Abstract
γ-Aminobutyric acid type A receptors (GABAARs) are ligand-gated chloride channels that exist in numerous distinct subunit combinations. At postsynaptic membrane specializations, different GABAAR isoforms colocalize with the tubulin-binding protein gephyrin. However, direct interactions of GABAAR subunits with gephyrin have not been reported. Recently, the GABAAR-associated protein GABARAP was found to bind to the γ2 subunit of GABAARs. Here we show that GABARAP interacts with gephyrin in both biochemical assays and transfected cells. Confocal analysis of neurons derived from wild-type and gephyrin-knockout mice revealed that GABARAP is highly enriched in intracellular compartments, but not at gephyrin-positive postsynaptic membrane specializations. Our data indicate that GABARAP–gephyrin interactions are not important for postsynaptic GABAAR anchoring but may be implicated in receptor sorting and/or targeting mechanisms. Consistent with this idea, a close homolog of GABARAP, p16, has been found to function as a late-acting intra-Golgi transport factor.
Keywords: p16, receptor clustering, postsynaptic density, Golgi transport, inhibitory synapse
Neuronal surface membranes contain numerous proteins that serve for intercellular communication. Among these, receptor ion channels, specialized for chemotransmission between neurons, are highly concentrated at postsynaptic densities (PSDs) apposed to the appropriate presynaptic nerve terminals. The targeting of neurotransmitter receptors to and clustering at PSDs is a complex process that requires receptor-associated proteins, cytoskeletal elements, and proteins involved in signal transduction. At excitatory synapses, PDZ domain (from postsynaptic density, disks large, zonula adherens)-mediated protein interactions generate a PSD that serves as a scaffold for glutamate receptors and regulatory enzymes involved in synaptic transmission (for reviews, see refs. 1 and 2). At inhibitory postsynaptic sites, the tubulin-binding protein gephyrin is known to play a crucial role in the synaptic localization of both glycine receptors (GlyRs) and γ-aminobutyric acid type A receptors (GABAARs) (for a review, see ref. 3).
Gephyrin (4, 5) was originally identified by copurification with the mammalian GlyR (6, 7). Gephyrin binds with high affinity to polymerized tubulin (8), and both microtubules and actin microfilaments are implicated in GlyR localization at postsynaptic sites (3, 9). Gephyrin also interacts with the GlyR β subunit (10, 11), thus serving as a receptor-cytoskeleton linker. At developing inhibitory PSDs, formation of a gephyrin scaffold precedes GlyR synaptic clustering (12, 13), and GlyR clusters do not form upon loss of gephyrin expression, either by antisense depletion in culture (12) or by gene knockout in mice (14).
Gephyrin is also found at many GABAergic synapses (15–20). Gene depletion and knockout experiments have revealed a crucial role of gephyrin in GABAAR clustering (21–23). Mice deficient for the GABAAR subunit γ2 display a strong reduction of postsynaptic gephyrin and GABAAR clusters, and the synaptic staining for the GABAAR α2 and γ2 subunits observed in cultured cortical neurons is reduced upon antisense oligonucleotide treatment (21). Neurons from gephyrin knockout mice show a total loss of postsynaptic GABAAR γ2 and α2 immunoreactivities, providing conclusive genetic evidence that gephyrin is essential for the postsynaptic localization of certain GABAARs (23). However, biochemical evidence for an association of gephyrin with GABAAR subunits is lacking at present (10), and cotransfection studies have reported only weak colocalization of the β3 but not other GABAAR subunits with gephyrin (24).
Recently, the GABAAR γ2 subunit was shown to bind the GABAAR-associated protein GABARAP, a 14-kDa polypeptide with sequence similarity to light chain 3 of microtubule-associated proteins (MAPs) 1A and 1B (25). Consistent with a potential receptor anchoring function, GABARAP harbors a tubulin-binding motif encompassing amino acids 1–36 (25, 26). In cultured cortical neurons, GABARAP displayed punctate immunoreactivity in both cell somata and neurites and colocalized with GABAARs (25). To clarify whether GABARAP may serve as a linker between GABAARs and gephyrin, we have now analyzed its interaction with gephyrin by genetic and biochemical methods. Moreover, we used immunocytochemistry to study the codistribution of these proteins in spinal cord and retinal sections. Our data show that GABARAP binds to gephyrin but is not found at GABAergic synapses and is localized intracellularly. These observations and GABARAP's close homology to p16, a late-acting intra-Golgi trafficking factor, suggest a role for GABARAP in intracellular receptor transport.
Materials and Methods
Binding Experiments.
The GABARAP cDNA and a GABARAP deletion mutant encoding amino acids 36–117 (GABARAP/36–117) were subcloned into pGEX-2T (Amersham Pharmacia) to generate glutathione S-transferase (GST) fusion proteins. Plasmids were transformed for isopropyl β-d-thiogalactopyranoside (IPTG)-induced protein production into Escherichia coli DH5α or BL21. Adult rat brains (3 g wet weight) were homogenized in 30 ml of homogenization buffer [100 mM NaCl/10 mM Tris⋅HCl (pH 7.5)/5 mM EDTA/10 mM MgCl2/0.5% Nonidet P-40/1% Triton X-100, including protease inhibitors] at 4°C for 60 min. After centrifugation at 10,000 × g for 30 min, 2.4 ml of the resulting supernatants was incubated with glutathione-agarose charged with 5 μg of GST, GST-GABARAP, or GST-GABARAP/36–117. After 2 h at 4°C, the beads were washed four times with homogenization buffer. Bound proteins were eluted with 5 mM glutathione/50 mM Tris (pH 8.0), separated by SDS/PAGE, and analyzed by Western blotting with anti-gephyrin (Transduction Laboratories, Lexington, KY, 1:250), as described (23).
Cell Culture and Transfection.
PC12 cells were cultured in Dulbecco's minimal essential medium supplemented with 10% (vol/vol) fetal calf serum and 5% (vol/vol) horse serum and seeded onto fibronectin-coated glass coverslips 1 day before transfection. Transfection was performed with 800 ng of DNA for 15 min at room temperature, using Lipofectamine Plus Reagent (Life Technologies, Eggenstein, Germany). Cells were then washed in phosphate-buffered saline and cultured in OptiMEM (Life Technologies) for 3–5 h at 37°C. Then the medium was exchanged for standard culture medium (see above), and cells were cultured for another 18 h at 37°C in an atmosphere of 5% CO2/95% air before being processed for immunostaining.
Cortical neurons were prepared as low-density cultures in coculture with astrocyte feeder layers as described (23). Cells were cultured for 7 days before being analyzed.
Cryostat Sections of Spinal Cord and Retina Tissue.
Spinal tissue of embryonic day 19.5 mice was cut into 5-mm blocks and fixed in 4% (wt/vol) paraformaldehyde for 10 min, followed by a short wash in phosphate-buffered saline. For cryoprotection, the sections were incubated in increasing concentrations [10% (wt/vol), 20% (wt/vol), 30% (wt/vol) + 0.01% sodium azide] of sucrose at 4°C for 1 h each. Cryostat sections were refixed for 5 min in 4% (wt/vol) paraformaldehyde and processed for immunofluorescence. For retina sections, tissue of adult (8–10 weeks) mice was prepared and processed for immunofluorescence as described (18).
Antibodies.
For Western blotting, a monoclonal anti-gephyrin antibody (Dianova, Hamburg, Germany) was used at a dilution of 1:250. For double-labeling experiments, the primary antibodies GABARAP 6402 (25) (1:100), anti-GABAAR γ2 subunit (27) (1:2000), anti-bassoon (28) (1:8000), and anti-gephyrin (1:100) were used.
Immunocytochemistry and Confocal Microscopy.
Coverslips carrying cortical neurons or spinal cord sections were prepared, cultured, and processed for immunofluorescence as described (23). Confocal microscopy was performed with a TCS-SP confocal laser-scanning microscope (Leica) equipped with the image software Leica tcs-nt (version 1.6.551).
Quantification of Immunoreactive Colocalization.
Three micrographs were taken from each double-labeled section with the 100× objective, using red and green fluorescence filters, and printed at a final magnification of 1,500×. The immunofluorescent puncta of the micrographs were transferred onto tracing paper. Micrograph images of double-labeled sections were superimposed at their correct position for counting the number of colocalized puncta. As a control for random overlap, images were also superimposed after a 180° turn to determine the number of coincidentally superimposed puncta. For each measurement, between 850 and 1,700 puncta, taken from at least three sections, were sampled (29).
Yeast Two-Hybrid Experiments.
Bait and prey plasmids were cotransfected with the lacZ reporter plasmid pSH18–34 into yeast strain EGY48 and assayed for lacZ and LEU2 expression. The cDNAs of the proteins to be analyzed were cloned into the plasmids pGILDA or pJG4–5 (Origene, Rockville, MD). The following amino acid residues, encoded by the partial cDNA fragments, were used in the binary interaction assays: rat GABAAR γ1/350–444, rat GABAAR γ2S/351–445, rat GABAAR γ3/333–446, rat GABAAR δ/327–429, rat gephyrin/2–733, rat gephyrin/2–152, rat gephyrin/153–348, rat gephyrin/349–736, GABARAP/37–117, and p16/37–117.
Results
GABARAP Is Homologous to p16, a Protein That Functions in Intra-Golgi Transport and Has Homologs in Different Invertebrate Species.
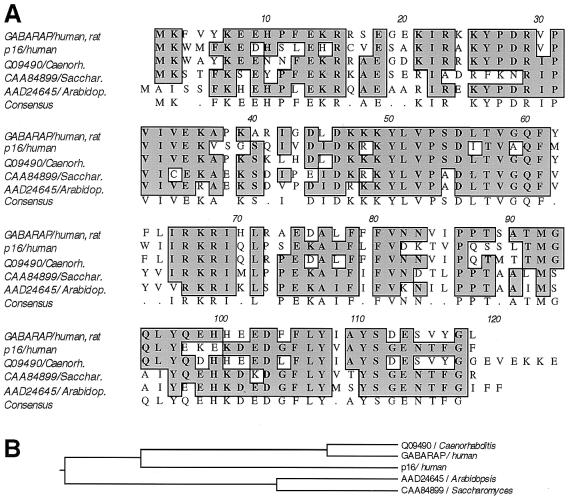
Database searches revealed a bovine homolog (GenBank accession no. AF020262; ref. 30), termed p16, that shares 57.3% amino acid identity with GABARAP (Fig. 1). The p16 protein was described as a late-acting 16-kDa intra-Golgi transport factor and was originally purified from bovine brain cytosol (30). The human p16 protein is termed ganglioside expression factor 2 (GenBank accession no. NP009216, unpublished) and shares 100% amino acid identity with the bovine protein. Additional homology searches revealed related sequences (Fig. 1) from Caenorhabditis elegans (GenBank accession no. Q09490), Saccharomyces cerevisiae (Genbank accession no. CAA84899), and Arabidopsis thaliana (GenBank accession no. AAD24645); the last has been classified as a putative microtubule-associated protein (31). These observations suggest a widespread function of p16 in many organisms that is not restricted to the nervous system, a finding that is consistent with GABARAP's ubiquitous mRNA expression (25). Searching mouse expressed sequence tag (EST) databases with the nucleotide sequence of p16 identified ESTs obtained from various tissues. These included hypothalamus (GenBank accession no. AA968244), liver (GenBank accession no. AI528168), kidney (GenBank accession no. AW318968), pancreas (GenBank accession no. AV056273), mammary gland (GenBank accession no. AA822987), lymph node (GenBank accession no. AA275333), tongue (GenBank accession no. AV082088), stomach (GenBank accession no. AV076945), and diaphragm (GenBank accession no. AA065794), suggesting that p16 is a ubiquitous protein with essential functions in all cells. Interestingly, ESTs corresponding to the p16 nucleotide sequence were also obtained from blastocysts (GenBank accession no. AA795530), indicating expression during early embryonic development.
Figure 1.
(A) Sequence alignment of GABARAP, p16, and their homologs from Caenorhabditis elegans, Saccharomyces cerevisiae, and Arabidopsis thaliana. The protein p16, recently described as a bovine late-acting intra-Golgi transport factor (30), shares 57.3% amino acid identity with GABARAP. Identities between GABARAP and its homologs are 78.9% (C. elegans), 54.7% (S. cerevisiae), and 54.2% (A. thaliana), respectively. (B) Phylogenetic tree of GABARAP, p16, and homolog proteins. Note that mammalian p16 is more distant from GABARAP than its homolog from C. elegans.
GABARAP, But Not p16, Binds Gephyrin.
In the yeast two-hybrid system, GABARAP gave strong positive signals for an interaction with the cytoplasmic loop regions of the GABAAR subunits γ1 and γ2 but not of the γ3 and δ subunits (Table 1). To investigate whether GABARAP might also bind to the receptor-anchoring protein gephyrin, we analyzed its interaction with either full-length gephyrin or partial gephyrin sequences, corresponding to amino acids 2–152, 153–348, or 349–736 (Table 1). Whereas full-length gephyrin did not give any β-galactosidase signal indicative of protein interaction, the fragment corresponding to amino acids 153–348 showed significant interaction (Table 1). In contrast, p16 did not interact with any of the analyzed GABAAR subunits (γ1, γ2, γ3, and δ), or with gephyrin or its fragments (Table 1).
Table 1.
Yeast two-hybrid analysis of protein–protein interactions
| Protein tested | GABARAP | p16 |
|---|---|---|
| GABAARγ1/350–444 | + | − |
| GABAARγ2/351–445 | + | − |
| GABAARγ3/333–446 | − | − |
| GABAARδ/327–429 | − | − |
| Gephyrin, full-length | − | − |
| Gephyrin/2–152 | − | − |
| Gephyrin/153–348 | + | − |
| Gephyrin/349–736 | − | − |
GABARAP and p16 bait constructs were used in binary interaction assays with GABAAR subunit and gephyrin sequences. Positive interactions revealed by lacZ and LEU2 expression are marked with a +, whereas − denotes white cells.
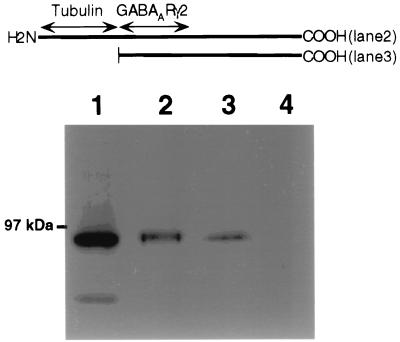
To further investigate the GABARAP–gephyrin interaction in vitro, we fused GABARAP to GST. Bacterially expressed fusion protein was incubated with rat brain homogenate. After washing, bound protein was eluted, subjected to SDS/PAGE, and probed with anti-gephyrin. Full-length GABARAP as well as a truncated protein containing amino acids 36–117 displayed efficient binding of the 93-kDa protein gephyrin (Fig. 2, lanes 2 and 3). In contrast, GST alone did not interact with gephyrin (Fig. 2, lane 4). Thus GABARAP, which binds the GABAAR subunit γ2 (25), also binds gephyrin.
Figure 2.
GABARAP interacts with gephyrin in vitro. GST-GABARAP or GST-GABARAP/36–117 (4 μg each) was bound to glutathione-agarose and incubated with brain homogenate. After washing, bound proteins were eluted with glutathione, separated by SDS/PAGE, and probed with anti-gephyrin. Brain homogenate input (lane 1), GST-GABARAP (lane 2), and GST-GABARAP/36–117 (lane 3) interact with gephyrin, whereas GST alone (lane 4) does not. The GABARAP sequences fused to GST are schematically given (bars) above the gel lanes; putative interaction domains are indicated by arrows.
GABARAP and Gephyrin Colocalize in PC12 Cells.
To unravel whether GABARAP and gephyrin interact in intact cells, we expressed both proteins in the neuron-like cell line PC12. GABARAP immunoreactivity was largely found at the plasma membrane (Fig. 3A), as revealed by confocal microscopy. Untransfected cells also displayed some endogenous GABARAP immunoreactivity, which again was mainly located at the membrane (not shown). This localization suggests the existence of membrane binding sites for GABARAP in PC12 cells. In contrast, single expression of gephyrin generated a largely diffuse cytoplasmic staining (Fig. 3B). Coexpression of both GABARAP and gephyrin caused a recruitment of cytoplasmic gephyrin to GABARAP-rich loci (Fig. 3C); this is indicative of a potential in vivo interaction between the two proteins, a finding consistent with the biochemical data.
Figure 3.
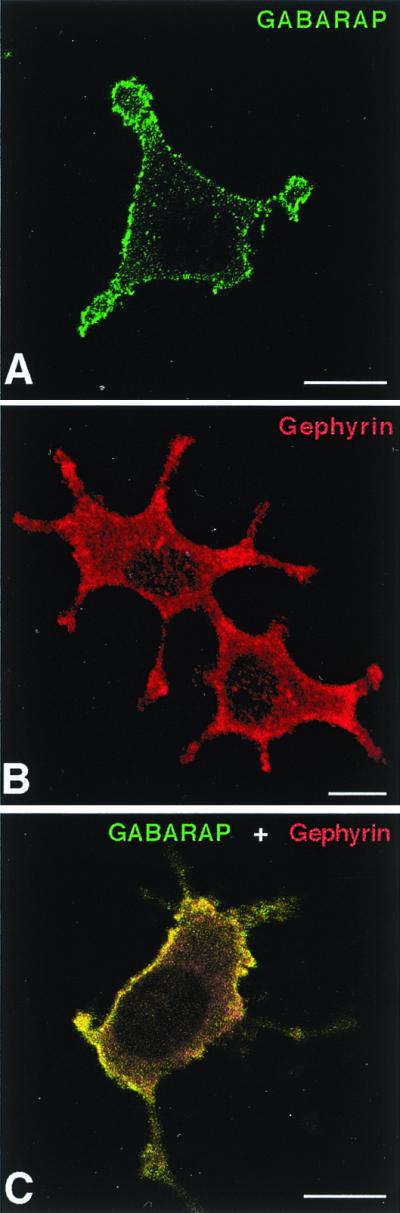
Colocalization of GABARAP and gephyrin upon heterologous expression in PC12 cells. Singly expressed GABARAP protein is located at the plasma membrane (A), whereas singly expressed gephyrin is diffusely distributed with some enrichment at submembranous compartments (B). Coexpression of GABARAP and gephyrin leads to a recruitment of gephyrin to GABARAP-rich loci (C). (Scale bars, 10 μm.)
Subcellular Distribution of GABARAP in Neurons of Wild-Type and Gephyrin-Knockout Mice.
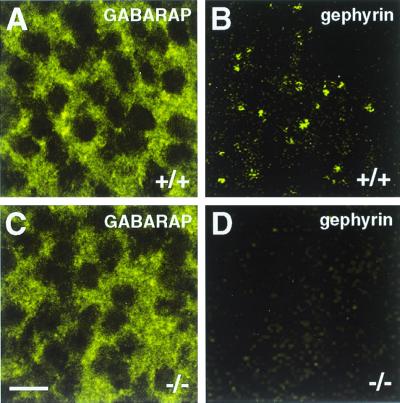
To unravel whether the interaction of GABARAP and gephyrin seen in vitro and in transfected cells may result in a synaptic colocalization of these proteins, we analyzed spinal cord sections and cultured cortical neurons from wild-type and gephyrin-deficient mice for GABARAP immunoreactivity. In spinal cord sections, the punctate distribution of GABARAP was less prominent than it was in cultured cortical neurons (25) but was still detectable (Fig. 4). Notably, sections derived from wild-type (+/+) and homozygous gephyrin-knockout mutants (−/−) did not significantly differ in the intensity and/or number of GABARAP immunoreactive puncta (Fig. 4) or show colocalization with gephyrin (not shown). Similarly, in cultured cortical neurons derived from wild-type and mutant mice, GABARAP puncta were not significantly different in number, size, and location between the different genotypes (Fig. 5), suggesting that GABARAP functions prior to or independently of gephyrin. More importantly, confocal images revealed that the majority of GABARAP puncta were seen in intracellular compartments, which according to their location represented putative endoplasmic reticulum (ER) and Golgi structures (Fig. 5). These data suggest that, in contrast to what is seen upon expression in transfected PC12 cells, in neurons GABARAP predominantly functions intracellularly.
Figure 4.
GABARAP immunoreactivity in embryonic day 19.5 spinal cord sections derived from wild-type (+/+) and gephyrin-knockout (−/−) mice. Anti-GABARAP stains punctate structures in spinal cells of wild-type tissue; in addition, a diffuse staining is seen (A). Gephyrin appears in synaptic clusters of wild-type tissue (B, and see ref. 14). In tissue sections derived from gephyrin-deficient mice, GABARAP immunoreactivity is unaltered (C), whereas gephyrin immunoreactivity is lost (D). (Scale bar, 10 μm.)
Figure 5.
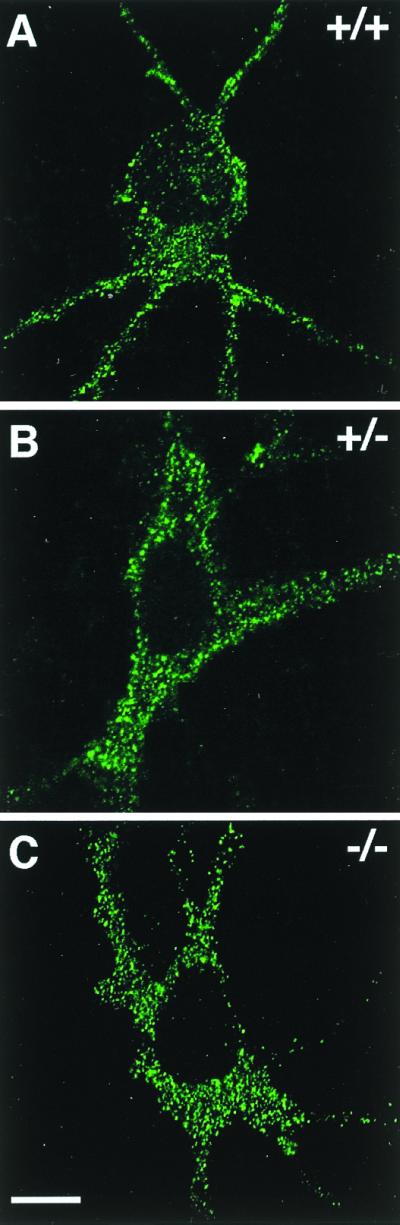
GABARAP staining of cultured cortical neurons derived from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) gephyrin-knockout mice. A punctate distribution of GABARAP (25) is seen throughout the cytoplasm of neurons derived from wild-type (A), heterozygous (B), and homozygous (C) gephyrin-knockout mice. Individual confocal sections are shown. (Scale bar, 10 μm.)
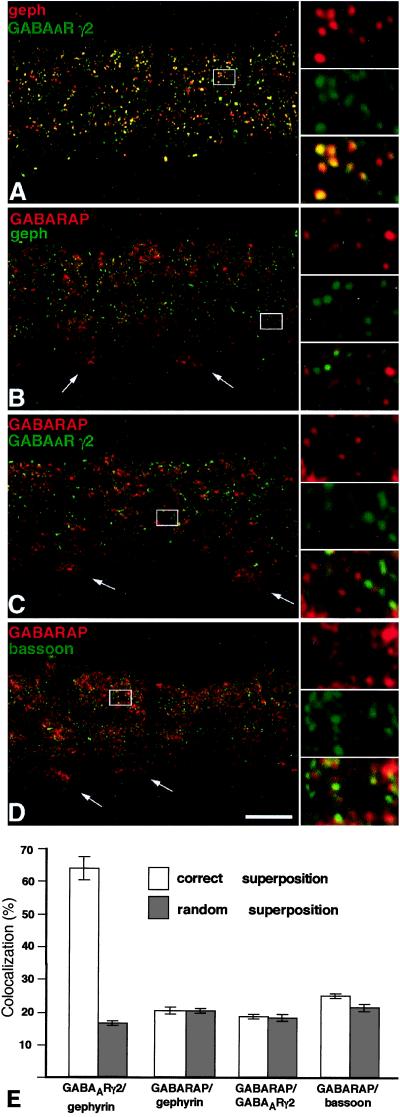
Quantitative Analysis of GABARAP, GABAAR, and Gephyrin Immunoreactivity in Retina.
To quantitatively evaluate whether GABAAR, GABARAP, and gephyrin colocalize at synaptic sites, we used retina sections, in which gephyrin is known to be exclusively synaptic, as revealed by electron microscopy (18). Such sections were stained for GABARAP, gephyrin, the GABAAR γ2 subunit, and the presynaptic protein bassoon (28). Notably, GABARAP and gephyrin immunoreactivities failed to show any significant colocalization above randomized values (Fig. 6 B and E). Furthermore, none of the GABAAR γ2 subunit (Fig. 6 C and E) or the bassoon (Fig. 6 D and E) immunoreactive puncta colocalized with GABARAP, whereas 64% of GABAAR γ2 subunit immunoreactive puncta colocalized with synaptic gephyrin (Fig. 6 A and E). These data indicate that GABARAP is not clustered at inhibitory synaptic sites, suggesting that GABARAP's interactions with the GABAAR γ2 subunit and gephyrin may be important for cellular functions other than receptor anchoring.
Figure 6.
Cofocal micrographs of vertical sections through the inner plexiform layer (IPL) of double-immunostained mouse retinae. Selected areas of the micrographs (frames) are shown at higher magnification, to the right. (A) Gephyrin (red) and the γ2 subunit of the GABAAR (green) are aggregated in synaptic hot spots, which are often colocalized. (B) GABARAP (red) shows diffuse, punctate distribution in the IPL but is not clustered with gephyrin (green) in synaptic hot spots. The white arrows point to the cell bodies of ganglion cells and show the expression of GABARAP in the cytoplasm. (C) GABARAP (red) and the γ2 subunit of the GABAAR (green) appear not to be aggregated within the same hot spots. (D) The presynaptic cytomatrix protein bassoon (green), which is clustered at both excitatory and inhibitory synapses, is not colocalized with GABARAP. (Scale bar, 10 μm.) (E) Quantifications of the colocalizations at their correct superpositions and at random superpositions. Only in the case of the γ2 subunit of the GABAAR and gephyrin was a significant colocalization of puncta observed.
Discussion
Here we have used yeast two-hybrid analysis as well as biochemical and anatomical methods to investigate whether GABARAP may mediate the specific localization of GABAARs at gephyrin-rich PSDs. In yeast, a partial gephyrin sequence, but not full-length gephyrin, gave a strong β-galactosidase signal, which is indicative of protein interaction with GABARAP. Furthermore, a recombinant GST-GABARAP fusion protein displayed high-affinity in vitro binding to gephyrin solubilized from brain. Upon heterologous coexpression in the neuron-like PC12 cell line, GABARAP and gephyrin were found to colocalize, indicating that these proteins can interact in a cellular environment. Because gephyrin has been shown to be essential for GABAAR clustering (21–23), these data are consistent with the proposal of Wang et al. (25) that GABARAP may be involved in the processes that underlie GABAAR targeting to and/or clustering at postsynaptic sites.
Despite the observed interaction of the two proteins, the comparison of spinal cord sections and cultured cortical neurons from wild-type and gephyrin-deficient mice did not reveal any differences in the number, size, or location of GABARAP immunoreactive puncta. Moreover, in wild-type retinae there was no significant colocalization of GABARAP with either the GABAAR γ2 subunit or gephyrin, whereas gephyrin-positive structures showed highly significant colocalization with the GABAAR. These observations indicate that GABARAP is not clustered at gephyrin-rich PSDs and is therefore unlikely to serve as a GABAAR-gephyrin linker molecule at differentiated synapses. Indeed, confocal sections of cultured cortical neurons revealed that the punctate GABARAP immunoreactivity is mainly found in intracellular compartments. According to their location, these GABARAP-positive sites are likely to represent ER/Golgi structures. This location is consistent with p16, a homolog of GABARAP, functioning as a late-acting intra-Golgi transport factor in mammalian brain (30). Thus GABARAP may be implicated in intracellular receptor sorting and targeting processes that precede and/or initiate receptor clustering at the synapse. Indeed, coexpression of GABARAP and GABAARs in QT6 cells has been found to increase the fraction of clustered GABAARs (L. Chen, H.W., S. Vicini, and R.W.O., unpublished results). In any case, the absence of GABARAP from GABAergic postsynaptic sites raises the question of whether there are yet-unidentified proteins that interact with both GABAARs and gephyrin and thus cause their synaptic colocalization.
Intracellular vesicular protein traffic includes different steps, such as budding, targeting, docking, and fusion of vesicles with their target membranes. Each step requires a different set of cytosolic factors. Its homology to p16 protein suggests that GABARAP may participate in a step preceding insertion of GABAARs into the plasma membrane. The tubulin cytoskeleton is known to be crucial for the organization of the Golgi complex (32) and the spatial distribution of peroxisomes (33). Furthermore, microtubules provide the structural basis for organelle transport via kinesin/dynein motor proteins, and a Golgi network-associated protein was recently found to directly bind to microtubules (34). The assignment of a putative tubulin-binding site to the N-terminal region of GABARAP (25, 26) is consistent with this protein having a role in the transport of GABAAR-containing Golgi vesicles before vesicle fusion with the plasma membrane. The yeast two-hybrid data obtained here show that GABARAP binds different GABAAR subunits and that only GABARAP and not its homolog p16 interacts with gephyrin. These data point to a role for GABARAP in receptor trafficking processes that may involve both different GABAAR isoforms and the receptor anchoring protein gephyrin. Whether gephyrin is transported to synaptic sites in vesicle-bound form, however, is not yet solved. Furthermore, it remains to be elucidated whether gephyrin can interact with inhibitory receptor subunits already at the level of the ER/Golgi compartment. If so, plasma membrane insertion of vesicles containing preassembled receptor–gephyrin complexes may provide a GABARAP-driven mechanism to allow for postnatal growth of inhibitory PSDs.
Acknowledgments
We thank Dagmar Magalei for expert technical assistance. The GABAAR γ2 subunit antibody was kindly provided by H. Möhler and J. M. Fritschy, and GABAAR subunit cDNAs were provided by P. Seeburg. This work was supported by grants from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie and the Fonds der Chemischen Industrie to H.B., the Deutsche Forschungsgemeinschaft (SFB 269), a Boehringer Ingelheim Fonds predoctoral fellowship to J.C.F., and National Institutes of Health Grant NS28772 to R.W.O.
Abbreviations
- PSD
postsynaptic density
- GABAAR
γ-aminobutyric acid type A receptor
- GABARAP
GABAAR-associated protein
- GlyR
glycine receptor
- GST
glutathione S-transferase
- EST
expressed sequence tag
- ER
endoplasmic reticulum
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sheng M. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- 2.Craven S E, Bredt D S. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- 3.Kneussel M, Betz H. J Physiol (London) 2000;525:1–9. doi: 10.1111/j.1469-7793.2000.t01-4-00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt B, Knaus P, Becker C M, Betz H. Biochemistry. 1987;26:805–811. doi: 10.1021/bi00377a022. [DOI] [PubMed] [Google Scholar]
- 5.Prior P, Schmitt B, Grenningloh G, Pribilla I, Multhaup G, Beyreuther K, Maulet Y, Werner P, Langosch D, Kirsch J, Betz H. Neuron. 1992;8:1161–1170. doi: 10.1016/0896-6273(92)90136-2. [DOI] [PubMed] [Google Scholar]
- 6.Pfeiffer F, Graham D, Betz H. J Biol Chem. 1982;257:9389–9393. [PubMed] [Google Scholar]
- 7.Graham D, Pfeiffer F, Simler R, Betz H. Biochemistry. 1985;12:990–994. doi: 10.1021/bi00325a027. [DOI] [PubMed] [Google Scholar]
- 8.Kirsch J, Langosch D, Prior P, Littauer U Z, Schmitt B, Betz H. J Biol Chem. 1991;266:22242–22245. [PubMed] [Google Scholar]
- 9.Kirsch J, Betz H. J Neurosci. 1995;15:4148–4156. doi: 10.1523/JNEUROSCI.15-06-04148.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer G, Kirsch J, Betz H, Langosch D. Neuron. 1995;15:563–572. doi: 10.1016/0896-6273(95)90145-0. [DOI] [PubMed] [Google Scholar]
- 11.Kneussel M, Hermann A, Kirsch J, Betz H. J Neurochem. 1999;72:1323–1326. doi: 10.1046/j.1471-4159.1999.0721323.x. [DOI] [PubMed] [Google Scholar]
- 12.Kirsch J, Wolters I, Triller A, Betz H. Nature (London) 1993;266:745–748. doi: 10.1038/366745a0. [DOI] [PubMed] [Google Scholar]
- 13.Bechade C, Colin I, Kirsch J, Betz H, Triller A. Eur J Neurosci. 1996;8:429–435. doi: 10.1111/j.1460-9568.1996.tb01226.x. [DOI] [PubMed] [Google Scholar]
- 14.Feng G, Tintrup H, Kirsch J, Nichol M C, Kuhse J, Betz H, Sanes J R. Science. 1998;282:1321–1324. doi: 10.1126/science.282.5392.1321. [DOI] [PubMed] [Google Scholar]
- 15.Triller A, Cluzeaud F, Korn H. J Cell Biol. 1987;104:947–956. doi: 10.1083/jcb.104.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohlhalter S, Möhler H, Fritschy J M. Brain Res. 1994;642:59–69. doi: 10.1016/0006-8993(94)90905-9. [DOI] [PubMed] [Google Scholar]
- 17.Cabot J B, Bushnell A, Alessi V, Mendell N R. J Comp Neurol. 1995;356:418–432. doi: 10.1002/cne.903560309. [DOI] [PubMed] [Google Scholar]
- 18.Sassoe-Pognetto M, Kirsch J, Grünert U, Greferath U, Fritschy J M, Möhler H, Betz H, Wässle H. J Comp Neurol. 1995;357:1–14. doi: 10.1002/cne.903570102. [DOI] [PubMed] [Google Scholar]
- 19.Todd A J, Watt C, Spike R C, Siegart W. J Neurosci. 1996;16:974–982. doi: 10.1523/JNEUROSCI.16-03-00974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giustetto M, Kirsch J, Fritschy J M, Cantino D, Sassoe-Pognetto M. J Comp Neurol. 1998;395:231–244. doi: 10.1002/(sici)1096-9861(19980601)395:2<231::aid-cne7>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Essrich C, Lorez M, Benson J A, Fritschy J M, Lüscher B. Nat Neurosci. 1998;7:563–571. doi: 10.1038/2798. [DOI] [PubMed] [Google Scholar]
- 22.Betz H. Nat Neurosci. 1998;7:541–543. doi: 10.1038/2777. [DOI] [PubMed] [Google Scholar]
- 23.Kneussel M, Brandstätter J H, Laube B, Stahl S, Müller U, Betz H. J Neurosci. 1999;19:9289–9297. doi: 10.1523/JNEUROSCI.19-21-09289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirsch J, Kuhse J, Betz H. Mol Cell Neurosci. 1995;6:450–461. doi: 10.1006/mcne.1995.1033. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Bedford F K, Brandon N J, Moss S J, Olsen R W. Nature (London) 1999;397:69–72. doi: 10.1038/16264. [DOI] [PubMed] [Google Scholar]
- 26.Wang, H. & Olsen, R. W. (2000) J. Neurochem., in press. [DOI] [PubMed]
- 27.Fritschy J M, Möhler H. J Comp Neurol. 1995;359:154–194. doi: 10.1002/cne.903590111. [DOI] [PubMed] [Google Scholar]
- 28.Richter K, Langnaese K, Kreutz M R, Olias G, Zhai R, Scheich H, Garner C G, Gundelfinger E D. J Comp Neurol. 1999;408:437–448. doi: 10.1002/(sici)1096-9861(19990607)408:3<437::aid-cne9>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 29.Fletcher E L, Hack I, Brandstätter J H, Wässle H. J Comp Neurol. 2000;420:98–112. [PubMed] [Google Scholar]
- 30.Legesse-Miller A, Sagiv Y, Porat A, Elazar Z. J Biol Chem. 1998;273:3105–3109. doi: 10.1074/jbc.273.5.3105. [DOI] [PubMed] [Google Scholar]
- 31.Lin X, Kaul S, Rounsley S D, Shea T P, Benito M-I, Town C D, Fujii C Y, Mason T M, Bowman C L, Barnstead M E, et al. Nature (London) 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 32.Thyberg J, Moskalewski S. Exp Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- 33.Wiemer E A C, Wenzel T, Deerinck T J, Ellisman M H, Subramani S. J Cell Biol. 1997;136:71–80. doi: 10.1083/jcb.136.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Infante C, Ramos-Morales F, Fedriani C, Bornens M, Rios R M. J Cell Biol. 1999;145:83–98. doi: 10.1083/jcb.145.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]