Abstract
Treatment of hematologic malignancies is evolving from a uniform approach to targeted therapies directed at the underlying molecular abnormalities of disease. The mixed lineage leukemia (MLL) proto-oncogene is a recurrent site of genetic rearrangements in acute leukemias; and since its discovery in 1992, many advances have been made in understanding its role in leukemogenesis. A variety of MLL translocation partners have been described, and detailed structure/function studies have identified functional domains that are required for transformation. Proteins associated with the MLL core complex or its fusion partners have been isolated and characterized for their critical roles in leukemia pathogenesis. Downstream mediators of MLL transcriptional regulation and multiple collaborating signaling pathways have been described and characterized. These advances in our understanding of MLL-related leukemogenesis provide a foundation for ongoing and future efforts to develop novel therapeutic strategies that will hopefully result in better treatment outcomes.
Introduction
The mixed lineage leukemia (MLL) gene is frequently rearranged in acute myeloid and lymphoblastic leukemias in adults and children1,2 and identifies a patient population with a particularly poor prognosis.3 Leukemogenic MLL rearrangements occur in a variety of forms, including reciprocal chromosomal translocations and partial tandem duplications of internal coding regions.4 The MLL gene encodes a histone methyltransferase (HMT) implicated in epigenetic regulation of transcription that is critical for normal embryonic development and hematopoiesis.5 Among the most widely studied target genes of MLL transcriptional regulation are HOX genes, which themselves are implicated in the malignant transformation of hematopoietic progenitors.6,7
In leukemias, chromosomal translocations fuse the amino-terminal part of MLL in-frame to one of more than 50 partner proteins.8 The most common translocations in acute lymphoblastic leukemia (ALL) are t(4;11) and t(11;19), resulting in expression of MLL-AF4 and MLL-ENL, respectively, whereas acute myeloid leukemia (AML) is frequently associated with t(9;11) and t(6;11) giving rise to MLL-AF9 and MLL-AF6, respectively. All MLL fusion proteins retain the amino-terminal portion containing AT hooks and the CxxC domain of MLL, thus preserving DNA-binding activity. In contrast, a region with transactivating potential, the plant homeodomain (PHD) fingers, and the suppressor of variegation-enhancer of zeste-trithorax (SET) domain, which mediates histone H3 lysine 4 (H3K4) methylation, are lost. Although loss of the carboxy-terminal regions of MLL in chimeric oncoproteins would be predicted to result in abrogation of transactivation and HMT functions, transforming MLL fusion proteins function as transcriptional regulators and induce aberrant expression of downstream MLL targets, including HOX genes.9,10 The precise mechanism for this aberrant transcriptional activity is not known but involves formation of a transcriptional core complex by the remaining N-terminal part of MLL and the fusion partner-portion of the chimeric MLL oncoprotein.
The partner proteins of MLL that normally reside in the nucleus commonly possess intrinsic transcriptional regulatory function and mediate aberrant recruitment of the transcription machinery to MLL target genes.11–14 Several MLL fusion partners, including AF4, AF9, AF10, and ENL, form a higher-order complex involved in transcriptional elongation and/or recruit the histone-modifying enzyme hDOT1L (human DOT1-like), ultimately allowing aberrant transcription of MLL target genes. In contrast, cytoplasmic MLL fusion partners do not appear to contain inherent transcriptional activation properties but allow self-association or dimerization of the N-terminal part of MLL, presumably enhancing the ability of MLL to bind to its traditional cofactors or promoting recruitment of additional factors by MLL or the partner protein moiety.15–18 Independent of the mechanistic differences, both types of MLL fusion proteins induce aberrant expression of downstream mediators, including HOX proteins, erythropoietin-producing hepatoma-amplified sequence A7 (EPHA7) and myeloid ecotropic viral integration site 1 (MEIS1). In addition, gene expression profiling of patient samples,19 as well as results from retroviral transduction studies,20 suggest a cooperative effect of activating mutation of FMS-like tyrosine kinase 3 (FLT3) with MLL fusion proteins. Other pathways that have recently been implicated in MLL-induced leukemogenesis include the glycogen synthase kinase 3 (GSK-3), heat shock protein 90 (HSP-90), and RAS pathways. This article reviews recent advances in MLL research with particular emphasis on the mechanisms of MLL-induced leukemogenesis and the identification of potential therapeutic targets (Figure 1).
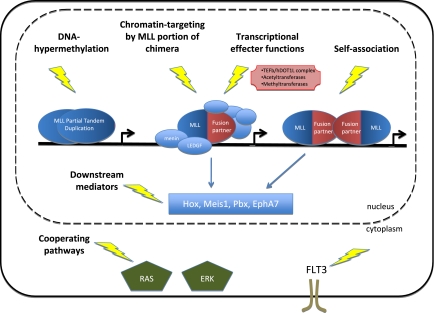
Figure 1.
Therapeutic targeting of MLL leukemia. Several key molecular functions that may provide potential therapeutic targets in MLL leukemia include chromatin association by the MLL portion of chimeric oncoproteins, nucleation of transcriptional effector complexes through the fusion partner moiety of MLL chimeras, downstream mediators of MLL, and pathways known to cooperate with MLL.
Chromatin targeting by the MLL portion of chimeric oncoproteins
MLL-menin-LEDGF interactions
Menin is one of several proteins that comprise the MLL core HMT complex. Originally identified as a product of the gene responsible for the heritable cancer syndrome multiple endocrine neoplasia type I,21 menin interacts with the N-terminal part of MLL through a conserved binding motif.22 Similar to loss of MLL, down-regulation of menin results in impaired maintenance of HOX gene expression.22,23 Consistent with this observation, menin is associated with the promoter regions of HOX genes and is essential for expression of Hoxa9.24 In addition, menin and MLL cooperatively regulate the expression of the cyclin-dependent kinase inhibitors (CDKI) p27Kip1 and p18Ink4c25 as well as the Hox cofactor Meis1. Genetic ablation of menin abrogates the differentiation arrest and oncogenic properties of MLL-transformed leukemic blasts.22 Similarly, disrupting the interaction between menin and MLL with a dominant negative inhibitor consisting of N-terminal MLL sequences results in down-regulation of Meis1 expression and inhibits proliferation of MLL-rearranged leukemic blasts, suggesting that targeting of the MLL-menin interaction may be a promising new approach for future drug development for MLL-induced leukemia,26 although the selectivity of this approach remains to be determined.
Recent studies27 further define the leukemogenic role of menin as a molecular adaptor linking MLL with lens epithelium-derived growth factor (LEDGF), a chromatin-associated protein previously implicated in transcriptional coactivation28 and leukemogenesis.29 Biochemical purification of the MLL-ENL/menin complex identified LEDGF as an associated protein, and chromatin immunoprecipitation demonstrated that LEDGF colocalizes with MLL-ENL/menin at crucial target sites within HOXA, MEIS1, and CDKI genes. Importantly, specific association with LEDGF is essential for MLL-dependent transcriptional regulation and leukemic transformation. Interestingly, engineered MLL oncoproteins in which the menin-binding motif was replaced with the chromatin-associating PWWP domain of LEDGF retained an ability to up-regulate Hoxa9 expression and efficiently transformed myeloid progenitors effectively bypassing menin, suggesting that menin's sole purpose is to tether MLL proteins with LEDGF promoting recruitment of the complex to transcriptionally active chromatin through the PWWP domain. These results establish LEDGF as an essential cofactor required for the leukemogenic function of the MLL fusion protein/menin complex and identify interactions between the components of the complex as potential therapeutic targets.
MLL-DNA interaction
CpG dinucleotide methylation is a critical epigenetic modification involved in the regulation of chromatin structure and gene transcription. MLL selectively binds to nonmethylated CpG DNA via its CxxC domain,30 which is retained in all leukemogenic MLL-fusion proteins and critically required for malignant transformation.31 Using chemical shift perturbation analysis and site-directed mutagenesis, the CxxC domain DNA binding interface has been identified and characterized.32 This structural information provides an important context for the design of therapeutics specifically targeting the interaction between the CxxC domain of MLL and nonmethylated CpG DNA.
Nucleation of transcriptional effector complexes through the fusion partner moiety of MLL chimeras
The nuclear fusion partners of MLL confer leukemogenic potential by providing domains with transcriptional effector activity and/or by recruiting transcriptional cofactors. Rarely, MLL is fused directly with known transcriptional activator proteins (eg, members of the forkhead transcription factor family) or coactivators (eg, CBP and p300). In most MLL leukemias, however, MLL is fused to proteins with previously unknown roles in transcriptional regulation. Immuno-purification and mass spectrometry studies have revealed that several of the more common MLL partner proteins associate with each other as components of a higher-order molecular complex. Molecular therapies directed at the interactions of these proteins or their functions may be an effective therapeutic strategy for leukemias with MLL rearrangements, although targeting of protein-protein interactions remains technically challenging. Alternatively, enzymes within the complexes that mediate critical posttranslational modifications represent potential therapeutic targets.
AF4 and AF9
AF4 and AF9 represent the most frequently observed MLL partner proteins in ALL and AML, respectively. Recent studies have revealed a critical role for AF4 and its associated proteins in the coordination of chromatin remodeling and maintenance of RNA polymerase II (Pol II) transcriptional elongation. Several MLL fusion proteins (including AF4, AF5, AF9, ENL, and AF10) normally comprise a higher-order complex that mediates histone H3 lysine 79 (H3K79) methylation by recruiting hDOT1L to elongating Pol II. In addition, the complex acts as a positive regulator of Pol II transcription elongation factor b (P-TEFb) kinase. In turn, the transactivation activity and protein stability of AF4, AF9, and ENL are tightly regulated by P-TEFb–dependent phosphorylation.33
An AF4-mimetic peptide, PFWT, has been engineered to competitively interfere with the interaction between MLL-AF4 and AF9,34 as demonstrated in vitro and in vivo. Furthermore, exposure of leukemic blasts carrying the t(4;11) translocation to PFWT, albeit at high concentrations, results in inhibition of proliferation and necrotic cell death, with little effect on normal hematopoietic progenitors,35 providing a promising rationale to develop therapeutic agents that target AF4-AF9 interactions.
Another possible therapeutic strategy in this context is the use of flavopiridol, a flavonoid derived from a medicinal plant in India. Flavopiridol is a potent inhibitor of cyclin-dependent kinase 9, a component of P-TEFb, and would therefore be expected to abrogate transcriptional elongation. However, detailed studies are needed to provide convincing support for the efficacy of flavopiridol in MLL-related leukemia.
hDOT1L
hDOT1L is an H3K79 methyltransferase that serves a critical role in regulating chromatin structure and transcription.36 In a yeast 2-hybrid screen, the MLL fusion partner AF10 was identified as an hDOT1L-interacting protein. The octapeptide motif-leucine zipper region of AF10, required for leukemic transformation,37 was found to mediate the interaction between AF10 and hDOT1L. This interaction is critical for leukemogenesis induced by MLL-AF10, and HMT activity of hDOT1L is necessary for transformation of hematopoietic progenitors by MLL-AF10 as well as a synthetic MLL-hDOT1L fusion protein. For both fusion proteins, leukemogenesis was dependent on up-regulation of Hoxa9 mediated through recruitment of hDOT1L and H3K79 methylation.38
Subsequent immuno-purification experiments suggested an association of hDOT1L with the MLL fusion partner ENL.39 Yeast 2-hybrid experiments confirmed a direct interaction between ENL and hDOT1L, and structure/function studies showed that the hDOT1L interaction domain in ENL coincides with a distinct C-terminal region that is critical for transformation by MLL-ENL. The binding affinity of ENL to hDOT1L was highly correlated with the transforming ability of MLL-ENL. In addition, transient knockdown of ENL diminished genome-wide as well as HOXA9 gene-specific H3K79 dimethylation, providing further circumstantial evidence of a critical role for ENL and hDOT1L interaction in leukemia related to MLL-ENL.
Taken together, the results of these studies establish hDOT1L as a candidate cancer drug target for treatment of MLL-induced leukemias. The interaction between hDOT1L and AF10 or ENL, respectively, may be a promising therapeutic target, although the recruitment of hDOT1L by both AF10 and ENL may complicate this strategy. Alternatively, compounds capable of directly inhibiting hDOT1L HMT activity may represent a more feasible approach. In this regard, a high-resolution structure of the catalytic domain of hDOT1L40 provides important information for rational drug design. Nevertheless, potential hDOT1L inhibitors will need to be used with caution as hDOT1L is the only known H3K79 methyltransferase and its genetic disruption in mice results in embryonic lethality.41 In murine embryonic stem cells, Dot1L deficiency leads to loss of H3K79 methylation and reduced levels of heterochromatic marks accompanied by telomere elongation, aneuploidy, and proliferation defects. These findings indicate a critical role for Dot1L in heterochromatin formation and embryonic development. Because the requirement of Dot1L for normal cellular function is probably not restricted to embryonic stem cells, careful studies are needed to ensure the safety of pharmacologic hDOT1L inhibition.
PRMT1
The protein arginine methyltransferase 1 (PRMT1) is an important epigenetic transcriptional regulator that maintains transcriptionally active chromatin through methylation of arginine 3 of histone 4 (H4R3).42 PRMT1 was recently shown to be an essential component of the leukemogenic MLL-EEN transcriptional core complex with both H4R3 methylation and histone acetylation activity.18 PRMT1 is recruited to MLL-EEN by the bridging molecule SAM68 (Src-associated in mitosis of 68 kDa), which binds directly to the SH3 domain of EEN. Chromatin immunoprecipitation assays demonstrated in vivo recruitment of PRMT1 by MLL-EEN to downstream targets, such as HOXA9, and confirmed specific H4R3 asymmetric dimethylation marks on the HOXA9 gene. Consistent with the role of H4R3 methylation in promoting histone acetylation by CBP/p300, increased H4 acetylation of the HOXA9 promoter was also observed. Further evidence supporting a critical function for the specific recruitment of PRMT1 by SAM68 in MLL leukemogenesis was provided by retroviral transduction and transplantation assays using synthetic constructs directly fusing MLL to PRMT1 and SAM68, respectively. In these assays, both MLL-PRMT1 and MLL-SAM68 were capable of transforming primary myeloid progenitors in vitro. In contrast, specific knockdown of PRMT1 or SAM68 expression suppressed MLL-mediated transformation. In addition, the catalytically inactive mutant MLL-PRMT1 (G80R) completely lost its transforming ability, confirming that H4R3 asymmetric dimethylation is critical for transformation induced by MLL-EEN.18
Although EEN is an exceedingly rare MLL fusion partner, this study establishes a critical role for protein arginine methylation in leukemic transformation and reveals its potential as a novel therapeutic target. Agents inhibiting the recruitment of PRMT1 to the transforming MLL transcriptional core complex by disrupting the interaction between SAM68 and PRMT1 or direct inhibitors of the catalytic activity of PRMT1 may allow to suppress the aberrant transcriptional activation property that PRMT1 confers to the MLL-EEN complex and possibly other MLL fusion proteins containing SH3 domains.
Self-association
MLL self-association mediated by the fusion partner domains of GAS7, AF1p, and gephyrin results in activation of the transcriptional activity of MLL and transformation.15,16 Although the mechanistic basis of the latter is not well understood, this process potentially involves recruitment of transcriptional cofactors by either the fusion partner or MLL itself. Interestingly, compared with MLL chimeras with a nuclear partner protein, self-associating MLL-fusion proteins bind to their target genes (eg, the HOXA9 locus) with a similar pattern and lead to increased histone acetylation and H3K4 methylation. However, unlike nuclear MLL fusion proteins, self-associating MLL fusion proteins do not induce H3K79 dimethylation, suggesting a different mechanism for transcriptional activation.43 Taken together, these results suggest that inhibition of the self-association process itself and inhibition of the currently unknown transcriptional cofactors recruited by self-associating MLL fusion proteins represent suitable approaches to interrupt key steps of leukemogenesis.
Downstream mediators of MLL
HOX genes
HOX proteins constitute a particular subgroup of homeodomain transcription factors that serve critical functions in patterning of the body axis. In addition, various members of the HOX gene family have been implicated in regulation of stem cell self-renewal and leukemic transformation. Gene expression profiling has consistently demonstrated up-regulation of a subset of major HOX genes, including HOXA4, HOXA5, HOXA7, HOXA9, HOXA10, and HOXC6 in leukemic blasts with MLL gene rearrangements.44,45 In mouse models of MLL-mediated transformation, most MLL oncoproteins tested demonstrate at least partial dependence on Hox function. MLL-ENL and MLL-AF10 are strongly dependent on Hoxa9, whereas the oncogenic potential of MLL-GAS7 is attenuated but not eliminated in the absence of Hoxa7 or Hoxa9.46,47 In contrast, mice with an MLL-AF9 knock-in mutation developed myeloid leukemia with a similar incidence in the presence or absence of Hoxa9.48 The observed variability probably reflects differences in experimental systems and functional redundancy between the multiple expressed HOX genes. Consistent with the latter, more severe abrogation of transformation is observed in vitro in cells deficient for 2 Hox genes (M.L.C., unpublished observations, 2007), which further supports a rate-limiting role for HOX in MLL-mediated transformation. Therefore, strategies based on broad down-regulation of HOXA gene expression or global inhibition of HOX protein activity might prove to be of benefit in the treatment of MLL-related leukemia.
EPHA7
EPH receptor tyrosine kinases and their ligands, the cell-surface bound ephrins, are part of a signaling system activated by cell-cell interaction. The EPH/ephrin signaling network plays an important role in neurodevelopmental processes and has also been implicated in malignant transformation. Recent work by Nakanishi et al49 has shown that MLL fusion proteins up-regulate EPHA7 by binding to the EPHA7 promoter, suggesting that it is a direct transcriptional target of MLL fusion proteins. Interestingly, subsequent studies have demonstrated that up-regulation of EPHA7 is associated with phosphorylation of the extracellular-signal-regulated kinase ERK. When treated with the potent ERK inhibitor 5-iodotubercidin, leukemic blasts expressing the MLL-AF4 fusion protein rapidly undergo apoptosis.49 Taken together, these results suggest a potential role for inhibition of EPH receptor tyrosine kinases and ERK in the treatment of MLL-induced leukemia.
MEIS and PBX
Homeodomain proteins of the PBX (pre-B-cell leukemia transcription factor) and MEIS families regulate gene expression as hetero-oligomeric complexes with HOX proteins.50 MEIS1 is consistently highly expressed in MLL-related leukemias44 and represents an important oncogenic collaborator. The collaborative activity of MEIS1 is mediated by 3 conserved domains that serve as the PBX interaction motif, the DNA-binding homeodomain, and the transactivating C-terminal region,51 respectively, suggesting that MEIS1 associates with PBX partners in a DNA-binding transcriptional complex to mediate MLL transformation.
Induction and maintenance of MLL-induced leukemogenesis in a murine model require Meis1 and are codependent on the redundant contributions of Pbx2 and Pbx3. In particular, Meis1 was found to regulate leukemia stem cell (LSC) potential and frequency by controlling the extent of self-renewal, differentiation arrest, and cell-cycle activity of MLL leukemic blasts as well as the rate of in vivo LSC generation from myeloid progenitors.52 The critical effects of Meis1 on LSC characteristics are in part mediated through the retinoblastoma (RB) pathway by epigenetic regulation of Ink4a expression. The results of this study indicate that Meis1 and Pbx proteins are essential for MLL-induced transformation and that Meis1 is a rate-limiting determinant of MLL LSC biology. Both families of homeodomain proteins, which form obligate heterodimers with each other, therefore represent promising therapeutic targets. In this context, targeting of Meis1 is of particular interest, as blocking its effect on the MLL LSC might represent a potentially curative intervention.
GSK-3
GSK-3 is a serine/threonine kinase that plays a role in numerous signaling pathways, including those governing stem cell maintenance, cell-cycle division, and apoptosis.53 In many malignancies, pathways that are usually inhibited by GSK-3, such as Wnt and Hedgehog, become aberrantly activated, in some cases resulting from loss of GSK-3 activity.54 In addition, GSK-3 inhibition enhances repopulation of normal hematopoietic stem cells after bone marrow transplantation.55
Interestingly, MLL-transformed cells are paradoxically dependent on GSK-3 for sustained proliferation and maintenance of their transformed phenotype.56 When exposed to GSK-3 inhibitors, human leukemia cell lines and primary mouse hematopoietic cells transformed by MLL fusion genes rapidly undergo proliferative arrest, whereas non-MLL leukemia cells and normal bone marrow progenitors are unaffected at similar concentrations of inhibitor. Furthermore, inhibition of GSK-3 by activation of AKT, a physiologic negative regulator of GSK-3, as well as genetic ablation of the GSK-3 isoforms α and β, resulted in marked suppression of MLL-transformed cell proliferation in vitro. Subsequent studies showed that the CDK inhibitor p27kip1 mediates the cell-cycle arrest of MLL-transformed cells observed with GSK-3 inhibition. In a preclinical mouse model of MLL leukemia, inhibition of GSK-3 showed promising evidence of efficacy. Using pharmacologic, physiologic, and genetic inhibition, this study establishes a critical role for GSK-3 in the proliferation of MLL-transformed cells and credentials GSK-3 as a candidate therapeutic target.56 Given the converse actions of GSK-3 in leukemia versus normal stem cell maintenance, GSK-3 inhibition could provide therapeutic selectivity in the treatment of MLL-induced leukemias if inhibitors with suitable pharmacologic properties are developed. A more detailed understanding of the mechanistic basis for the paradoxical response to GSK-3 inhibition may provide additional targets.
Cooperating mutations and pathways
FLT3
Gene expression profiling of leukemic blasts isolated from patients with MLL-related ALL has revealed a high expression level of FLT3, and mutations resulting in activation of FLT3 are frequently observed.19 Similarly, MLL rearrangements, particularly MLL-AF9 and MLL PTD, are frequent in AML patients with an FLT3 mutation.57 Additional evidence for cooperation between MLL rearrangements and activating FLT3 mutations is provided by an experimental retroviral transduction model. Although retroviral transfer of the MLL fusion protein MLL-SEPTIN6 resulted in a myeloproliferative syndrome with long latency in mice, the addition of activated FLT3 induced acute biphenotypic or myeloid leukemia with short latency in vivo.20
It therefore appears plausible that inhibitors of FLT3 might have activity in MLL-related leukemia. Indeed, the FLT3 inhibitor PKC412 has been shown to differentially kill leukemic blasts carrying MLL translocations and activated or mutated FLT3. PKC412 was also shown to have significant activity in a murine model of MLL leukemia.19,58 Furthermore, sensitivity to the potent and selective FLT3-inhibitor CEP-701 was very high in patient samples with MLL rearrangements, and apoptosis of the leukemic blasts in vitro was observed in up to 82% of cases.59 Importantly, subsequent studies evaluating the combination of CEP-701 with chemotherapy in MLL-rearranged cell lines and patient samples showed a strong synergistic effect when CEP-701 was given after chemotherapy but an antagonistic effect when it was given before chemotherapy. This sequence dependence is thought to be the result of the effect of CEP-701 on cell-cycle progression and is mediated specifically by FLT3 inhibition, as it is not observed in control cells that do not express activated FLT3.60
In contrast to these studies, work by Lavau et al questions the significance of targeting FLT3 in leukemias associated with MLL fusion proteins.61 When injected into recipient mice, Flt3−/− and Flt3+/+ bone marrow progenitors transduced with MLL-ENL or MLL-CBP gave rise to rapidly progressing AML with similar phenotype and latency. Importantly, Flt3−/− cells expressing MLL-ENL were equally sensitive to growth inhibition by PKC412 as Flt3+/+ leukemic cells. These results indicate that Flt3 is dispensable for leukemic transformation induced by MLL fusion proteins and suggest that the efficacy of FLT3 inhibitors may be mediated by an off-target effect involving a different pathway. Nevertheless, ongoing trials should help establish whether FLT3 inhibitors, regardless of their targets, hold promise for treatment of MLL leukemia.
RAS
The RAS proteins belong to a large family of guanosine triphosphate (GTP)–binding proteins that play crucial roles in controlling the activity of signaling pathways that govern normal cellular growth and proliferation. In malignant cells, constitutively activated RAS proteins contribute to transformation by deregulating cell growth and apoptosis and by enhancing angiogenesis and metastatic potential.62 A retrospective analysis of 130 children with de novo AML and 313 children with B-precursor ALL showed a strong association between RAS mutations (50%), particularly K-RAS mutations, and MLL-related B-precursor ALL.63 These results suggest the possibility that RAS mutations might play a role in the pathogenesis of MLL-induced B-precursor ALL.
Several therapeutic agents have been developed that target RAS proteins or components of their upstream or downstream signaling pathways. Farnesyltransferase inhibitors prevent the posttranslational attachment of the farnesyl isoprenoid group to RAS and thereby inhibit its correct localization to the plasma membrane that is essential to initiate signal transduction. In addition, several kinase inhibitors have been developed that act downstream of RAS targeting the RAF/MAPK pathway or the PI3K/AKT pathway, respectively.64 Given the presence of the underlying MLL translocation, RAS pathway inhibitors probably do not fully block the transformed phenotype of MLL leukemia when given alone. However, these agents may significantly enhance the effectiveness of other chemotherapeutic agents when used in combination by abrogating the proliferative advantage conferred by the constitutive activation of the RAS pathway.
HSP-90
Proteomics-based strategies have been used to discover pharmacologic targets in leukemias with t(4;11) translocations.65 Initial screening experiments using 2-dimensional differential in-gel electrophoresis demonstrated that heat shock protein 90 alpha (HSP90α) is a potential target for pharmacologic inhibition in MLL-related leukemia. Treatment of a cell line expressing the MLL-AF4 fusion protein with the HSP90-inhibitor 17-demethoxygeldanamycin (17-AAG) resulted in a significant decrease of HSP90α protein expression, increase in apoptosis, and decreased expression of the disease biomarker nucleoside diphosphate kinase (NM23). Further studies showed a synergistic effect of combining 17-AAG with the FLT3 inhibitor GTP14564. In MLL-AF4 cell lines expressing either the FLT3-ITD or amplified wild-type FLT3, combined treatment with 17-AAG and GTP14564 reduced the levels of activated phosphorylated FLT3 and increased G0/G1 arrest and apoptosis. Responses to GTP14564 were restricted to cell lines with activated FLT3 and correlated with the level of STAT5 phosphorylation in these cells, suggesting a critical role of the STAT5 signal transduction pathway in the response to FLT3 inhibition.66
MCL-1
Myeloid cell leukemia sequence-1 (MCL-1) is an antiapoptotic protein related to BCL-2 that plays an important role in survival of multiple myeloma cells and is associated with resistance to chemotherapeutic intervention, particularly prednisone.67 Compared with ALL without MLL rearrangements, MLL leukemic blasts show markedly increased MCL-1 expression.68 SNS-032, a selective inhibitor of cyclin-dependent kinases 2, 7, and 9, triggers cell death in multiple myeloma cells by down-regulation of MCL-1.69 Furthermore, subtherapeutic doses of the MCL-1 inhibitor R-etodolac restored sensitivity of multiple myeloma cells to steroids, suggesting that it might be possible to overcome steroid-resistance by down-regulation of MCL-1.70 Targeting of MCL-1 therefore appears to be a potential therapeutic strategy for MLL-induced leukemia, particularly in cases of disease resistant to induction with steroid-containing regimens.
Approved drugs with activity against MLL leukemia
All-trans retinoic acid and vitamin D3
The retinoid all-trans retinoic acid (ATRA) is able to overcome the differentiation block induced by chimeric promyelocytic leukemia proteins in acute promyelocytic leukemia and constitutes the standard of care for induction therapy in this disease. The ATRA analog 9-cis retinoic acid has been shown to induce differentiation of a myeloid cell line (SN-1) expressing the MLL-CBP fusion protein. Interestingly, the maturation process of SN-1 cells was associated with down-regulation of the MLL-CBP fusion protein.71 Similarly, the MLL-AF9 expressing cell line MOLM-14 undergoes differentiation when exposed to ATRA or 1β,25-dihydroxyvitamin D3. The combination of ATRA with the histone deacetylase inhibitor trichostatin enhances the differentiating effect on MOLM-14 cells, suggesting a synergistic effect between the 2 drugs.72
This hypothesis is further supported by the finding that the DNA-methyltransferase inhibitor 5-azacytidine restores sensitivity to ATRA-induced differentiation in MLL-rearranged cell lines that are resistant to ATRA alone.71 The restoration of ATRA sensitivity in the resistant cell lines is associated with expression of p16, a CDK inhibitor. Taken together, these results suggest that combined treatment with ATRA and histone deacetylase inhibitors or DNA methyltransferase inhibitors may be beneficial in leukemia with MLL translocations.
Histone deacetylase and DNA methyltransferase inhibitors
Recent evidence also demonstrates a role for histone deacetylase and DNA methyltransferase inhibitors in leukemia mediated by the MLL partial tandem duplication (PTD). In contrast to leukemic cells with MLL translocations, blasts with the MLL PTD do not express the wild-type MLL protein from the contralateral unaffected allele. Interestingly, exposure of MLL PTD leukemic cells to histone deacetylase and DNA methyltransferase inhibitors resulted in reversal of the MLL wild-type allele silencing. Furthermore, the expression of MLL wild-type protein induced selective sensitivity to apoptosis, suggesting that the loss of MLL wild-type function is critical for leukemic transformation induced by MLL PTD.73 However, it remains unclear whether the effect of histone deacetylase and DNA methyltransferase inhibitors is specifically mediated through up-regulation of wild-type MLL or through another of the pleiotropic effects of these compounds.
Additional evidence for the use of histone deacetylase inhibitors in MLL leukemia is provided by studies using valproic acid. Inhibition of histone deacetylase by valproic acid induced G1 cell-cycle arrest and apoptosis in cell lines and patient samples expressing MLL-AF9.74 This effect is mediated by p21.
The DNA methyltransferase inhibitor decitabine induces apoptosis of MLL leukemia cells in vitro. Although demethylating agents are able to restore expression of a variety of different genes, one confirmed candidate gene of decitabine is the tumor suppressor Fragile Histidine Triad gene (FHIT). Gene expression profiling demonstrated that expression of FHIT is down-regulated in samples from pediatric patients with MLL-rearranged ALL compared with ALL carrying unmutated MLL. On exposure to decitabine, expression of FHIT is restored, and the exposed leukemic blasts undergo apoptosis. In addition, restoration of FHIT expression by genetic means induces cell death, confirming a critical role for FHIT in MLL-associated leukemogenesis and supporting it as a potential mediator for therapy.75 In summary, these results provide evidence for a potential role of histone deacetylase and DNA methyltransferase inhibitors in the treatment of MLL-rearranged leukemia. Additional studies are necessary to conclusively confirm their activity and further delineate targets of their therapeutic effect.
Cytosine arabinoside
Cytosine arabinoside (Ara-C) is a pyrimidine analog that is widely used in the treatment of acute leukemia. The hydrophilic drug gains entry into cells by the membrane transport protein human equilibrative nucleoside transporter 1 (hENT1) and is subsequently metabolized into its active form cytosine arabinoside triphosphate (Ara-CTP). Ara-CTP competes with deoxycytidine triphosphate for incorporation into DNA and inhibits DNA polymerase, resulting in decreased DNA synthesis and repair and ultimately leading to apoptosis. Interestingly, MLL gene-rearranged infant ALL cells, although generally considered resistant to chemotherapeutic agents, are highly sensitive to Ara-C.76 On exposure to Ara-C in vitro, infant ALL cells were 3.3-fold more sensitive and accumulated 2.3-fold more Ara-CTP compared with ALL cells with germline MLL from older children. A corresponding oligonucleotide microarray screen found a 2.7-fold higher mRNA expression of the transport protein hENT1 in leukemia cells with MLL gene rearrangements compared with nonrearranged ALL. Moreover, a strong correlation between hENT1 mRNA expression and Ara-C sensitivity was observed, suggesting that elevated expression of hENT1 contributes to increased sensitivity of MLL gene-rearranged infant ALL by enhanced uptake of Ara-C into the cell. These important findings provide a scientific rationale for the use of Ara-C in infant ALL and suggest that hENT1 mRNA expression may be a valuable predictor of Ara-C sensitivity in ALL patients.76
Conclusions and future directions
In conclusion, the treatment of leukemias with MLL rearrangements constitutes a major challenge in the adult as well as the pediatric population. Improvement of existing therapies will depend on further elucidation of the molecular mechanisms involved in transformation by MLL fusion proteins and the subsequent targeting of the identified pathways. Many important advances have recently been made in our understanding of MLL-induced leukemogenesis (Figure 1) and will hopefully result in meaningful improvements in our patient's lives once they are translated into the clinic. In the future, molecular studies will continue to be instrumental in identifying candidate genes or pathways critical for MLL-induced leukemogenesis, providing the basis for the development of novel targeted treatment strategies. Furthermore, molecular profiling will also be helpful in stratifying MLL leukemias into subclasses of disease with respect to prognosis and expected sensitivity to specific chemotherapy regimens.
At least equally important, the identification and specific targeting of MLL leukemia stem cells remain a fundamental goal of ongoing research efforts.77,78 The leukemia stem cell is characterized by its ability to self-renew and proliferate and is probably responsible for maintenance and relapse of disease. Unlike the majority of leukemic blasts, leukemia stem cells are often resistant to standard chemotherapy, allowing the leukemia to relapse after chemotherapy has resulted in apparent remission. Based on these characteristics, the leukemia stem cell is critical for leukemogenesis and represents the most relevant target for leukemia therapy. Consequently, to achieve the highest chance of curing the disease, it is essential to identify and understand unique characteristics of the leukemia stem cell population that can be exploited to facilitate its efficient and specific eradication.
Acknowledgments
The authors thank Dr Kevin Smith for insightful discussions in preparation of this review.
The authors apologize to those authors whose work is not appropriately cited because of the limitations of space and/or knowledge.
This work was supported by grants from Hope Street Kids (Alexandria, VA; M.L.), the Leukemia & Lymphoma Society (White Plains, NY; M.L.C.), the American Society of Clinical Oncology Foundation (M.L.), and the National Institutes of Health (Bethesda, MD; grant K08 CA120349 to M.L. and M.L.C.).
Authorship
Contribution: M.L. and M.L.C. wrote the review.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michaela Liedtke, Stanford University School of Medicine, 300 Pasteur Dr, Stanford, CA 94305; e-mail: mliedtke@stanford.edu; or Michael L. Cleary, Stanford University School of Medicine, 300 Pasteur Dr, Stanford, CA 94305; e-mail: mcleary@stanford.edu.
References
- 1.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 2.Gu Y, Nakamura T, Alder H, et al. The (4;11) chromosome translocations of human acute leukemia's involves fusion between the ALL-1 gene encoding a protein with homologies to Drosophila trithorax gene and a gene AF-4 on chromosome 4. Cell. 1992;71:701. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 3.Pui CH, Gaynon PS, Boyett JM, et al. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet. 2002;359:1909–1915. doi: 10.1016/S0140-6736(02)08782-2. [DOI] [PubMed] [Google Scholar]
- 4.Schichman SA, Caligiuri MA, Gu Y, et al. ALL-1 partial tandem duplication in acute leukemia. Proc Natl Acad Sci U S A. 1994;21:6236–6239. doi: 10.1073/pnas.91.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90:1799–1806. [PubMed] [Google Scholar]
- 6.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 7.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 8.Daser A, Rabbitts TH. The versatile mixed lineage leukaemia gene MLL and its many associations in leukaemogenesis. Semin Canc Biol. 2005;15:175–188. doi: 10.1016/j.semcancer.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Rozovskaia T, Feinstein E, Mor O, et al. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemia's with the t(4:11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- 10.Zeisig BB, Milne T, Garcia-Cuellar MP, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slany RK, Lavau C, Cleary ML. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavau C, Du C, Thirman M, Zeleznik-Le N. Chromatin-related properties of CBP fused to MLL generate a myelodysplastic-like syndrome that evolves into myeloid leukemia. EMBO. 2000;19:4655–4664. doi: 10.1093/emboj/19.17.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasad R, Yano T, Sorio C, et al. Domains with transcriptional regulatory activity within the ALL-1 and AF-4 proteins involved in acute leukemia. Proc Natl Acad Sci U S A. 1995;92:12160–12164. doi: 10.1073/pnas.92.26.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiner SA, Garcia-Cuellar MP, Fey GH, Slany RK. The leukemogenic fusion of MLL with ENL creates a novel transcriptional transactivator. Leukemia. 1999;13:1525–1533. doi: 10.1038/sj.leu.2401534. [DOI] [PubMed] [Google Scholar]
- 15.Martin ME, Milne TA, Bloyer S, et al. Dimerization of MLL-fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4:197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- 16.So CW, Lin M, Ayton PA, Chen EH, Cleary ML. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4:99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- 17.Eguchi M, Eguchi-Ishimae M, Graves M. The small oligomerization domain of gephyrin converts MLL to an oncogene. Blood. 2004;103:3876–3882. doi: 10.1182/blood-2003-11-3817. [DOI] [PubMed] [Google Scholar]
- 18.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 19.Armstrong SA, Kung AL, Mabon ME, et al. Inhibition of FLT3 in MLL: validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 20.Ono R, Nakajima H, Ozaki K, et al. Dimerization of MLL-fusion proteins and FLT3 activation synergize to induce multiple-lineage leukemogenesis. J Clin Invest. 2005;115:919–929. doi: 10.1172/JCI22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrasekharappa SC, Guru SC, Manickam P, et al. Positional cloning of the gene for multiple endocrine neoplasia-type 1. Science. 1997;276:404–407. doi: 10.1126/science.276.5311.404. [DOI] [PubMed] [Google Scholar]
- 22.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama A, Wang Z, Wysocka J, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes CM, Rozenblatt-Rosen O, Milne TA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 25.Milne TA, Hughes CM, Lloyd R, et al. Menin and MLL cooperatively regulate expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci U S A. 2005;102:749–754. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67:7275–7283. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge H, Si Y, Roeder RG. Isolation of cDNA encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 1998;17:6723–6729. doi: 10.1093/emboj/17.22.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahuja HG, Hong J, Aplan PD, Tcheurekdjian L, Forman SJ, Slovak ML. t(9;11)(p22;p15) in acute myeloid leukemia results in a fusion between NUP98 and the gene encoding transcriptional coactivators p52 and p75-lens epithelium-derived growth factor (LEDGF). Cancer Res. 2000;60:6227–6229. [PubMed] [Google Scholar]
- 30.Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 2002;30:958–965. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen MD, Grummit CG, Hilcenko C, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukemia-associated MLL histone methyltransferase. EMBO J. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 34.Srinivasan RS, Nesbit JB, Marrero L, Erfurth F, LaRussa VF, Hemenway CS. The synthetic peptide PFWT disrupts AF4-AF9 protein complexes and induces apoptosis in t(4;11) leukemia cells. Leukemia. 2004;18:1364–1372. doi: 10.1038/sj.leu.2403415. [DOI] [PubMed] [Google Scholar]
- 35.Palermo CM, Bennett CA, Winters AC, Hemenway CS. The AF4-mimetic peptide, PFWT, induces necrotic cell death in MV4-11 leukemia cells. Leuk Res. 2007;32:633–642. doi: 10.1016/j.leukres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Q, Wang H, Ng HH, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 37.DiMartino JF, Ayton PM, Chen EH, Naftzger CC, Young BD, Cleary ML. The AF10 leucine zipper is required for leukemic transformation of myeloid progenitors by MLL: AF10. Blood. 2002;99:3780–3785. doi: 10.1182/blood.v99.10.3780. [DOI] [PubMed] [Google Scholar]
- 38.Okada Y, Feng Q, LinY, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 39.Mueller D, Bach C, Zeisig D, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Min J, Feng Q, Zhizhong L, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 41.Jones B, Su H, Bhat A, et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLOS Genet. 2008;4:e1000190. doi: 10.1371/journal.pgen.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang S, Litt M, Felsenfeld G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 2005;19:1885–1893. doi: 10.1101/gad.1333905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL-fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer. 2005;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- 44.Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 45.Ferrando AA, Armstrong SA, Neuberg DS, et al. Gene expression signatures in MLL rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102:262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 46.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17:2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.So CW, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- 48.Kumar AR, Hudson WA, Chen W, et al. Hoxa9 influences the phenotype but not the incidence of Mll-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- 49.Nakanishi H, Nakamura T, Canaani E, Croce CM. ALL1 fusion proteins induce deregulation of EphA7 and ERK phosphorylation in human acute leukemias. Proc Natl Acad Sci U S A. 2007;104:14442–14447. doi: 10.1073/pnas.0703211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mann RS. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- 51.Mamo A, Krosl J, Kroon E, et al. Molecular dissection of Meis1 reveals 2 domains required for leukemia induction and a key role for Hoxa gene activation. Blood. 2006;108:622–629. doi: 10.1182/blood-2005-06-2244. [DOI] [PubMed] [Google Scholar]
- 52.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller JR. The Wnts. Genome Biol. 2002;3 doi: 10.1186/gb-2001-3-1-reviews3001. reviews 3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trowbridge JJ, Xenocostas A, Moon RT, Bhatia M. Glycogen synthase kinase-3 is an in vivo regulator of hematopoietic stem cell repopulation. Nat Med. 2006;12:89–98. doi: 10.1038/nm1339. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TCP, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carnicer MJ, Nomdedeu JF, Lasa A, et al. FLT3 mutations are associated with other molecular lesions in AML. Leuk Res. 2004;28:19–23. doi: 10.1016/s0145-2126(03)00125-5. [DOI] [PubMed] [Google Scholar]
- 58.Stubbs MC, Kim YM, Krivtsov AV, et al. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment. Leukemia. 2008;22:66–77. doi: 10.1038/sj.leu.2404951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown P, Levis M, Shurtleff S, Campana D, Downing J, Small D. FLT3 inhibition selectively kills childhood acute lymphoblastic leukemia cells with high levels of FLT3 expression. Blood. 2005;105:812–820. doi: 10.1182/blood-2004-06-2498. [DOI] [PubMed] [Google Scholar]
- 60.Brown P, Levis M, McIntyre E, Griesemer M, Small D. Combinations of the FLT3 inhibitor CEP-701 and chemotherapy synergistically kill infant and childhood MLL rearranged ALL cells in a sequence-dependent manner. Leukemia. 2006;20:1368–1376. doi: 10.1038/sj.leu.2404277. [DOI] [PubMed] [Google Scholar]
- 61.Lavau C, Libura M, Morgado E, Albouhair S. The leukemogenic properties of MLL fusion genes do not require FLT3 signaling. Blood. 2005;106:1201. [Google Scholar]
- 62.Shields JM, Pruitt K, McFall A, Shaub A, Der CJ. Understanding Ras: ‘it ain't over ‘til it's over.’. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 63.Liang DC, Shih LY, Fu JF, et al. K-Ras mutations and N-Ras mutations in childhood acute leukemias with or without mixed-lineage leukemia gene rearrangements. Cancer. 2006;106:950–956. doi: 10.1002/cncr.21687. [DOI] [PubMed] [Google Scholar]
- 64.Sebolt-Leopold JS. Advances in the development of cancer therapeutics directed against the RAS-mitogen-activated protein kinase pathway. Clin Cancer Res. 2008;14:3651–3655. doi: 10.1158/1078-0432.CCR-08-0333. [DOI] [PubMed] [Google Scholar]
- 65.Yocum AK, Busch CM, Felix CA, Blair IA. Proteomics-based strategy to identify biomarkers and pharmacological targets in leukemias with t(4;11) translocations. J Proteome Res. 2006;5:2743–2753. doi: 10.1021/pr060235v. [DOI] [PubMed] [Google Scholar]
- 66.Yao Q, Nishiuchi R, Kitamura T, Kersey JH. Human leukemias with mutated FLT3 kinase are synergistically sensitive to FLT3 and Hsp90 inhibitors: the key role of the STAT5 signal transduction pathway. Leukemia. 2005;19:1605–1612. doi: 10.1038/sj.leu.2403881. [DOI] [PubMed] [Google Scholar]
- 67.Puthier D, Thabard W, Rapp M, et al. Interferon alpha extends the survival of human myeloma cells through an upregulation of the Mcl-1 anti-apoptotic molecule. Br J Haematol. 2001;112:358–363. doi: 10.1046/j.1365-2141.2001.02575.x. [DOI] [PubMed] [Google Scholar]
- 68.Stam RW, den Boer ML, Pieters R. Towards targeted therapy for infant acute lymphoblastic leukaemia. Br J Haematol. 2006;132:539–551. doi: 10.1111/j.1365-2141.2005.05909.x. [DOI] [PubMed] [Google Scholar]
- 69.Hawtin E, Cohen R, Haas N, et al. SNS-032 exhibits dose-dependent mechanism-based inhibition of CDK7 and cyclin-dependent kinase 9 in peripheral blood mononuclear cells from patients with advanced cancers treated in an ongoing phase 1 trial. Haematologica. 2007;93(suppl 2):276. [Google Scholar]
- 70.Yasui H, Hideshima T, Hamasaki M, et al. SDX-101, the R-enantiomer of etodolac, induces cytotoxicity, overcomes drug resistance, and enhances the activity of dexamethasone in multiple myeloma. Blood. 2005;106:706–712. doi: 10.1182/blood-2005-02-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niitsu N, Hayashi Y, Sugita K, Honma Y. Sensitization by 5-aza-2′-deoxycytidine of leukaemia cells with MLL abnormalities to induction of differentiation by all-trans retinoid acid and 1alpha,25-dihydroxyvitamin D3. Br J Haematol. 2001;112:315–326. doi: 10.1046/j.1365-2141.2001.02523.x. [DOI] [PubMed] [Google Scholar]
- 72.Iijima K, Honma Y, Niitsu N. Granulocytic differentiation of leukemic cells with t(9;11)(p22;q23) induced by all-trans-retinoic acid. Leuk Lymphoma. 2004;45:1017–1024. doi: 10.1080/1042819031000163887. [DOI] [PubMed] [Google Scholar]
- 73.Whitman SP, Liu S, Vukosavljevic T, et al. The MLL partial tandem duplication: evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular targeted therapy. Blood. 2005;106:345–352. doi: 10.1182/blood-2005-01-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tonelli R, Sartini R, Fronza R, et al. g1 cell-cycle arrest and apoptosis by histone deacetylase inhibition in MLL AF9 acute myeloid leukemia cells is p21 dependent and MLL-AF9 independent. Leukemia. 2006;20:1307–1310. doi: 10.1038/sj.leu.2404221. [DOI] [PubMed] [Google Scholar]
- 75.Stam RW, den Boer ML, Passier MM, et al. Silencing of the tumor suppressor gene FHIT is highly characteristic for MLL gene rearranged infant acute lymphoblastic leukemia. Leukemia. 2006;20:264–271. doi: 10.1038/sj.leu.2404074. [DOI] [PubMed] [Google Scholar]
- 76.Stam RW, den Boer ML, Meijerink JPP, et al. Differential mRNA expression of Ara-C-metabolizing enzymes explains Ara-C sensitivity in MLL gene-rearranged infant acute lymphoblastic leukemia. Blood. 2003;101:1270–1276. doi: 10.1182/blood-2002-05-1600. [DOI] [PubMed] [Google Scholar]
- 77.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 78.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]