Abstract
Polarity of the microtubule (MT) cytoskeleton is essential for many cell functions. Cytoplasmic linker–associated proteins (CLASPs) are MT-associated proteins thought to organize intracellular MTs and display a unique spatiotemporal regulation. In migrating epithelial cells, CLASPs track MT plus ends in the cell body but bind along MTs in the lamella. In this study, we demonstrate that glycogen synthase kinase 3β (GSK3β) directly phosphorylates CLASPs at multiple sites in the domain required for MT plus end tracking. Although complete phosphorylation disrupts both plus end tracking and association along lamella MTs, we show that partial phosphorylation of the identified GSK3β motifs determines whether CLASPs track plus ends or associate along MTs. In addition, we find that expression of constitutively active GSK3β destabilizes lamella MTs by disrupting lateral MT interactions with the cell cortex. GSK3β-induced lamella MT destabilization was partially rescued by expression of CLASP2 with mutated phosphorylation sites. This indicates that CLASP-mediated stabilization of peripheral MTs, which likely occurs in the vicinity of focal adhesions, may be regulated by local GSK3β inactivation.
Introduction
The microtubule (MT) network is important for many cell functions such as intracellular transport, cell division, and cell migration during development or tissue remodeling. In addition to actin-driven lamella/lamellipodia protrusion in the direction of migration (Small and Resch, 2005), migrating cells are characterized by orientation of the MT-organizing center and a large population of MTs toward the leading edge. This polarity of the MT network is thought to be important for directional migration (Wittmann and Waterman-Storer, 2001). MTs are dynamic polymers that stochastically switch between phases of polymerization and depolymerization (Howard and Hyman, 2003). Stabilization of lamella MTs is mediated by a bias of MT polymerization dynamics toward net growth downstream of intracellular signaling pathways (Wittmann et al., 2003), which may result from spatiotemporally regulated interactions of these MTs with the cortical cytoskeleton in the cell's lamella (Gundersen et al., 2004; Lansbergen and Akhmanova, 2006).
Plus end tracking proteins (+TIPs) are a heterogeneous class of proteins that specifically accumulate at or near the plus ends of growing MTs in cells (Galjart, 2005; Akhmanova and Steinmetz, 2008). Three different +TIP complexes have been suggested to participate in MT linkage to cortical sites. CLIP-170 may mediate lamella MT attachments through interactions with activated Rac1, IQGAP1, and components of the dynein–dynactin complex (Fukata et al., 2002; Lansbergen et al., 2004). Adenomatous polyposis coli protein may link lamella MTs to the actin cytoskeleton through EB1-mediated binding to the formin mDia1 (Wen et al., 2004), and cytoplasmic linker–associated proteins (CLASPs) may link lamella MTs to adhesion sites by interactions with the spectraplakin ACF7 and the phosphatidylinositol (3,4,5)-trisphosphate sensor LL5β (Drabek et al., 2006; Lansbergen et al., 2006; Wu et al., 2008). However, it is not understood how such MT attachment is regulated to specifically stabilize lamella MTs. In directionally migrating cells, CLASPs display a spatiotemporal gradient of intracellular distribution that correlates with the cell's direction of migration. In fibroblasts, CLASPs accumulate in cortical patches near the cell's leading edge (Drabek et al., 2006; Lansbergen et al., 2006). In epithelial cells, CLASPs associate along lamella MTs independently of their plus end tracking activity in the cell body (Wittmann and Waterman-Storer, 2005). In addition, CLASPs may anchor MTs at the Golgi apparatus (Efimov et al., 2007).
Many MT-associated proteins are negatively regulated by phosphorylation (Cassimeris and Spittle, 2001), and expression of constitutively active glycogen synthase kinase 3β (GSK3β) disrupts CLASP–MT association in cells (Akhmanova et al., 2001; Wittmann and Waterman-Storer, 2005). GSK3β substrates include several MT-associated proteins such as the adenomatous polyposis coli (APC) protein, the von Hippel-Lindau (VHL) protein, and neuronal MT-associated proteins (Zumbrunn et al., 2001; Cho and Johnson, 2003; Trivedi et al., 2005; Hergovich et al., 2006; Ferrarese et al., 2007), as well as other MT regulatory proteins (Yoshimura et al., 2005; Fumoto et al., 2006). Thus, GSK3β may be a master regulator of the MT cytoskeleton. In contrast to most other protein kinases, GSK3β activity is high in nonstimulated cells and is inhibited in the leading edge of migrating astrocytes and developing neurons (Etienne-Manneville and Hall, 2003; Yoshimura et al., 2006). Because this proposed gradient of GSK3β activity correlates well with the spatiotemporal distribution of CLASP–MT association in migrating cells, we investigated the regulation of CLASP2 by GSK3β in greater detail. In this study, we identify a cluster of GSK3β phosphorylation sites in the CLASP–MT plus end tracking domain and show that phosphorylation of a specific GSK3β motif switches CLASP activity between plus end tracking and association along MTs. In addition, our data indicate that GSK3β-mediated CLASP phosphorylation plays an important role in the cortical attachment and stabilization of lamella MTs in collectively migrating epithelial cells.
Results
GSK3β regulates lamella MT persistence in migrating epithelial cells
To analyze whether local GSK3β inactivation regulates lamella MT dynamics in migrating epithelial cells, we used an immortalized human keratinocyte cell line (HaCaT; Boukamp et al., 1988) because localization of endogenous CLASP along lamella MTs in these HaCaT cells was very similar to what we previously observed in PtK1 cells (Fig. 1 A; Wittmann and Waterman-Storer, 2005). In addition, cells at the edge of a HaCaT monolayer robustly polarized, formed extensive lamella, and migrated as an epithelial sheet retaining contacts with neighboring cells (Fig. S5 and Video 1). For the purpose of this paper, we define the lamella as the flat cell region in which CLASP is localized along MTs as opposed to the cell body in which CLASP is only associated with growing MT plus ends.
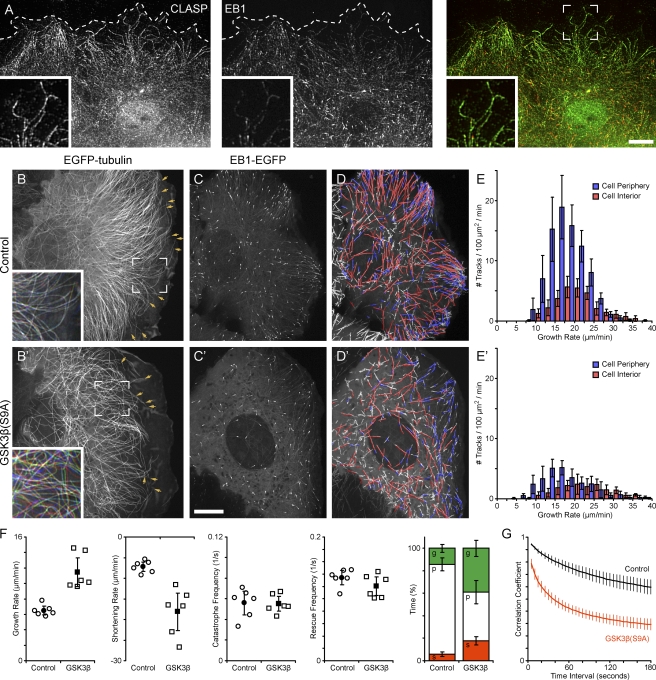
Figure 1.
Ectopic GSK3β activation increases lamella MT dynamics. (A) Immunofluorescence of endogenous CLASP and EB1 in migrating HaCaT keratinocytes. Dashed lines indicate the cell's leading edge. Insets show a peripheral cell region at a higher magnification. (B) Migrating HaCaT keratinocyte expressing EGFP-tubulin. (B and B′) Arrows indicate MTs that are part of the quantification in F, and insets show an overlay of images from the original time-lapse sequence acquired at 0, 30, and 60 s. The colors indicate lateral movement of MTs during this time period. (C) HaCaT cell expressing EB1-EGFP. (D) Computer-generated tracks of EB1 comets over 38 s (99 frames) overlaid on a maximum intensity projection of the entire sequence. Red indicates growth rates above and blue below the median. Only tracks with a lifetime of >4 s (10 frames) are shown. (E) Histogram of growth rates in the cell periphery (blue) and cell interior (red). (B–E) Control HaCaT cells. (B′–E′) HaCaT cells expressing constitutively active mRFP-GSK3β(S9A). (F) Analysis of lamella MT polymerization dynamics in control and GSK3β(S9A)-expressing cells. Open symbols indicate parameters calculated from individual cells, and closed symbols indicate means. The bar graph summarizes time MTs spent growing (g), pausing (p), or shortening (s). (G) Plot of the correlation coefficient between image regions in the lamella of EGFP-tubulin–expressing cells as a function of the time interval between images. Error bars indicate 95% confidence intervals; n = 6 cells. Bars, 10 µm.
MTs in the lamella of migrating HaCaT cells formed a dense array, were oriented roughly perpendicular to the cell edge, and displayed very little lateral movement (Fig. 1 B and Video 2). The number of these MTs seemed large compared with other epithelial cell types (Waterman-Storer and Salmon, 1997; Wittmann et al., 2003), indicating that HaCaT keratinocytes are a good model to study this lamella-specific MT population in truly migratory epithelial cells. Manual MT end tracking in EGFP-tubulin–expressing cells revealed that lamella MTs were relatively nondynamic and spent a large amount of time in pause with mostly short excursions of growth or shortening (Fig. 1 F and Table S1).
HaCaT cells transiently expressing constitutively active monomeric RFP (mRFP)–GSK3β(S9A) displayed significant morphological differences as compared with control cells (Fig. S1). These cells were generally bigger with a flatter lamella and migrated significantly slower than control cells (control, 13.4 ± 3.8 µm/h; GSK3β(S9A), 9.0 ± 4.1 µm/h; Fig. S5 and see Fig. 7 E). Although several aspects of cytoskeleton organization likely caused this phenotype, the most striking difference was a dramatically decreased density of lamella MTs in GSK3β(S9A)-expressing cells (Fig. 1 B and Fig. S1 A) and the absence of stable acetylated MTs (Fig. S1 B). This decrease in lamella MT density was accompanied by altered MT polymerization dynamics. Although transition frequencies between phases of growth and shortening were only decreased mildly in GSK3β(S9A)-expressing cells, both growth and shortening rates of lamella MTs were significantly increased, and MTs spent less time in pause and more time growing and shortening (Fig. 1 F and Table S1). In addition, lamella MTs in GSK3β(S9A)-expressing cells displayed increased lateral movement, buckling, and looping (Fig. 1, B and B′, compare insets; and Video 3). We confirmed this by image cross-correlation analysis in which the correlation coefficient between images at increasing time intervals decreased significantly more rapidly in GSK3β(S9A)-expressing cells than in control cells (Fig. 1 G).
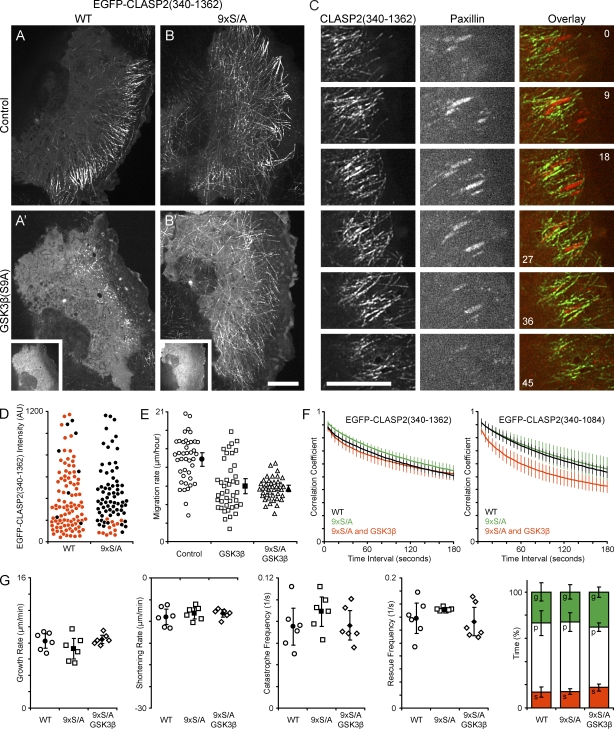
Figure 7.
CLASP2–MT interactions mediated by GSK3β inactivation contribute to lamella MT dynamics. (A) Migrating HaCaT keratinocyte expressing EGFP-CLASP2(340–1,362). (B) HaCaT cell expressing 9×S/A EGFP-CLASP2(340–1,362) containing mutated GSK3β phosphorylation sites. (A and B) Control HaCaT cells. (A′ and B′) HaCaT cells additionally expressing constitutively active mRFP-GSK3β(S9A) (insets). (C) Dynamics of EGFP-CLASP2(340–1,362) and mCherry-paxillin in focal adhesions in the leading lamella of a migrating HaCaT cell. Elapsed time is indicated in minutes. (D) Quantification of ectopic EGFP-CLASP2(340–1,362) association along cell body MTs of WT or the nonphosphorylatable 9×S/A construct as a function of cytoplasmic EGFP fluorescence intensity. Red symbols represent cells with plus end tracking only, and black symbols represent cells in which the CLASP construct is clearly detectable along cell body MTs. (E) Quantification of migration rates of control HaCaT cells and cells expressing GSK3β(S9A) alone or in combination with 9×S/A CLASP2(340–1,362). Open symbols represent individual cells, and closed symbols represent means. n = 42 cells. (F) Plot of the correlation coefficient between image regions in the lamella of cells expressing the indicated EGFP-CLASP2(340–1,362) or EGFP-CLASP(340–1,084) constructs as a function of the time interval between images. n = 6 cells. (G) Analysis of lamella MT polymerization dynamics by direct manual tracking of EGFP-CLASP2(340–1,362)–decorated MTs. Open symbols represent parameters calculated from individual cells, and closed symbols represent means of measurements from all six cells analyzed. The bar graph summarizes time MTs spent growing (g), pausing (p), or shortening (s). AU, arbitrary unit. Error bars indicate 95% confidence intervals. Bars, 10 µm.
Because we were not able to distinguish individual MT ends in the cell body of EGFP-tubulin–expressing cells, we used EB1-EGFP as a marker for growing MT plus ends to compare MT polymerization dynamics in the cell body and lamella of control and mRFP-GSK3β(S9A)–expressing cells (Videos 4 and 5). Computer vision tracking of a large population of EB1-labeled MT ends revealed that the density of growing MTs was significantly higher in the lamella of control cells as compared with the cell body (Fig. 1, C–E). In GSK3β(S9A)-expressing cells, MT density was significantly reduced 1.7-fold in the cell body and threefold in the lamella (Fig. 1, E and E′). In both control and GSK3β(S9A)-expressing cells, the growth rate of lamella MTs (control, 18.0 ± 0.8 µm/min; GSK3β(S9A), 17.3 ± 2.9 µm/min) was reduced as compared with MTs in the cell body (control, 21.4 ± 1.4 µm/min; GSK3β(S9A), 24.1 ± 1.6 µm/min), which is similar to what we previously observed in Ptk1 cells (Wittmann and Waterman-Storer, 2005). However, the growth rate of lamella MTs in control and GSK3β(S9A)-expressing cells was not significantly different. Because the sampling frequency used for EB1 tracking was more than 10-fold faster than for direct MT end tracking, this indicates that the increase in growth rate observed by MT end tracking (Fig. 1 F) is likely a result of the decreased amount of time lamella MTs spend in the pause state in GSK3β(S9A)-expressing cells.
GSK3β phosphorylates CLASP2 in the MT plus end tracking domain
CLASP knockdown by RNA interference results in decreased MT density in the cell periphery in both HeLa and HaCaT cells (Mimori-Kiyosue et al., 2005; Fig. S2). Because this phenotype is similar to cells expressing constitutively active GSK3β(S9A), and because CLASP2–MT association is disrupted in GSK3β(S9A)-expressing cells (Akhmanova et al., 2001; Wittmann and Waterman-Storer, 2005), we next examined GSK3β-dependent CLASP phosphorylation. We focused our analysis on CLASP2 phosphorylation because this is the main CLASP expressed in HaCaT cells (Fig. S3).
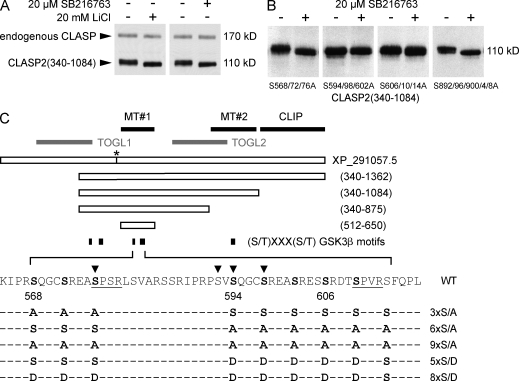
We used a gel shift assay in which we compared the relative migration of CLASP in control HeLa cells and cells treated with two different GSK3β inhibitors. Both LiCl and SB216763 induced a downshift of the endogenous CLASP band and of EGFP-CLASP2(340–1,084), indicating GSK3β-mediated CLASP phosphorylation and high GSK3β activity in nonstimulated cells (Fig. 2 A). Because CLASP2 contains several potential GSK3β phosphorylation motifs (Fig. 2 C), we used truncated CLASP2 constructs to define the parts of the CLASP molecule phosphorylated by GSK3β. We found that only constructs containing the MT plus end tracking domain (MT#1; Fig. 2 C) between amino acid 512 and 650 displayed a GSK3β-dependent gel shift (unpublished data). This region contains nine serine residues constituting GSK3β consensus motifs (Fig. 2 C), which we replaced by nonphosphorylatable alanines in groups of three. S594/598/602A or S606/610/614A abolished the GSK3β inhibitor–induced shift (Fig. 2 B). In contrast, the gel shift was not affected by mutation of S568/572/576A or of a potential GSK3β motif in the domain required for lamella MT binding (MT#2; Fig. 2 C; Wittmann and Waterman-Storer, 2005).
Figure 2.
Identification of physiological CLASP2 GSK3β phosphorylation sites. (A) Immunoblot of HeLa cell lysates probed with a polyclonal CLASP antibody (Hannak and Heald, 2006). Treatment with GSK3β inhibitors (20 µM SB216763 or 10 mM LiCl) results in dephosphorylation (downshift) of endogenous CLASP and transfected EGFP-CLASP2(340–1,084) that contains all potential GSK3β sites. (B) Mutation of GSK3β motifs within the MT plus end tracking domain identifies the motif between S594 and S614 as responsible for the GSK3β inhibitor–induced gel shift. (C) Diagram of constructs and phosphorylation site mutants used in this study. Bolded letters indicate serine residues identified to be phosphorylated by GSK3β. Predicted Cdk consensus motifs are underlined. Arrowheads indicate phosphorylated residues identified by mass spectrometry phosphoproteomics (Trinidad et al., 2006; Matsuoka et al., 2007; Dephoure et al., 2008; Imami et al., 2008). The numbering of amino acid positions corresponds to the old National Center for Biotechnology Information reference sequence XP_291057.5. This record has been replaced by NP_055912, which has a longer N terminus. However, we have kept the numbering for consistency with our previous paper (Wittmann and Waterman-Storer, 2005). The asterisk indicates an eight–amino acid difference between the CLASP2 clone used in this study and XP_291057.5. Black bars indicate functionally defined domains: MT#1, MT plus end tracking and EB1-binding domain; MT#2, region required for binding along lamella MTs; CLIP, C-terminal domain required for association with CLIP-170 and LL5β. Gray bars indicate putative TOG-like domains (Slep and Vale, 2007).
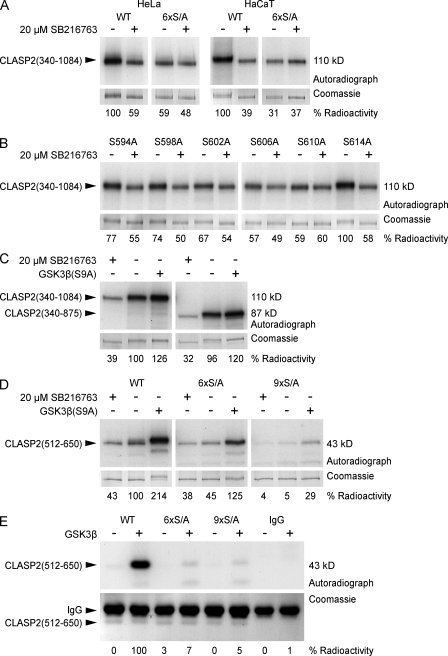
To more directly analyze phosphorylation of GSK3β motifs in MT#1, we performed metabolic labeling experiments. In both HeLa and HaCaT cells, SB216763 dramatically decreased EGFP-CLASP2(340–1,084) phosphorylation (Fig. 3 A). In contrast, phosphorylation of mutated EGFP-CLASP2(340–1,084) in which S594/598/602/606/610/614 were replaced with alanines (6× serine to alanine mutation [S/A]) was severely reduced and did not decrease further in response to SB216763. Because this construct contains all other potential GSK3β motifs, it indicates that this is the main physiologically relevant GSK3β-dependent phosphorylation. Because residual phosphorylation of the 6×S/A construct was GSK3β inhibitor independent, it is likely caused by phosphorylation by other kinases.
Figure 3.
Analysis of CLASP2 phosphorylation. (A–D) Metabolic labeling of tissue culture cells with [32P]-labeled phosphate. EGFP-tagged CLASP2 constructs were immunoprecipitated and analyzed by SDS-PAGE. Top panels show autoradiograph, and bottom panels show the corresponding Coomassie-stained gel as loading control. Quantification of radioactivity incorporation by densitometry is shown below the gel images. (A) In both HeLa and HaCaT cells, GSK3β inhibition with 20 µM SB216763 decreases CLASP2(340–1,084) phosphorylation. Mutation of the GSK3β motif between S594 to S614 (6×S/A) eliminates GSK3β-dependent phosphorylation. (B) Mutation of individual serine residues between S594 and S614 shows that S614 is not part of the motif and reveals hierarchical phosphorylation by GSK3β. (C) The domain required for CLASP2 association along lamella MTs (875–1,084) is not required for efficient phosphorylation by GSK3β. (D) Combined mutation of the GSK3β motifs between S568 to S576 and S594 to S614 (9×S/A) is required to completely abolish phosphorylation of the MT plus end tracking domain CLASP2(512–650) by constitutively active GSK3β(S9A). (E) In vitro phosphorylation of immunoprecipitated EGFP-CLASP2(512–650) by purified GSK3β in the presence of γ-[32P]ATP.
To further dissect phosphorylation of this motif, we replaced each individual serine with alanine and analyzed phosphorylation with and without SB216763 (Fig. 3 B). S614A, which is not conserved in CLASP1, did not affect GSK3β-dependent phosphorylation, indicating that it is not part of the motif. In addition, mutation of serines at the C-terminal end of the motif had a larger impact on GSK3β-dependent phosphorylation than mutation of more N-terminal serine residues consistent with the proposed sequential mechanism of GSK3β phosphorylation (Frame et al., 2001).
We next analyzed phosphorylation of EGFP-CLASP2(340–875) that specifically lacks MT#2 required for CLASP binding along lamella MTs. EGFP-CLASP2(340–875) was phosphorylated to a similar extent as EGFP-CLASP2(340–1,084), phosphorylation was inhibited by SB216763, and coexpression of mRFP-GSK3β(S9A) led to a small increase in phosphorylation of both constructs. This indicates that association along lamella MTs is not required for GSK3β-dependent phosphorylation (Fig. 3 C).
Although our initial gel shift experiments indicated that the GSK3β motif at S568 was not phosphorylated under physiological conditions (Fig. 2 B), this sequence is an exact duplication of the N-terminal part of the motif at S594 (Fig. 2 C). We therefore tested whether this motif can be phosphorylated in cells expressing constitutively active GSK3β(S9A) (Fig. 3 D). A construct containing only the plus end tracking domain (MT#1), EGFP-CLASP2(512–650), was less efficiently phosphorylated by endogenous GSK3β than the longer constructs. However, coexpression of mRFP-GSK3β(S9A) boosted phosphorylation more than twofold. The 6×S/A construct was still phosphorylated to a reduced extent in the presence of mRFP-GSK3β(S9A), indicating that GSK3β can phosphorylate additional sites in the plus end tracking domain. However, additional S568/572/576A mutation (9×S/A) abolished phosphorylation even in mRFP-GSK3β(S9A)–expressing cells.
Finally, we tested whether CLASP2 is a direct GSK3β substrate. To retain potential priming phosphorylation, instead of using bacterially expressed protein, we immunoprecipitated EGFP-tagged CLASP2(512–650) from HeLa cell lysates and performed in vitro kinase assays with purified GSK3β (Fig. 3 E). As compared with the wild-type (WT) protein, phosphorylation of both the 6×S/A and 9×S/A mutant was greatly reduced. Together, these results demonstrate that the S594/598/602/606/610 motif is a physiological target of GSK3β and is highly phosphorylated in nonstimulated cells. However, a partial duplication of this motif at S568/572/576 can be phosphorylated by high levels of intracellular GSK3β activity and may contribute to the regulation of CLASP function.
GSK3β phosphorylation modulates CLASP2–MT binding in cells
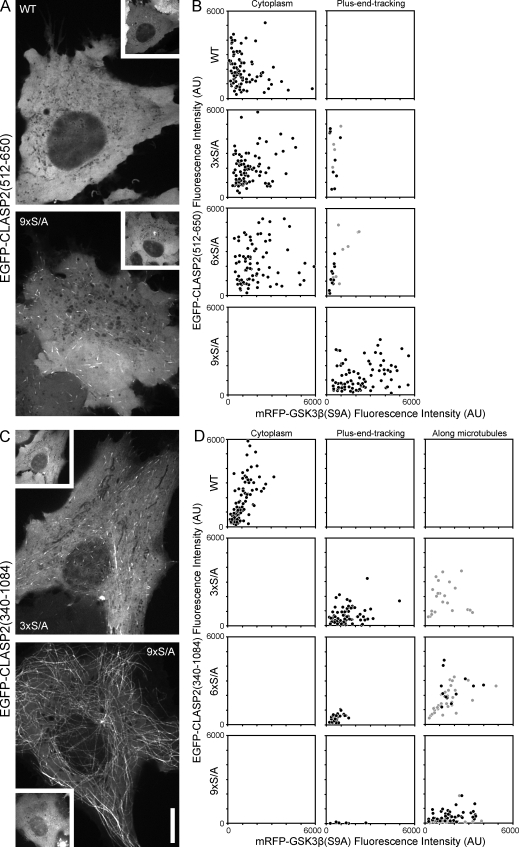
The CLASP2 domain between amino acid 512 and 650 containing the GSK3β phosphorylation sites identified in the previous section is sufficient for MT plus end tracking in cells (Mimori-Kiyosue et al., 2005; Fig. 2 C, MT#1). To test the functional significance of these sites for CLASP2–MT interactions in vivo, we analyzed whether EGFP-CLASP2(512–650) constructs with mutated phosphorylation sites rescued CLASP2 plus end tracking in HeLa cells expressing constitutively active mRFP-GSK3β(S9A). We analyzed cells in a wide range of fluorescence intensity levels to correct for variations in CLASP2 phosphorylation as a result of relative differences of GSK3β(S9A) and EGFP-CLASP(512–650) expression in individual transfected cells (Fig. 4, A and B). Even at the detection limit of GSK3β(S9A) expression, MT plus end association of WT CLASP2(512–650) was completely disrupted. In contrast, the 9×S/A construct in which all GSK3β sites are replaced with nonphosphorylatable alanine residues always tracked MT plus ends even at the highest GSK3β(S9A) expression levels (Fig. 4 A). Both of the constructs with only one of the GSK3β motifs mutated, 3×S/A and 6×S/A, rescued MT plus end association only at very low GSK3β(S9A) expression levels. This indicates that GSK3β phosphorylation of either motif is sufficient to inhibit the CLASP2 plus end tracking domain.
Figure 4.
Phosphorylation site mutations rescue CLASP2–MT binding in cells expressing constitutively active GSK3β(S9A). (A) Representative examples of HeLa cells expressing WT or mutated EGFP-CLASP2(512–650) containing only the plus end tracking domain MT#1 in combination with mRFP-GSK3β(S9A) (insets). WT EGFP-CLASP2(512–650) is completely cytoplasmic in cells expressing constitutively active GSK3β(S9A), whereas the 9×S/A construct robustly tracks MT plus ends. (B) Scatter plots of cells with different expression levels of EGFP-CLASP2(512–650) phosphorylation site mutants and of constitutively active mRFP-GSK3β(S9A) categorized by whether the CLASP2 construct was detectable on MT plus ends or not. Each symbol represents the mean cytoplasmic EGFP and mRFP fluorescence intensities of an individual HeLa cell. Gray symbols indicate cells in which plus end tracking was barely detectable. (C) Representative examples of HeLa cells expressing mutated EGFP-CLASP2(340–1,084) containing both MT#1 as well as MT#2 required for CLASP association along lamella MTs in combination with mRFP-GSK3β(S9A) (insets). Mutation of the first GSK3β motif, 3×S/A, completely rescues plus end tracking but has little effect on binding along MTs. Mutation of all GSK3β sites, 9×S/A, is necessary to rescue binding along MTs, and this construct shows almost no preference for plus ends. (D) Scatter plots of cells with different expression levels of EGFP-CLASP2(340–1,084) phosphorylation site mutants and of constitutively active mRFP-GSK3β(S9A) categorized by whether the CLASP2 construct was only in the cytoplasm, on MT plus ends, or bound along MTs. Each symbol represents the mean cytoplasmic EGFP and mRFP fluorescence intensities of an individual HeLa cell. Gray symbols indicate cells with only weak binding along MTs. All images were taken at identical illumination and exposure settings. The axes on all graphs are scaled identically. AU, arbitrary unit. Bar, 10 µm.
Because constitutively active GSK3β(S9A) also disrupts CLASP2 binding along lamella MTs (Wittmann and Waterman-Storer, 2005), we next analyzed how partial GSK3β phosphorylation affected MT association of EGFP-CLASP2(340–1,084), which also contains MT#2 required for CLASP association along lamella MTs (Fig. 4, C and D). As expected, GSK3β(S9A) expression completely disrupted MT association of WT CLASP2(340–1,084). Surprisingly, mutation of either the motif at S568 (3×S/A) or S594 (6×S/A), which had little effect on the plus end tracking domain alone (Fig. 4 B), completely restored plus end tracking even at high mRFP-GSK3β(S9A) expression levels, indicating that MT#2 contributes to plus end tracking. Complete mutation of the GSK3β motif 9×S/A rescued binding along MTs (Fig. 4 D). In most cells, 9×S/A EGFP-CLASP2(340–1,084) decorated the entire MT cytoskeleton, and there was no discernable difference between CLASP2 association to MT plus ends or along MTs (Fig. 4 C). Such binding along MTs was never observed with the short EGFP-CLASP2(512–650) construct containing only the plus end tracking domain MT#1. In addition, a construct specifically lacking MT#2 did not rescue binding along MTs and behaved identical to the plus end tracking domain alone (Fig. S4).
These results are summarized in Table S2 and demonstrate that both MT#1 and MT#2 function synergistically and that the phosphorylation state of the GSK3β motif at S594-S610, which is highly phosphorylated by endogenous GSK3β, determines whether CLASP2 tracks MT plus ends or binds along MTs. Additional phosphorylation of the GSK3β motif at S568-S576, which is not phosphorylated by endogenous GSK3β levels, completely inhibits MT association.
GSK3β phosphorylation directly inhibits the CLASP2 plus end tracking domain
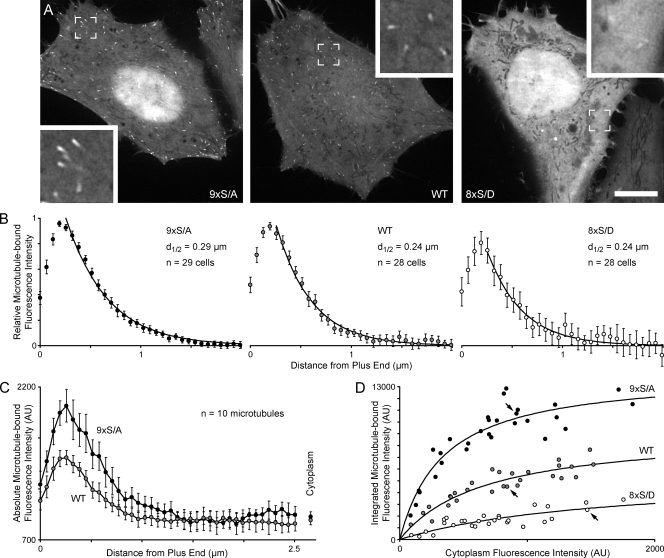
To test whether endogenous levels of GSK3β regulate the CLASP2 plus end tracking domain, we compared WT EGFP-CLASP2(512–650), the 9×S/A construct, and a construct in which the GSK3β sites were replaced with phosphomimetic aspartate residues (8× serine to aspartate mutation [S/D]) in HeLa cells. Both WT CLASP2(512–650) and the 9×S/A construct clearly tracked MT plus ends in all cells observed (Fig. 5 A). In contrast, the 8×S/D construct largely reproduced the phenotype observed in GSK3β(S9A)-expressing cells. 8×S/D only bound very weakly to MT ends and in many cells could not be detected on MTs at all.
Figure 5.
GSK3β phosphorylation modulates MT affinity of the CLASP2 plus end tracking domain. (A) Representative examples of HeLa cells expressing WT or mutated EGFP-CLASP2(512–650) as indicated. Images were taken at identical illumination and exposure settings. Insets show MT plus ends at a higher magnification. (B) Normalized mean EGFP-CLASP2(512–650) fluorescence profile along MT plus ends showing that mutation of GSK3β phosphorylation sites does not affect the decay of CLASP2(512–650) binding with increasing distance from plus ends. Fluorescence intensities are normalized to the maximum MT-bound fluorescence intensity (1) and the background in the surrounding cytoplasm (0). Three MTs were measured per cell. The number of cells per condition is indicated on the graphs. The measurement error for the 8×S/D construct is large because MT binding of this construct was very weak. d1/2 indicates the distance from the MT plus end at which half of the EGFP-CLASP construct has dissociated from the growing plus end as determined by least-square curve fitting to a single exponential decay function (solid lines). (C) Absolute EGFP-CLASP2(512–650) fluorescence profile along MT plus ends in cells with similar expression levels (the same cells as in A). Although the fluorescence decay constant is similar, the absolute amount of plus end–bound 9×S/A is significantly larger than of WT EGFP-CLASP2(512–650). (D) Integrated fluorescence intensity from absolute fluorescence profiles as in C plotted against the mean cytoplasmic fluorescence, indicating quantitative differences in MT binding of the different mutants. This represents the same dataset as in B, but profiles were not normalized. Each circle represents the mean of three MTs per cell. Arrows indicate the cells shown in A. Solid lines show least-square fits to a hyperbolic binding isotherm. AU, arbitrary unit. Error bars indicate 95% confidence intervals. Bar, 10 µm.
Because phosphorylation has been proposed to regulate +TIP release from growing MT ends (Vaughan, 2004), we measured the decay of the EGFP-CLASP2(512–650) fluorescence intensity profile as a function of increasing distance from the MT end (Fig. 5 B). To account for different expression levels, we normalized these intensity profiles relative to the maximum brightness on the MT end. A least-square fit of an exponential decay function to the mean fluorescence profile of many MT ends (∼85 MTs per condition) showed that the decay constants were very similar for WT, nonphosphorylatable, and pseudophosphorylated CLASP2(512–650), indicating that phosphorylation does not directly regulate CLASP2 release from growing plus ends.
Because plus-end comets of the 9×S/A construct consistently appeared brighter than WT, we next analyzed the absolute amount of fluorescence on MT ends. Absolute intensity decay profiles of a mean of 10 MTs revealed significantly more 9×S/A on plus ends compared with WT EGFP-CLASP2(512–650) in cells expressing similar levels of these constructs (Fig. 5 C). In addition, plotting of the integrated intensity of such profiles as a function of expression level showed that plus end association of WT CLASP2(512–650) was in between that of the 9×S/A and the 8×S/D constructs (Fig. 5 D). The binding curves of all three constructs were fitted reasonably well by a single-site hyperbolic binding equation, although both the apparent Kd and maximal binding decreased with increasing phosphorylation state.
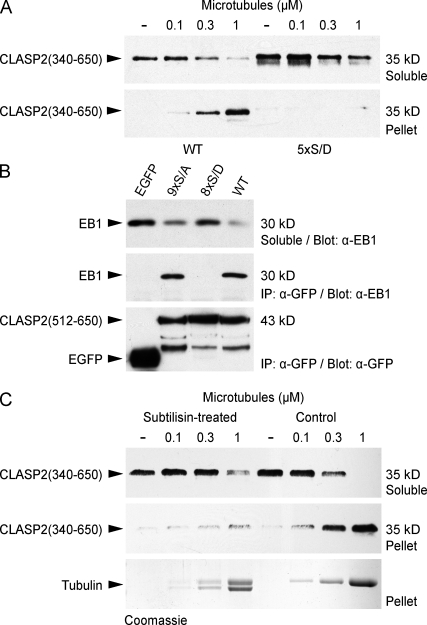
To test whether phosphorylation of the CLASP2 plus end tracking domain directly affected MT affinity, we performed in vitro MT sedimentation assays with bacterially expressed purified protein. Because we were not able to achieve sufficient GSK3β phosphorylation of CLASP2 protein in vitro, we instead compared MT binding of WT 6×His-CLASP2(340–650) and a construct in which the GSK3β motif between S594 and S610 was replaced with phosphomimetic aspartate residues (5×S/D; Fig. 2 C). WT CLASP2(340–650) bound to taxol-stabilized MTs with high affinity (apparent Kd < 0.3 µM), which is similar to what we observed previously (Wittmann and Waterman-Storer, 2005). In contrast, the 5×S/D mutant was not recovered in the MT pellet up to a MT concentration of 1 µM, indicating that binding affinity was decreased by at least 10-fold (Fig. 6 A).
Figure 6.
Binding of the CLASP2 plus end tracking domain to the tubulin C terminus and EB1 is directly inhibited by GSK3β phosphorylation. (A) Sedimentation assay of 6×His-CLASP2(340–650) at constant concentration (0.5 µM) with an increasing concentration of taxol-stabilized MTs. Comparison of the WT with a mutated protein in which the GSK3β sites between S594 and S610 were replaced with phosphomimetic aspartate residues (5×S/D) shows that the phosphomimetic mutant does not bind MTs. (B) Immunoprecipitation (IP) using GFP antibodies from HeLa cells expressing EGFP-tagged CLASP2–MT plus end tracking domain constructs. Endogenous EB1 only immunoprecipitates with WT EGFP-CLASP2(512–650) and the nonphosphorylatable mutant 9×S/A but not with pseudophosphorylated 8×S/D or EGFP alone. (C) Sedimentation assay using MTs treated with subtilisin, which removes the flexible tubulin C terminus, resulting in a downshift of the α/β-tubulin bands on the Coomassie-stained gel. 6×His-CLASP2(340–650) does not bind to subtilisin-treated MTs.
The CLASP2 plus end tracking domain also interacts with EB1 (Mimori-Kiyosue et al., 2005). To test whether phosphorylation affects EB1 binding, we immunoprecipitated WT and mutant EGFP-CLASP2(512–650) from HeLa cell lysates (Fig. 6 B). Endogenous EB1 immunoprecipitated with WT CLASP2(512–650) and with the 9×S/A construct in which all GSK3β sites were replaced with nonphosphorylatable alanine residues. In contrast, no EB1 was recovered with a construct in which GSK3β sites were replaced with phosphomimetic aspartate residues (8×S/D), indicating that CLASP2 phosphorylation also inhibits EB1 binding. Consistent with these data, we were unable to immunoprecipitate EB1 with longer CLASP2 constructs (not depicted), which may be more efficiently phosphorylated by endogenous GSK3β levels than EGFP-CLASP2(512–650) (Fig. 3, C and D).
Because the C termini of tubulin and EB1 are strikingly similar (Komarova et al., 2005) and CLASPs do not bind C-terminally truncated EB1 (Mimori-Kiyosue et al., 2005), we hypothesized that the CLASP2 plus end tracking domain interacts with the tubulin C terminus. To test this, we removed tubulin C termini by limited subtilisin proteolysis of taxol-stabilized MTs, which was accompanied by a characteristic downshift of the tubulin band by SDS–PAGE (Knipling et al., 1999). MTs were still recovered in the pellet after centrifugation, confirming that MTs remained assembled during subtilisin treatment. However, 6×His-CLASP2(340–650) no longer bound to subtilisin-treated MTs (Fig. 6 C). Together, these results indicate that the CLASP2 plus end tracking domain binds to the negatively charged C termini of MTs and EB1 and that this interaction is directly inhibited by GSK3β multisite phosphorylation.
GSK3β inactivation is required for CLASP2-mediated lamella MT attachment
Because GSK3β may be specifically inactivated in the leading edge of migrating cells (Etienne-Manneville and Hall, 2003), we hypothesized that such a GSK3β gradient may be the reason for the observed spatiotemporal gradient of CLASP2–MT association. CLASP2 plus end tracking in the cell body may be the result of high GSK3β activity. In contrast, CLASP2 association along lamella MTs may be the result of local GSK3β inactivation. To test this, we compared the dynamics of WT and 9×S/A EGFP-CLASP2(340–1,362) in migrating HaCaT keratinocytes. Similar to endogenous CLASP (Fig. 1 A), WT CLASP2(340–1,362), which contains the complete CLASP2 C terminus, tracked MT plus ends in the cell body and associated along MTs in the cell's protruding lamella (Fig. 7 A and Video 6). In contrast, nonphosphorylatable 9×S/A CLASP2(340–1,362) also bound along cell body MTs (Fig. 7, B and D), indicating that GSK3β phosphorylation is required for the spatiotemporal gradient of CLASP–MT association in migrating epithelial cells.
We next tested whether mutation of the GSK3β phosphorylation sites was sufficient to rescue defects mediated by expression of constitutively active GSK3β(S9A). As expected, MT binding of WT EGFP-CLASP2(340–1,362) was completely disrupted in mRFP-GSK3β(S9A)–expressing cells (Fig. 7 A′; Akhmanova et al., 2001; Wittmann and Waterman-Storer, 2005). In contrast, mutation of the GSK3β phosphorylation sites in 9×S/A CLASP2(340–1,362) rescued MT binding (Fig. 7 B′). However, even in these cells in which a hypothetical CLASP2 phosphorylation gradient is doubly disrupted, we often observed a residual gradient of CLASP2–MT association. This indicates that mechanisms in addition to GSK3β phosphorylation contribute to the spatiotemporal regulation of CLASP2–MT association in migrating epithelial cells. In addition, MT density was still decreased in these cells, and cell migration rate was not rescued, which is consistent with GSK3β phosphorylation of other targets important for cell migration and MT regulation (Fig. 7 E and Fig. S5).
However, we found that MTs decorated with 9×S/A CLASP2(340–1,362) in GSK3β(S9A)-expressing cells displayed less lateral movement than MTs in GSK3β(S9A)-expressing cells (Video 7). Consistent with this, image cross-correlation analysis revealed no significant difference in the time-dependent decay of the correlation coefficient between cells expressing WT or 9×S/A EGFP-CLASP2(340–1,362) or cells expressing both mRFP-GSK3β(S9A) and 9×S/A EGFP-CLASP2(340–1,362) (Fig. 7 F). In contrast, 9×S/A EGFP-CLASP2(340–1,084) did not rescue GSK3β(S9A)-induced lateral MT movement, indicating that the CLASP2 C terminus is required to anchor lamella MTs to the cortical cytoskeleton.
In addition, in the lamella of migrating HaCaT keratinocytes, we often observed clusters of EGFP-CLASP2(340–1,362), which remained stationary relative to the substrate reminiscent of adhesion dynamics in migrating cells (Video 8). Such clusters were largely absent in cells expressing EGFP-CLASP2(340–1,084) (Video 9). Time-lapse analysis of EGFP-CLASP2(340–1,362) in combination with mCherry-paxillin as a focal adhesion marker further revealed that CLASP-decorated MTs engulf focal adhesions (Fig. 7 C and Video 10) and that focal adhesion and CLASP dynamics are spatiotemporally correlated during epithelial cell migration.
To test how such CLASP-mediated MT attachment to cortical sites affects MT polymerization dynamics, we directly tracked EGFP-CLASP2–decorated lamella MT ends. MT dynamic instability parameters determined in this way were very similar to what we measured in EGFP-tubulin–expressing cells, and both 9×S/A EGFP-CLASP2(340–1,362) and 9×S/A EGFP-CLASP2(340–1,084) largely rescued GSK3β(S9A)-induced MT polymerization dynamics defects (Fig. 7 G and Table S1). This indicates that nonphosphorylated CLASP2(340–1,084) has a stabilizing effect on MTs, which is independent of MT interactions with the cell cortex mediated by the C-terminal domain.
Discussion
CLASP phosphorylation by GSK3β
In this study, we show that CLASPs are physiological GSK3β substrates. We used a candidate approach in which we replaced GSK3β consensus motifs with nonphosphorylatable alanine residues to identify sites that are phosphorylated by endogenous levels of GSK3β. Although CLASP2 contains several potential GSK3β motifs, mutation of five serine residues between S594 and S610 abolished GSK3β-dependent phosphorylation in different cell types. Furthermore, we found that a partial duplication of the N-terminal half of this motif between S568 and S576 can be phosphorylated by elevated GSK3β activity levels but may only have a small contribution to endogenous CLASP2 phosphorylation. These GSK3β phosphorylation motifs are highly conserved in all mammalian CLASPs, and phosphorylation of some of these sites has been observed by mass spectrometry proteomic approaches (Fig. 2 C).
Phosphorylation of most GSK3β substrates depends on priming phosphorylation by a different protein kinase (Jope and Johnson, 2004). We currently do not know whether CLASP2 phosphorylation by GSK3β requires priming phosphorylation, but it is intriguing that both C-terminal residues of the functional GSK3β motifs (S610 and S576) are near perfect matches to the Cdk consensus (S/T)PX(K/R) (Fig. 2 C). In addition, inefficient phosphorylation of bacterially produced CLASP2 protein in vitro suggests that priming phosphorylation is required (unpublished data). Although Cdk5 may act as a priming kinase for other MT regulatory proteins (Cho and Johnson, 2003; Yoshimura et al., 2005), different Cdk inhibitors did not decrease CLASP phosphorylation in HeLa cells (unpublished data).
Regulation of CLASP–MT interactions
All functional GSK3β phosphorylation sites reside within the CLASP2 domain required for MT plus end tracking and association with EB1 (Mimori-Kiyosue et al., 2005), and we found that both of these activities are inhibited by GSK3β-mediated phosphorylation. Our experiments using subtilisin-treated MTs further indicate that MT binding of the CLASP2 plus end tracking domain requires the negatively charged tubulin C terminus. However, CLASP plus end tracking does not require the very C-terminal tubulin tyrosine residue (Peris et al., 2006). Thus, because the CLASP plus end tracking domain contains numerous strongly positively charged arginine and lysine residues, it is possible that the interaction of this short domain with MTs is primarily electrostatic.
Multisite phosphorylation by GSK3β could gradually disrupt this electrostatic glue by introducing multiple strongly negatively charged phosphate groups. Because a single phosphorylation may not be sufficient to disrupt the interaction of many charged residues, such a mechanism could explain the role of GSK3β as an amplifier of an initial priming kinase-dependent signaling event. The positive feedback achieved by hierarchical phosphorylation may contribute to a steeper response of such an electrostatic switch (Roach, 1991; Ferrarese et al., 2007). The role of GSK3β as a signal amplifier rather than a signal transducer could further explain why GSK3β is involved in such a wide variety of seemingly unrelated intracellular processes (Jope and Johnson, 2004).
Intracellular dynamics of full-length CLASPs are complex, and CLASPs can directly or indirectly interact with MTs through at least three different domains. Although TOG-like domains, which may mediate interactions with MTs, only partially overlap with functionally defined domains involved in MT interactions (Mimori-Kiyosue et al., 2005; Wittmann and Waterman-Storer, 2005; Slep and Vale, 2007; Fig. 2 C), differential regulation of multiple weak interactions may determine whether CLASPs track MT plus ends or bind along MTs. We propose a model in which partial phosphorylation of the motif at S594 inhibits MT#2, the domain required for association along MTs, through a long-range conformational change before directly affecting the plus end tracking domain (MT#1). Because MT#2 has very weak MT-binding activity on its own (Wittmann and Waterman-Storer, 2005), it may not be involved directly in MT binding. Only complete phosphorylation of both GSK3β motifs in the presence of elevated GSK3β activity disrupted CLASP2(340–1,084) plus end tracking, and it remains to be determined whether this occurs under physiological conditions.
Consistent with a model that partial phosphorylation may distinguish between different modes of CLASP–MT association, complete phosphorylation of all eight GSK3β sites disrupted both plus end tracking and binding along lamella MTs, and mutation of all GSK3β sites rescued both activities in cells expressing elevated GSK3β levels. However, most importantly, mutation of the first GSK3β motif that is not phosphorylated by physiological GSK3β levels preferentially rescued plus end tracking. This strongly indicates that in cells, the physiologically relevant GSK3β motif at S594 is responsible for regulating CLASP–MT association in the lamella. Thus, the observed gradient of CLASP–MT association is likely the result of different CLASP phosphorylation states as a result of GSK3β inactivation in the protruding lamella (Etienne-Manneville and Hall, 2003), although additional mechanisms may be involved.
It is possible that the two CLASP–MT-binding domains recognize slightly different tubulin conformations (Nogales and Wang, 2006), resulting in different affinities of these domains for MT plus ends and MT lattices. In this case, CLASP plus end tracking would simply be an equilibrium binding process depending on a structural property of growing MT ends. This is entirely consistent with the observed rapid exchange of plus end–bound and soluble +TIPs in cells (Wittmann and Waterman-Storer, 2005; Dragestein et al., 2008), recent in vitro studies (Bieling et al., 2008; Wittmann, 2008), and our observation that the EGFP-CLASP2(512–650) plus end comet length does not depend on the phosphorylation state.
CLASP function in migrating cells
In migrating HaCaT keratinocytes, CLASP2-decorated lamella MTs often persisted in immobile clusters as the cell migrated across. We show that these clusters of CLASP-decorated lamella MTs are closely associated with focal adhesions. In addition, CLASP and focal adhesion dynamics were spatiotemporally correlated, and focal adhesion turnover preceded the disappearance of CLASP-decorated MTs. Thus, GSK3β and CLASPs may play a role in adhesion site turnover consistent with the observation that GSK3β inhibition freezes focal adhesion dynamics (Kobayashi et al., 2006).
Because cortical interactions are mediated by the CLASP C terminus, CLASP-interacting proteins such as LL5β or ACF7 (Drabek et al., 2006; Lansbergen et al., 2006) are likely involved. We speculate that interactions of CLASP-decorated MTs with focal adhesions contribute to lamella MT stability, although our results suggest that CLASPs also influence MT polymerization dynamics directly, independent of interactions of the C terminus. Because the spectraplakin ACF7 guides MTs along actin cables in keratinocytes (Wu et al., 2008), it is likely that CLASPs and ACF7 cooperate to organize the MT cytoskeleton in migrating epithelial cells.
Punctate cortical structures often remained in cells with elevated levels of GSK3β activity, suggesting that cortical interactions may not be regulated directly by GSK3β phosphorylation. In cells expressing constitutively active GSK3β, MTs cannot interact with phosphorylated CLASP at these cortical sites, resulting in increased lateral MT movements, altered MT polymerization dynamics, and decreased MT stability as indicated by decreased acetylation and detyrosination (Eng et al., 2006). Consistent with this hypothesis, both CLASP RNA interference and ACF7-null cells displayed defects in peripheral MT density and dynamics that were qualitatively similar to the expression of constitutively active GSK3β(S9A) (Kodama et al., 2003; Mimori-Kiyosue et al., 2005). Interactions of nonphosphorylated CLASPs with the cell cortex likely occur along the sides of lamella MTs, which represent a more robust mechanism of lamella MT stabilization than direct capture of highly dynamic MT ends. Additional stabilization of CLASP lamella MT binding through interactions of the CLASP C terminus with the cell cortex may also explain the residual gradient of CLASP–MT interactions in cells in which the GSK3β activity gradient was disrupted. Alternatively, gradients of other signaling molecules may affect CLASP–MT binding, for example, by modulating priming phosphorylation (Ridley et al., 2003).
Materials and methods
Constructs and adenoviral vectors
pEGFP-CLASP2 and other constructs were used as described previously (Wittmann and Waterman-Storer, 2005). Phosphorylation site point mutations were generated with QuikChange II (Agilent Technologies) and verified by sequencing. Replication-deficient adenovirus particles for transient HaCaT transfection experiments were produced using the AdEasy adenoviral vector system (Agilent Technologies). EGFP-CLASP2 constructs, EGFP-tubulin, and mRFP-GSK3β(S9A) were amplified by PCR using Vent polymerase (New England Biolabs, Inc.) and cloned into pShuttle-CMV (Agilent Technologies) between KpnI and NotI restriction sites. Homologous recombination with pAdEasy-1 and production of adenovirus particles was performed according to the manufacturer's instructions. Adenovirus particles were purified by CsCl2 density gradient ultracentrifugation following published procedures (Tollefson et al., 2007). Purified virus was stored in aliquots in 10 mM Tris-HCl, pH 8.0, 1 mM MgCl2, 5% sucrose, 1% glycine, and 0.05% Tween-80 at −80°C. Virus particle concentration was estimated from absorbance at 260 nm (Sweeney and Hennessey, 2002).
Cell culture and immunofluorescence
HeLa cells and HaCaT keratinocytes were cultured in DME, 10% fetal bovine serum (Invitrogen) at 37°C, and 5% CO2 (Boukamp et al., 1988). To induce epithelial sheet migration, HaCaT cells were grown to confluency on #1.5 glass coverslips, washed with PBS without calcium or magnesium, and half of the cell monolayer was removed by scratching with a razor blade. Dead cells at the wound edge were removed by extensive washing with medium. Wounded HaCaT monolayers were immediately infected with recombinant adenovirus for 1–2 h, washed, mounted in culture medium in sealed observation chambers (Wittmann et al., 2004), and allowed to polarize and migrate overnight. For double infections, both viruses were added simultaneously. Immunostaining of endogenous CLASP and EB1 was performed as described previously (Wittmann and Waterman-Storer, 2005).
Image acquisition and analysis
Intracellular EGFP- and mCherry-tagged protein dynamics were imaged at 37°C with a 100× NA 1.49 objective lens (CFI APO TIRF; Nikon) on an inverted microscope system (TE2000 Perfect Focus System; Nikon) equipped with a spinning-disk confocal unit (CSU10; Yokogawa) with 200-mW, 488-nm, and 561-nm solid-state lasers (LMM5; Spectral Applied Research), electronic shutters, a cooled charge-coupled device camera (CoolSNAP HQ2; Photometrics), and controlled by NIS-Elements software (Nikon).
MT ends were tracked manually in time-lapse sequences of EGFP-tubulin–expressing cells acquired every 5 s using NIS-Elements software. These tracks were analyzed in Excel (Microsoft) using a custom-written macro as described previously (Wittmann et al., 2003). Alternatively, EB1-EGFP at growing MT ends was tracked in time-lapse sequences acquired every 400 ms using custom-written software in MatLab (The MathWorks), which will be described elsewhere. Image cross-correlation was performed in ImageJ (National Institutes of Health) using a plug-in by Chinga and Syverud (2007). Image regions with comparable lamella MT density were selected in different cells, and all possible combinations of these regions in a time-lapse sequence were compared, and the correlation coefficient was plotted as a function of increasing time interval.
To measure the amount of MT plus end–bound EGFP-CLASP2(512–650), we performed background-corrected line scans along the MT end using NIS-Elements software. The integrated fluorescence intensity along this line was used as an estimate of total plus end–bound protein. The relative expression level of cells was estimated by measuring the fluorescence intensity in homogenous regions in the cytoplasm, and excitation laser intensities and acquisition times were kept constant in these experiments. All quantification of fluorescence intensities was performed on raw 16-bit images. Images for presentation in figures and videos were processed with a 3 × 3 Gaussian low pass filter to reduce high frequency camera noise and an unsharp mask (7 × 7 kernel size; scaling factor 0.5) to enhance dim features using MetaMorph (MDS Analytical Technologies; Wittmann et al., 2004).
Phosphorylation assays and immunoprecipitation
To analyze intracellular CLASP phosphorylation by metabolic labeling, HeLa cells were transfected with EGFP-tagged CLASP constructs using Fugene 6 (Roche) and incubated in serum- and phosphate-free DME (Invitrogen) in the presence of 0.1 mCi/ml 32[PO4]3− for 2 h at 37°C in 5% CO2. Cells were washed with ice-cold PBS, lysed in 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 10% glycerol, 1% NP-40, containing phosphatase (1 mM EGTA, 50 mM NaF, 10 mM Na4P2O7, 1 mM β-glycerophosphate, and 1 mM Na3VO4), and protease inhibitors and were centrifuged for 3 min at 14,000 g to remove cell debris. EGFP-tagged proteins were immunoprecipitated using a polyclonal GFP antibody (provided by J. Huppa, Stanford University, Palo Alto, CA) immobilized on protein A beads (Affi-Prep; Bio-Rad Laboratories). After several washes with lysis buffer, bound proteins were eluted by boiling in SDS sample buffer containing 5% β-mercaptoethanol and analyzed by gel electrophoresis and autoradiography. To analyze in vitro phosphorylation by GSK3β, transfected HeLa cells were incubated with 20 µM SB216763 for 2 h, and EGFP-tagged protein was isolated by immunoprecipitation. After extensive washing in lysis buffer, the beads containing EGFP-CLASP2(512–650) were resuspended in 20 µl 20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 5 mM DTT, 200 µM ATP containing 10 µCi γ-[32P]ATP, and 100 U GSK3β (New England Biolabs, Inc.), incubated at 30°C for 30 min, and analyzed by autoradiography.
To analyze EB1 binding, transfected HeLa cells were washed with ice-cold PBS and lysed in 30 mM K-Hepes, pH 7.5, 100 mM KCl, 1% NP-40 containing phosphatase, and protease inhibitors. EGFP-tagged CLASP was immunoprecipitated, washed, and eluted as described in the previous paragraph. Bound proteins were analyzed by SDS gel electrophoresis and immunoblot probed with monoclonal antibodies against GFP (JL-8; Clontech Laboratories, Inc.) or EB1 (BD). Polyclonal CLASP antibodies were used as described previously (Hannak and Heald, 2006).
MT sedimentation assay
6×His-tagged CLASP2(340–650) was prepared, and in vitro MT-binding assays were performed as previously described in 80 mM K-Pipes, pH 6.8, 70 mM NaCl, 1 mM EGTA, and 1 mM MgCl2 (Wittmann and Waterman-Storer, 2005). Equivalent amounts of supernatant and pellet were analyzed by immunoblot with a monoclonal His-tag antibody (27E8; Cell Signaling Technology). Cleavage of the unstructured tubulin C terminus was done by limited proteolysis of taxol-stabilized MTs with subtilisin (Sigma-Aldrich; Knipling et al., 1999). The proteolysis reaction was stopped by adding freshly prepared 20 mM PMSF. Subtilisin-treated MTs were pelleted by centrifugation at 60,000 g and resuspended in MT-stabilizing buffer containing 20 mM PMSF to ensure complete removal of active subtilisin and cleaved C-terminal tubulin fragments.
Online supplemental material
Table S1 shows a summary of MT dynamic instability parameters. Table S2 shows a summary of MT-binding data of the different CLASP2 phosphorylation mutants. Fig. S1 shows that constitutively active GSK3β(S9A) disrupts cytoskeletal organization. Fig. S2 shows CLASP knockdown by RNA interference in HaCaT cells. Fig. S3 shows analysis of CLASP expression in HaCaT cells. Fig. S4 shows phosphorylation site mutations rescue CLASP2(340–875) plus end tracking in cells expressing constitutively active GSK3β(S9A). Fig. S5 shows HaCaT migration at the edge of a cell monolayer. Video 1 shows HaCaT keratinocyte sheet migration at the edge of a confluent monolayer. Video 2 shows EGFP-tubulin in a control HaCaT cell at the edge of a migrating cell sheet. Video 3 shows EGFP-tubulin in a HaCaT cell expressing constitutively active GSK3β(S9A). Video 4 shows EB1-EGFP in a control HaCaT cell. Video 5 shows EB1-EGFP in a HaCaT cell expressing constitutively active GSK3β(S9A). Video 6 shows EGFP-CLASP2(340–1,362) in a migrating HaCaT cell. Video 7 shows nonphosphorylatable 9×S/A EGFP-CLASP2(340–1,362) in a HaCaT cell expressing mRFP-GSK3β(S9A) (inset). Video 8 shows EGFP-CLASP2(340–1,362) in a migrating HaCaT cell. Video 9 shows EGFP-CLASP2(340–1,084) in a migrating HaCaT cell. Video 10 shows EGFP-CLASP2(340–1,362) and mCherry-paxillin in a HaCaT migrating cell.
Acknowledgments
We thank Diane Barber and members of the Barber laboratory for constructive discussions and chocolate cake.
This work was supported by the University of California San Francisco Academic Senate Committee on Research, the American Heart Association Scientist Development Grant (0730032N), and National Institutes of Health grants (R01GM079139 to T. Wittmann and U01GM067230 to G. Danuser). This research was conducted in a facility constructed with support from the Research Facilities Improvement Program (grant C06 RR16490) from the National Center for Research Resources of the National Institutes of Health.
Footnotes
Abbreviations used in this paper: CLASP, cytoplasmic linker–associated protein; GSK3β, glycogen synthase kinase 3β; mRFP, monomeric RFP; MT, microtubule; WT, wild type.
References
- Akhmanova A., Steinmetz M.O. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips.Nat. Rev. Mol. Cell Biol. 9:309–322 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., Hoogenraad C.C., Drabek K., Stepanova T., Dortland B., Verkerk T., Vermeulen W., Burgering B.M., De Zeeuw C.I., Grosveld F., Galjart N. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts.Cell. 104:923–935 [DOI] [PubMed] [Google Scholar]
- Bieling P., Kandels-Lewis S., Telley I.A., van Dijk J., Janke C., Surrey T. 2008. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites.J. Cell Biol. 183:1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P., Petrussevska R.T., Breitkreutz D., Hornung J., Markham A., Fusenig N.E. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line.J. Cell Biol. 106:761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L., Spittle C. 2001. Regulation of microtubule-associated proteins.Int. Rev. Cytol. 210:163–226 [DOI] [PubMed] [Google Scholar]
- Chinga G., Syverud K. 2007. Quantification of paper mass distributions within local picking areas.Nordic Pulp and Paper Res. J. 22:441–446 [Google Scholar]
- Cho J.H., Johnson G.V. 2003. Glycogen synthase kinase 3beta phosphorylates tau at both primed and unprimed sites. Differential impact on microtubule binding.J. Biol. Chem. 278:187–193 [DOI] [PubMed] [Google Scholar]
- Dephoure N., Zhou C., Villen J., Beausoleil S.A., Bakalarski C.E., Elledge S.J., Gygi S.P. 2008. A quantitative atlas of mitotic phosphorylation.Proc. Natl. Acad. Sci. USA. 105:10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabek K., van Ham M., Stepanova T., Draegestein K., van Horssen R., Sayas C.L., Akhmanova A., Ten Hagen T., Smits R., Fodde R., Grosveld F., Galjart N. 2006. Role of CLASP2 in microtubule stabilization and the regulation of persistent motility.Curr. Biol. 16:2259–2264 [DOI] [PubMed] [Google Scholar]
- Dragestein K.A., van Cappellen W.A., van Harren J., Tsibidis G.D., Akhmanova A., Knoch T.A., Grosveld F., Galjart N. 2008. Dynamic behavior of GFP–CLIP-170 reveals fast protein turnover on microtubule plus ends.J. Cell Biol. 180:729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A., Kharitonov A., Efimova N., Loncarek J., Miller P.M., Andreyeva N., Gleeson P., Galjart N., Maia A.R., McLeod I.X., et al. 2007. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network.Dev. Cell. 12:917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng C.H., Huckaba T.M., Gundersen G.G. 2006. The formin mDia regulates GSK3beta through novel PKCs to promote microtubule stabilization but not MTOC reorientation in migrating fibroblasts.Mol. Biol. Cell. 17:5004–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. 2003. Cdc42 regulates GSK-3β and adenomatous polyposis coli to control cell polarity.Nature. 421:753–756 [DOI] [PubMed] [Google Scholar]
- Ferrarese A., Marin O., Bustos V.H., Venerando A., Antonelli M., Allende J.E., Pinna L.A. 2007. Chemical dissection of the APC repeat 3 multistep phosphorylation by the concerted action of protein kinases CK1 and GSK3.Biochemistry. 46:11902–11910 [DOI] [PubMed] [Google Scholar]
- Frame S., Cohen P., Biondi R.M. 2001. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation.Mol. Cell. 7:1321–1327 [DOI] [PubMed] [Google Scholar]
- Fukata M., Watanabe T., Noritake J., Nakagawa M., Yamaga M., Kuroda S., Matsuura Y., Iwamatsu A., Perez F., Kaibuchi K. 2002. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170.Cell. 109:873–885 [DOI] [PubMed] [Google Scholar]
- Fumoto K., Hoogenraad C.C., Kikuchi A. 2006. GSK-3beta-regulated interaction of BICD with dynein is involved in microtubule anchorage at centrosome.EMBO J. 25:5670–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galjart N. 2005. CLIPs and CLASPs and cellular dynamics.Nat. Rev. Mol. Cell Biol. 6:487–498 [DOI] [PubMed] [Google Scholar]
- Gundersen G.G., Gomes E.R., Wen Y. 2004. Cortical control of microtubule stability and polarization.Curr. Opin. Cell Biol. 16:106–112 [DOI] [PubMed] [Google Scholar]
- Hannak E., Heald R. 2006. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle.J. Cell Biol. 172:19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Lisztwan J., Thoma C.R., Wirbelauer C., Barry R.E., Krek W. 2006. Priming-dependent phosphorylation and regulation of the tumor suppressor pVHL by glycogen synthase kinase 3.Mol. Cell. Biol. 26:5784–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J., Hyman A.A. 2003. Dynamics and mechanics of the microtubule plus end.Nature. 422:753–758 [DOI] [PubMed] [Google Scholar]
- Imami K., Sugiyama N., Kyono Y., Tomita M., Ishihama Y. 2008. Automated phosphoproteome analysis for cultured cancer cells by two-dimensional nanoLC-MS using a calcined titania/C18 biphasic column.Anal. Sci. 24:161–166 [DOI] [PubMed] [Google Scholar]
- Jope R.S., Johnson G.V. 2004. The glamour and gloom of glycogen synthase kinase-3.Trends Biochem. Sci. 29:95–102 [DOI] [PubMed] [Google Scholar]
- Knipling L., Hwang J., Wolff J. 1999. Preparation and properties of pure tubulin S.Cell Motil. Cytoskeleton. 43:63–71 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Hino S., Oue N., Asahara T., Zollo M., Yasui W., Kikuchi A. 2006. Glycogen synthase kinase 3 and h-prune regulate cell migration by modulating focal adhesions.Mol. Cell. Biol. 26:898–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama A., Karakesisoglou I., Wong E., Vaezi A., Fuchs E. 2003. ACF7: an essential integrator of microtubule dynamics.Cell. 115:343–354 [DOI] [PubMed] [Google Scholar]
- Komarova Y., Lansbergen G., Galjart N., Grosveld F., Borisy G.G., Akhmanova A. 2005. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules.Mol. Biol. Cell. 16:5334–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Akhmanova A. 2006. Microtubule plus end: a hub of cellular activities.Traffic. 7:499–507 [DOI] [PubMed] [Google Scholar]
- Lansbergen G., Komarova Y., Modesti M., Wyman C., Hoogenraad C.C., Goodson H.V., Lemaitre R.P., Drechsel D.N., van Munster E., Gadella T.W.J., et al. 2004. Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization.J. Cell Biol. 166:1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G., Grigoriev I., Mimori-Kiyosue Y., Ohtsuka T., Higa S., Kitajima I., Demmers J., Galjart N., Houtsmuller A.B., Grosveld F., Akhmanova A. 2006. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta.Dev. Cell. 11:21–32 [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., III, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage.Science. 316:1160–1166 [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Grigoriev I., Lansbergen G., Sasaki H., Matsui C., Severin F., Galjart N., Grosveld F., Vorobjev I., Tsukita S., Akhmanova A. 2005. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex.J. Cell Biol. 168:141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Wang H.W. 2006. Structural intermediates in microtubule assembly and disassembly: how and why? Curr. Opin. Cell Biol. 18:179–184 [DOI] [PubMed] [Google Scholar]
- Peris L., Thery M., Faure J., Saoudi Y., Lafanechere L., Chilton J.K., Gordon-Weeks P., Galjart N., Bornens M., Wordeman L., et al. 2006. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends.J. Cell Biol. 174:839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. 2003. Cell migration: integrating signals from front to back.Science. 302:1704–1709 [DOI] [PubMed] [Google Scholar]
- Roach P.J. 1991. Multisite and hierarchal protein phosphorylation.J. Biol. Chem. 266:14139–14142 [PubMed] [Google Scholar]
- Slep K.C., Vale R.D. 2007. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1.Mol. Cell. 27:976–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J.V., Resch G.P. 2005. The comings and goings of actin: coupling protrusion and retraction in cell motility.Curr. Opin. Cell Biol. 17:517–523 [DOI] [PubMed] [Google Scholar]
- Sweeney J.A., Hennessey J.P., Jr 2002. Evaluation of accuracy and precision of adenovirus absorptivity at 260 nm under conditions of complete DNA disruption.Virology. 295:284–288 [DOI] [PubMed] [Google Scholar]
- Tollefson A.E., Kuppuswamy M., Shashkova E.V., Doronin K., Wold W.S. 2007. Preparation and titration of CsCl-banded adenovirus stocks.Methods Mol. Med. 130:223–235 [DOI] [PubMed] [Google Scholar]
- Trinidad J.C., Specht C.G., Thalhammer A., Schoepfer R., Burlingame A.L. 2006. Comprehensive identification of phosphorylation sites in postsynaptic density preparations.Mol. Cell. Proteomics. 5:914–922 [DOI] [PubMed] [Google Scholar]
- Trivedi N., Marsh P., Goold R.G., Wood-Kaczmar A., Gordon-Weeks P.R. 2005. Glycogen synthase kinase-3{beta} phosphorylation of MAP1B at Ser1260 and Thr1265 is spatially restricted to growing axons.J. Cell Sci. 118:993–1005 [DOI] [PubMed] [Google Scholar]
- Vaughan K.T. 2004. Surfing, regulating and capturing: are all microtubule-tip-tracking proteins created equal? Trends Cell Biol. 14:491–496 [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Salmon E.D. 1997. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling.J. Cell Biol. 139:417–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Eng C.H., Schmoranzer J., Cabrera-Poch N., Morris E.J., Chen M., Wallar B.J., Alberts A.S., Gundersen G.G. 2004. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration.Nat. Cell Biol. 6:820–830 [DOI] [PubMed] [Google Scholar]
- Wittmann T. 2008. EBs clip CLIPs to growing microtubule ends.J. Cell Biol. 183:1183–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T., Waterman-Storer C.M. 2001. Cell motility: can Rho GTPases and microtubules point the way? J. Cell Sci. 114:3795–3803 [DOI] [PubMed] [Google Scholar]
- Wittmann T., Waterman-Storer C.M. 2005. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3β in migrating epithelial cells.J. Cell Biol. 169:929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T., Bokoch G.M., Waterman-Storer C.M. 2003. Regulation of leading edge microtubule and actin dynamics downstream of Rac1.J. Cell Biol. 161:845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T., Littlefield R., Waterman-Storer C.M. 2004. Fluorescent speckle microscopy of cytoskeletal dynamics in living cells. Live Cell Imaging: A Laboratory Manual. Goldman R.D., Spector D.L., Cold Spring Harbor Press, Cold Spring Harbor, NY: 187–204 [Google Scholar]
- Wu X., Kodama A., Fuchs E. 2008. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity.Cell. 135:137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T., Kawano Y., Arimura N., Kawabata S., Kikuchi A., Kaibuchi K. 2005. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity.Cell. 120:137–149 [DOI] [PubMed] [Google Scholar]
- Yoshimura T., Arimura N., Kaibuchi K. 2006. Signaling networks in neuronal polarization.J. Neurosci. 26:10626–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbrunn J., Kinoshita K., Hyman A.A., Nathke I.S. 2001. Binding of the adenomatous polyposis coli protein to microtubules increases microtubule stability and is regulated by GSK3 beta phosphorylation.Curr. Biol. 11:44–49 [DOI] [PubMed] [Google Scholar]