SUMMARY
I-TevI is a modular intron-encoded endonuclease, consisting of an N-terminal catalytic and a C-terminal DNA-binding domain, joined by a 75-amino acid linker. This linker can be divided into three regions, from N- to C-terminal: the deletion intolerant (DI) region; the deletion tolerant (DT) region; and a zinc finger, which acts as a distance determinant for cleavage. To further explore linker function, we generated deletion and substitution mutants that were tested for their preference to cleave at distance, or at the correct sequence. Our results demonstrate that the I-TevI linker is multi-functional, a property which sets it apart from junction sequences in most other proteins. First, the linker DI region plays a role in I-TevI cleavage activity. Second, the DT linker region participates in distance determination, as evident from some DT mutants that display a phenotype similar to that of the zinc-finger mutants in their cleavage-site selection. Finally, NMR analysis of a freestanding 56-residue linker segment showed an unstructured stretch corresponding to the DI and a portion of the DT region, followed by a β-strand corresponding to the remainder of the DT region and containing a key distance-determining arginine, R129. Mutation of this arginine to alanine abolished distance determination and disrupted the β-strand, indicating that structure of the DT linker region plays a role in cleavage at a fixed distance.
Keywords: intron endonuclease, GIY-YIG enzyme, NMR spectroscopy, linker function, protein ruler
INTRODUCTION
Linkers in proteins of modular architecture frequently promote communication between domains and/or they facilitate the specific functions of multi-domain proteins 1; 2. The homing endonucleases of the GIY-YIG family are modular enzymes, composed of distinct domains with a functional linker 3. These endonucleases recognize lengthy DNA targets and make double-strand breaks. A well-conserved N-terminal domain of approximately 90 amino acids includes the defining GIY-YIG motif and catalytic residues, whereas the C-terminal domain is more variable in length, and shows less overall similarity among endonucleases 4; 5; 6.
The best characterized GIY-YIG endonuclease, I-TevI, encoded by the thymidylate synthase (td) intron of the bacteriophage T4, cleaves intronless DNA at sites 23 and 25 nt upstream of the intron insertion site (IS) and creates a double-strand break with a 2-nt 3′ extension 7; 8. I-TevI interacts with two regions of its 38-bp homing site on the intronless td allele 9. The DNA-binding domain contacts the primary binding region of 20 bp, which is centered on the intron IS 10. This domain is joined via the lengthy linker to the catalytic domain, which interacts with the cleavage site (CS), 23–25 nt upstream of the IS.
I-TevI uses both sequence and distance determinants in selecting its cleavage site 11. If the natural sequence is displaced from the optimal distance of 23 and 25 nt, then I-TevI searches bi-directionally from its cleavage position to locate a preferred site, 5′-CX↑XX↓G-3′, and cleaves at alternative distances, albeit with reduced efficiency 11; 12. The cleavage window extends from 5 bp upstream to 16 bp downstream of the normal cleavage site 11. When a preferred site is not within the window, the enzyme defaults to the optimal distance and cleaves with reduced efficiency 11; 13.
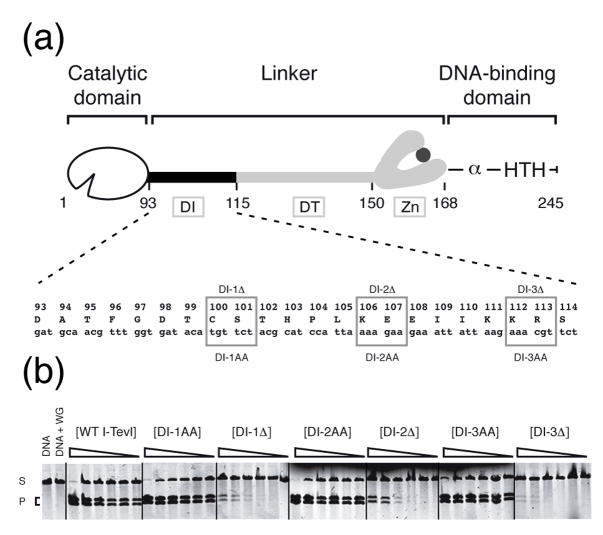
The catalytic domain of I-TevI ends at residue 92, as determined by informatics, NMR spectroscopy and X-ray crystallography 4; 14. The DNA-binding domain begins at residue 168, as shown biochemically by deletion analysis and crystallography 10; 13. The interdomain linker is extraordinary in that it is 75 amino acids long, constituting almost one-third of I-TevI (245 amino acids), and it is functionally complex. This complexity became apparent in deletion analysis which showed that the linker is organized into three regions (Fig. 1(a)): the 22-amino acid region from residue 93 to 114 is the DI region (deletion intolerant); the central 35 amino acids, from residue 115 to 149, constitute the DT region (deletion tolerant); and the last 18 amino acids form a zinc finger 4; 13.
Figure 1. Length rather than sequence of the DI region is needed for cleavage.
(a) Representation of I-TevI domains. The I-TevI linker between the catalytic and DNA-binding domains is differentiated into three regions: the DI region (deletion intolerant); the DT region (deletion tolerant); and the zinc finger (Zn) 4. The α and HTH in the cartoon indicate the α-helix and helix-turn-helix motifs of the DNA binding domain. Numbers on the cartoon (except the last) correspond to the beginning of a domain or segment. The mutated residues are enclosed within boxes. (b) Cleavage assays on linearized pTZtdΔI DNA. Cleavage assays were performed with proteins synthesized in vitro with [35S]-Met as described in Materials and Methods and the reaction products were resolved on agarose gels. S, substrate; P, cleavage products. WG = unprogrammed wheat germ extract. The cleavage reaction mixtures were adjusted to equivalent 35S -labeled protein and diluted step-wise 1/3 from 1 (undiluted) to 1:243.
In our previous work we demonstrated that the zinc finger serves as a distance determinant to constrain the catalytic domain, through putative intramolecular protein-protein interactions, such that it is proximal to the cleavage site, thereby promoting catalysis 13. Interestingly, I-BmoI, a GIY-YIG endondonuclease encoded by the thymidylate synthase intron of Bacillus mojavensis 15, is capable of some degree of distance determination, despite the absence of a zinc finger, suggesting that there must be other molecular features that are important in these “protein rulers” 5. Although the structure of the I-TevI zinc finger was solved with that of the DNA-binding domain 10, the structure of the remainder of the linker remains undefined.
To probe the function of the linker and to address a possible role of the DI and DT regions in cleavage efficiency and distance determination, we created deletions and point mutations. Our data suggest that the DI region is important for the functional integrity of the catalytic domain, while demonstrating the essential role of the DT linker region in I-TevI distance determination. Moreover, NMR analysis indicates that a freestanding linker segment comprising the DI and DT regions preferentially forms a β-strand over a portion of its length, and that this structure is disrupted by mutation of a single residue in the DT region that is critical for distance determination.
RESULTS
In a prior study, which involved artificial hybrid endonucleases created between I-TevI and I-BmoI, we hypothesized that linker components aside from the I-TevI zinc finger are involved in distance determination 5. To test this hypothesis and to define the functions of different components of the I-TevI linker, we undertook a mutational analysis of the linker.
A role for the DI region in promoting cleavage
The DI and DT regions were defined by 2-residue deletions 4. To distinguish whether the phenotype of the deletions resulted from a shorter DI linker that structurally disturbs the enzyme’s proper conformation for catalysis, or from a loss of specific side chains that participate in enzyme function, we performed Ala-substitution analysis in the DI region. We made three mutants: DI-1AA (C100A, S101A), DI-2AA (K106A, E107A) and DI-3AA (K112A, R113A) (Fig. 1(a)). Because of the toxicity of I-TevI, we constructed all linker mutants in a pSP65 vector background and the mutant proteins were synthesized in vitro 16. Cleavage efficiency was estimated by comparison to wild-type I-TevI activity on wild-type I-TevI homing site, using linearized pTZtdΔI substrate, which yields two cleavage products on agarose gels (Fig. 1(b)). All three DI-Ala-substitution mutants were able to cleave the wild-type I-TevI homing site with similar accuracy (Supplementary Fig. 1(a)) and efficiency to wild-type I-TevI, whereas the corresponding deletion mutants were at least 100-fold less active (Fig. 1(b)). These observations suggest that a fixed linker length rather than amino acid composition in the DI region is required for efficient cleavage of the wild-type substrate by I-TevI.
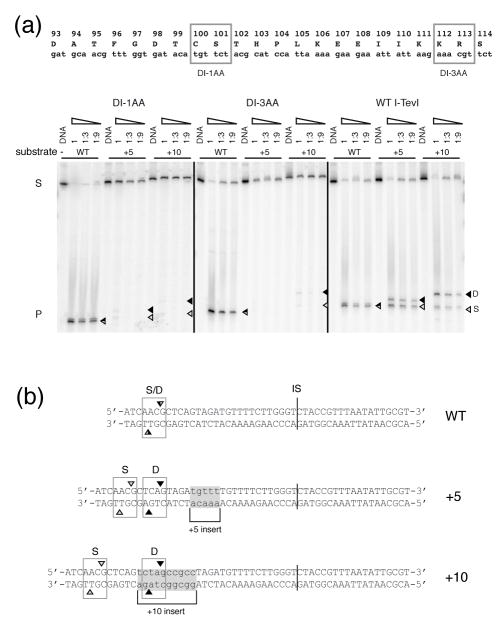
To probe linker flexibility, we further investigated the catalytic activities of the DI Ala-substitution mutants (Fig. 2(a)) on homing sites containing a 5-nt (+5) or a 10-nt (+10) insertion between the intron IS and the cleavage site (CS) (Fig. 2(b)) 11. We measured cleavage by DI Ala-substitution mutants DI-1AA and DI-3AA on a 5′-end 32P-labeled 300-bp PCR fragment containing wild-type, +5 or +10 I-TevI homing-site. The two DI Ala-substitution mutants retained cleavage activity only slightly lower than that of the native enzyme on wild-type substrate, as judged by substrate disappearance and product appearance, but they effected barely perceptible cleavage on the +5 and +10 homing sites (Fig. 2(a)). However, the weak cleavages observed on the +5 and +10 homing sites displayed a cleavage-site preference similar to that of the wild-type I-TevI. Together these observations indicate that the Ala substitutions severely impair I-TevI’s ability to extend its catalytic domain to reach shifted cleavage sequences, whether in or out of phase on the B-form DNA helix, but that the individual side chains are not involved in distance determination.
Figure 2. Ala-substitution mutants in the DI region reduced linker flexibility.
(a) Cleavage activity on wild-type, +5 and +10 mutant substrates. DI-1AA, DI-3AA and wild-type I-TevI cleavage activity is shown on each of the three DNA substrates. In vitro-synthesized enzymes were incubated with 5′ end-labeled double-strand substrates (S) to yield products (P), and resolved on a 6% sequencing gel. The cleavage reaction mixtures were adjusted to equivalent 35S -labeled protein and diluted step-wise 1/3 from 1 (undiluted) to 1:9. D = cleavage at distance (black triangles), S = cleavage at sequence (gray triangles). (b) WT, +5 and +10 substrate sequences. The inserted sequences are shaded. D and S are as in (a). The intron IS is indicated by a vertical line.
The DT linker region has a role in I-TevI distance determination for remote cleavage
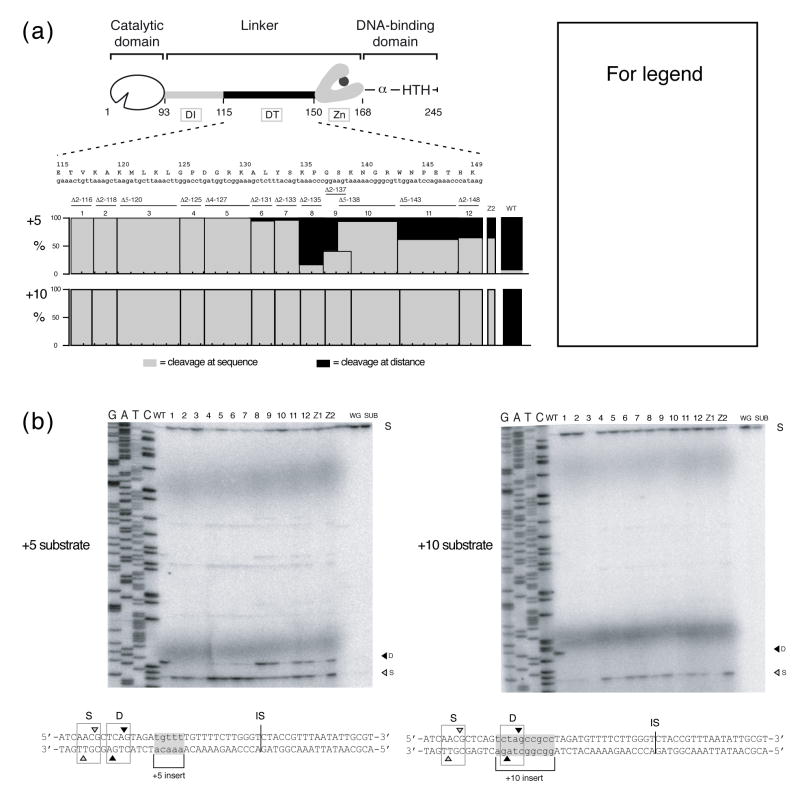
To evaluate the hypothesis that the DT region of the linker participates in distance determination 5, we tested the propensities of the cleavage-competent linker deletion derivatives to cleave at sequence or at distance. The deletion mutants that were used to define the DT region (Fig. 3(a)) 4, as well as zinc-finger mutants 13, were synthesized in vitro. After confirming the cleavage activity of all the mutants on linearized pTZtdΔI DNA 4, we mapped the cleavage sites of the DT and zinc-finger mutants on +5 and +10 substrates (Fig. 3(b)). On +5 substrate, wild-type I-TevI cleaved the DNA predominantly at the optimal distance, 23 nt from the IS on the top strand. However, the DT mutants showed distinct phenotypes: mutants from residue 116 to 134 cleaved almost exclusively at the correct sequence on the +5 substrate, 28 nt upstream from the IS, whereas mutants from the remainder of the DT region (residues 135 to 148) behaved similarly to zinc-finger mutants, cleaving at both distance and sequence (Fig. 3(b), top). Further, cleavage-site mapping on a +10 substrate showed that all of the DT linker mutants had an identical phenotype to the zinc-finger mutants, cleaving strictly at sequence, 33 nt upstream from the IS, whereas the wild-type enzyme cleaves the +10 substrate strictly at the optimal distance (Fig. 3(b), bottom). The data were quantitated and are shown in Figure 3(a). Cleavage patterns on the bottom strand were similar to those on the top strand (data not shown). The increased flexibility of the interdomain linker in these deletion mutants of the DT region relieves the constraint of wild-type I-TevI, and enables the mutant enzymes to extend further to cleave at the natural sequence, as for the zinc-finger mutants, despite being shorter proteins than wild-type I-TevI.
Figure 3. DT linker deletion mutants affect cleavage-site selection.
(a) Schematic representation of mutations in the DT region. The deletion mutants were constructed with two-, four- or five-amino acid deletions, represented below the linker sequence by bars numbered 1–12. Mutants are named by indicating the size of the deletion followed by the location of the starting amino acid. For example, Δ2-125 is a two-amino acid deletion starting at residue 125 and Δ5-138 is a five-amino acid deletion starting from residue 138 4. Cleavage assay data are represented as % cleavage at distance (black bars) over sequence (gray bars) for each deletion mutant and WT control, and are averaged over three independent experiments. (b) Top strand cleavage-site mapping on +5 and +10 I-TevI homing site (HS). Numbers correspond to constructs in (a). WT, wild-type I-TevI; 1, Δ2-116; 2, Δ2-118; 3, Δ5-120; 4, Δ2-125; 5, Δ4-127; 6, Δ2-131; 7, Δ2-133; 8, Δ2-135; 9, Δ2-137; 10, Δ5-138; 11, Δ5-143; 12, Δ2-148; Z1, ΔZn; Z2, CZnA; WG, wheat germ extract; and SUB, DNA substrate. Substrate sequences and labeling as in Figure 2. Similar ratios of cleavage at sequence versus distance were observed on the bottom strand (data not shown).
The roles of charged residues of the DT linker region in I-TevI distance determination
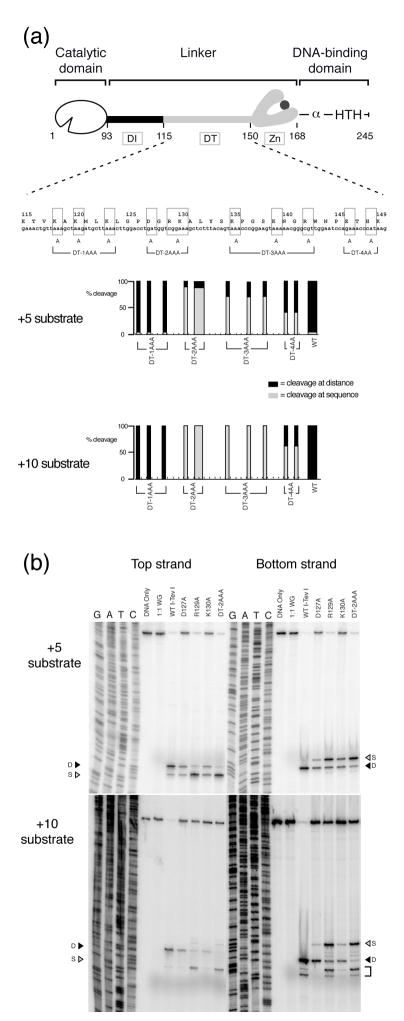
We wished to distinguish if the loss of specific residues in the DT region or shorter protein length impaired the distance determination for cleavage in the deletion mutants. Furthermore, if specific residues were responsible, we wanted to evaluate the extent of their involvement in distance determination. We therefore modified charged residues, which are often involved in electrostatic interactions and salt-bridge formation. From residues 116 to 134, six residues are either acidic or basic (K118, K120, K123, D127, R129, K130) and in the remaining DT region, residues 135 to 149, four amino acids are basic (K135, K139, R142, K149). We constructed four DT Ala-substitution mutants in the pSP65 vector background: DT-1AAA (K118A, K120A, K123A), DT-2AAA (D127A, R129A, K130A), DT-3AAA (K135A, K139A, R142A) and DT-4AA (E146A, H148A) (Fig. 4(a)). The cleavage activities of the mutants synthesized in vitro were first tested on a wild-type homing site. The DT Ala-substitution mutants each cleaved the wild-type homing site with a similar efficiency and at the same sites on top and bottom strands, 23 and 25 nt from the IS, as for wild-type I-TevI (Supplementary Fig. 1(a) and (b)).
Figure 4. Defining R129 as a key residue in distance determination.
(a) Sequence of DT region. Ala-substitution mutations are indicated by boxes and brackets. Cleavage assay data for +5 and +10 substrates are represented as % cleavage at distance (black bars) over sequence (gray bars) for each substitution mutant and WT control, and are an average over three independent experiments. (b) Representative cleavage reactions on +5 and +10 substrates. Cleavage reactions and gels are as in Figure 2(a). Cleavage on WT substrate is shown in Supplementary Fig. 2. The square bracket represents spurious cleavage on the +10 substrate, as previously reported 11.
To determine whether the Ala-substitutions may affect distance discrimination, we mapped the cleavage sites of these mutants on +5 and +10 homing sites on both strands. The data were quantitated as shown in Figure 4(a), with representative data for the DT-2AAA mutant shown in Figure 4(b). The four substitution mutants showed different preferences for distance or sequence in selecting their cleavage sites. DT-1AAA behaved similarly to wild-type I-TevI and cleaved predominantly or exclusively at distance on the +5 and +10 substrates, respectively (Fig. 4(a)). Thus, Ala substitutions at positions K118, K120 and K123 did not affect the protein’s distance determination ability, although the deletions at these positions completely abolished this function (Fig. 3(a)). This observation suggests that protein length, rather than the nature of the charged side chains at these positions is important for distance determination.
The two point mutants DT-2AAA and DT-3AAA showed a different phenotype than that of the wild-type enzyme, cleaving preferentially at sequence on the +5 homing site, and exclusively at sequence on the +10 homing site, with the DT-2AAA mutant exhibiting the more dramatic phenotype (Fig. 4(a) and (b)). Finally, the DT-4AA mutant displayed an intermediate phenotype among the Ala-substitution mutants, with approximately equivalent preference for sequence and distance on the +5 and +10 homing sites, suggesting that the roles of individual amino acids are not as important as those of DT-2AAA or DT-3AAA.
In the DT-2AAA mutant, the substitution of only three charged residues switched the cleavage behavior of I-TevI from a preference to cleave at distance to a preference to cleave at sequence. Mutations of D127, R129 and K130 to Ala thus increased the linker flexibility and extended the reach of the catalytic domain by 5 or 10 additional nucleotides to enable the enzyme to find the correct sequence. Like the zinc-finger mutants, but with perhaps an even more dramatic phenotype, these charged residues seem to be crucial for imparting distance determination capability to I-TevI. While the data confirm a role for a portion of the DT segment of the linker in I-TevI distance sensing (residues 127-149), as suggested by the hybrid endonuclease study 5, they divide the DT portion of the linker with respect to phenotype, and they implicate residues 127-130 as most critical to distance determination among the mutants tested.
Residue R129 in the DT linker segment is a strong distance determinant
Because the DT-2AAA triple Ala-substitution mutant was distinctive in its phenotype, almost completely abolishing cleavage at the correct distance, we wished to tease apart the individual contributions of D127, R129 and K130. We therefore made single mutants and, after testing them on wild-type substrate (Supplementary Fig. 2), we determined sequence versus distance preferences for cleavage, alongside the triple mutant and wild-type enzyme (Fig. 4(b)). Whereas D127A had slight distance preferences for cleavage on both strands on +5 and +10 substrates, akin to the behavior of the wild-type enzyme, and K130A had little bias toward distance or sequence, the R129A mutant had strong sequence preferences on both strands of the +5 and +10 substrates. Indeed R129A could account for most if not all the phenotypic properties of the triple mutant, indicating that this single residue, like the zinc finger, is a critical distance determinant.
Linker structure by NMR and the role of R129
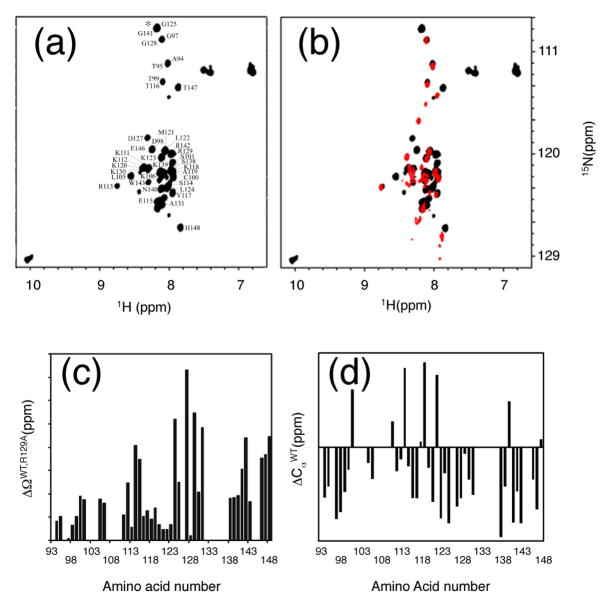
Some linkers fold autonomously 2; 17. To determine if the freestanding I-TevI linker has an autonomous fold, as well as to explore a potential structural basis for the phenotype of the R129A mutant, and to begin to correlate phenotypic with physical properties of the linker, we performed NMR spectroscopy on a segment of the I-TevI linker. Two linker segments, consisting of amino acids 93 to 167 (I-TevI93-167), including the zinc finger, and 93 to 148 (I-TevI93-148), encompassing only the DI and DT linker regions, were overexpressed and purified. Only the shorter linker, I-TevI93-148, was monodispersed in solution. The zinc finger-containing I-TevI93-167 was prone to degradation and quickly aggregated during NMR experiments. Therefore I-TevI93-148, containing either the wild-type arginine or mutant alanine residue at position 129 (R129A-I-TevI93-148) were analyzed.
The 1H{15N}-HSQC spectrum of I-TevI93-148 exhibited a limited spectral dispersion in the amide proton region, from 7.5 ppm to 8.7 ppm, that suggested that this segment of the linker has a minimally defined tertiary structure (Fig. 5(a)). The R129A mutation led to further narrowing of the amide proton resonances (Fig. 5(b)) and significant changes in chemical shifts as compared to the wild type I-TevI93-148.
Figure 5. R129A mutation changes the secondary structure of I-TevI93-148.
(a) NMR 1H{15N} HSQC spectrum of I-TevI93-148. Chemical shift assignments of I-TevI93-148 are indicated. A peak marked by an asterisk is from the uncut thrombin restriction site. (b). Overlay of the 1H{15N}-HSQC spectra of I-TevI93-148 (black) and R129A-I-TevI93-148 (red). (c). Chemical shift changes, ΔΩ, due to the R129A mutation. Chemical shift changes were calculated by using , where ΔΩH and ΔΩN are changes in amide proton and nitrogen chemical shifts expressed in ppm, respectively. (d) Differences in 13Cα chemical shifts of I-TevI93-148 from the random coil values, . Consistent negative values of indicate the preference of the protein 123–148 region to form a β-strand.
To identify which area of the linker was affected by the R129A mutation, we assigned 82% of the I-TevI93-148 resonances. Surprisingly, the R129A mutation resulted in relatively small changes in chemical shifts in the region 93–122 (Fig. 5(c)). The most significant differences occurred after amino acid L122. Since unstructured parts of the protein would usually be unaffected by the mutation, the global change in chemical shifts of the 123–148 region indicates that the preexisting structure of this region was altered by the R129A mutation.
We analyzed the secondary structure of I-TevI93-148 using chemical shift indices (CSIs). CSIs provide a well-established tool for identifying the elements of protein secondary structure 18. They are based on the difference between the protein proton and carbon chemical shifts and amino acid random coil chemical shifts. CSIs were also used to define regions of the protein that did not have a stable tertiary structure but had preference towards forming secondary structure elements 19. Consistently negative values of the difference between I-TevI93-148 and random coil α-carbon chemical shifts are indicative of the predominantly β-strand structure in the 123–148 region (Fig. 5(d)). Therefore, large changes in the chemical shift of the amide protons and nitrogens far away from the mutation site of Arg-to-Ala at position 129 indicate substantial structural changes that are associated with this mutation in the 123-148 amino acid region, suggesting dissolution of the β-strand structure.
Identification of I-TevI linker-like sequences in environmental database
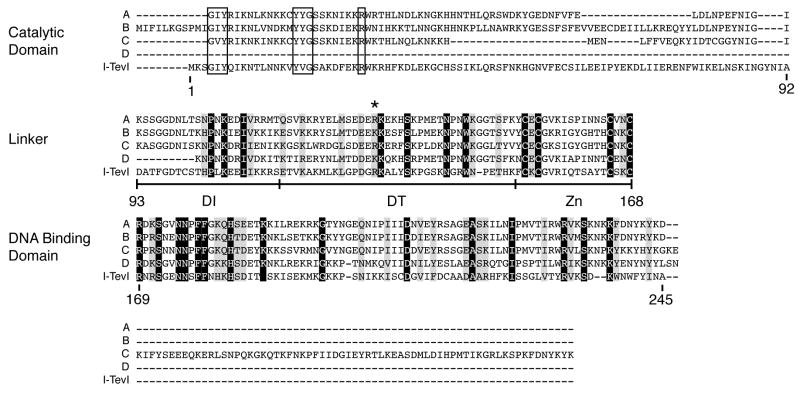
To generalize our findings, sequence similarity searches were performed. A BLAST search of the NCBI non-redundant protein sequences database (nr) using the full-length I-TevI sequence revealed a significant number of known GIY-YIG family endonucleases, including I-BmoI. A BLAST search of the environmental non-redundant sequences database (env_nr) using the full-length I-TevI sequence again revealed many sequences with substantial homology to I-TevI. When the search was confined to the linker sequence (residues 93-167), four striking matches were found. These sequences and neighboring residues on the respective contigs were used in a multiple sequence alignment with I-TevI using MAFFT (version 6, http://align.bmr.kyushu-u.ac.jp/mafft/software/) (Fig. 6). All four sequences are highly conserved with I-TevI in the linker and DNA-binding domains. Three of these have the signature GIY-YIG motifs and a conserved arginine corresponding to a catalytic residue of I-TevI, and they are presumably also homing endonucleases (Fig. 6, sequences A–C). The fourth sequence begins 9 residues into the linker, at the start of the contig, and therefore yields no information on N-terminal residues (Fig. 6, sequence D). The zinc-finger motif of the linker is conserved in all four sequences, and the amino acids corresponding to R129 are arginine in two cases and lysine in the other two.
Figure 6. I-TevI linker sequence alignment.
Sequences A-D were recovered from a BLAST search of the environmental subdivision database using the I-TevI linker as query sequence (amino acids 93-167). Residue numbering of I-TevI is below the sequence. Sequence alignments are shown as blocks that correspond to the I-TevI catalytic domain, the linker, the DNA-binding domain, and residues C-terminal to the DNA binding domain. Black shading represents residues that are absolutely conserved among the five sequences and gray shading represents conserved residues with conservative changes. Residues that correspond to the GIY-YIG signature sequences are boxed. GenBank IDs to the hypothetical proteins (Marine Metagenome ID) are: A, 140852551 (Gos_3956749); B, 136178932 (Gos_8413892); C, 136165525 (Gos_8427889); D, 140538853 (Gos_4034147).
We also ran secondary structure prediction algorithims on the linker sequences. They all have a low-probability α-helix in the 93–122 region, corresponding to the unstructured stretch of I-TevI linker, and a putative beta sheet structure in the region homologous to amino acids 123–148 of I-TevI. These results suggest that these linker sequences have a similar fold.
DISCUSSION
Although some linkers are simply connectors between discrete functional domains, for example in the RhaS and RhaR proteins 20, many have adapted and assume additional roles related to the catalytic or binding properties of the domains that they join. They have thus evolved to act as communication devices 1, molecular rulers and other dynamic regulatory devices 2. Domain movement is thought to be conducted by “soft”, flexible linkers, whereas there is an absence of motion in ruler-type, rigid linkers 2. Interestingly, the I-TevI linker has a measurement device and yet exhibits flexibility. Whatever the mechanism, we postulate that the I-TevI linker has evolved the metric device to meet the protein’s two functions, DNA endonuclease and transcription repressor 21. Thus efficient cleavage of the homing site is achieved because the site is at optimal distance, whereas repression on the operator occurs with a minimum of cleavage, because potential cut-sites are out of the preferred distance range 5.
Structural and functional segmentation of the I-TevI linker
Our mutational analysis of the DI region of the linker indicates that the length or register that maintains spacing of residues in the DI linker region, rather than specific amino acids, is essential for the I-TevI catalytic function (Fig. 1) 4. Moreover, specific amino acids in the DI region are essential for protein flexibility, required to search for a shifted cleavage site along the DNA helix. Because no structure was observed in the DI region of the free linker by NMR analysis, one might hypothesize that the linker only folds if it is joined to the catalytic and/or DNA-binding domains of the protein. Alternatively, structure may be acquired upon DNA binding, dictating subsequent enzyme conformation required for catalysis.
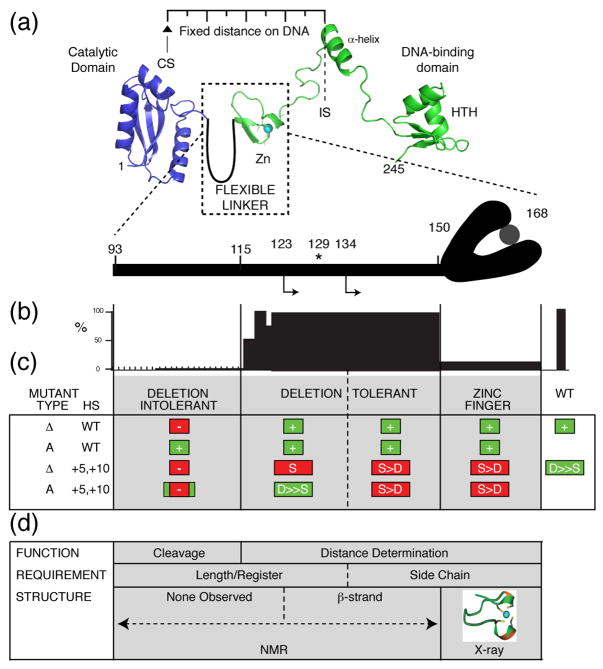
The DT linker region contributes to cleavage at a fixed distance, resembling the zinc finger in function. However, the DT region can be divided into subsegments on the basis of both the NMR and biochemical data (Fig. 7; Supplementary Fig. 3). The N-terminal portion of the DT region forms an extension of the DI region in two respects. First, protein length or register of residues up to approximately residue 134, and not the nature of amino acid side chains, is important for function, which in this case is distance determination. The length requirement for distance determination is manifest in the strict sequence preference of deletion mutants, in sharp contrast to the ability, and indeed most commonly, the preference of the point mutants to cleave at a distance. Second, no autonomous structure was observed in the DI region or first portion of the DT region of the linker, which assumes a β-strand configuration from approximately residue 123 (Figs. 5 and 6). In a satisfying correspondence between structure and function, the R129A mutant, which has the strongest phenotype of any other linker mutant in abolishing distance determination, has lost the β-strand structure in the free linker. Like the zinc finger, the β-strand is therefore an important structural element in distance-directed cleavage.
Figure 7. Summary of function and structure of the I-TevI linker.
(a) I-TevI linker between the catalytic and DNA-binding domains. The linker is cartooned as a loop beside the zinc finger (Zn), the structure of which was solved along with the DNA binding domain 10. The catalytic domain structure is from ref 13. Where the protein sits on the DNA is marked by the intron insertion site (IS) and I-TevI cleavage site (CS), which are a fixed distance apart. The asterisk corresponds to R129, which disrupts distance determination function and NMR structure when mutated. The rightward arrows correspond to the beginning of the β-strand (residue 123) and the distance determination transition (residue 134) of the DT region. (b) Summary of activity of deletion mutants 4; 13. (c) Summary of activity of mutant types with respect to linker segments: Δ = deletion; A = Ala-substitution. HS = homing site DNA substrate (WT DNA or +5, +10 insertions). In colored boxes (green = WT phenotype, red = mutant phenotype): + = active; - = inactive; S = cleavage at sequence; S > D = preferred cleavage at sequence over distance, D ≫ S = greatly preferred cleavage at distance over sequence. Green box indicates mutant’s general preference for cleavage at distance, like WT. Red box indicates mutant’s general preference for cleavage at sequence. (d) Requirements for function corresponding to structure.
Similarities to the linker in other GIY-YIG proteins in public databases
Database searches identified four proteins with striking sequence similarities to the I-TevI linker. Since these are from the environmental metagenomics database of GenBank, their identities are unknown, but they are likely to encode I-TevI-like homing endonucleases. Interestingly, the three sequences that have N-terminal domains with GIY-YIG motifs are all downstream of coding sequences that match multiple bacterial thymidylate synthase genes (E values in the 1e-29 range; data not shown). This finding, considered in the context of I-TevI being encoded in the phage T4 thymidate synthase intron, suggests a tantalizing relationship between thymidylate synthase genes and I-TevI-like proteins.
The linker may interact broadly within the I-TevI molecule
The loss of cleavage activity of the DI mutants indicates a role for the linker in the integrity or positioning of the catalytic domain. Furthermore, we noted previously that a hybrid endonuclease of the I-BmoI DNA-binding domain with the full-length I-TevI linker plus catalytic domain had lost distance determination on a +10 substrate 5. This observation implies that either the presence of the I-BmoI DNA binding domain disrupts the overall hybrid protein conformation required for I-TevI distance determination, or that the I-TevI DNA-binding domain is required for distance determination. A preliminary intramolecular protein-protein chemical crosslinking study suggests that this may indeed be the case, with interaction between the C-terminal DNA binding domain and the linker (Liu, unpublished) 22. There is also preliminary evidence that I-TevI split at various positions in the linker can reassemble into a functional enzyme to cleave DNA, much as artificially split I-DmoI can reassemble 23, suggesting intra-linker and/or inter-domain interactions (K. Lehtonen and Belfort, unpublished). We propose that the I-TevI zinc finger mediates protein-protein interactions, as do similar 4-Cys zinc fingers (ref 24 and references therein), possibly in concert with the DT linker region, to effect distance-dependent cleavage. However, exactly how the I-TevI linker serves its metric functions remains a mystery, particularly since rigid peptides have previously been associated with protein rulers 2.
Possible relationship to other modular GIY-YIG enzymes
The overall role of the I-TevI linker is to act as a communication device between the DNA-binding and catalytic GIY-YIG domains, such that they act in concert for DNA cleavage, but the DNA-binding domain acts independently when serving as a transcriptional repressor 5. GIY-YIG proteins have been found in all three domains of life, as DNA repair enzymes, restriction enzymes and endonucleases encoded by other mobile DNAs, such as the Penelope-like elements of Drosophila 6. Linkers are apparent in some of these proteins. Of particular interest are the Penelope-like retroelements, in which the GIY-YIG domains are joined to a module of different function, namely reverse transcriptase 25. The reverse transcriptase is thought to make a cDNA copy of the element to be inserted into the cut created by the GIY-YIG module. As with I-TevI, the linkers of Penelope-like elements are in the 70-80 amino acid range. What the role of the linker might be in directing physical interaction or functional communication between the endonuclease and polymerization domains is not known. However, the adaptability of the I-TevI linker to accommodate the different functions of endonuclease and repressor stimulates one to ask what the specific principles might be that govern the interplay between discrete functions of other modular GIY-YIG enzymes.
MATERIALS AND METHODS
I-TevI linker mutagenesis
The expression plasmid pSP65-716, a pSP65 vector with the wild-type I-TevI sequence inserted between the EcoRI and HindIII sites under the control of the SP6 promoter 7; 13, was used as a template for the PCR to generate DI and DT linker deletion and Ala-substitution mutants, as previously described 4. Briefly, mutagenic oligonucleotide primer pairs complementary to sequences flanking a single location introduced a unique restriction site and the desired deletions or Ala substitutions. The resulting PCR products were gel purified, digested with the appropriate restriction enzyme and self-ligated. These plasmids were sequenced to verify the mutations.
In vitro transcription and translation
All DI and DT deletion and Ala-substitution mutant proteins were expressed in vitro. HindIII-linearized plasmid DNA (~1 μg) was used as template for in vitro transcription with SP6 RNA polymerase (Invitrogen). The resulting mRNA was purified by Qiagen RNeasy mini kit, and translated in vitro using wheat germ extract (Promega) in the presence of [35S]-Met. Aliquots of the reaction mixtures were fractionated on 12% SDS polyacrylamide gels and the relative amounts of the I-TevI linker mutants were determined by comparison of radioactive counts of each protein using a Typhoon 9400 (GE Healthcare) phosphorimager and ImageQuant software (Molecular Dynamics).
Cleavage assays
To compare the cleavage efficiency of I-TevI linker derivatives, the same relative concentration of each protein, determined by the [35S]-Met signal on the Typhoon, was used in each reaction. Cleavage reactions (20 μl) were performed at 37°C by incubating 250 ng of ScaI-linearized pTZtdΔI, containing a wild-type td homing site, with decreasing amounts of protein for 15 min in 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, and 100 mM NaCl 13. The reactions were quenched with 4 μl 10X stop-load buffer (50 μM EDTA, 25% glycerol, 5% SDS and 0.025% bromophenol blue), followed by an incubation at 37°C with 1 μl RNase A (500 μg/ml) for 15 min, then with 1 μl proteinase K (20 mg/ml) for 1 h. Samples were fractionated on 1% agarose gels in TAE buffer (40 mM Tris-acetate, 1 mM EDTA), and the gels were stained with SYBR-Gold (Molecular Probes). The cleavage products were visualized with a Typhoon 9400 phosphorimager, and the extent of cleavage was determined using ImageQuant.
Cleavage-site mapping
Double-stranded, [32P]-labeled wild-type (304 bp), +5 (309bp) and +10 (314 bp) I-TevI homing-site substrates were generated by the PCR using pTZtdΔI, pTZ18UtdIC+5 or pTZtdIC+10 as templates, respectively 13. Twenty picomoles of sequencing primers, W311 (5′-TATTGATCG-TATTAAAAAACTGCC-3′) and W312 (5′-ACATTGTTCTACGTGATTC-3′), 5′ end-labeled with 16.8 pmol (50 μCi) of γ[32P]ATP by T4 polynucleotide kinase (New England Biolabs), were used for substrate amplification. Each labeled primer was used in PCR reactions with a non-isotopically labeled primer partner. Cleavage-site mapping was performed as previously described 26. Briefly, the cleavage reactions were conducted at 37°C for 15 min by incubating 5,000 cpm of 5′ end-labeled double-stranded DNA substrate with in vitro synthesized I-TevI derivatives in 20 μl of 50 mM Tris-HCl (pH 8.0), 10mM MgCl2 and 100 mM NaCl. The reaction mixtures were phenol extracted and ethanol precipitated, resuspended in 5 μl stop solution (95% formamide, 20 mM EDTA, 0.05% bromophenol blue and 0.05% xylene cyanol) and fractionated on 6% denaturing polyacrylamide (8.3 M urea) sequencing gels. Samples were fractionated alongside a dideoxynucleotide sequencing ladder generated with the same end-labeled primer using a Thermo Sequenase Cycle Sequencing Kit (USB).
Linker cloning and purification for NMR
DNA corresponding to the wild-type linker region of I-TevI (amino acids 93-148, I-TevI93-148) or R129A mutant of the linker region of I-TevI (R129A-I-TevI93-148) was cloned into the EcoRI/SalI restriction sites of pMAL-c2x (New England Biolabs), in which the Factor Xa cleavage site had been replaced with a thrombin cleavage site (LVPR↓GS, arrow = cleavage site), and a 6-His tag had been added N-terminal to the maltose binding protein. The resulting fusion protein was thus 6-His/maltose binding protein/LVPRGSEF/I-TevI93-148, or 6-His/maltose binding protein/LVPRGSEF/R129A-I-TevI93-148.
To obtain [U-15N, 13C]-I-TevI93-148 and [U-15N, 13C]-R129A-I-TevI93-148 for the NMR studies, the plasmid constructs were grown at 37°C in E. coli ER2566 in M9 minimal medium containing 1 g per liter [U-15N] NH4Cl (Sigma Aldrich) and 2 g per liter of [U-13C] dextrose (Sigma Aldrich) as sole nitrogen and carbon sources to OD600 0.4, induced with 1 mM IPTG for 3 h at 20°C and harvested by centrifugation. Cell pellets were sonicated in 20 ml 50 mM Tris pH 8.0, 500 mM NaCl, and 20 mM imidazole supplemented with 1 tablet Complete Mini EDTA-free protease inhibitors (Roche) per 10 ml, and the supernatant was applied to a 5 ml Hi-Trap chelating column (GE Healthcare) that was previously charged with NiSO4 and equilibrated in the same buffer without protease inhibitors. Thrombin (20 U, Novagen) was added to the column in 5 ml buffer and the protein was left to digest at 25°C overnight. The cleaved protein was eluted in 1 ml fractions and visualized by SDS-PAGE. Select fractions were dialyzed overnight at 4°C against 50 mM Tris pH 8.0, 20 mM NaCl, 1 mM DTT, 0.01% NaN3 and 1 mM EDTA supplemented with 1 tablet protease inhibitors per liter. The sample was then applied to a Hi-Trap Q-HP anion exchange column (GE Healthcare) in sequence with a Hi-Trap SP-HP cation exchange column (GE Healthcare), with the protein fragment passing through the Q column and binding to the SP column. The protein was then eluted from the SP column over a 1 M NaCl gradient using an AKTA FPLC (GE Healthcare). After SDS-PAGE, select fractions were concentrated to 1 ml final volume with an Amicon Ultra spin concentrator. The sample was then run on a Superdex 75 (GE Healthcare) size exclusion column equilibrated in 50 mM Tris pH 8.0, 250 mM NaCl, 1 mM DTT, 0.01% NaN3, and 1 mM EDTA with 1 tablet per liter protease inhibitors. Select fractions were dialyzed against 10 mM KPO4 pH 7.0, 100 mM NaCl, 1 mM DTT, 0.01% NaN3, and 1 mM EDTA and concentrated to 500 μl final volume using an Amicon Ultraspin concentrator for the NMR analysis.
NMR Spectroscopy
NMR experiments analyzing linker segments were performed on an Avance Bruker spectrometer, operating at a 1H frequency of 700 MHz and equipped with a z-axis gradient cryoprobe. All NMR data were collected at 25°C. Protein samples of [U-15N, 13C] I-TevI93-148 and [U-15N, 13C] R129A-I-TevI93-148 with concentrations ranging from 0.3–0.5 mM were dissolved in the NMR buffer (10 mM KPO4 pH 7.0, 100 mM NaCl, 0.02% NaN3, and 90%/10% H2O/D2O.). An 1H{15N}-HSQC experiment was used to optimize NMR buffer conditions and monitor structural integrity of the NMR samples. The triple resonance experiments HNCA, HNCOCA, 15N-edited NOESY and 15N-edited TOCSY experiments 27 were collected using previously described sequences. All spectra were processed using TOPSPIN (Bruker, Inc), and NMR chemical shift assignments were made using CARA 28.
Computational analysis
The amino acid sequence of the linker region of I-TevI (residues 93 to 167) was used as a query sequence to search the NCBI Environmental database using protein-protein BLAST. The top four results (with e-values ranging from 2e −5 to 0.12) were then aligned with the full-length I-TevI sequence using the EMBL-EBI program MAFFT (version 6, http://align.bmr.kyushu-u.ac.jp/mafft/software/). Sequences used to align the N-terminal portions of hypothetical proteins A and C (Fig. 6) were obtained from the NCBI env_nr database contigs. For secondary structure prediction, the PredictProtein Server was used 29.
Supplementary Material
(a) Cleavage-site mapping. Position of the top- and bottom-strand cleavage sites are depicted by open (gray) and black triangles, respectively. DNA substrates containing the wild-type I-TevI homing site 5′-end labeled on the top or bottom strand were incubated with in vitro synthesized DI or DT Ala-substitution mutants, and cleavage products were fractionated on a 6% denaturing gel alongside a sequencing ladder generated with the same labeled primer and template DNA used to generate the DNA substrates. Lane 1, DI-1AA; lane 2, DI-2AA; lane 3, DI-3AA; lane 4, DT-1AAA; lane 5, DT-2AAA; lane 6, DT-3AAA; lane 7, DT-4AA; lane 8, wild-type I-TevI. The wild-type I-TevI homing site sequence is shown below. (b) Cleavage efficiency of the I-TevI DT Ala-substitution mutants. Cleavage assays were performed as described in Materials and Methods on linearized pTZtdΔI DNA and the reaction products were resolved on agarose gels. S, substrate; P, cleavage products. The proteins were synthesized in vitro, and the cleavage reactions mixtures contained decreasing amounts of I-TevI mutants as described in the legend to Figure 1(b).
The gel is labeled as in Figure 4(b). The spurious cleavages demarcated by square brackets on the bottom strand are thought to result from a CXXXG motif 5 nt downstream of the wild-type cleavage site (overscored).
(a) Schematic of cleavage activity of deletion mutants taken from ref 4. Data for ΔZn are from ref 12. The relative cleavage activity of the I-TevI deletion derivatives, expressed as % cleavage of substrate, is shown in the bar chart. Columns on the right refer to mutation type (Mut: Δ = deletion, P = point mutation) and substrate (Sub: WT, +5 or +10). (b) Cleavage efficiency of DI and DT Ala-substitution mutants on wild-type I-TevI HS. The cleavage activities are plotted as in (a). Data for CZnA are from ref 12. (c) – (f). Graphical representation of percent cleavage at the natural sequence (gray bars) versus at fixed distance (black bars). Data in the compilation are taken from Figures 2–4 in the text.
Acknowledgments
We are thankful to Karla Lehtonen and Matt Stanger for generating the data shown in Figure 4(b). We are grateful to lab members for their critical suggestions, and particularly to Justin Robbins, Jeanne DiChiara and David Edgell for their thoughtful comments on the MS. We thank Justin Robbins and Gil Amitai for their help with the computational analyses, and an anonymous reviewer for suggesting that we perform these studies. Thanks also to Maryellen Carl for preparing the manuscript, and to the Wadsworth Center Molecular Genetics Core for DNA sequencing. This work was supported by grants from the NIH to MB (GM39422 and GM44844) and a Career Development Award from the American Diabetes Association to AS (1-06-CD-23).
References
- 1.Gokhale RS, Khosla C. Role of linkers in communication between protein modules. Cur Opin Chem Biol. 2000;4:22–27. doi: 10.1016/s1367-5931(99)00046-0. [DOI] [PubMed] [Google Scholar]
- 2.Wriggers W, Chakravarty S, Jennings PA. Control of protein functional dynamics by peptide linkers. Biopolymers. 2005;80:736–46. doi: 10.1002/bip.20291. [DOI] [PubMed] [Google Scholar]
- 3.Van Roey P, Derbyshire V. GIY-YIG homing endonucleases - beads on a string. In: Belfort M, Derbyshire V, Stoddard BL, Wood DW, editors. Homing Endonucleases and Inteins. Springer-Verlag; 2005. pp. 67–83. [Google Scholar]
- 4.Kowalski JC, Belfort M, Stapleton MA, Holpert M, Dansereau JT, Pietrokovski S, Baxter SM, Derbyshire V. Configuration of the catalytic domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu QQ, Derbyshire V, Belfort M, Edgell DR. Distance determination by GIY-YIG intron endonucleases: discrimination between repression and cleavage functions. Nucleic Acids Res. 2006;34:1755–1764. doi: 10.1093/nar/gkl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunin-Horkawicz S, Feder M, Bujnicki JM. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics. 2006;7:98. doi: 10.1186/1471-2164-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell-Pedersen D, Quirk S, Clyman J, Belfort M. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res. 1990;18:3763–3770. doi: 10.1093/nar/18.13.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu FK, Maley G, Pedersen-Lane J, Wang AM, Maley F. Characterization of the restriction site of a prokaryotic intron-encoded endonuclease. Proc Natl Acad Sci USA. 1990;87:3574–3578. doi: 10.1073/pnas.87.9.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell-Pedersen D, Quirk SM, Bryk M, Belfort M. I-TevI, the endonuclease encoded by the mobile td intron, recognizes binding and cleavage domains on its DNA target. Proc Natl Acad Sci USA. 1991;88:7719–7723. doi: 10.1073/pnas.88.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Roey P, Waddling CA, Fox KM, Belfort M, Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryk M, Belisle M, Mueller JE, Belfort M. Selection of a remote cleavage site by I-TevI, the td intron-encoded endonuclease. J Mol Biol. 1995;247:197–210. doi: 10.1006/jmbi.1994.0133. [DOI] [PubMed] [Google Scholar]
- 12.Edgell DR, Stanger MJ, Belfort M. Coincidence of cleavage sites of intron endonuclease I-TevI and critical sequences of the host thymidylate synthase gene. J Mol Biol. 2004;343:1231–1241. doi: 10.1016/j.jmb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Dean AB, Stanger MJ, Dansereau JT, Van Roey P, Derbyshire V, Belfort M. Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI. Proc Natl Acad Sci USA. 2002;99:8554–8561. doi: 10.1073/pnas.082253699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Roey P, Meehan L, Kowalski J, Belfort M, Derbyshire V. Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nat Struct Biol. 2002;9:806–811. doi: 10.1038/nsb853. [DOI] [PubMed] [Google Scholar]
- 15.Edgell DR, Shub DA. Related homing endonucleases I-BmoI and I-TevI use different strategies to cleave homologous recognition sites. Proc Natl Acad Sci USA. 2001;98:7898–7903. doi: 10.1073/pnas.141222498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derbyshire V, Kowalski JC, Dansereau JT, Hauer CR, Belfort M. Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J Mol Biol. 1997;265:494–506. doi: 10.1006/jmbi.1996.0754. [DOI] [PubMed] [Google Scholar]
- 17.Richter CD, Stanmore DA, Miguel RN, Moncrieffe MC, Tran L, Brewerton S, Meersman F, Broadhurst RW, Weissman KJ. Autonomous folding of interdomain regions of a modular polyketide synthase. FEBS J. 2007;274:2196–2209. doi: 10.1111/j.1742-4658.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- 18.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 19.Sung YH, Eliezer D. Residual structure, backbone dynamics, and interactions within the synuclein family. J Mol Biol. 2007;372:689–707. doi: 10.1016/j.jmb.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolin A, Jevtic V, Swint-Kruse L, Egan SM. Linker regions of the RhaS and RhaR proteins. J Bacteriol. 2007;189:269–271. doi: 10.1128/JB.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgell DR, Derbyshire V, Van Roey P, LaBonne S, Stanger MJ, Li Z, Boyd TM, Shub DA, Belfort M. Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor. Nat Struct Mol Biol. 2004;11:936–944. doi: 10.1038/nsmb823. [DOI] [PubMed] [Google Scholar]
- 22.Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc Natl Acad Sci USA. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva GH, Belfort M, Wende W, Pingoud A. From monomeric to homodimeric endonucleases and back: engineering novel specificity of LAGLIDADG enzymes. J Mol Biol. 2006;361:744–754. doi: 10.1016/j.jmb.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 24.Ye J, Cho SH, Fuselier J, Li W, Beckwith J, Rapoport TA. Crystal structure of an unusual thioredoxin protein with a zinc finger domain. J Biol Chem. 2007;282:34945–34959. doi: 10.1074/jbc.M704044200. [DOI] [PubMed] [Google Scholar]
- 25.Evgen’ev MB, Arkhipova IR. Penelope-like elements--a new class of retroelements: distribution, function and possible evolutionary significance. Cytogenet Genome Res. 2005;110:510–521. doi: 10.1159/000084984. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q, Belle A, Shub DA, Belfort M, Edgell DR. SegG endonuclease promotes marker exclusion and mediates co-conversion from a distant cleavage site. J Mol Biol. 2003;334:13–23. doi: 10.1016/j.jmb.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 27.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR Spectroscopy: Principles and practice. Academic Press; San Diego: 1996. [Google Scholar]
- 28.Masse JE, Keller R. AutoLink: automated sequential resonance assignment of biopolymers from NMR data by relative-hypothesis-prioritization-based simulated logic. J Mag Res. 2005;174:133–151. doi: 10.1016/j.jmr.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 29.Rost B, Yachdav G, Liu J. The PredictProtein Server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Cleavage-site mapping. Position of the top- and bottom-strand cleavage sites are depicted by open (gray) and black triangles, respectively. DNA substrates containing the wild-type I-TevI homing site 5′-end labeled on the top or bottom strand were incubated with in vitro synthesized DI or DT Ala-substitution mutants, and cleavage products were fractionated on a 6% denaturing gel alongside a sequencing ladder generated with the same labeled primer and template DNA used to generate the DNA substrates. Lane 1, DI-1AA; lane 2, DI-2AA; lane 3, DI-3AA; lane 4, DT-1AAA; lane 5, DT-2AAA; lane 6, DT-3AAA; lane 7, DT-4AA; lane 8, wild-type I-TevI. The wild-type I-TevI homing site sequence is shown below. (b) Cleavage efficiency of the I-TevI DT Ala-substitution mutants. Cleavage assays were performed as described in Materials and Methods on linearized pTZtdΔI DNA and the reaction products were resolved on agarose gels. S, substrate; P, cleavage products. The proteins were synthesized in vitro, and the cleavage reactions mixtures contained decreasing amounts of I-TevI mutants as described in the legend to Figure 1(b).
The gel is labeled as in Figure 4(b). The spurious cleavages demarcated by square brackets on the bottom strand are thought to result from a CXXXG motif 5 nt downstream of the wild-type cleavage site (overscored).
(a) Schematic of cleavage activity of deletion mutants taken from ref 4. Data for ΔZn are from ref 12. The relative cleavage activity of the I-TevI deletion derivatives, expressed as % cleavage of substrate, is shown in the bar chart. Columns on the right refer to mutation type (Mut: Δ = deletion, P = point mutation) and substrate (Sub: WT, +5 or +10). (b) Cleavage efficiency of DI and DT Ala-substitution mutants on wild-type I-TevI HS. The cleavage activities are plotted as in (a). Data for CZnA are from ref 12. (c) – (f). Graphical representation of percent cleavage at the natural sequence (gray bars) versus at fixed distance (black bars). Data in the compilation are taken from Figures 2–4 in the text.