Abstract
The half-a-tetratricopeptide (HAT) repeat motif is of interest because it is found exclusively in proteins that are involved in RNA metabolism. Little is known about structure–function relationships in this class of repeat motif. Here, we present the results of a combined bioinformatics, modeling and mutagenesis study of the HAT domain of Utp6. We have derived a new HAT consensus, delineated its structure-defining residues and, by homology modeling, have placed these residues in a structural context. By considering only HAT motifs from Utp6 proteins, we identified residues that are shared by, and unique to, only this subset of HAT motifs, suggesting a key functional role. Employing both random and directed mutagenesis of the HAT domain in yeast Utp6, we have identified residues whose mutation results in loss of function. By examining these residues in the context of the homology model, we have delineated those that act by perturbing structure and those that more likely have a direct effect on function. Importantly, the residues we predict to have a direct effect on function map together on the tertiary structure, thus defining a potential functional interaction surface.
Keywords: HAT, homology model, RNA processing, Utp6, yeast
Introduction
A protein's sequence determines its structure, and its structure determines its function. Nevertheless, it remains a fundamental challenge in protein research to decipher the function of a protein knowing only its sequence. Significant progress has been made in our ability to build homology models of a protein's structure from its amino acid sequence, but understanding a protein's function based on the predicted structure is still no easy task. Additional data, e.g. from mutagenesis studies, can be extremely useful.
Repeat proteins represent a particularly tractable system because typically the underlying repeat motif and the interactions between repeat units are extremely well conserved. Superimposed upon the basic structural motif are functional residues, which can vary to accommodate different functions. Thus, one can delineate residues that specify the structure and those which are functionally important. The abundance of repeat protein sequences has made such analyses increasingly feasible, and significant inroads have been made with regard to our understanding of structure–function relationships in the ankyrin repeat, HEAT repeat, leucine-rich repeat and the tetratricopeptide repeat (TPR) (Andrade et al., 2000, 2001; Main et al., 2003; Cortajarena et al., 2004, 2008; Magliery and Regan, 2004, 2005) There are, however, limitations to statistical approaches, not the least of which is that they require large data sets.
Here, we focus on the structure–function relationships in the HAT motif (half-a-TPR), and in particular the three tandem HAT motifs in the Saccharomyces cerevisiae protein Utp6. It is less ubiquitous than the ankyrin and TPR repeats, being found in only eight proteins in humans, compared with TPRs, which are found in over 100 proteins in humans. Although nearly 10 000 TPR sequences are available in Pfam (Bateman et al., 2004), there are fewer than 1000 HAT sequences. This relative rarity and the observation that HAT repeats occur exclusively in proteins involved in RNA processing make them of particular interest. The HAT repeat, until recently, was little studied. The exclusivity of HAT repeats in RNA-processing proteins prompted early speculation that the HAT domain may bind RNA (Vincent et al., 2003; Liu et al., 2006; Bai et al., 2007; Legrand et al., 2007); however, a peptide ligand for the HAT domain of Utp6 has recently been identified (Champion et al., 2008). Typically, proteins containing HAT motifs are part of larger RNA-processing complexes and are hypothesized to play a scaffolding role in assembling these protein complexes (Ben-Yehuda et al., 2000; Vincent et al., 2003; Liu et al., 2006; Bai et al., 2007). In yeast, the HAT-containing proteins Prp6, Prp39, Prp42, Clf1 and Syf1 are involved in pre-mRNA splicing (McLean and Rymond, 1998; Vincent et al., 2003; Ben-Yehuda et al., 2000; Liu et al., 2006); Utp6 and Rrp5 are involved in pre-rRNA processing (Torchet et al., 1998; Dragon et al., 2002); and Rna14 is involved in polyadenylation (Minvielle-Sebastia et al., 1991; Bai et al., 2007). Human homologs exist for each of the yeast proteins except Prp42, and an additional human protein, SART3, contains HAT repeats and is involved in pre-mRNA processing (Stanek et al., 2003).
The recently published crystal structures of a HAT-containing protein, CstF-77 (Bai et al., 2007; Legrand et al., 2007), have made modeling of the HAT motifs of different proteins feasible. Because the HAT-containing protein Utp6 is essential for yeast viability, it is possible to harness the power of genetics to gain insights into functionally important residues.
Here, we present the results of a combined bioinformatics, modeling and mutagenesis study of the HAT domain of Utp6, an essential protein involved in ribosome biogenesis. We initially identify HAT structural residues by creating a HAT alignment and consensus. Using a Utp6 homolog alignment, we identify potential functional residues as those conserved only in Utp6 HATs. We next create a homology model of the HAT domain of yeast Utp6, and identify both types of residues in this model. We use random and directed mutagenesis of yeast UTP6 to identify residues whose mutation results in loss of function and use the homology model to determine which of these residues are likely to participate in the protein's function. This tertiary mapping of the predicted functional residues reveals a potential interaction surface for the Utp6 HAT domain.
Materials and methods
Bioinformatics
An alignment of 742 HAT motifs was downloaded from Pfam (Bateman et al., 2004). This alignment included gaps so that many of the 107 amino acid positions consisted of gaps in greater than 90% of the sequences. The alignment was therefore edited to contain 33 positions with each position filled in greater than 10% of sequences (>10% occupancy). Of these 33 positions, 32 had greater than 90% occupancy, and position 17 had 57% occupancy. It should be noted that the convention with the TPR consensus puts a highly conserved tryptophan at position 4 in helix A, followed by helix B, whereas the convention with the HAT consensus begins with helix B, followed by helix A, containing the conserved tryptophan at position 24.
Relative entropy analysis is described in Magliery and Regan (2004, 2005). Briefly, it compares the usage of a particular amino acid at a given motif position within a multiple sequence alignment to the overall usage of that amino acid in the yeast genome. The relative entropy calculation, therefore, is a measure of the conservation of an amino acid at a given position, weighted by its usage. To calculate this in the HAT alignment, the usage of each amino acid was divided by that amino acid's fractional usage in the yeast genome. The relative entropy of an amino acid at a given position is its usage multiplied by the natural log of that ratio, or HAT usage × ln(HAT usage/yeast usage). The total relative entropy at a given position in the HAT sequence is a sum of the relative entropies of each amino acid at that position. The HAT consensus sequence was determined using the HAT alignment and its relative entropy calculations. These calculations were done using Microsoft Excel.
Utp6 HAT motif homology model
The CLUSTAL W (Thompson et al., 1994) multiple sequence alignment program at the Network Protein Sequence Analysis website (Combet et al., 2000) was used to create two amino acid sequence alignments between residues 87–194 of yeast Utp6 and either residues 47–158 (#1) or 80–200 (#2) of murine CstF-77. The sequence alignment #2 and 2OOE PDB file of murine CstF-77 HAT motifs crystal structure were then used in ‘alignment mode’ of the SWISS-MODEL program (Peitsch, 1995; Guex and Peitsch, 1997; Schwede et al., 2003; Kopp and Schwede, 2004; Arnold et al., 2006) to create a homology model of the yeast Utp6 HAT motif. The Utp6 HAT motif homology model and the CstF-77 structure were viewed using PyMol (DeLano, 2002).
Yeast manipulations
The genetic screen for cold-sensitivity (CS), site-directed mutagenesis of the UTP6 gene and growth assays were performed as described (Champion et al., 2008).
Results
Recent studies have shown that not all HAT motifs are functionally equivalent, although they share certain structural residues (Vincent et al., 2003). Although the structure-determining residues are conserved in all HATs, each has a unique function. We hypothesized, therefore, that by using sequence analysis of the HAT domain, we could distinguish structural residues from functional residues. Until now, no universally accepted consensus sequence for the HAT repeat has existed.
We first determined the consensus residues for all HAT sequences currently available (13 December 2007). Previous alignments were limited because they were based on the HAT repeats from a single protein, rather than combining data from several proteins of different functions (Zhang et al., 1991; McLean and Rymond, 1998; Preker and Keller, 1998; Torchet et al., 1998; Chung et al., 1999, 2002; Ben-Yehuda et al., 2000; Amada et al., 2003). The Pfam database (Bateman et al., 2004) alignment of 742 HAT motifs is based on the seed of HAT motifs from two crooked neck homologs (Drosophila melanogaster CRN, amino acids 191–222 and Schizosaccharomyces pombe CLF1, amino acids 185–216) and one from a hypothetical Caenorhabditis elegans protein presumed to be an ortholog of Clf1 (O16376; Chung et al., 1999; amino acids 201–233). Despite our initial concern that this seed may skew the results toward the structure and function of the crooked neck subset of HAT proteins, we found very similar results using a more complete, but redundant, set of sequences (data not shown). We therefore chose to work with the universally available Pfam-derived alignment.
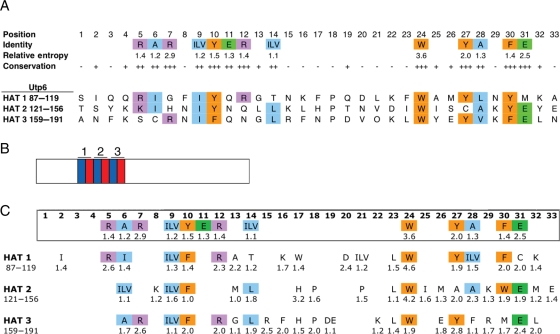
Starting with the alignment of the 742 HAT motifs downloaded from the Pfam database, we first defined new HAT consensus residues (Fig. 1A). The Pfam alignment contained 107 positions, but many of those positions were due to gaps in greater than 90% of the sequences. Positions that were occupied in fewer than 10% of sequences were omitted, leaving 33 positions. The degree of amino acid conservation at each position in the HAT motif was assessed using a relative entropy calculation. A higher relative entropy value reflects higher conservation of a given amino acid at a particular position in the HAT consensus. Because it is likely that a given position in the HAT consensus tolerates a family of similar amino acids rather than a single amino acid, similar amino acids were grouped and the sum of their individual relative entropies calculated. Specifically, at positions 9 and 14, no individual amino acid has a relative entropy value >1, but the combined relative entropies of aliphatic residues isoleucine, leucine and valine at each position are >1, and thus are treated as a ‘consensus’ residue type (medium-sized hydrophobic).
Fig. 1.
A homology-derived HAT consensus sequence. (A) Annotated HAT consensus sequence aligned with the HAT motifs in Utp6. Positions at which a single amino acid has a relative entropy >1.4 are ‘conserved’, and marked by ‘+++’. Positions at which an individual amino acid or amino acid family has a combined relative entropy >1 are ‘invariable’, and marked by ‘++’. Positions at which an amino acid family has a combined relative entropy >0.6 are ‘variable’, and marked by ‘+’. Finally, positions at which no individual amino acid and no amino acid family have a relative entropy value greater than 0.6 are ‘hypervariable’, and marked by ‘-’. Amino acids are color-coded by family: aliphatic residues (alanine, glycine, isoleucine, leucine and valine) are blue, aromatic residues (phenylalanine, tryptophan and tyrosine) are orange, positively charged residues (arginine and lysine) are purple, negatively charged residues (aspartic acid and glutamic acid) are green. The sequences of the Utp6 HAT motifs are shown aligned with the HAT consensus residues and matching amino acids are highlighted using the same color code. (B) Cartoon of the HAT domain organization of Utp6. Helix B (blue) and helix A (red) of the HAT motifs are shown and numbered. The N-terminus is on the left. (C) An alignment of 31 Utp6 homologs was used to calculate the relative entropy and determine a consensus sequence for each of the three HAT motifs in Utp6. Relative entropy values are listed under each residue if the value is greater than 1. Amino acids are colored as described in (A) if they match the HAT consensus. Amino acid numbers for each HAT motif in yeast Utp6 are given.
Threshold values were used to classify each position of the HAT sequence. A position was designated as ‘conserved’ if the relative entropy of a single amino acid is >1.4, ‘invariable’ if the relative entropy of an individual amino acid or amino acid family is >1 but <1.4, ‘variable’ if the relative entropy of an individual amino acid or amino acid family is >0.6 but <1 and ‘hypervariable’ if the relative entropy of an individual amino acid or amino acid family is <0.6. This analysis identified residues, or sets of similar residues, that are conserved at certain positions, and it is this pattern that defines the HAT fold.
The Utp6 protein in S. cerevisiae contains three tandem HAT motifs (Fig. 1A) spanning amino acids 87–191 of the 440 amino acid protein (Fig. 1B). We created an alignment of the three tandem HAT motifs from yeast Utp6 and its homologs and for each position in the alignment we calculated relative entropy values (Fig. 1C; Table I). As expected, the Utp6 HAT consensus fits the overall HAT consensus. In particular, the most conserved HAT residue, tryptophan at HAT position 24, has similarly high relative entropy values in Utp6 HATs 1 and 2, and a relatively high relative entropy value in HAT 3. The other aromatic HAT-conserved residues, tyrosine at positions 10 and 27 and phenylalanine at position 30, are still aromatic in most Utp6 consensus positions, but not necessarily the same amino acid as in the HAT consensus. The degenerate nature of the HAT consensus is also evident from the Utp6 alignment. Although at least eight of the HAT-conserved residues are also conserved in each of the Utp6 HATs, there are notable exceptions. For instance, tyrosine at position 27 of the HAT consensus is a conserved tyrosine in Utp6 HATs 1 and 3, but is a conserved alanine in HAT 2. Similarly, although the HAT consensus prescribes a small aliphatic alanine at position 28, this position in Utp6 HAT 3 is occupied by a large aliphatic phenylalanine. The significance of these differences between the Utp6 consensus and the HAT consensus remains to be resolved.
Table I.
HAT-conserved residues and Utp6-conserved residues
| HAT consensus |
Utp6 HAT 1 |
Utp6 HAT 2 |
Utp6 HAT 3 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Position | Amino acid | Position | Consensus amino acid | Amino acid | Position | Consensus amino acid | Amino acid | Position | Consensus amino acid | Amino acid |
| 1 | 87 | S | 124 | T | 159 | A | ||||
| 2 | 88 | I | I | 125 | S | 160 | N | |||
| 3 | 89 | Q | 126 | Y | 161 | F | ||||
| 4 | 90 | Q | 127 | K | 162 | K | ||||
| 5 | R | 91 | R | R | 128 | K | 163 | S | ||
| 6 | A | 92 | I | I | 129 | ILV | I | 164 | A | C |
| 7 | R | 93 | G | 130 | H | 165 | R | R | ||
| 8 | 94 | F | 131 | K | N | 166 | N | |||
| 9 | ILV | 95 | ILV | I | 132 | ILV | I | 167 | ILV | I |
| 10 | Y | 96 | F | Y | 133 | F | Y | 168 | F | F |
| 11 | E | 97 | Q | 134 | N | 169 | Q | |||
| 12 | R | 98 | R | R | 135 | Q | 170 | R | N | |
| 13 | 99 | A | G | 136 | M | L | 171 | G | G | |
| 14 | ILV | 100 | T | T | 137 | L | L | 172 | L | L |
| 15 | 101 | N | 138 | K | 173 | R | R | |||
| 16 | 102 | K | K | 139 | L | 174 | F | F | ||
| 17 | 103 | W | F | 140 | H | H | 175 | H | N | |
| 18 | 104 | P | 141 | P | P | 176 | P | P | ||
| 19 | 105 | Q | 142 | T | 177 | DE | D | |||
| 20 | 106 | D | D | 143 | N | 178 | V | |||
| 21 | 107 | ILV | L | 144 | P | V | 179 | P | ||
| 22 | 108 | K | 145 | D | 180 | K | K | |||
| 23 | 109 | L | F | 146 | L | I | 181 | L | L | |
| 24 | W | 110 | W | W | 147 | W | W | 182 | W | W |
| 25 | 111 | A | 148 | I | I | 183 | Y | |||
| 26 | 112 | M | 149 | M | S | 184 | E | E | ||
| 27 | Y | 113 | Y | Y | 150 | A | C | 185 | Y | Y |
| 28 | A | 114 | ILV | L | 151 | A | A | 186 | F | V |
| 29 | 115 | N | 152 | K | K | 187 | R | K | ||
| 30 | F | 116 | F | Y | 153 | W | Y | 188 | M | F |
| 31 | E | 117 | C | M | 154 | E | E | 189 | E | E |
| 32 | 118 | K | K | 155 | M | Y | 190 | L | L | |
| 33 | 119 | A | 156 | E | E | 191 | N | |||
Positions that are conserved or invariable in both the alignment of the 742 HAT sequences and in the alignment of the 31 Utp6-specific HAT sequences are in bold. Positions that are conserved or invariable only in the alignment of the 31 Utp6-specific HAT sequences are in italic.
Importantly, the Utp6 HAT alignment shows strong conservation of particular residues that are not conserved in all HATs, including several in the loop regions between helices B and A of each HAT motif. Position 17 of Utp6 HAT 2 contains histidine with a relative entropy value of 3.2; this position in Utp6 HATs 1 and 3 is also relatively well conserved and occupied by a tryptophan (HAT 1) or histidine (HAT 3). Additionally, the loop region of Utp6 HAT 1 contains conserved charged residues lysine and aspartic acid. Although it is common to find a conserved proline in this region in many HAT sequences, its position is not static (for instance in Utp6 HAT 2, it can be found in both positions 18 and 21), and therefore it does not appear conserved in the HAT consensus. Overall, the alignment shows that while the HAT motifs in Utp6 fit the HAT consensus, each motif has unique features (deviations from the HAT consensus sequence or particularly well-conserved residues in positions that are not conserved in the HAT consensus) that may affect the functional role of Utp6.
Homology model of the HAT domain of yeast Utp6 based on the crystal structure of the murine CstF-77 HAT domain
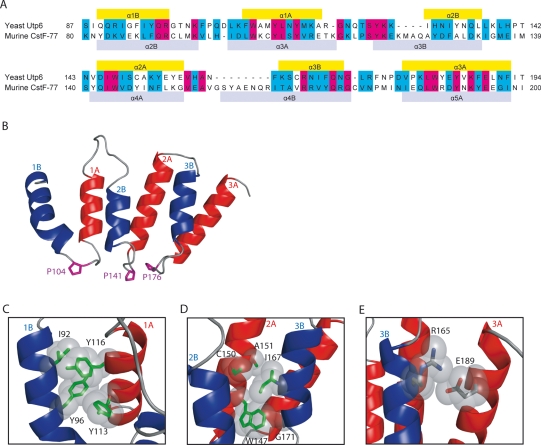
We built a homology model of the HAT domain of Utp6 based on the crystal structure of the murine CstF-77 HAT domain (pdbID=2OOE) (Bai et al., 2007). CstF-77 contains two HAT domains: HAT-N and HAT-C. Each HAT motif encodes two α-helices: helix A is identified by a highly conserved tryptophan (position 24), helix B is identified by a well-conserved tyrosine (position 10). Because HAT repeats are found in tandem, the order of the helices, A followed by B or B followed by A, is somewhat arbitrary, and both nomenclatures have been used (Preker and Keller, 1998; Bai et al., 2007). The first identified α-helix in Utp6 fits the consensus for helix B and is followed by a regular length loop, which contains a proline residue. We therefore describe the Utp6 HAT domain as three tandem HAT repeat motifs, each of which is comprised of helix B, followed by helix A (Fig. 1A and B). In contrast, Bai et al.'s nomenclature for the HAT repeats of CstF-77 starts with helix A followed by helix B, such that the proline occurs between adjacent repeats, rather than between helices in the same repeat. Using our definition, the HAT-N domain of CsfF-77 has four HAT repeats, thus permitting two different alignments with the three Utp6 HAT repeats. Two multiple sequence alignments were made between Utp6's three HAT repeats (residues 87–194) and either helices 1B through 4A (#1: residues 47–158) or helices 2B through 5A (#2: residues 80–200) of CstF-77. Alignment #2 is preferred (Fig. 2A) because in the homology model derived from alignment #1, the second helix of HAT1 is poorly formed. The sequences in alignment #2 are 18% identical and 37% similar. The 2B helix of CstF-77 (Bai et al., 2007, nomenclature) is aligned with the 1B helix of the HATs of Utp6, which is equivalent to position 87 of Utp6 matched with position 80 of CstF-77. Key hydrophobic-conserved residues align well, notably hydrophobic residues at positions 6 (residues 62, 129, 164), 9 (residues 95, 132, 167) and 10 (residues 96, 133, 168) of helix B and larger hydrophobic residues at positions 24 (residues 110, 147, 182), 27 (residues 113, 150, 185) and 30 (residues 116, 153, 188) of helix A (Fig. 2A). Although the overall sequence identity and similarity are not high, the alignment of the key HAT residues indicates that the homology model based on this sequence alignment is reliable.
Fig. 2.
Utp6 HAT motif homology model. (A) Sequence alignment of the three HAT repeats of yeast Utp6 and HAT repeats 2B–5A of murine CstF-77 (Thompson et al., 1994). Residues that are identical in the two proteins are highlighted in pink, and residues that are similar in the two proteins are highlighted in teal. CstF-77 helices (based on the crystal structure; Bai et al., 2007) are indicated as gray bars, and helices predicted in the model are indicated as yellow bars. (B) Ribbon representation of the model of the HAT domain of Utp6. Helix B of each HAT repeat is colored blue, and helix A of each HAT repeat is colored red. The helices are numbered according to which HAT repeat they are found in, and whether they are in helix A or B. The proline residues between helices B and A of each repeat are shown in purple to aid orientation. (C) Intra-HAT helix–helix packing. Key hydrophobic interactions between residues in helices 1B and 1A of HAT motif 1 are shown. Conserved hydrophobic residues are drawn as sticks (green) and their van der Waals radii are colored gray. (D) Inter-HAT helix–helix packing. Key hydrophobic interactions between helices 2A and 3B of HAT motifs 2 and 3 are shown. Key as in (C). (E) Electrostatic interaction between residues distant sequence in helices 3B and 3A of HAT 3. Residues are drawn as sticks (colored by atom) and their van der Waals radii are colored gray. Figures B–E were prepared using PyMol (DeLano, 2002).
The working homology model of the Utp6 HAT motif consisting of three HAT repeats is shown in Fig. 2B. To assess the plausibility of this model, we mapped HAT-conserved residues onto the structure. As expected, most of these residues are buried in the interfaces between helices. There are two different types of helix–helix interfaces: the intra-HAT interface between B and A helices of one HAT motif and the inter-HAT interface between A and B helices of adjacent HAT motifs. Examples of the packing interactions between conserved residues at the intra-HAT helix interface are shown in Fig. 2C. Residues I92 and Y96 of helix 1B pack with Y116 and Y113 of helix 1A. I92 is equivalent to HAT position 6, which has a conserved hydrophobic residue in all HAT motifs (the three Utp6 HAT motifs have isoleucine, isoleucine and cysteine at this position). Y96 is equivalent to HAT position 10; Y114 to HAT position 27 and Y116 to HAT position 30. These conserved, aromatic, hydrophobic residues pack with each other, thus stabilizing the intra-HAT interface.
The inter-HAT interface, shown in Fig. 2D, includes HAT position 24 (W147) which is the most highly conserved residue of the HAT motif. It shows complete conservation in HATs, and all three HAT repeats in Utp6 have tryptophan at this position. In our model, the side chain of the tryptophan residue on helix 2A packs against G171 on helix 3B that is equivalent to position 13 in the HAT motif. Position 13 shows some preference for smaller hydrophobic residues with glycine, leucine and glycine at this position in the three HATs of yeast Utp6. The remainder of this inter-HAT packing interface is formed by I167 on helix 3B and C150 and A151 on helix 2A. I167 is equivalent to HAT position 9 with conserved aliphatic residues in HATs and isoleucine in each of the three Utp6 HATs. C150 and A151 are conserved hydrophobic positions 27 and 28 in HATs. The side chains of residues at position 27 of helices 1A (Y113) and 2A (C150) are oriented in such a way that they can pack with both intra- and inter-HAT hydrophobic interfaces. Positions R7 and E31 are both well conserved in the HAT consensus. In the HATs of Utp6, only the HAT3 motif has the typical R7–E31 pair (R165–E189). Examination of the model (Fig. 2E) shows that these residues are well-positioned to make an electrostatic interaction. Using MolProbity (Davis et al., 2007) to assess the overall stereochemistry of the model, we find over 95% of the residues have φ–ψ angles that fall in the allowed regions of Ramachandran backbone conformational space.
Genetic screen for mutations in Utp6 that confer cold sensitivity
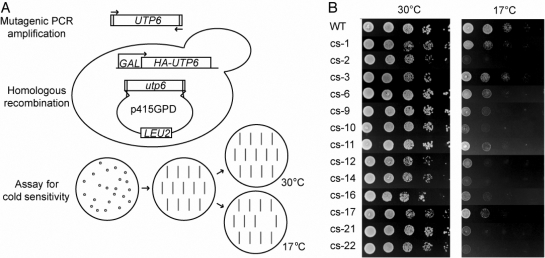
We employed a genetic approach to identify, without bias, residues that are essential for the function of Utp6 in ribosome biogenesis. If the HAT domain is indeed essential to ribosome biogenesis, we would expect to find mutations within the HAT domain that cause a defect in ribosome biogenesis, which we can measure as a defect in growth. Because our goal was to identify conditional mutations in Utp6 that cause defects in ribosome assembly, a process that is strongly inhibited by lowered temperature (Guthrie et al., 1969), we employed a screen for mutations in Utp6 that confer CS. This work has been previously summarized in Champion et al. (2008), but will be described in more detail here.
First, in a strain where the essential endogenous Utp6 protein can be conditionally depleted, we used mutagenic PCR of the UTP6 gene to create a library of randomly mutagenized utp6 (Fig. 3A; Muhlrad et al., 1992). Colonies expressing randomly mutated Utp6 proteins were re-streaked onto selective media and tested for CS by comparing growth to that of wild-type cells at 17°C. Of 1500 colonies screened, 13 were CS, and utp6 was sequenced to identify mutations (Fig. 3B; Table II). Of these 13 strains, four utilized utp6 with mutations that encode a premature stop (two due to frameshifts, the other two due to nonsense mutations) between amino acids 211 and 242, just C-terminal to the HAT motif. That these four strains grow normally at 30°C (Fig. 3B), while expressing only the N-terminal half of Utp6 suggests that the C-terminal half of Utp6 is not essential for growth. Several non-synonymous mutations were found in the utp6 of each of the remaining nine strains, and some of these were chosen for individual analysis to determine which mutations were responsible for causing the CS phenotype as described in detail in the next section. Only 6 of the 34 mutations found were in the C-terminal half of the protein, whereas 15 mutations were found in the HAT motif, 3 mutations were found in the N-terminus and 6 mutations were found within 50 amino acids of the end of the HAT motif.
Fig. 3.
Genetic screen for mutations in Utp6 that confer CS. (A) Schematic for the design of the genetic screen for mutations in Utp6. Gap repair using a mutagenized library of utp6 with the p415GPD expression vector was performed in yeast with the endogenous UTP6 under a galactose-inducible promoter. Double-transformants were selected by growth on glucose plates lacking leucine. Colonies were re-struck onto new plates, then replica plated to test for growth at 17°C. Colonies that grew normally at 30°C, but not at 17°C, were further validated as described in Champion et al., 2008. (B) Results of the genetic screen for mutations in Utp6. Indicated are strains derived from YPH499 GAL::3HA-UTP6, each containing a plasmid expressing either wild type (WT) or mutated (cs-1 to cs-22) Utp6. Cells were plated in serial dilutions on glucose plates lacking leucine and grown at either 30°C or 17°C to test for CS. Plasmid inserts were sequenced, and mutations found in Utp6 are listed in Table II.
Table II.
Utp6 mutations isolated in the screen for CS
| Strain | Mutations | Conservation | Location |
|---|---|---|---|
| cs-1 | I88T | I in 61% of Utp6 | HAT1, position 2 |
| Y96C | Y in HATs | HAT2, position 10 | |
| N191D | None | HAT3, position 33 | |
| Q211Z | None | C-term | |
| cs-2 | S84P | S in 39% of Utp6 | N-term |
| G99E | A/G in all Utp6 | HAT1, position 13 | |
| cs-3 | R165G | R in HATs | HAT3, position 7 |
| E218D | E in 52% of HATs | C-term | |
| cs-6 | I12N | I in 45% of Utp6 | N-term |
| I146N | I in 68% of HATs | HAT2, position 23 | |
| D177N | E/D in 65% of Utp6 | HAT2, position 19 | |
| S438R | None | C-term | |
| cs-9 | K34E | K in 65% of Utp6 | N-term |
| H289R | None | C-term | |
| cs-10 | L136P | L in 84% of Utp6 | HAT2, position 16 |
| K195R | K in 84% of Utp6 | C-term | |
| S236P | None | C-term | |
| cs-11 | L139S | L in 29% of Utp6 | HAT2, position 6 |
| F192L | None | C-term | |
| cs-12 | Frameshift 238 | None | C-term |
| cs-14 | E210V | E in 26% of Utp6 | C-term |
| N217D | None | C-term | |
| K290R | None | C-term | |
| I381T | None | C-term | |
| cs-16 | Q219Z | None | C-term |
| cs-17 | T100A | T in 55% of Utp6 | HAT1, position 14 |
| N175S | N in 23% of Utp6 | HAT3, position 17 | |
| Frameshift 232 | None | C-term | |
| cs-21 | S87T | S in 42% of Utp6 | HAT1, position 1 |
| K138E | None | HAT2, position 15 | |
| N336S | None | C-term | |
| K389E | K in 26% of Utp6 | C-term | |
| cs-22 | L136P | L in 35% of Utp6 | HAT2, position 13 |
| K187R | K/R in 87% of Utp6 | HAT3, position 29 |
Thirteen strains were isolated in the screen for mutations in Utp6 that confer CS (cs-1 through cs-22). Sequencing of the utp6 gene from each strain revealed the mutations listed. For each mutated amino acid, its conservation in HATs or in Utp6 homologs is listed, if any. Also for each mutated amino acid, its position in Utp6 is listed: N-term indicates the residue lies N-terminal to the HAT domain, within amino acids 1–86; C-term indicates the residue lies C-terminal to the HAT domain, within amino acids 192–440; residues within the HAT domain are noted by their position within each HAT motif. This table was revised from Champion et al. (2008) based on the new Utp6 alignment.
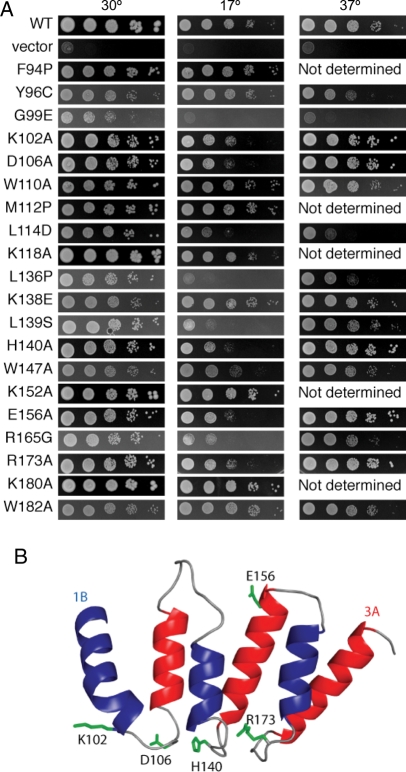
Because many of the strains isolated in the screen for CS contained multiple mutations in Utp6, and because we wanted to examine the role of particular amino acids in the Utp6 HAT motif function, we used directed mutagenesis to make some mutations separately in Utp6 (Table III). Several mutations identified in the screen were chosen for study as single mutations based on educated guesses that they were causing the phenotype: Y96C, G99E, L136P, K138E and L139S. Of these, only G99E, L136P and L139S were found to contribute to the CS phenotype (Fig. 4A; Table III).
Table III.
The effect of single point mutations in the HAT motif of Utp6 on function
| Mutations | Conservation | Location | Phenotype |
|---|---|---|---|
| F94P | none | HAT1, position 8 | No effect at 17°C |
| Y96C | Y in HATs | HAT1, position 10 | TS |
| G99E | A/G in all Utp6 | HAT1, position 13 | CS and TS |
| K102A | K in 74% of Utp6 | HAT1, position 16 | CS |
| D106A | D in 87% of Utp6 | HAT1, position 20 | CS |
| W110A | W in HATs | HAT1, position 24 | No effect on growth |
| M112P | none | HAT2, position 26 | No effect at 17°C |
| L114D | A in HATs | HAT1, position 28 | CS and TS |
| K118A | K in 65% of Utp6 | HAT1, position 32 | No effect on growth |
| L136P | L/M/A/V in 97% of Utp6 | HAT2, position 13 | CS |
| K138E | none | HAT2, position 15 | No effect on growth |
| L139S | none | HAT2, position 16 | CS |
| H140A | H in 87% of Utp6 | HAT2, position 17 | CS |
| W147A | W in HATs | HAT2, position 24 | CS |
| K152A | K in 60% of Utp6 | HAT2, position 29 | No effect at 17°C |
| E156A | E in 61% of Utp6 | HAT2, position 33 | CS |
| R165G | R in HATs | HAT3, position 7 | CS |
| R173A | R in 84% of Utp6 | HAT3, position 15 | CS |
| K180A | K in 58% of Utp6 | HAT3, position 22 | No effect at 17°C |
| W182A | W in HATs | HAT3, position 24 | No effect on growth |
Positions that are conserved or invariable in the alignment of the 742 HAT sequences are emboldened. Positions that are conserved or invariable only in the alignment of the 31 Utp6-specific HAT sequences are italicized. Conservation noted as ‘none’ indicates less than 50% conservation at that position in both HAT and Utp6 alignments; these positions are in roman text.
Fig. 4.
Point mutations in Utp6 that confer growth defects. (A) Yeast harboring the indicated mutations in the HAT motif of Utp6 were serially diluted and grown on glucose plates lacking leucine for 3 days at 30°C or 37°C or for 6 days at 17°C. Strains expressing wild-type Utp6 or no Utp6 (vector) were used as positive and negative controls, respectively. (B) Ribbon representation of the model of the HAT motif of Utp6. Residues that are conserved only in Utp6 HAT motifs and whose mutation causes a defective phenotype are shown as green sticks. The figure was prepared using PyMol (DeLano, 2002).
Additional mutations were designed with the aim of disrupting the structure of the HAT motif: F94P, W110A, M112P, L114D, W147A and W182A. Insertion of a proline at position 94 or 112 was intended to disrupt the α-helical structures of helix 1B or 1A, respectively. However, for reasons that remain unclear, neither of these proline substitutions resulted in a CS phenotype. Position L114 is a conserved aliphatic position of helix 1B and was mutated to a charged residue, aspartic acid, resulting in strong CS and temperature sensitive (TS) phenotypes. Finally, the tryptophan at positions 110, 147 and 183, which correspond to the most highly conserved position (24) in the HAT motif, were mutated to the much smaller hydrophobic residue, alanine. The W147A mutation (in HAT 2) resulted in CS, whereas the analogous mutation in HAT 1 or HAT 3 had no observed effect (W110A and W182A, Fig. 4A; Table III). Other mutations were made in residues that are conserved only in the HAT motifs of Utp6 homologs: K102A, D106A, H140A, E156A and R173A each exhibited a CS phenotype. Three other mutations in conserved Utp6 residues, K118A, K152A and K180A, had no observable effect on growth at 17°C (Fig. 4A; Table III).
Utp6-specific residues in the HAT motif
Certain predicted surface residues were identified as common only to Utp6 HATs, suggesting they are important for Utp6-specific HAT function (italicized residues in Table I). Several of these conserved residues were mutated individually to alanine, as described above. One would not expect mutation to a surface alanine to have a significant effect on protein stability, unless the residue is involved in a charge–charge interaction, which these are not. The observation that the point mutations K102A (position 16 of HAT 1), D106A (position 20 of HAT1), H140A (position 17 of HAT 2), E156A (position 33 of HAT2) and R173A (position 15 of HAT3) all give rise to a CS phenotype (Fig. 4A; Table III) strongly suggests that they play a key functional role. We hypothesize that the phenotype associated with the alanine substitutions occurs because a key Utp6-specific functional interaction of the HAT motif with a protein partner is lost. Figure 4B shows the Utp6 HAT-specific residues whose mutation gives rise to a CS phenotype. It is clear that four of the five residues map together in a cluster at one end of the molecule. Because these residues are highly conserved in Utp6 homologs, but not in all HAT sequences, because the mutation of these residues confers a growth defect and because these residues are predicted by our model to cluster on the surface of the molecule, we predict that this region of the HAT domain likely represents an interaction surface.
Discussion
The HAT motif is a relatively rare and understudied repeat motif that is hypothesized to play a scaffolding role in the formation of RNA-processing protein complexes. Here, we used a bioinformatics approach to analyze all available HAT sequences and identified the highly conserved residues that specify the structure of the HAT fold. We performed similar analyses on 31 Utp6 sequences and identified residues conserved only in the HAT domains of Utp6 proteins. We hypothesized that these residues are related to the specific function of Utp6 HAT domains. To further analyze the structure and function of Utp6 HAT domains, we performed genetic studies and assessed the effect of mutations, either random or directed, on Utp6 cellular function. Lastly, we built a homology model of the yeast Utp6 based on the recently solved crystal structure of CstF-77 (Bai et al., 2007). We then analyzed the homology model of the Utp6 HAT domain in terms of the bioinformatic and genetic data.
In the screen for mutations that affect Utp6 function, many single mutations of conserved residues yielded a CS and/or TS phenotype, indicating that the mutations of structural HAT fold-determining residues that we identified have a major impact on the function of the protein. A single mutation, Y96C, at position 10 in the first HAT repeat gave a weak TS phenotype. This result is consistent with the expectation predicted by our model that a decrease in protein stability would occur as a consequence of mutating a large buried hydrophobic residue. The L114D mutation (equivalent to position 28 in the first HAT repeat) was identified in the genetic screen, and when present alone it confers both cold and temperature sensitivity. L114 is buried in the inter-HAT helix–helix interface. Mutating L114 to aspartic acid would be expected to destabilize the protein, because of the energetically unfavorable consequences of introducing a charged residue into a hydrophobic environment. Likewise, position 24 is the most conserved in the HAT motif, and in our model of the structure of the HAT domain of Utp6, it interacts with other hydrophobic residues at the helix–helix interface. The W147A mutation at this position in the second HAT repeat does give the expected CS phenotype, albeit weakly. We were surprised, however, that the analogous mutation in either the first or the third HAT repeat of Utp6 (W110A and W182A) resulted in no observable growth defect, neither CS nor TS. We speculate that the helices in HATs 1 and 3 may be able to move to maintain close-packing around the small, hydrophobic alanine side chain in the inter-helix hydrophobic cluster, but that this cannot happen in HAT 2. The complementary charge pair, R7–E31, is a conserved feature of HATs. The R7–E31 pair is found only in the third HAT repeat of Utp6, and the homology model shows that these residues are well-positioned to make a stabilizing charge–charge interaction. The mutation R165G (R7 of the third HAT) was identified in the screen and confers a CS phenotype. This result lends support to the idea of a structurally stabilizing role for the R7–E31 interaction.
G99 is an Utp6-conserved residue, not a conserved HAT consensus residue. However, the homology model and mutational analysis suggest that it is structurally important. When present alone, the G99E mutation gave rise to both CS and TS phenotypes. Pfam identifies only three HAT motifs in Utp6, as described here. However, the identification of additional HAT motifs in its homologs suggests that Utp6 may include additional HAT motifs N-terminal or C-terminal to these three, but that they are too degenerate to be identified by the Pfam algorithm. G99 is at position 13 in the first helix of HAT 1, and we propose that it is likely interacting with a preceding stretch of amino acids, perhaps an additional HAT motif, as we have seen in the homology model of the equivalent glycine at position 171 of HAT 3, which packs against W147 of the preceding helix. Thus, a glycine to glutamic acid mutation would be expected to destroy such an interaction.
While most of the conserved structural residues in HAT motifs were, as we expected, found at the helix–helix interfaces of our Utp6 HAT model, thus validating its reliability, genetic studies show that mutations of a few residues did not affect the in vivo function of Utp6. For example, substituting a proline within a helix at positions 94 and 112 or replacing the conserved tryptophan at position 24 with alanine did not result in the CS or TS phenotype. The significance of these repeatable observations is unknown, but they may suggest that the HAT fold is functionally flexible and local structural disruptions do not significantly destroy the protein's function. Indeed, the mutational flexibility reflected in this study may explain why the HAT consensus is so poorly conserved.
In addition to the G99E and L136P mutations (Table III), we have identified five Utp6-conserved residues that are sensitive to mutation. These are surface residues and we believe they are most likely directly involved in the function rather than structure of Utp6 HAT motifs. The observation that four of these residues cluster on the loops at one end of the molecule strongly suggests that the loop region forms an interaction surface. There are two possible interaction partners for these loops. The first is another region of the Utp6 protein which folds against this side of the HAT domain, stabilizing the overall tertiary structure of the protein. The second and equally plausible interacting partner is another protein. HAT motifs are hypothesized to mediate protein–protein interactions in RNA-processing complexes, and the identified interaction surface on the Utp6 HAT could be binding another component of the Utp6-containing complex. Further studies will be required to distinguish between these two possibilities. The probable interacting loop surface we identified on the Utp6 HAT domain is structurally plausible because loops provide a good scaffold for binding interactions (Skerra, 2000). Recently, a peptide segment of Utp21, a component of the UtpB subcomplex involved in small ribosomal subunit maturation, has been identified as a ligand of the Utp6 HAT domain (Champion et al., 2008) and is thus a candidate for the interaction partner.
Funding
These studies were supported by the following grants from the National Institutes of Health: National Research Service Award T32 HD07149 (E.A.C.), a subcontract from GM080515 (L.R.), and National Institute of General Medical Sciences grant number 52581 (S.J.B.).
Acknowledgements
We are grateful to Chris Wilson, Tom Magliery and Ajit Divakaruni for discussions and guidance with creating a consensus sequence and calculating relative entropy.
Footnotes
Edited by Lars Baltzer
References
- Amada N., Tezuka T., Mayeda A., Araki K., Takei N., Todokoro K., Nawa H. J. Biochem. 2003;133:615–623. doi: 10.1093/jb/mvg079. [DOI] [PubMed] [Google Scholar]
- Andrade M.A., Ponting C.P., Gibson T.J., Bork P. J. Mol. Biol. 2000;298:521–537. doi: 10.1006/jmbi.2000.3684. [DOI] [PubMed] [Google Scholar]
- Andrade M.A., Perez-Iratxeta C., Ponting C.P. J. Struct. Biol. 2001;134:117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Bai Y., Auperin T.C., Chou C.Y., Chang G.G., Manley J.L., Tong L. Mol. Cell. 2007;25:863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S., Dix I., Russell C.S., McGarvey M., Beggs J.D., Kupiec M. Genetics. 2000;156:1503–1517. doi: 10.1093/genetics/156.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion E.A., Lane B.H., Jackrel M.E., Regan L., Baserga S.J. Mol. Cell. Biol. 2008;28:6547–6556. doi: 10.1128/MCB.00906-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., McLean M.R., Rymond B.C. RNA. 1999;5:1042–1054. doi: 10.1017/s1355838299990635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Zhou Z., Huddleston K.A., Harrison D.A., Reed R., Coleman T.A., Rymond B.C. Biochim. Biophys. Acta. 2002;1576:287–297. doi: 10.1016/s0167-4781(02)00368-8. [DOI] [PubMed] [Google Scholar]
- Combet C., Blanchet C., Geourjon C., Deleage G. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Cortajarena A.L., Kajander T., Pan W., Cocco M.J., Regan L. Protein Eng. Des. Sel. 2004;17:399–409. doi: 10.1093/protein/gzh047. [DOI] [PubMed] [Google Scholar]
- Cortajarena A.L., Yi F., Regan L. ACS Chem. Biol. 2008;3:161–166. doi: 10.1021/cb700260z. [DOI] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., 3rd, Snoeyink J., Richardson J.S., et al. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. Palo Alto, CA, USA: DeLano Scientific; 2002. http://www.pymol.org . [Google Scholar]
- Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Bernstein K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y.N., et al. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Nashimoto H., Nomura M. Proc. Natl Acad. Sci. USA. 1969;63:384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp J., Schwede T. Nucleic Acids Res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand P., Pinaud N., Minvielle-Sebastia L., Fribourg S. Nucleic Acids Res. 2007;35:4515–4522. doi: 10.1093/nar/gkm458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Rauhut R., Vornlocher H.P., Luhrmann R. RNA. 2006;12:1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliery T.J., Regan L. J. Mol. Biol. 2004;343:731–745. doi: 10.1016/j.jmb.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Magliery T.J., Regan L. BMC Bioinformatics. 2005;30:240–250. doi: 10.1186/1471-2105-6-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main E.R., Xiong Y., Cocco M.J., D'Andrea L.D., Regan L. Structure. 2003;11:497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- McLean M.R., Rymond B.C. Mol. Cell. Biol. 1998;18:353–360. doi: 10.1128/mcb.18.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Winsor B., Bonneaud N., Lacroute F. Mol. Cell. Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Hunter R., Parker R. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Peitsch M.C. Biotechnology. 1995;13:658–660. [Google Scholar]
- Preker P.J., Keller W. Trends Biochem. Sci. 1998;23:15–16. doi: 10.1016/s0968-0004(97)01156-0. [DOI] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N., Peitsch M.C. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerra A. J. Mol. Recognit. 2000;13:167–187. doi: 10.1002/1099-1352(200007/08)13:4<167::AID-JMR502>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Stanek D., Rader S.D., Klingauf M., Neugebauer K.M. J. Cell. Biol. 2003;160:505–516. doi: 10.1083/jcb.200210087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet C., Jacq C., Hermann-Le Denmat S. RNA. 1998;4:1636–1652. doi: 10.1017/s1355838298981511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent K., Wang Q., Jay S., Hobbs K., Rymond B.C. Genetics. 2003;164:895–907. doi: 10.1093/genetics/164.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Smouse D., Perrimon N. Genes Dev. 1991;5:1080–1091. doi: 10.1101/gad.5.6.1080. [DOI] [PubMed] [Google Scholar]