Abstract
Skin morphogenesis, maintenance, and healing after wounding require complex epithelial–mesenchymal interactions. In this study, we show that for skin homeostasis, interleukin-1 (IL-1) produced by keratinocytes activates peroxisome proliferator–activated receptor β/δ (PPARβ/δ) expression in underlying fibroblasts, which in turn inhibits the mitotic activity of keratinocytes via inhibition of the IL-1 signaling pathway. In fact, PPARβ/δ stimulates production of the secreted IL-1 receptor antagonist, which leads to an autocrine decrease in IL-1 signaling pathways and consequently decreases production of secreted mitogenic factors by the fibroblasts. This fibroblast PPARβ/δ regulation of the IL-1 signaling is required for proper wound healing and can regulate tumor as well as normal human keratinocyte cell proliferation. Together, these findings provide evidence for a novel homeostatic control of keratinocyte proliferation and differentiation mediated via PPARβ/δ regulation in dermal fibroblasts of IL-1 signaling. Given the ubiquitous expression of PPARβ/δ, other epithelial–mesenchymal interactions may also be regulated in a similar manner.
Introduction
Adult epidermis is a stratified epithelium in which keratinocytes from the basal and suprabasal layers cease to divide concomitantly with their outward movement and terminal differentiation all through enucleation and keratinization. After injury, the restoration of its functional integrity is of utmost importance to the survival of the organism. The maintenance of epithelium and its regeneration to close the wound is orchestrated with the contribution of the underlying dermal tissue. This synchrony is key to preventing either insufficient or excess wound repair. The regulation of wound repair is dictated by epithelial–mesenchymal interactions and purportedly mediated by the action of central players such as growth factors. This complex interplay demands the expression of soluble factors exerting autocrine and paracrine activities and, importantly, the integration of such diverse signals, which culminate in appropriate cellular responses (Fusenig, 1994). Although the importance of the epithelial–mesenchymal communication is well recognized, the mechanism underlying this process needs in-depth study.
Of the numerous cytokines produced by skin cells, interleukin-1 (IL-1) has a pronounced influence on skin homeostasis and wound repair, and there are multiple mechanisms to regulate IL-1 signaling (Schroder, 1995). Both human and mouse keratinocytes constitutively produce biologically active IL-1α/β (Arend et al., 1998). The two distinct forms of IL-1, termed IL-1α and IL-1β, are capable of binding to the same receptor with similar affinities to trigger biological effects. For any cells to respond productively to IL-1α/β, they must possess corresponding cell surface receptors. Two distinct IL-1 receptors (IL-1Rs) have been cloned and characterized. The type I IL-1R (IL-1R1) mediates all known responses of IL-1. In contrast, the type 2 IL-1R (IL-1R2) is incapable of participating in signal transduction by virtue of its short cytoplasmic tail and thus is thought to neutralize the action of the bound IL-1α/β. Interestingly, the predominant IL-1R species expressed by keratinocytes, both constitutively and after activation of the cells, is the IL-1R2. Its production by keratinocytes would represent an efficient mean for these cells to escape autocrine IL-1α/β signaling (Rauschmayr et al., 1997). In contrast, the dermal fibroblasts express mainly the IL-1R1 isotype and thus are very sensitive to IL-1α/β signaling.
The effects of IL-1α/β on target cells can also be inhibited via the binding of the IL-1R antagonist (IL-1ra) to IL-1R1. There are two structural variants of IL-1ra, secreted IL-1ra (sIL-1ra) and intracellular IL-1ra (icIL-1ra). Both isoforms are transcribed from the same gene via two different promoters and alternative first exons. IL-1ra binds to IL-1R1 with an affinity that is 100–500 times higher than for IL-1R2 and competitively inhibits IL-1α/β binding to the receptor. Although icIL-ra may be released from keratinocytes under some conditions to compete for receptor binding, it can inhibit IL-1α/β–induced cytokine production intracellularly via the COP9 signalosome (Banda et al., 2005). Fibroblasts produce both IL-1ra isoforms when appropriately stimulated, whereas keratinocytes only constitutively produce the icIL-1ra (Arend et al., 1998).
Peroxisome proliferator–activated receptors (PPARs) are nuclear hormone receptors involved in the control of chronic diseases such as diabetes and obesity (Kersten et al., 2000). The PPAR subgroup comprises three related members named PPARα (NR1C1), PPARβ/δ (NR1C2), and PPARγ (NR1C3; Nuclear Receptors Nomenclature Committee, 1999). Ligand-activated PPARs form obligate heterodimers with the retinoid X receptor and bind to defined peroxisome proliferator response elements (PPREs) in the regulatory regions of target genes. Several works have shown that ligand-activated PPARβ/δ can induce terminal differentiation of keratinocytes (Tan et al., 2001; Burdick et al., 2006). Other studies comparing PPARβ/δ-null (KO) with wild-type (WT) mice showed that inflammatory signals increase the expression and activity of PPARβ/δ, which confer a pronounced antiapoptotic capacity to the keratinocytes, mediated in part via the increased activity of the PKBα survival pathway (Tan et al., 2001; Di Poi et al., 2002). Nonetheless, it is paradoxical that in the KO mice, epidermal hyperproliferation was observed either in early wound repair, upon hair plucking, or in response to a topical challenge with the phorbol ester (TPA; Peters et al., 2000; Michalik et al., 2001). Thus, the role of PPARβ/δ on cell proliferation remains debatable, and its mechanism of action is not sufficiently known (Michalik et al., 2001; Kim et al., 2005; Man et al., 2008).
Herein, we provide evidence for a novel paracrine effect of PPARβ/δ on epithelial cell growth. We found that PPARβ/δ in the fibroblasts attenuates keratinocyte proliferation by directly increasing the expression of sIL-1ra, thereby repressing IL-1 signaling. Epithelial–mesenchymal communications and IL-1 signaling are responsible for a wide range of biological events such as wound repair and tumorigenesis. Our results underscore the paracrine role of PPARβ/δ in the control of cell proliferation involving epithelial–mesenchymal interactions.
Results
PPARβ/δ knockdown in dermal fibroblasts results in increased keratinocyte proliferation
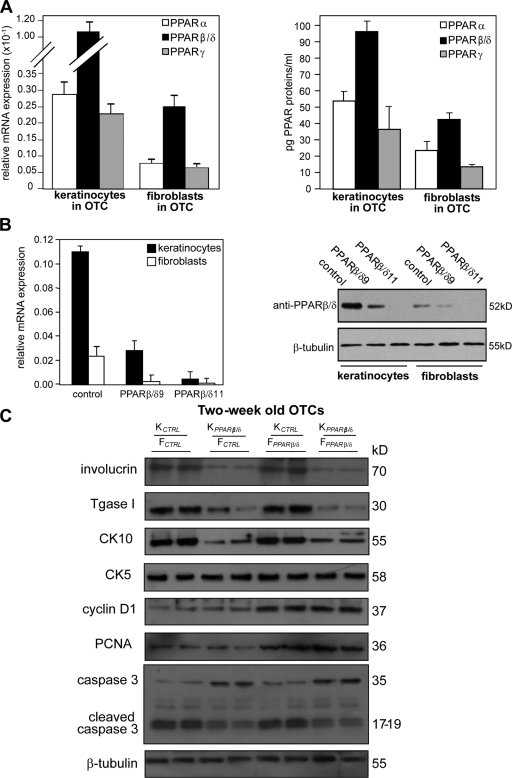
Earlier studies have shown that ligand-activated PPARβ/δ induced the differentiation of human keratinocytes in monolayer cultures (Tan et al., 2001; Schmuth et al., 2004). Herein, we examined the autocrine and paracrine consequences of PPARβ/δ deficiency during human epidermis formation using the organotypic skin culture (OTC) model. We first examined the expression pattern of the three PPAR isotypes in fibroblasts and keratinocytes of control OTC. Both quantitative real-time PCR (qPCR) and ELISA analyses performed on mechanically separated dermis and epidermis equivalents of OTCs revealed that PPARβ/δ is the predominant isotype in the keratinocytes and dermal fibroblasts, whereas the lower levels of PPARα and PPARγ are comparable (Fig. 1 A). We next assessed the knockdown efficiency of PPARβ/δ expression in human keratinocytes and fibroblasts by lentivirus-mediated siRNAs using qPCR and immunoblot analyses. qPCR revealed a >95% reduction of PPARβ/δ expression in cells transduced with the siRNA PPARβ/δ11 sequence (Fig. 1 B, left). Immunoblot analysis also showed negligible levels of PPARβ/δ protein in the transduced cells (Fig. 1 B, right). These cells were used for subsequent experiments.
Figure 1.
PPARβ/δ-deficient fibroblasts increase epidermal proliferation. (A) Expression profile of PPARs in OTC keratinocytes and fibroblasts. Total RNA and protein were extracted from keratinocytes and fibroblasts in OTC. Expression levels of PPAR mRNA (left) and protein (right) were monitored by qPCR and PPAR transcription factor assay kit, respectively. PPARβ/δ mRNA was normalized with control ribosomal protein P0 mRNA. (B) Human keratinocytes or fibroblasts were transduced with a lentiviral vector harboring a control or two different PPARβ/δ (PPARβ/δ9 and PPARβ/δ11) siRNAs. (C) Immunoblot analysis of epidermis from 2-wk-old OTCs constructed using KCTRL or KPPARβ/δ and FCTRL or FPPARβ/δ. Involucrin and transglutaminase I (Tgase I) are terminal differentiation markers, and keratin 10 (CK10) is an early differentiation marker. Keratin 5 (CK5) identifies the basal keratinocytes. Cell proliferation was measured using PCNA and cyclin D1. Apoptosis was detected using caspase 3. β-Tubulin showed equal loading and transfer. Representative immunoblots of epidermis from two OTCs are shown. Data are mean ± SD, n = 3.
Next, OTCs were reconstructed using control keratinocytes (KCTRL), PPARβ/δ knockdown keratinocytes (KPPARβ/δ), control fibroblasts (FCTRL), and PPARβ/δ knockdown fibroblasts (FPPARβ/δ) in various combinations. Immunoblot analysis and immunofluorescence staining of the 2-wk-old KCTRL/FCTRL OTCs showed the expected keratinocyte differentiation markers keratin 5, keratin 10, and involucrin (Fig. 1 C and Fig. S1). OTCs with KPPARβ/δ showed a reduced expression of terminal markers, which is consistent with the known prodifferentiation role of PPARβ/δ (Fig. 1 C and Fig. S1; Schmuth et al., 2004). No difference in keratin 5 expression, localized to the basal layer of the epidermis, was observed among the various OTCs (Fig. 1 C and Fig. S1). This provided evidence for a cell-autonomous action of PPARβ/δ in keratinocyte differentiation. Interestingly, OTC with FPPARβ/δ led to increased keratinocyte proliferation as indicated by an increase in expression of cyclin D1 and proliferating cell nuclear antigen (PCNA; Fig. 1 C) and in Ki67-positive cells with respect to the control OTC (Fig. S1). Notably, the KPPARβ/δ/FPPARβ/δ OTC similarly displayed this enhanced proliferation of keratinocytes regardless of the impaired differentiation of the KPPARβ/δ (Fig. 1 C and Fig. S1). Furthermore, OTC with KPPARβ/δ showed threefold more apoptotic cells (TUNEL positive) and a higher level of cleaved caspase 3 as compared with OTC with KCTRL (Fig. 1 C and Fig. S1). A dynamic epithelial–mesenchymal interaction is essential for the proper formation of the basement membrane (Smola et al., 1998). The reduced laminin 5 staining in OTCs with either KPPARβ/δ or FPPARβ/δ suggested a dysregulated epithelial–mesenchymal communication (Fig. S1). Altogether, these results are consistent with the observations from KO mice (Peters et al., 2000; Michalik et al., 2001) and revealed a potent proliferation stimulatory effect of FPPARβ/δ on cocultured keratinocytes, providing evidence for an important noncell autonomous PPARβ/δ-dependent mechanism regulating keratinocyte proliferation.
Organotypic cultures with FPPARβ/δ show increased expression of mitogenic factors
The paracrine effect of FPPARβ/δ on epidermal proliferation is likely mediated by changes in the secretion of mitogenic or antimitogenic factors by the fibroblasts. To understand the mechanism underlying the enhanced epidermal proliferation in OTC with FPPARβ/δ, an unbiased protein array was performed. Inflammatory cytokine and growth factor arrays were used to compare conditioned media from OTCs reconstructed using WT keratinocytes with either FCTRL or FPPARβ/δ. A total of 76 distinct proteins were screened, and the results showed that the protein expression of several mitogenic factors and cytokines were increased in OTC with FPPARβ/δ (Table I). Notably, most of the proteins whose expression was increased have a known mitogenic action on keratinocytes. The expression of TGF-β1, which exerts an antiproliferative effect on keratinocytes, and the abundant IL-1α, constitutively produced by keratinocytes, remained unchanged. The expression of two proangiogenic factors, namely VEGF and placental growth factor (PIGF), were reduced in OTC with FPPARβ/δ when compared with FCTRL.
Table I.
Relative fold change of protein expression in OTCs with FPPARβ/δ compared with FCTRL
| Cytokines/growth factors | Fold increasea |
| KGFb | 4.01 ± 0.11c |
| IL-6b | 5.56 ± 0.02c |
| IL-8b | 2.94 ± 0.14d |
| IL-10 | 2.13 ± 0.07c |
| IL-16 | 2.86 ± 0.11d |
| GMCSFb | 5.26 ± 0.10c |
| I-309 | 3.33 ± 0.16c |
| Eotaxin-2 | 3.03 ± 0.12c |
| VEGF | 0.25 ± 0.011d |
| PIGF | 0.33 ± 0.014d |
The intensities of signals were quantified by densitometry. Positive control included in the array blot was used to normalize and compare the results from different membranes. Values obtained from the control KCTRL/FCTRL OTC were arbitrarily assigned a value of 1. Values represent the mean fold increase as compared with control OTC (n = 3).
mRNA expression levels of these genes were further verified by qPCR.
P < 0.01.
P < 0.1.
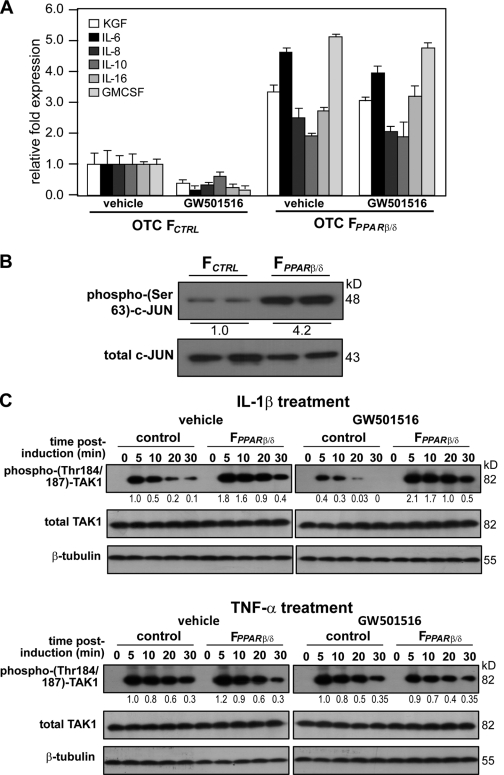
To verify that a reduced PPARβ/δ expression leads to a transcriptional up-regulation of mitogenic factor expression, we performed qPCR on selected mitogenic genes from OTC FCTRL and FPPARβ/δ in the absence or presence of the PPARβ/δ agonist GW501516. Consistent with the protein array results, FCTRL exposed to GW501516 for 12 h showed a decrease in the expression of these genes (Fig. 2 A). Importantly, FPPARβ/δ expressed higher basal levels of these mitogenic factors and, as expected, ligand treatment had no effect. Altogether, fibroblasts deficient in PPARβ/δ have an increased expression of mitogenic factors (Fig. 2 A).
Figure 2.
Reduced fibroblast PPARβ/δ expression increases IL-1β activation of TAK1 and up-regulation of AP-1–controlled mitogenic target genes. (A) Expression of mitogenic factor mRNAs in 2-wk old OTC FPPARβ/δ and FCTRL treated with PPARβ/δ agonist (500 nM GW501516 for 24 h) or vehicle. The expression levels of the indicated mitogenic factors were analyzed by qPCR and normalized to control ribosomal protein P0. Results are represented in fold induction as compared with OTC FCTRL. (B) Immunoblot analysis of phosphorylated c-Jun from FPPARβ/δ and FCTRL extracted from KCTRL/FPPARβ/δ and KCTRL/FCTRL OTCs (n = 2), respectively. Total c-Jun and TAK1 protein expression level, which remains unchanged, showed equal loading and transfer. (C) Immunoblot analysis of IL-1β and TNF-α activation of TAK1 in FCTRL or FPPARβ/δ. Cells were treated with either vehicle (DMSO) or 800 nM GW501516 for 24 h prior to exposure to 10 ng/ml IL-1β (top) or TNF-α (bottom). At the indicated time points, total cell lysates were extracted. Equal amounts of total protein (50 µg) were resolved, electrotransferred, and probed for phosphorylated TAK1 (Thr184/187), total TAK1, and β-tubulin. Values below each band represent the mean fold differences (n = 3) in expression level with respect to vehicle-treated FCTRL at 5 min, which was assigned the value of one. Data are mean ± SEM, n = 3.
Increased expression of mitogenic factors by FPPARβ/δ results from attenuated IL-1β/TAK1/AP-1 activation
Next, we sought to identify the signaling cascade that culminates in the up-regulation of the mitogenic growth factors in FPPARβ/δ. The expression of most of these genes is known to be regulated by the transcription factor activation protein-1 (AP-1; Szabowski et al., 2000; Florin et al., 2004). Although direct regulation of eotaxin-2 by AP-1 has not been reported, the expression of this gene is stimulated by IL-1 (Watanabe et al., 2002; Heiman et al., 2005). The increased expression of I-309 is likely the consequence of increased IL-8 level (Wiener et al., 2008). We first addressed the hypothesis that AP-1 belongs to the pathway through which PPARβ/δ regulates the expression of mitogenic factors in FPPARβ/δ. We examined the levels of phosphorylated (activated) c-Jun in FPPARβ/δ and FCTRL fibroblasts extracted from KCTRL/FPPARβ/δ and KCTRL/FCTRL OTCs, respectively. Immunoblot analysis showed that FPPARβ/δ isolated from the KCTRL/FPPARβ/δ OTCs had enhanced phosphorylated c-Jun, which is consistent with an increased activity of the AP-1 complex (Fig. 2 B) and increased production of growth factors.
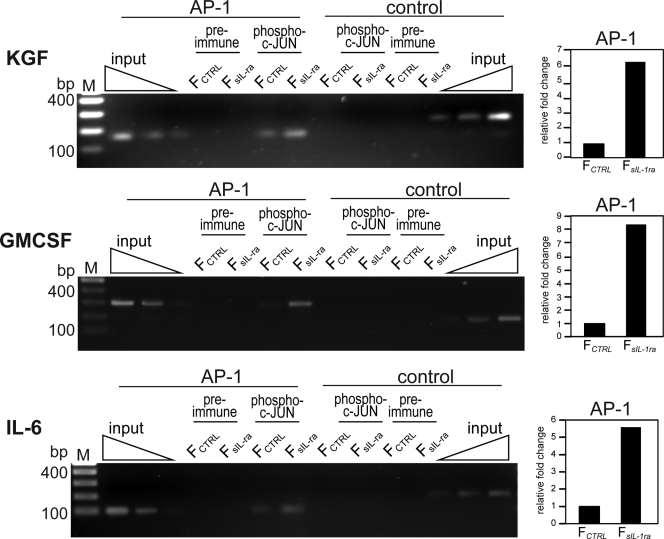
The TGF-activated kinase 1 (TAK1) plays a pivotal role in the activation of many genes, including genes encoding mitogenic factors, via activation of AP-1 (Florin et al., 2004, 2005). The IL-1α/β released by keratinocytes and TNF-α present in wound sites activate TAK1 (Shim et al., 2005). To investigate the effect of PPARβ/δ on TAK1 activation, we examined the expression of phosphorylated TAK1 in FCTRL and FPPARβ/δ after IL-1β and TNF-α stimulations. FCTRL and FPPARβ/δ were treated with either vehicle (DMSO) or PPARβ/δ agonist (GW501516) for 24 h prior to stimulation by either IL-1β or TNF-α. As expected, there was an increase in phospho-TAK1 in vehicle-treated FCTRL exposed to either IL-1β or TNF-α (Fig. 2 C). Cotreatment of these fibroblasts with IL-1β and GW501516 significantly prevented the increase in phospho-TAK1 levels in a dose-dependent manner (Fig. 2 C and Fig. S2 A). In the FPPARβ/δ, a more robust increase in phospho-TAK1 was observed, which was only marginally affected by GW501516, showing that the ligand effect in FCTRL was PPARβ/δ specific (Fig. 2 C and Fig. S2 A). In contrast, neither ligand-activated PPARβ/δ nor PPARβ/δ deficiency had an effect on TNF-α–mediated TAK1 activation (Fig. 2 C and Fig. S2 A). To further this observation, we performed chromatin immunoprecipitation (ChIP) using phospho–c-Jun antibodies on the keratinocyte growth factor (KGF) and granulocyte macrophage colony-stimulating factor (GMCSF) gene promoters in fibroblasts from KCTRL/FCTRL and KCTRL/FPPARβ/δ OTCs (Fig. S2 B). There was more phospho–c-Jun immunoprecipitated chromatin from FPPARβ/δ when compared with FCTRL, pointing to enhanced AP-1 binding and activation of the KGF and GMCSF genes in FPPARβ/δ (Fig. S2 B). Together, these results indicate that PPARβ/δ specifically attenuates IL-1β–mediated TAK1 activation and thus AP-1 activity.
Neutralization of IL-1α/β signaling abolishes the mitogenic action of FPPARβ/δ
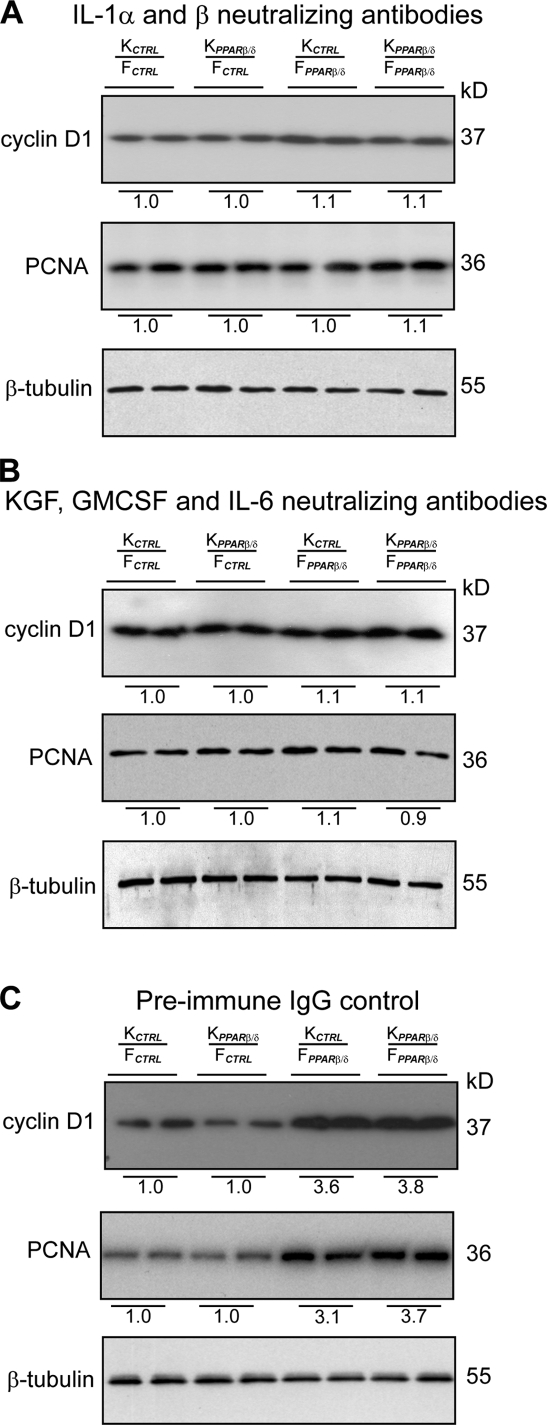
Our results showed that IL-1β–mediated but not TNF-α–mediated TAK1 activation and AP-1–dependent expression of mitogenic factors were increased in FPPARβ/δ. This result suggested that the IL-1α/β pathway is responsible for the mitogenic effect of PPARβ/δ knockdown in fibroblasts. Conceivably, negating the effect of IL-1α/β with neutralizing antibodies against IL-1α/β would counteract the effect of PPARβ/δ knockdown in FPPARβ/δ. As anticipated, OTCs exposed to anti–IL-1α/β antibodies displayed a reduction in Ki67-positive keratinocytes, and the mitogenic action of FPPARβ/δ was completely abolished (Fig. S3, compare A with C), indicating that IL-1α/β signaling is a major pathway in the control of keratinocyte proliferation. This was further confirmed by immunoblot analysis of cyclin D1 and PCNA (Fig. 3, compare A with C). Similarly, inhibiting the action of the mitogenic factors should replicate the neutralizing effect of anti–IL-1α/β antibodies. To test this possibility, we treated the OTCs with neutralizing antibodies against KGF, GMCSF, and IL-6, which are the three most abundant mitogenic factors in our model. To avoid possible compensatory effects, we neutralized all three growth factors simultaneously. Consistent with the aforementioned results, all OTCs showed thinner epidermis and reduced keratinocyte proliferation when compared with the corresponding OTC treated with preimmune IgG (Fig. 3, B and C and Fig. S3, B and C). Altogether, our observations indicated that the mitogenic action of the FPPARβ/δ was caused by an increase in IL-1α/β–mediated expression of mitogenic factors via TAK1 and AP-1.
Figure 3.
Neutralizing antibodies against IL-1α/β or KGF, GMCSF, and IL-6 abolished the mitogenic effect of FPPARβ/δ. (A–C) Immunoblot analysis of epidermis from indicated OTCs treated with 400 ng/ml IL-1α/β (A), KGF, GMCSF, and IL-6 (B) neutralizing antibodies (each at 400 ng/ml) or 400 ng/ml preimmune IgG (C). Antibodies were added to OTC medium at each change of medium. Cell proliferation was measured using cyclin D1 and PCNA. Values below each band represent the mean fold differences in expression level with respect to KCTRL from KCTRL/FCTRL OTC, which was assigned the value of 1. β-Tubulin served as a loading control. Representative immunoblots of epidermis from two indicated OTCs are shown.
Human sIL-1ra gene is a direct target of PPARβ/δ in fibroblasts
In vivo, the release of IL-1α/β by basal keratinocytes is sufficient to maintain tissue homeostasis and to initiate cutaneous inflammation (Groves et al., 1996; Maas-Szabowski et al., 2000). Although we did not detect a change in the expression of IL-1α/β in the protein array (Table I), we clearly showed that PPARβ/δ specifically attenuated IL-1β response, including the activation of TAK1. This suggested that the attenuation of IL-1β signaling by PPARβ/δ occurs upstream of TAK1 activation. To examine whether PPARβ/δ has a regulatory effect in IL-1α/β signaling cascade, we measured the mRNA levels of IL-1α, IL-1β, IL-1R1, IL-1R2, icIL-1ra, sIL-1ra, and IL-1R–associated kinase, which are all key players of IL-1 signaling, in OTC epidermal keratinocytes (KCTRL and KPPARβ/δ) and dermal fibroblasts (FCRTL and FPPARβ/δ). With the exception of a higher level of sIL-1ra in the OTC FCRTL, we detected no change in the expression of the other mediators of IL-1α/β signaling (Fig. 4 A and not depicted). Consistently, elevated sIL-1ra was detected in the medium of OTC with FCTRL as compared with FPPARβ/δ, but no difference was observed in icIL-1ra protein within the fibroblasts (Fig. 4 A).
Figure 4.
Human sIL-1ra is encoded in a direct PPARβ/δ target gene in fibroblasts. (A) Expression of sIL-1ra and icIL-1ra mRNA (left) and protein (right) in OTC keratinocytes (KCTRL and KPPARβ/δ) and fibroblasts (FCRTL and FPPARβ/δ). sIL-1ra and icIL-1ra mRNA were analyzed by qPCR and normalized to ribosomal protein P0. The sIL-1ra level was determined by ELISA from medium of KCTRL/FCTRL and KCTRL/FPPARβ/δ OTCs. The icIL-1ra levels were measured by ELISA from cell lysates. (B) PPRE1 and PPRE3 of the human sIL-1ra gene are functional. Transactivation assay in fibroblasts cotransfected with a luciferase (luc) promoter driven by the human sIL-1ra promoter and pEF1–β-galactosidase as control of transfection efficiency. Relative positions of the three putative PPREs (PPRE 1–3) and their mutants (mPPRE 1–3) are represented in closed and open ovals, respectively. Cells were treated with either 500 nM GW501516 (GW) and/or 10 ng/ml IL-1β for 24 h. Luciferase activity was measured, and normalized reporter activity is shown as fold induction as compared with untreated fibroblasts. (C) EMSA of human sIL-1ra PPRE1 and PPRE3. Radiolabeled PPRE1 (left) and PPRE3 (middle) were incubated either with RXRα, PPARβ/δ, or both. NSC denotes nonspecific competitor, the nonfunctional MEd DR1 element in the malic enzyme promoter. SC denotes nonradiolabeled consensus PPRE (conPPRE). As positive control, conPPRE was used. Mutated consensus PPRE is denoted by mconPPRE. PPARβ/δ did not bind to PPRE2 and mutated PPRE probes (mconPPRE, mPPRE1, mPPRE2, and mPPRE3; right). (D) PPARβ/δ binds to PPRE1 and PPRE3 of the human sIL-1ra gene in fibroblasts. ChIP assays were conducted using preimmune IgG or antibodies against PPARβ/δ (AB) in FCTRL (WT) and FPPARβ/δ (kd) fibroblasts extracted from two independent OTCs (OTC1 and OTC2). The regions spanning PPRE1 and PPRE2 of the sIL-1ra gene were amplified using appropriate primers (Table S1). A control region between PPRE1 and PPRE2 served as negative control. *, P < 0.05; **, P < 0.01. Data are mean ± SEM, n = 4.
To determine whether the sIL-1ra promoter is directly regulated by PPARβ/δ in the fibroblasts, a 4.4-kb fragment of the promoter region was subcloned into a luciferase reporter vector and analyzed in transactivation assays. Three putative PPRE sequences were identified in the sIL-1ra promoter (GenBank no. X64532), PPRE1 at position −1,038/−1,050, PPRE2 at −1,072/−1,084, and PPRE3 at −4,067/−4,079, using NUBIScan (Fig. S4 A; Podvinec et al., 2002). Transfected primary fibroblasts treated with the PPARβ/δ ligand GW501516 alone showed only a modest approximately threefold increase in reporter activity (Fig. 4 B). As expected, treatment with IL-1β increased the reporter activity by ∼20-fold, which is consistent with an earlier study (Smith et al., 1994). Importantly, this IL-1β–mediated transactivation was further enhanced by treatment with GW501516 (to ∼50-fold; Fig. 4 B). To identify the functional PPREs responsible for the PPAR-mediated up-regulation, site-directed mutagenesis of the PPREs was performed. Transactivation assays with the various mutated promoters clearly showed that the stimulatory effects of PPARβ/δ were mediated by two PPREs, i.e., PPRE1 and PPRE3. To investigate whether PPARβ/δ was truly bound to these identified PPREs, electrophoretic mobility shift assay (EMSA) and ChIP were performed. As expected, specific protein–DNA complexes were detected in EMSA for PPRE1 and PPRE3, which were effectively competed by a consensus PPRE oligonucleotide (Fig. 4 C). As a positive control, the consensus PPRE oligonucleotide was used as a probe, which was specifically competed by the unlabeled consensus PPRE but not by a nonspecific competitor containing a nonfunctional MEd (malic enzyme distal PPRE) DR1 element from the malic enzyme promoter (Fig. S4 B; IJpenberg et al., 2004). Similarly, ChIP performed on OTC FCTRL using a monoclonal anti-PPARβ/δ antibody showed the binding of PPARβ/δ to both PPRE1 and PPRE3 (Fig. 4 D). This effect was not observed in FPPARβ/δ, and no signal was seen with preimmune serum either. In addition, no binding was detected to a control sequence residing between PPRE1 and PPRE3. These data clearly showed that PPARβ/δ specifically binds to the identified functional PPREs in the human sIL-1ra promoter in OTC FCTRL. They indicated that the human sIL-1ra promoter is a direct PPARβ/δ target in primary human dermal fibroblasts.
Altogether, we showed that IL-1α/β secreted by the keratinocytes activates TAK1/AP-1 signaling, which up-regulates the expression of mitogenic factors in the fibroblasts and thus enhances epidermal proliferation. We also showed that PPARβ/δ in the fibroblasts plays a homeostatic role in which it stimulates the expression of the sIL-1ra, modulates IL-1α/β–mediated mitogenic factors gene expression, and thus attenuates epidermal proliferation.
sIL-1ra knockdown fibroblasts increase keratinocyte proliferation
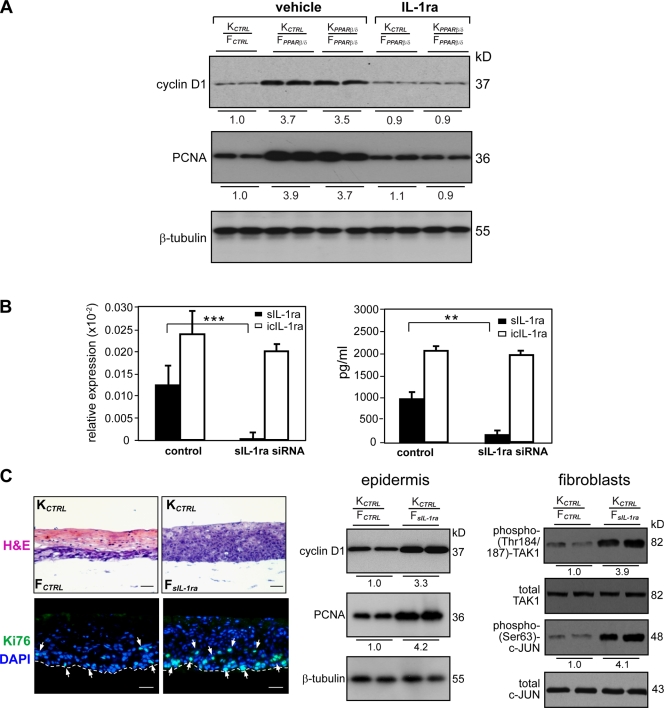
We showed that the sIL-1ra gene in fibroblasts is a direct target of PPARβ/δ and that accordingly, PPARβ/δ knockdown results in a reduced production of sIL-1ra. To test whether the reduced production of sIL-1ra by FPPARβ/δ triggers keratinocyte proliferation, we examined the effect of exogenous IL-1ra supplemented into the medium of OTCs with FPPARβ/δ. Importantly, exogenous IL-1ra abolished the mitogenic effect of FPPARβ/δ (Fig. S5 A). This was further confirmed by immunoblot analysis of cyclin D1 and PCNA on the epidermis from the various OTCs (Fig. 5 A). Thus, inhibition of IL-1α/β signaling using IL-1ra can rescue the pro-proliferating effect that PPARβ/δ deficiency in the fibroblasts has on keratinocyte proliferation.
Figure 5.
Reduced sIL-1ra in fibroblasts potentiates epidermal keratinocyte proliferation. (A) Immunoblot analysis of epidermis from indicated OTCs treated with either vehicle (PBS) or 50 ng/ml exogenous IL-1ra. Cell proliferation was measured using PCNA and cyclin D1. Values below each band were derived as described in Fig. 3 C. (B) Specific knockdown of sIL-1ra in human fibroblasts. (left) Knockdown efficiency was monitored by qPCR and normalized with control ribosomal protein P0. Specificity of knockdown was assessed by the relative expression level of icIL-1ra. (right) Protein expression of sIL-1ra and icIL-1ra as determined by ELISA. (C) Reduced fibroblasts sIL-1ra increase epidermal proliferation. OTCs were constructed using KCTRL with either control (FCTRL) or sIL-1ra knockdown (FsIL-1ra) fibroblasts. H&E, hematoxylin and eosin staining. (top) Ki67, cell proliferation (white arrows); DAPI, nuclear staining. Bars, 40 µm. (bottom) Bars, 20 µm. (left) Mean numbers of proliferating cells were derived as described in Fig. S1. Broken lines denote epidermal–dermal junction. (right) Immunoblot analysis of keratinocytes and fibroblasts extracted from two independent indicated OTCs. Equal amounts of total protein (50 µg) were resolved, electrotransferred, and probed for the indicated proteins. Values below each band represent the mean fold differences in expression level with respect to KCTRL or FCTRL extracted from KCTRL/FCTRL OTC. **, P < 0.01; ***, P < 0.001. Data are mean ± SD, n = 3.
To strengthen these results, we examined the role of sIL-1ra on epidermal proliferation using RNA interference. An siRNA designed to target sIL-1ra was introduced into the human primary fibroblasts. qPCR and ELISA analyses revealed >95% reduction in sIL-1ra mRNA and protein in the transduced fibroblasts (Fig. 5 B). The knockdown showed high specificity for the targeted sIL-1ra mRNA as the level of icIL-1ra mRNA was unchanged (Fig. 5 B, left). The sIL-1ra knockdown fibroblasts (FsIL-1ra) were used in OTC. Consistent with the aforementioned results, reducing the sIL-1ra production in fibroblasts resulted in increased keratinocyte proliferation as indicated by the large increase in Ki67-positive cells and cyclin D1 and PCNA expression (Fig. 5 C, left and middle). This marked increase in epidermal cell proliferation is accompanied by enhanced activation of TAK1 and c-Jun in the FsIL-1ra when compared with FCTRL (Fig. 5 C, right).
To provide further evidence that reduced sIL-1ra in fibroblasts results in the increased expression of mitogenic factors, protein arrays were used to compare conditioned media from OTCs using WT keratinocytes with either FCTRL or FsIL-1ra (Table II). In addition to those listed in Table I, the expression of two additional factors, chemokine ligand 1 (CXCL1)/growth-related oncogene-α and stromal-derived growth factor-1 (SDF-1), were increased in OTC FsIL-1ra compared with FCTRL. The expression of CXCL1 and SDF-1 is c-Jun dependent, and their expression increased keratinocyte proliferation (Steude et al., 2002; Florin et al., 2005). Similarly, when comparing OTCs with FCTRL and FPPARβ/δ, there was a trend for higher CXCL1 and SDF-1 expression in OTCs FPPARβ/δ, although the fold increase was not significant (unpublished data).
Table II.
Relative fold change of protein expression in OTCs with FsIL1ra compared with FCTRL
| Cytokines/growth factors | Fold increasea |
| KGFb | 5.47 ± 0.22c |
| IL-6b | 6.11 ± 0.31c |
| IL-8 | 3.47 ± 0.06c |
| IL-10 | 3.88 ± 0.15c |
| IL-16 | 3.78 ± 0.41c |
| GMCSFb | 7.74 ± 0.31c |
| CXCL1/GRO-α | 2.67 ± 0.16d |
| SDF-1 | 2.33 ± 0.07d |
| I-309 | 4.57 ± 0.20c |
| Eotaxin-2 | 6.19 ± 0.62c |
| VEGF | 0.22 ± 0.030d |
| PIGF | 0.25 ± 0.021d |
GRO-α, growth-related oncogene-α.
The intensities of signals were quantified by densitometry. Positive control included in the array blot was used to normalize and compare the results from different membranes. Values obtained from the control KCTRL/FCTRL OTC were arbitrarily assigned a value of 1. Values represent the mean fold increase as compared with control OTC (n = 3).
ChIP was performed on these genes using phospho–c-Jun antibody.
P < 0.01.
P < 0.1.
Finally, we also performed ChIP using phospho–c-Jun antibodies on the KGF, GMCSF, and IL-6 gene promoters in fibroblasts extracted from KCTRL/FCTRL and KCTRL/FsIL-1ra OTCs. The sequences spanning the AP-1–binding site were significantly enriched in the immunoprecipitates obtained from FsIL-1ra compared with FCTRL (Fig. 6, left). This was further confirmed by qPCR normalized to chromatin before immunoprecipitation, i.e., input (Fig. 6, right). Altogether, we concluded that reduced sIL-1ra production in the fibroblasts, as observed in FPPARβ/δ, had a pronounced mitogenic effect on the epidermis.
Figure 6.
Increased c-Jun binding to AP-1 site of human KGF, GMCSF, and IL-6 gene promoter in FsIL-1ra. ChIP of AP-1–binding site of human KGF (top), GMCSF (middle), and IL-6 (bottom) genes using phospho–c-Jun antibodies. The gene sequence spanning the AP-1 site and a random control sequence were analyzed by PCR in the immunoprecipitated chromatin of FCTRL and FsIL-1ra fibroblasts extracted from K/FCTRL and K/FsIL-1ra OTC, respectively. Preimmune serum was used as a control. qPCR was performed on immunoprecipitates of phospho–c-Jun antibodies and normalized to input (chromatin before immunoprecipitation). Results are represented in fold change as compared with FCTRL extracted from K/FCTRL OTC. M, 100 bp DNA ladder.
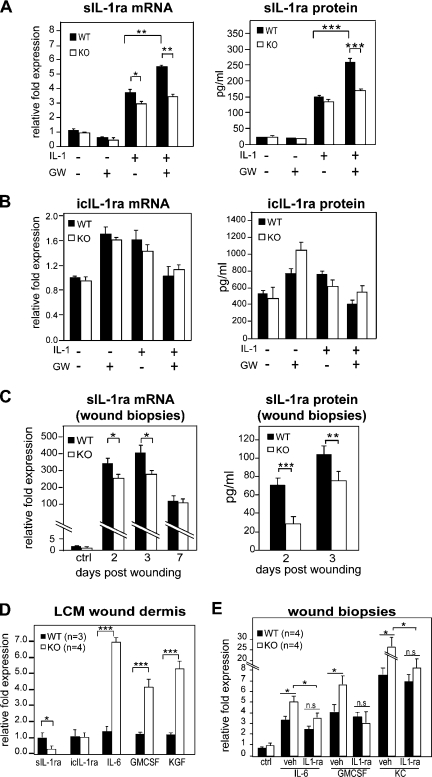
PPARβ/δ-KO mice show reduced sIL-1ra expression during early wound healing
Next, we evaluated the relevance of our findings during mouse skin wound healing, an in vivo model that involves the IL-1α/β pathway. We first showed that activated PPARβ/δ increased the IL-1β–induced sIL-1ra expression in WT but not in KO primary fibroblasts (Fig. 7, A and B). Next, we examined the expression of IL-1ra in skin wound biopsies collected during early wound healing. qPCR analysis of early wound biopsies (days 1–3) showed a significantly lower sIL-1ra expression in KO tissue when compared with WT (Fig. 7 C), which is consistent with the enhanced cell proliferation previously reported during early wound healing (Tan et al., 2001). Notably, this difference in sIL-1ra expression was no longer detected in later wound biopsy specimens (Fig. 7 C, day 7). The analysis of wound fluids collected from day 2 and 3 wound beds showed reduced sIL-1ra protein in KO mice when compared with WT mice (Fig. 7 C). The expression of icIL-1ra was unchanged between WT and KO mice (unpublished data). Because keratinocytes only produced icIL-1ra, the change in sIL-1ra expression was attributed to the fibroblasts, a hypothesis that was verified by qPCR analysis of laser microdissected epithelium and dermis sections from day 4 wound biopsies of WT and KO mice (Fig. S5 B). Consistent with our aforementioned results, wound KO dermis had reduced sIL-1ra expression and concomitantly increased levels of IL-6, GMCSF, and KGF when compared with wound WT dermis (Fig. 7 D). These results confirmed that the expression of sIL-1ra was also modulated by PPARβ/δ in mouse fibroblasts. We next examined the effect of topical application of exogenous IL-1ra on the expression of IL-6, GMCSF, and mouse chemokine ligand 1 (KC; also known as CXCL1) in early wound biopsies. As expected, exogenous IL-1ra reduced IL-6, GMCSF, and KC expression in KO wounds (Fig. 7 E). In summary, in vivo wound biopsy and fluid analyses revealed lower sIL-1ra mRNA and protein in the KO mice. Our results showed that PPARβ/δ-deficient fibroblasts had reduced sIL-1ra expression compared with WT cells and that this reduced sIL-1ra production increased epidermal proliferation.
Figure 7.
Regulation of IL-1β signaling via PPARβ/δ during wound repair in KO mice. (A and B) Expression level of sIL-1ra (A) and icIL-1ra (B) in WT and KO fibroblasts. WT and KO primary fibroblasts were treated with PPARβ/δ ligand (100 nM GW501516 [GW]) and/or 10 ng/ml IL-1β. The mRNA (left) and protein (right) expression levels were measured by qPCR and ELISA, respectively. (C) Expression level of sIL-1ra in early wound biopsies of KO mice. Wound fluids and biopsies were collected at the indicated day post-wounding (days 1–3, n = 7; day 7, n = 4). Unwounded skin was used as control (ctrl; n = 4). sIL-1ra mRNA and protein levels were determined by qPCR and ELISA, respectively. (D) Expression of sIL-1ra, icIL-1ra, KGF, IL-6, and GMCSF mRNAs from laser capture microdissected (LCM) dermis of WT (n = 3) and KO (n = 4) wound biopsies. Indicated mitogenic factor mRNA levels were analyzed by qPCR and normalized to control ribosomal protein P0. (E) Expression levels of indicated factor mRNAs in vehicle (veh; carboxymethylcellulose)- and IL-1ra–treated (3 × 2.5 µg) wounds of KO and WT mice as compared with corresponding unwounded (control) skin. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data are mean ± SEM, n = 4.
Discussion
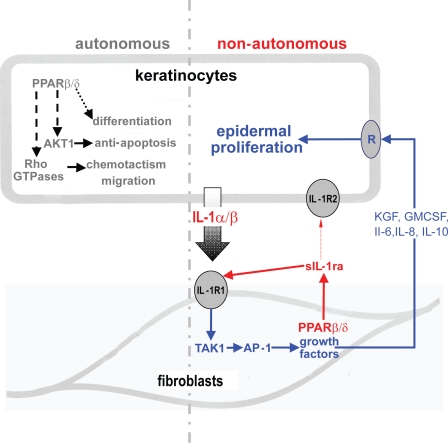
Wound healing is a complex process that consists of a cascade of overlapping events, including inflammation reepithelialization and remodeling, directed at the restoration of the epidermal barrier. Throughout the whole healing process, interaction between different cell types provides coordination of the individual events, allowing for a temporal and spatial control. Reepithelialization is accomplished by increased keratinocyte proliferation and guided migration over the granulation tissue. Such processes require ordered changes in keratinocyte behavior that is dictated by the interplay of keratinocytes with dermal fibroblasts, i.e., epithelial–mesenchymal communication. The results presented in this study characterize a novel unsuspected role of PPARβ/δ in fibroblasts, which ensure a balanced functional interaction between dermis and epidermis. We show that keratinocyte proliferation is controlled by the dermal fibroblasts that produce AP-1–dependent mitogenic factors under the control of the IL-1/TAK1 signaling cascade whose activity level is modulated by PPARβ/δ via a direct control sIL-1ra production (Fig. 8).
Figure 8.
The autonomous and nonautonomous actions of PPARβ/δ. PPARβ/δ confers antiapoptotic properties to keratinocytes and potentiates their migration via AKT1 and Rho GTPases in a cell-autonomous manner (Di Poi et al., 2002; Tan et al., 2007). PPARβ/δ also influences keratinocyte differentiation in a yet unknown mechanism. PPARβ/δ regulates epidermal proliferation in a nonautonomous fashion via a paracrine mechanism. IL-1 constitutively produced by the adjacent keratinocytes activates c-Jun, an obligate partner of AP-1 transcription factor via TAK1, and consequently increases the production of several mitogenic factors. The expression of PPARβ/δ in the underlying fibroblasts attenuates this IL-1 signaling via the production of sIL-1ra. sIL-1ra has little affinity for IL-1R2, which is highly expressed in keratinocytes. Thus, sIL-1ra acts in an autocrine fashion onto the fibroblasts, which expressed the predominant functional IL-1R1. The binding of sIL-1ra to IL-1R1 modulates the IL-1–mediated signaling events and consequently decreases the production of several AP-1–mediated mitogenic factors. The mitogenic factors exert a reduced paracrine effect on the epithelial proliferation via their cognate receptors (R). Therefore, PPARβ/δ in the fibroblasts plays an important homeostatic role in maintaining epidermal homeostasis, the absence of PPARβ/δ resulting in significant epidermal proliferation.
Epidermal homeostasis requires a continuous exchange of signals with the underlying dermal compartment. Fibroblasts are a major cell type in the dermis, and their function in regulating extracellular matrix deposition and growth factor expression determines the differentiation states of the epidermis. Previous studies have showed that gene expression changes in the dermal fibroblasts significantly influence epithelial cell fate (Cheng et al., 2005). In the presence of IL-1–producing keratinocytes, PPARβ/δ was the predominant PPAR isotype in fibroblasts. Interestingly, PPARβ/δ directly increased the expression of sIL-1ra in the fibroblasts that acted in an autocrine manner to modulate the expression of several IL-1–dependent mitogenic factors (Fig. 8). This autocrine action is possible because IL-1R1 is the predominant IL-1R subtype in fibroblasts, whereas keratinocytes exhibit IL-1R2 that has very low affinity for sIL-1ra and protects them from an exacerbated autocrine action of the IL-1α/β that they constitutively produce. This study underscores the fact that PPARβ/δ in the dermal fibroblasts fulfils a homeostatic role during epidermal formation, balancing cell proliferation via a control on IL-1α/β signaling.
It was reported that the enhanced TPA-induced hyperplasia in KO mice was a result of reduced ubiquitin C, whose expression is regulated by PPARβ/δ (Kim et al., 2004). The impaired ubiquitin-dependent proteosome-mediated protein turnover in KO mice resulted in a higher PKCα level, increased Raf1, and MAPK/extracellular signal-regulated kinase activities, which led to an increase in cell proliferation (Kim et al., 2005). Other studies have shown that PKCα overexpression in two different models did not affect the proliferation of primary human keratinocytes and of PKCα transgenic mouse keratinocytes (Wang and Smart, 1999; Cataisson et al., 2003). In fact, PKCα transgenic mouse keratinocytes treated with TPA underwent apoptosis (Cataisson et al., 2003). This is in apparent discrepancy with the delayed wound healing kinetics observed in KO mice. Enhanced keratinocyte proliferation was also observed in many situations not involving TPA exposure (Michalik et al., 2001; Tan et al., 2001). Furthermore, no difference was observed in phosphorylated MEK-1/2 and ERK-1/2 expression between untreated KO and WT mice (Kim et al., 2005). This raises the question of how epidermal hyperproliferation occurs at wound edges early after injury of KO mice (Michalik et al., 2001). These observations point to a complex role of PPARβ/δ in cell proliferation. Our results showing that PPARβ/δ deficiency in fibroblasts increases their mitogenic effect on epithelial cells unveil part of this complexity. The underlying mechanism involves reduced sIL-1ra production by the fibroblasts, allowing a stronger IL-1 signaling, resulting in increased TAK1 activity with a consequent rise in mitogenic growth factor production. The in vivo relevance of this mechanism was observed during early wound repair in KO mice with reduced sIL-1ra in early but not late wound biopsies. This pattern is consistent with the early epidermal hyperproliferation reported in KO mice during the inflammatory phase of wound healing (Tan et al., 2001).
This study and earlier studies suggest different roles of PPARβ/δ in the dermis and adjacent epidermis (Tan et al., 2001; Man et al., 2008). The activation of PPARβ/δ conferred antiapoptotic properties to keratinocytes, which are mediated in part by the increased activation of the PKBα pathway (Di Poi et al., 2002). This antiapoptotic role was similarly observed in mouse tubular renal epithelial cells (Letavernier et al., 2005) and in human keratinocyte HaCaT cells (Schug et al., 2007). Our data now clearly show that PPARβ/δ signaling in fibroblasts has a pronounced effect on the growth potential of adjacent epithelia. Such differential roles were previously observed for TGF-β1 signaling in wound repair (Beanes et al., 2003). Interestingly, an in vivo cross talk between TGF-β1/Smad3 and PPARβ/δ signaling in keratinocytes during wound repair has been reported (Tan et al., 2004). The development of a vascular supply is important for wound healing. Interestingly, we also observed a reduced expression of VEGF and PIGF in OTC with FPPARβ/δ. Although it is tempting to speculate that both VEGF and PIGF may be target genes of PPARβ/δ, their regulation are complex and context dependent, involving numerous factors such as pVHL (von Hippel-Lindau tumor suppressor), Sp-1, NF-κB (nuclear factor κ light chain enhancer of activated B cells), hypoxia-inducible factor α, and AP-1, among others (Mukhopadhyay et al., 1997; Green et al., 2001). Importantly, epithelial–mesenchymal interactions can also modulate VEGF expression (Ong et al., 2007). During wound repair, an impaired angiogenesis is in line with the delayed wound healing observed in KO mice (Michalik et al., 2001).
The tissue microenvironment plays a pivotal role in modulating gene expression and cellular behavior, including epithelial–mesenchymal communication. Likewise, the microenvironment milieu is constituted by factors produced and released by these various cell types as communication signals. Of interest, the activation of IL-1 signaling was reported to coerce the growth of tumors, whereas its repression by IL-1ra has an antitumor effect (Lewis et al., 2006). The role of PPARβ/δ in tumorigenesis remains debatable because there is evidence that this PPAR isotype potentiates or attenuates epithelial cancers (Gupta et al., 2000; Barak et al., 2002; Harman et al., 2004). Although this question is not directly addressed in this study, our preliminary data showed that FPPARβ/δ can increase the proliferation of human squamous carcinoma cells (unpublished data).
Materials and methods
Reagents
The following antibodies and suppliers were used: anti-Ki67, antikeratin 10, anti-involucrin, antikeratin 6 (Novacastra), anti-PPARα, anti-PPARβ/δ, and anti-PPARγ (Millipore and Thermo Fisher Scientific); all cytokines and neutralizing antibodies against IL-1α, IL-1β, KGF, IL-6, and GMCSF (PeproTech); anti–c-Jun, antiphospho(Ser63)–c-Jun, anti-TAK1, and antiphospho-TAK1 (Cell Signaling Technology); and Alexa Fluor 488–conjugated goat anti–mouse antibody (Invitrogen). Real-time reagent SYBR GreenER (Invitrogen), rat tail collagen type I (BD), and transfection reagent Superfect (QIAGEN) were used. All restriction enzymes and DNA/RNA modifying enzymes were obtained from Fermentas. Double-promoter lentivirus-based siRNA vector and pFIV Packaging kit (System Biosciences) was used. The PPARα, β/δ, γ Complete Transcription Factor Assay kit was obtained from Cayman Chemical. Primary neonatal human fibroblasts and keratinocytes were obtained from Invitrogen. GW501516 was obtained from Enzo Biochem, Inc. Otherwise, chemicals were obtained from Sigma-Aldrich.
Total RNA isolation, reverse transcription PCR, and qPCR
Total RNA was extracted from fresh tissues using Aurum total RNA kit (Bio-Rad Laboratories) following the supplier’s protocol. 5 μg total RNA was reverse transcribed with oligo-dT primers using RevertAid H Minus M-MuLV (Fermentas). After reverse transcription, the RNAs were removed by RNase H digestion. qPCR was performed with platinum Taq polymerase and SYBR GreenER super mixes using a PCR machine (MiniOpticon; Bio-Rad Laboratories). Melt curve analysis was included to assure that only one PCR product was formed. Primers were designed to generate a PCR amplification product of 100–250 bp. Only primer pairs yielding unique amplification products without primer dimer formation were subsequently used for real-time PCR assays. Expression was related to the control gene ribosomal protein P0 (RPLP0), which did not change under any of the experimental conditions studied. The sequence of primers is available in Table S1. For each wound biopsies, 10 8-µm-thick tissues were microdissected and pooled. RNA was isolated from microdissected paraformaldehyde-fixed paraffin-embedded sections using RecoverAll Total Nucleic Acid Isolation kit (Applied Biosystems). 5 ng RNA was subjected to Full Spectrum Complete Transcriptome RNA Amplification kit (System Biosciences) prior to qPCR.
Lentivirus-mediated knockdown of PPARβ/δ and sIL-1ra
Two siRNAs targeting the human PPARβ/δ, one against sIL-1ra and one unrelated control siRNA, were subcloned into the lentiviral-based siRNA vector pFIV-H1/U6-puro. The correct pFIV siRNA constructs were verified by sequencing using H1 primer. The sequence of the siRNAs was as given in Table S1. Transduction-ready pseudoviral particles (System Biosciences) were produced and harvested as described by the manufacturer. Transduced cells were enriched by puromycin selection for 1 wk. Western blot analysis, ELISA, or qPCR were used to assess the efficiency of knockdown.
Transactivation assay
An ∼4.4-kb (−1572 to −5980) promoter of the human sIL-1ra was PCR amplified from human genomic DNA using Pfu polymerase. The primer pairs used are given in Table S1. The resulting fragment was used for a second PCR amplification step introducing SacI and BglII sites that were used for subcloning into the pGL-3 Basic vector (Promega). Site-directed mutagenesis of the three putative PPREs (PPRE1 at −1038/−1050, PPRE2 at −1072/−1084, and PPRE3 at −4067/−4079) were achieved using QuikChange site-directed mutagenesis kit (Agilent Technologies). Human fibroblasts were routinely grown in medium 106 (Invitrogen). Fibroblasts were cotransfected with a luciferase reporter driven by the various sIL-1ra promoter constructs and pEF1–β-galactosidase as a control of transfection efficiency using Superfect. After transfection, cells were incubated in the presence or absence of 500 nM PPARβ/δ ligand GW501516 and IL-1β for 24 h before lysis. Transfections with the positive control reporter pGL-3xPPRE-tk-Luc were included (provided by R.M. Evans, The Salk Institute for Biological Studies, University of California, San Diego, La Jolla, CA). Luciferase activity was measured using the luciferase assay (Promega) on a Microbeta Trilux (PerkinElmer). β-Galactosidase activity was measured in the cell lysate by a standard assay using 2-nitrophenyl-βD-galactopyranoside as a substrate.
ChIP
ChIP was performed as described previously (Tan et al., 2004) with minor modifications. In brief, epidermis from OTC was physically separated from the collagen-embedded human fibroblasts after 2.5 U/ml dispase treatment at 37°C for 20 min. The dermal equivalents were thoroughly washed with PBS. The collagen gel was cut into small pieces prior to digestion with 0.5% collagenase I at 37°C. The fibroblasts were retrieved and cross-linked with 1% formaldehyde for 15 min at 37°C prior to sonication in lysis buffer. Monoclonal anti-PPARβ/δ antibody was used. The immunoprecipitates were reverse cross-linked for PCR by heating at 65°C for 6 h. The primer pairs used are shown in Table S1.
OTC
Primary human keratinocytes and fibroblasts were routinely maintained in defined keratinocyte medium (EpiLife; Invitrogen) and medium 106, respectively, as described by the manufacturer. OTCs were performed as previously described (Maas-Szabowski et al., 2000) in serum-free OTC medium with some modifications. 1 vol 10× HBSS containing phenol red was mixed with 8 vol 4 mg/ml rat tail type I collagen (BD). The acetic acid was neutralized with 1 N NaOH on ice. Fibroblasts were resuspended in 1 vol 1× HBSS and added dropwise to the neutralized collagen to achieve a final cell density of 105 cells/ml of collagen. 3 ml of this fibroblast/collagen mixture was dispensed into a 24-mm diameter Transwell culture insert (3-µm pore size; BD). The culture insert was placed in a 6-well Deep-Well Plate (BD) and allowed to gel in a 37°C, 5% CO2, 70% humidity incubator (Thermo Fisher Scientific). Glass rings (outer diameter, 20 mm; thickness, 1.2 mm; height, 12 mm) were centrally placed onto the collagen. The glass rings serve to compress the collagen and delimit the area for the seeding of keratinocytes. The fibroblast-embedded collagen was submerged and cultured in serum-free OTC medium overnight. The serum-free OTC contains basal medium, 3:1 (vol/vol) DME (low glucose), Ham’s F-12 nutrient mixture, basal supplements 5 µg/ml insulin, 1 ng/ml epidermal growth factor, 0.4 µg/ml hydrocortisone, 100 nM adenine, 10 µM serine, 100 nM cholera toxin, 10 µM carnitine, 1 mg/ml fatty acid–free albumin, lipid supplements 0.05 mM ethanolamine, 1 µM isoproterenol, 1 µM α-tocopherol, and 50 µg/ml ascorbic acid. The following day, the medium was removed and 106 keratinocytes were seeded into the center of the glass ring. OTC medium was added, and the setup was once again incubated at 37°C with 5% CO2 for 24 h to allow the keratinocytes to attach. The next day, the glass ring was removed, and culture was maintained at air liquid interface. Medium was changed every 3 d for a total of 2 wk. Neutralizing antibodies (each at 400 ng/ml) were freshly supplemented at every change of medium throughout the 2-wk culture period.
Immunofluorescence
OTCs were fixed with 4% paraformaldehyde in PBS for 2 h at 25°C. The fixed OTCs were washed twice with PBS and embedded in Tissue-Tek OCT compound medium (Sakura) overnight at 4°C. The skin cultures were subsequently frozen at −70°C for cryosectioning. 10-µm cryostat tissue sections were mounted on SuperFrost Plus slides (Menzel-Gläser). The sections were processed for immunofluorescence as described previously (Michalik et al., 2001) except that Alexa Fluor 488–conjugated goat anti–mouse secondary antibody was used. The apoptotic keratinocytes were detected using the TUNEL assay according to the manufacturer’s protocol (Roche). As positive control for TUNEL assay, the section was pretreated with DNase I. The slides were mounted with antifade reagent (ProLong Gold; Invitrogen) with DAPI. Images were taken with an inverted microscope (ECLIPSE TE2000-U; Nikon) using a Plan Fluor 20×/0.45 objective (Nikon), Retiga EXi FAST cooled mono 12-bit camera (QImaging), and Image-Pro Plus software (Media Cybernetics).
Wounding experiment
Wounding of the mice dorsal skin was performed as previously described (Michalik et al., 2001). Wound biopsies and fluids were isolated as previously described (Tan et al., 2004) and were immediately snap frozen in liquid nitrogen for further analysis. Topical application of exogenous IL-1ra treatment was performed as previously described (Tan et al., 2005) except that three applications of 2.5 µg recombinant IL-1ra were added over 3 d.
Measurement of cytokines by ELISA
The concentration of sIL-1ra and icIL-1ra measured using sandwich ELISA (R&D Systems). In brief, 2 × 105 cells were cultured in a 35-mm culture dish with 750 µl of medium and subjected to the indicated treatments. After 24 h, the media were harvested and treated with the addition of Complete protease inhibitors (Roche). The level of sIL-1ra was measured in culture supernatants accordingly to the manufacturer’s instruction. To measure icIL-1ra, the cells were washed thoroughly, harvested by treatment with trypsin, and collected by centrifugation in fresh serum-free culture medium. The cell pellet was resuspended in 750 µl of medium, disrupted by sonication, and the debris was removed by centrifugation. The concentration of icIL-ra in the supernatant was measured by ELISA.
Protein arrays
Human Inflammation Antibody Array 3 and Growth Factor Antibody Array membranes (RayBiotech) were processed according to the manufacturer’s protocol. Protein spots were detected by chemiluminescence. Signal intensities were quantified using ImageJ (National Institutes of Health) analysis software and were normalized with the mean intensity of the positive controls on each membrane.
Western blot analysis
Epidermis was physically separated from OTC after a 20-min treatment with dispase. Fibroblasts embedded in collagen were isolated after collagenase treatment. For Western blotting, protein extracts were made in ice-cold lysis buffer (20 mM Na2H2PO4, 250 mM NaCl, 1% Triton X-100, and 0.1% SDS). Equal amounts of protein extracts (50 µg) were resolved by SDS-PAGE and electrotransferred onto PVDF membranes. Membranes were processed as described by the manufacturer of antibodies, and proteins were detected by chemiluminescence (Millipore). Coomassie blue–stained membrane or tubulin was used to check for equal loading and transfer.
Online supplemental material
Fig. S1 shows immunofluorescence staining of 2-wk old OTCs. Fig. S2 shows immunoblot analysis of IL-1β and TNF-α activation of TAK1 in FCTRL or FPPARβ/δ fibroblasts and ChIP of AP-1–binding site of human KGF and GMCSF genes using phospho–c-Jun antibodies. Fig. S3 shows immunofluorescence staining of 2-wk-old OTCs treated with neutralizing antibodies against IL-1α and β or against KGF, GMCSF, and IL-6. Fig. S4 shows the alignment of the two functional PPREs in human sIL-1ra promoter to consensus PPRE. Fig. S5 shows immunofluorescence staining of 2-wk old OTCs derived using PPARβ/δ-deficient fibroblasts (FPPARβ/δ) treated with either vehicle or exogenous IL-1ra. Table S1 shows the sequence of primers used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200809028/DC1.
Acknowledgments
We thank Dr. Samuel Ko and Anna Teo (Carl Zeiss, Inc.) for their expertise in laser capture microdissection and R.M. Evans for the positive PPRE reporter construct.
This work was supported by grants from the Academic Research Fund, the Ministry of Education (ARC8/06 to N.S. Tan), Supplementary Equipment Purchase (RG127/05 to N.S. Tan), and the Swiss National Science Foundation (W. Wahli).
Footnotes
Abbreviations used in this paper: AP-1, activation protein-1; ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; GMCSF, granulocyte macrophage colony-stimulating factor; icIL-1ra, intracellular IL-1ra; IL, interleukin; IL-1R, IL-1 receptor; IL-1ra, IL-1R antagonist; KGF, keratinocyte growth factor; OTC, organotypic skin culture; PCNA, proliferating cell nuclear antigen; PIGF, placental growth factor; PPAR, peroxisome proliferator–activated receptor; PPRE, peroxisome proliferator response elements; qPCR, quantitative real-time PCR; SDF-1, stromal-derived growth factor-1; sIL-1ra, secreted IL-1ra; TAK1, TGF-activated kinase 1; WT, wild type.
References
- Arend W.P., Malyak M., Guthridge C.J., Gabay C. 1998. Interleukin-1 receptor antagonist: role in biology.Annu. Rev. Immunol. 16:27–55 [DOI] [PubMed] [Google Scholar]
- Banda N.K., Guthridge C., Sheppard D., Cairns K.S., Muggli M., Bech-Otschir D., Dubiel W., Arend W.P. 2005. Intracellular IL-1 receptor antagonist type 1 inhibits IL-1-induced cytokine production in keratinocytes through binding to the third component of the COP9 signalosome.J. Immunol. 174:3608–3616 [DOI] [PubMed] [Google Scholar]
- Barak Y., Liao D., He W., Ong E.S., Nelson M.C., Olefsky J.M., Boland R., Evans R.M. 2002. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer.Proc. Natl. Acad. Sci. USA. 99:303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanes S.R., Dang C., Soo C., Ting K. 2003. Skin repair and scar formation: the central role of TGF-beta.Expert Rev. Mol. Med. 5:1–22 [DOI] [PubMed] [Google Scholar]
- Burdick A.D., Kim D.J., Peraza M.A., Gonzalez F.J., Peters J.M. 2006. The role of peroxisome proliferator-activated receptor-beta/delta in epithelial cell growth and differentiation.Cell. Signal. 18:9–20 [DOI] [PubMed] [Google Scholar]
- Cataisson C., Joseloff E., Murillas R., Wang A., Atwell C., Torgerson S., Gerdes M., Subleski J., Gao J.L., Murphy P.M., et al. 2003. Activation of cutaneous protein kinase C alpha induces keratinocyte apoptosis and intraepidermal inflammation by independent signaling pathways.J. Immunol. 171:2703–2713 [DOI] [PubMed] [Google Scholar]
- Cheng N., Bhowmick N.A., Chytil A., Gorksa A.E., Brown K.A., Muraoka R., Arteaga C.L., Neilson E.G., Hayward S.W., Moses H.L. 2005. Loss of TGF-beta type II receptor in fibroblasts promotes mammary carcinoma growth and invasion through upregulation of TGF-alpha-, MSP- and HGF-mediated signaling networks.Oncogene. 24:5053–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Poi N., Tan N.S., Michalik L., Wahli W., Desvergne B. 2002. Antiapoptotic role of PPARbeta in keratinocytes via transcriptional control of the Akt1 signaling pathway.Mol. Cell. 10:721–733 [DOI] [PubMed] [Google Scholar]
- Florin L., Hummerich L., Dittrich B.T., Kokocinski F., Wrobel G., Gack S., Schorpp-Kistner M., Werner S., Hahn M., Lichter P., et al. 2004. Identification of novel AP-1 target genes in fibroblasts regulated during cutaneous wound healing.Oncogene. 23:7005–7017 [DOI] [PubMed] [Google Scholar]
- Florin L., Maas-Szabowski N., Werner S., Szabowski A., Angel P. 2005. Increased keratinocyte proliferation by JUN-dependent expression of PTN and SDF-1 in fibroblasts.J. Cell Sci. 118:1981–1989 [DOI] [PubMed] [Google Scholar]
- Fusenig N.E. 1994. Epithelial-mesenchymal interactions regulate keratinocyte growth and differentiation in vitro. The Keratinocyte Handbook. Leigh I.M., Lane E.B., Watt F.M., Cambridge University Press, Cambridge/New York: 71–94 [Google Scholar]
- Green C.J., Lichtlen P., Huynh N.T., Yanovsky M., Laderoute K.R., Schaffner W., Murphy B.J. 2001. Placenta growth factor gene expression is induced by hypoxia in fibroblasts: a central role for metal transcription factor-1.Cancer Res. 61:2696–2703 [PubMed] [Google Scholar]
- Groves R.W., Rauschmayr T., Nakamura K., Sarkar S., Williams I.R., Kupper T.S. 1996. Inflammatory and hyperproliferative skin disease in mice that express elevated levels of the IL-1 receptor (type I) on epidermal keratinocytes. Evidence that IL-1-inducible secondary cytokines produced by keratinocytes in vivo can cause skin disease.J. Clin. Invest. 98:336–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.A., Tan J., Krause W.F., Geraci M.W., Willson T.M., Dey S.K., DuBois R.N. 2000. Prostacyclin-mediated activation of peroxisome proliferator-activated receptor delta in colorectal cancer.Proc. Natl. Acad. Sci. USA. 97:13275–13280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman F.S., Nicol C.J., Marin H.E., Ward J.M., Gonzalez F.J., Peters J.M. 2004. Peroxisome proliferator-activated receptor-delta attenuates colon carcinogenesis.Nat. Med. 10:481–483 [DOI] [PubMed] [Google Scholar]
- Heiman A.S., Abonyo B.O., Darling-Reed S.F., Alexander M.S. 2005. Cytokine-stimulated human lung alveolar epithelial cells release eotaxin-2 (CCL24) and eotaxin-3 (CCL26).J. Interferon Cytokine Res. 25:82–91 [DOI] [PubMed] [Google Scholar]
- IJpenberg A., Tan N.S., Gelman L., Kersten S., Seydoux J., Xu J., Metzger D., Canaple L., Chambon P., Wahli W., Desvergne B. 2004. In vivo activation of PPAR target genes by RXR homodimers.EMBO J. 23:2083–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S., Desvergne B., Wahli W. 2000. Roles of PPARs in health and disease.Nature. 405:421–424 [DOI] [PubMed] [Google Scholar]
- Kim D.J., Akiyama T.E., Harman F.S., Burns A.M., Shan W., Ward J.M., Kennett M.J., Gonzalez F.J., Peters J.M. 2004. Peroxisome proliferator-activated receptor beta (delta)-dependent regulation of ubiquitin C expression contributes to attenuation of skin carcinogenesis.J. Biol. Chem. 279:23719–23727 [DOI] [PubMed] [Google Scholar]
- Kim D.J., Murray I.A., Burns A.M., Gonzalez F.J., Perdew G.H., Peters J.M. 2005. Peroxisome proliferator-activated receptor-beta/delta inhibits epidermal cell proliferation by down-regulation of kinase activity.J. Biol. Chem. 280:9519–9527 [DOI] [PubMed] [Google Scholar]
- Letavernier E., Perez J., Joye E., Bellocq A., Fouqueray B., Haymann J.P., Heudes D., Wahli W., Desvergne B., Baud L. 2005. Peroxisome proliferator-activated receptor beta/delta exerts a strong protection from ischemic acute renal failure.J. Am. Soc. Nephrol. 16:2395–2402 [DOI] [PubMed] [Google Scholar]
- Lewis A.M., Varghese S., Xu H., Alexander H.R. 2006. Interleukin-1 and cancer progression: the emerging role of interleukin-1 receptor antagonist as a novel therapeutic agent in cancer treatment.J. Transl. Med. 4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas-Szabowski N., Stark H.J., Fusenig N.E. 2000. Keratinocyte growth regulation in defined organotypic cultures through IL-1-induced keratinocyte growth factor expression in resting fibroblasts.J. Invest. Dermatol. 114:1075–1084 [DOI] [PubMed] [Google Scholar]
- Man M.Q., Barish G.D., Schmuth M., Crumrine D., Barak Y., Chang S., Jiang Y., Evans R.M., Elias P.M., Feingold K.R. 2008. Deficiency of PPARbeta/delta in the epidermis results in defective cutaneous permeability barrier homeostasis and increased inflammation.J. Invest. Dermatol. 128:370–377 [DOI] [PubMed] [Google Scholar]
- Michalik L., Desvergne B., Tan N.S., Basu-Modak S., Escher P., Rieusset J., Peters J.M., Kaya G., Gonzalez F.J., Zakany J., et al. 2001. Impaired skin wound healing in peroxisome proliferator–activated receptor (PPAR)α and PPARβ mutant mice.J. Cell Biol. 154:799–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D., Knebelmann B., Cohen H.T., Ananth S., Sukhatme V.P. 1997. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity.Mol. Cell. Biol. 17:5629–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee 1999. A unified nomenclature system for nuclear receptor superfamily.Cell. 97:161–163 [DOI] [PubMed] [Google Scholar]
- Ong C.T., Khoo Y.T., Tan E.K., Mukhopadhyay A., Do D.V., Han H.C., Lim I.J., Phan T.T. 2007. Epithelial-mesenchymal interactions in keloid pathogenesis modulate vascular endothelial growth factor expression and secretion.J. Pathol. 211:95–108 [DOI] [PubMed] [Google Scholar]
- Peters J.M., Lee S.S., Li W., Ward J.M., Gavrilova O., Everett C., Reitman M.L., Hudson L.D., Gonzalez F.J. 2000. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta).Mol. Cell. Biol. 20:5119–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podvinec M., Kaufmann M.R., Handschin C., Meyer U.A. 2002. NUBIScan, an in silico approach for prediction of nuclear receptor response elements.Mol. Endocrinol. 16:1269–1279 [DOI] [PubMed] [Google Scholar]
- Rauschmayr T., Groves R.W., Kupper T.S. 1997. Keratinocyte expression of the type 2 interleukin 1 receptor mediates local and specific inhibition of interleukin 1-mediated inflammation.Proc. Natl. Acad. Sci. USA. 94:5814–5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuth M., Haqq C.M., Cairns W.J., Holder J.C., Dorsam S., Chang S., Lau P., Fowler A.J., Chuang G., Moser A.H., et al. 2004. Peroxisome proliferator-activated receptor (PPAR)-beta/delta stimulates differentiation and lipid accumulation in keratinocytes.J. Invest. Dermatol. 122:971–983 [DOI] [PubMed] [Google Scholar]
- Schroder J.M. 1995. Cytokine networks in the skin.J. Invest. Dermatol. 105:20S–24S [PubMed] [Google Scholar]
- Schug T.T., Berry D.C., Shaw N.S., Travis S.N., Noy N. 2007. Opposing effects of retinoic Acid on cell growth result from alternate activation of two different nuclear receptors.Cell. 129:723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J.H., Xiao C., Paschal A.E., Bailey S.T., Rao P., Hayden M.S., Lee K.Y., Bussey C., Steckel M., Tanaka N., et al. 2005. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo.Genes Dev. 19:2668–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.F., Jr., Eidlen D., Arend W.P., Gutierrez-Hartmann A. 1994. LPS-induced expression of the human IL-1 receptor antagonist gene is controlled by multiple interacting promoter elements.J. Immunol. 153:3584–3593 [PubMed] [Google Scholar]
- Smola H., Stark H.J., Thiekötter G., Mirancea N., Krieg T., Fusenig N.E. 1998. Dynamic of basement membrane formation by keratinocyte-fibroblast interactions in organotypic skin culture.Exp. Cell Res. 239:399–410 [DOI] [PubMed] [Google Scholar]
- Steude J., Kulke R., Christophers E. 2002. Interleukin-1-stimulated secretion of interleukin-8 and growth-related oncogene-alpha demonstrates greatly enhanced keratinocyte growth in human raft cultured epidermis.J. Invest. Dermatol. 119:1254–1260 [DOI] [PubMed] [Google Scholar]
- Szabowski A., Maas-Szabowski N., Andrecht S., Kolbus A., Schorpp-Kistner M., Fusenig N.E., Angel P. 2000. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal-epidermal interaction in skin.Cell. 103:745–755 [DOI] [PubMed] [Google Scholar]
- Tan N.S., Michalik L., Noy N., Yasmin R., Pacot C., Heim M., Fluhmann B., Desvergne B., Wahli W. 2001. Critical roles of PPAR beta/delta in keratinocyte response to inflammation.Genes Dev. 15:3263–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan N.S., Michalik L., Di-Poi N., Ng C.Y., Mermod N., Roberts A.B., Desvergne B., Wahli W. 2004. Essential role of Smad3 in the inhibition of inflammation-induced PPARbeta/delta expression.EMBO J. 23:4211–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan N.S., Michalik L., Desvergne B., Wahli W. 2005. Genetic- or transforming growth factor-beta1-induced changes in epidermal peroxisome proliferator-activated receptor beta/delta expression dictate wound repair kinetics.J. Biol. Chem. 280:18163–18170 [DOI] [PubMed] [Google Scholar]
- Tan N.S., Icre G., Montagner A., Bordier-ten-Heggeler B., Wahli W., Michalik L. 2007. The nuclear hormone receptor peroxisome proliferator-activated receptor beta/delta potentiates cell chemotactism, polarization, and migration.Mol. Cell. Biol. 27:7161–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Q., Smart R.C. 1999. Overexpression of protein kinase C-alpha in the epidermis of transgenic mice results in striking alterations in phorbol ester-induced inflammation and COX-2, MIP-2 and TNF-alpha expression but not tumor promotion.J. Cell Sci. 112(Pt 20):3497–3506 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Jose P.J., Rankin S.M. 2002. Eotaxin-2 generation is differentially regulated by lipopolysaccharide and IL-4 in monocytes and macrophages.J. Immunol. 168:1911–1918 [DOI] [PubMed] [Google Scholar]
- Wiener Z., Pocza P., Racz M., Nagy G., Tolgyesi G., Molnar V., Jaeger J., Buzas E., Gorbe E., Papp Z., et al. 2008. IL-18 induces a marked gene expression profile change and increased Ccl1 (I-309) production in mouse mucosal mast cell homologs.Int. Immunol. 20:1565–1573 [DOI] [PubMed] [Google Scholar]