid Res. 48:2547–2559 [DOI] [PubMed] [Google Scholar]
- Bray S.J. 2006. Notch signalling: a simple pathway becomes complex.Nat. Rev. Mol. Cell Biol. 7:678–689 [DOI] [PubMed] [Google Scholar]
- Cermelli S., Guo Y., Gross S.P., Welte M.A. 2006. The lipid-droplet proteome reveals that droplets are a protein-storage depot.Curr. Biol. 16:1783–1795 [DOI] [PubMed] [Google Scholar]
- Chung H.Y., Morita E., von Schwedler U., Muller B., Krausslich H.G., Sundquist W.I. 2008. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains.J. Virol. 82:4884–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli F.D., Proukakis C., Patel H., Cross H., Azam S., Patton M.A., Bork P., Crosby A.H. 2003. The identification of a conserved domain in both spartin and spastin, mutated in hereditary spastic paraplegia.Genomics. 81:437–441 [DOI] [PubMed] [Google Scholar]
- Cohen A.W., Razani B., Schubert W., Williams T.M., Wang X.B., Iyengar P., Brasaemle D.L., Scherer P.E., Lisanti M.P. 2004. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation.Diabetes. 53:1261–1270 [DOI] [PubMed] [Google Scholar]
- Eastman S.W., Martin-Serrano J., Chung W., Zang T., Bieniasz P.D. 2005. Identification of human VPS37C, a component of endosomal sorting complex required for transport-I important for viral budding.J. Biol. Chem. 280:628–636 [DOI] [PubMed] [Google Scholar]
- Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., Yang H. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast.J. Cell Biol. 180:473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn P. 1999. Neuropathy target esterase.Biochem. J. 344:625–631 [PMC free article] [PubMed] [Google Scholar]
- Greenberg A.S., Obin M.S. 2008. Many roads lead to the lipid droplet.Cell Metab. 7:472–473 [DOI] [PubMed] [Google Scholar]
- Guo Y., Walter T.C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J.S., Vale R.D., Walter P., Farese R.V. 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization.Nature. 453:657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H., Yang D. 2008. MIT domainia.Dev. Cell. 14:6–8 [DOI] [PubMed] [Google Scholar]
- Ingham R.J., Gish G., Pawson T. 2004. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture.Oncogene. 23:1972–1984 [DOI] [PubMed] [Google Scholar]
- Ito D., Suzuki N. 2007. Molecular pathogenesis of seipin/BSCL2-related motor neuron diseases.Ann. Neurol. 61:237–250 [DOI] [PubMed] [Google Scholar]
- Kienesberger P.C., Lass A., Preiss-Landl K., Wolinski H., Kohlwein S.D., Zimmermann R., Zechner R. 2008. Identification of an insulin-regulated lysophospholipase with homology to neuropathy target esterase.J. Biol. Chem. 283:5908–5917 [DOI] [PubMed] [Google Scholar]
- Laine A., Ronai Z. 2007. Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1.Oncogene. 26:1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger L.L., Ostermeyer-Fay A.G., Goldberg E.B., Brown W.J., Brown D.A. 2007. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover.J. Lipid Res. 48:2751–2761 [DOI] [PubMed] [Google Scholar]
- Londos C., Sztalryd C., Tansey J.T., Kimmel A.R. 2005. Role of PAT proteins in lipid metabolism.Biochimie. 87:45–49 [DOI] [PubMed] [Google Scholar]
- Lu J., Rashid F., Byrne P.C. 2006. The hereditary spastic paraplegia protein spartin localises to mitochondria.J. Neurochem. 98:1908–1919 [DOI] [PubMed] [Google Scholar]
- Marchese A., Raiborg C., Santini F., Keen J.H., Stenmark H., Benovic J.L. 2003. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4.Dev. Cell. 5:709–722 [DOI] [PubMed] [Google Scholar]
- Martin S., Parton R.G. 2005. Caveolin, cholesterol, and lipid bodies.Semin. Cell Dev. Biol. 16:163–174 [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P.D. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress.Nat. Med. 7:1313–1319 [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P.D. 2003. Role of ESCRT-I in retroviral budding.J. Virol. 77:4794–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J., Eastman S.W., Chung W., Bieniasz P.D. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway.J. Cell Biol. 168:89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moren A., Imamura T., Miyazono K., Heldin C.H., Moustakas A. 2005. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases.J. Biol. Chem. 280:22115–22123 [DOI] [PubMed] [Google Scholar]
- Neil S.J., Eastman S.W., Jouvenet N., Bieniasz P.D. 2006. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane.PLoS Pathog. 2:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita T., Saksena S., Ghazi-Tabatabai S., Gill D.J., Perisic O., Emr S.D., Williams R.L. 2007. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4.Nature. 449:735–739 [DOI] [PubMed] [Google Scholar]
- Ohsaki Y., Maeda T., Maeda M., Tauchi-Sato K., Fujimoto T. 2006. Recruitment of TIP47 to lipid droplets is controlled by the putative hydrophobic cleft.Biochem. Biophys. Res. Commun. 347:279–287 [DOI] [PubMed] [Google Scholar]
- Ozeki S., Cheng J., Tauchi-Sato K., Hatano N., Taniguchi H., Fujimoto T. 2005. Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane.J. Cell Sci. 118:2601–2611 [DOI] [PubMed] [Google Scholar]
- Patel H., Cross H., Proukakis C., Hershberger R., Bork P., Ciccarelli F.D., Patton M.A., McKusick V.A., Crosby A.H. 2002. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia.Nat. Genet. 31:347–348 [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification.Methods. 24:218–229 [DOI] [PubMed] [Google Scholar]
- Puri V., Ranjit S., Konda S., Nicoloro S.M., Straubhaar J., Chawla A., Chouinard M., Lin C., Burkart A., Corvera S., et al. 2008. Cidea is associated with lipid droplets and insulin sensitivity in humans.Proc. Natl. Acad. Sci. USA. 105:7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainier S., Bui M., Mark E., Thomas D., Tokarz D., Ming L., Delaney C., Richardson R.J., Albers J.W., Matsunami N., et al. 2008. Neuropathy target esterase gene mutations cause motor neuron disease.Am. J. Hum. Genet. 82:780–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E., Connell J., Edwards T.L., Duley S., Brown S.E., Sanderson C.M. 2005. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B.Hum. Mol. Genet. 14:19–38 [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration.Nat. Biotechnol. 17:1030–1032 [DOI] [PubMed] [Google Scholar]
- Robay D., Patel H., Simpson M.A., Brown N.A., Crosby A.H. 2006. Endogenous spartin, mutated in hereditary spastic paraplegia, has a complex subcellular localization suggesting diverse roles in neurons.Exp. Cell Res. 312:2764–2777 [DOI] [PubMed] [Google Scholar]
- Row P.E., Liu H., Hayes S., Welchman R., Charalabous P., Hofmann K., Clague M.J., Sanderson C.M., Urbe S. 2007. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation.J. Biol. Chem. 282:30929–30937 [DOI] [PubMed] [Google Scholar]
- Sato S., Fukasawa M., Yamakawa Y., Natsume T., Suzuki T., Shoji I., Aizaki H., Miyamura T., Nishijima M. 2006. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein.J. Biochem. 139:921–930 [DOI] [PubMed] [Google Scholar]
- Scott A., Gaspar J., Stuchell-Brereton M.D., Alam S.L., Skalicky J.J., Sundquist W.I. 2005. Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A.Proc. Natl. Acad. Sci. USA. 102:13813–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Goldberg E.B., Makarova K.S., Lin L., Brown W.J., Jackson C.L. 2006. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells.EMBO Rep. 7:106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton M.D., Skalicky J.J., Kieffer C., Karren M.A., Ghaffarian S., Sundquist W.I. 2007. ESCRT-III recognition by VPS4 ATPases.Nature. 449:740–744 [DOI] [PubMed] [Google Scholar]
- Szymanski K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., Goodman J.M. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology.Proc. Natl. Acad. Sci. USA. 104:20890–20895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turró S., Ingelmo-Torres M., Estanyol J.M., Tebar F., Fernández M.A., Albor C.V., Gaus K., Grewal T., Enrich C., Pol A. 2006. Identification and characterization of associated with lipid droplet protein 1: a novel membrane-associated protein that resides on hepatic lipid droplets.Traffic. 7:1254–1269 [DOI] [PubMed] [Google Scholar]
- Usami Y., Popov S., Popova E., Gottlinger H.G. 2008. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase.J. Virol. 82:4898–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U.K., Stuchell M., Muller B., Ward D.M., Chung H.Y., Morita E., Wang H.E., Davis T., He G.P., Cimbora D.M., et al. 2003. The protein network of HIV budding.Cell. 114:701–713 [DOI] [PubMed] [Google Scholar]
- Welte M.A. 2007. Proteins under new management: lipid droplets deliver.Trends Cell Biol. 17:363–369 [DOI] [PubMed] [Google Scholar]
- Wolins N.E., Rubin B., Brasaemle D.L. 2001. TIP47 associates with lipid droplets.J. Biol. Chem. 276:5101–5108 [DOI] [PubMed] [Google Scholar]
- Wolins N.E., Quaynor B.K., Skinner J.R., Schoenfish M.J., Tzekov A., Bickel P.E. 2005. S3-12, Adipophilin, and TIP47 package lipid in adipocytes.J. Biol. Chem. 280:19146–19155 [DOI] [PubMed] [Google Scholar]
- Xu G., Sztalryd C., Lu X., Tansey J.T., Gan J., Dorward H., Kimmel A.R., Londos C. 2005. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway.J. Biol. Chem. 280:42841–42847 [DOI] [PubMed] [Google Scholar]
- Yuan B., Campbell S., Bacharach E., Rein A., Goff S.P. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses.J. Virol. 74:7250–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccheo O., Dinsdale D., Meacock P.A., Glynn P. 2004. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells.J. Biol. Chem. 279:24024–24033 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chang C., Gehling D.J., Hemmati-Brivanlou A., Derynck R. 2001. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase.Proc. Natl. Acad. Sci. USA. 98:974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang H.R., Wrana J.L. 2004. Smurf1: a link between cell polarity and ubiquitination.Cell Cycle. 3:391–392 [DOI] [PubMed] [Google Scholar]