Abstract
Cell death is of broad physiological and pathological importance, making quantification of biochemical events associated with cell demise a high priority for experimental pathology. Fibrosis is a common consequence of tissue injury involving necrotic cell death. Using tissue specimens from experimental mouse models of traumatic brain injury, cardiac fibrosis, and cancer, as well as human tumor specimens assembled in tissue microarray (TMA) format, we undertook computer-assisted quantification of specific immunohistochemical and histological parameters that characterize processes associated with cell death. In this study, we demonstrated the utility of image analysis algorithms for color deconvolution, colocalization, and nuclear morphometry to characterize cell death events in tissue specimens: (a) subjected to immunostaining for detecting cleaved caspase-3, cleaved poly(ADP-ribose)-polymerase, cleaved lamin-A, phosphorylated histone H2AX, and Bcl-2; (b) analyzed by terminal deoxyribonucleotidyl transferase–mediated dUTP nick end labeling assay to detect DNA fragmentation; and (c) evaluated with Masson's trichrome staining. We developed novel algorithm-based scoring methods and validated them using TMAs as a high-throughput format. The proposed computer-assisted scoring methods for digital images by brightfield microscopy permit linear quantification of immunohistochemical and histochemical stainings. Examples are provided of digital image analysis performed in automated or semiautomated fashion for successful quantification of molecular events associated with cell death in tissue sections. (J Histochem Cytochem 57:649–663, 2009)
Keywords: cell death, automated image analysis, algorithm, autoscore, virtual pathology
Inhibition or promotion of cell demise may have a curative or deleterious effect in a range of human diseases. The therapeutic induction of cell death is sought in cancer (reviewed in Reed 2006), whereas tilting the cell death–survival equilibrium toward the preservation of cell survival constitutes a therapeutic approach to stroke, infarction, and neurodegeneration (Bredesen et al. 2006).
One of the possible consequences of cell death events is structural remodeling of different organs, such as heart, liver, and lung, which often leads to irreversible loss of their normal function. Cardiac fibrosis, attributed both to necrotic (Weber 2004) and apoptotic (Takemura and Fujiwara 2004) processes, reduces myocardial distensibility and contractility.
The physiological importance and pathological consequences of cell demise have spurred attempts to define and classify cell death modalities, but simple categorization does not adequately reflect the complexity of endogenous cell death mechanisms. Thus, the Nomenclature Committee on Cell Death (NCCD) recommended mechanism-based morphological criteria that define death modalities such as apoptosis, autophagy, necrosis, mitotic catastrophe, anoikis, and paraptosis (Kroemer et al. 2009), emphasizing the stochastic, as opposed to synchronous, character of death and the importance of discriminating between dying (as a process) and death (as a point of no return). Other proposed cell death classification systems are based on biochemical criteria, functional features, or immunological attributes, arguing against a clear-cut distinction between different modes of cell demise (Galluzzi et al. 2007). Recent developments in automated image analysis technologies enable more objective and accurate assessment of diverse immunohistochemical markers and histochemical stains with relevance to cell death.
Dysregulation of apoptosis makes an important contribution to pathogenesis, progression, and treatment of most cancers (reviewed in Reed 2004). Although the various identified apoptotic pathways follow initially different tracks, they converge to activate the aspartic acid–specific proteases, caspases (reviewed in Meier and Vousden 2007). Caspase-3 is activated by most apoptotic stimuli (Kumar 1997). Numerous substrates of caspase-3 have been discovered, including poly(ADP-ribose) polymerase (PARP-1) (Lazebnik et al. 1994). Apart from its role in repair of damaged DNA (Berger 1985), several studies implicate PARP-1 in necrotic death (Ha and Snyder 1999). Lamins A and C are also cleaved by caspases (Takahashi et al. 1996; Kivinen et al. 2005).
Because most cytotoxic therapeutics commonly used in cancer therapy target DNA, the assessment of cellular responses to DNA damage may assist in designing new strategies for therapeutic interventions. DNA damage activates cell cycle checkpoint mechanisms that arrest cell growth, allowing time for damage repair. If DNA repair is unsuccessful, the cell often dies (Plesca et al. 2008). Posttranslational modifications play an essential role in regulating the cellular response to DNA damage. Phosphorylation of histone H2AX at serine 139 (Rogakou et al. 1998) is an early step in response to DNA double-strand breaks, resulting in discrete γH2AX foci at the DNA damage sites, which may initiate recruitment of repair or damage-signaling factors to the break (Paull et al. 2000). In response to cellular stress, p53 protein can initiate DNA repair, cell cycle arrest, senescence, and apoptosis (reviewed in (Vazquez et al. 2008). In addition to its roles as a transcription factor, the p53 protein participates directly in the intrinsic apoptosis pathway by interacting with the multidomain members of the Bcl-2 family to induce mitochondrial outer-membrane permeabilization, an event inhibited by Bcl-2 and other anti-apoptotic proteins (reviewed in Yip and Reed 2008).
Embracing the NCCD's recommendation to abandon poorly defined terms such as “the percentage of apoptosis, necrosis” (Kroemer et al. 2009), we propose computer-assisted quantification of specific parameters that characterize processes associated with cell death. In this study, we aim to provide automated image analysis solutions to gauging biochemical events associated with cell death in tissue specimens.
Materials and Methods
The experimental protocols were approved by the Burnham Institute for Medical Research Institutional Animal Care and Use Committee (#07-049, 07-144) and comply with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Tumor Xenograft Experiments
Experimental Design
Tumor xenograft experiments have been described in detail elsewhere (Hyer et al. 2005). In brief, MDA-MB-468 breast cancer cells were injected subcutaneously into 4-week-old female Balb/c nu/nu mice. When tumor volumes reached 25 mm3 (∼14 days), animals were treated daily for 14 days with intraperitoneal injections of 1-(2-cyano-3,12-dioxooleana-1,9-dien-28-oyl) imidazole (CDDO-Im). Six hours later, a subgroup was given intraperitoneal injections of 5 mg/kg/day of tumor necrosis factor–related apoptosis-inducing ligand (TRAIL).
Tissue Preparation and IHC
Mice were anesthetized (n=5/group) using Avertin (Sigma-Aldrich; St. Louis, MO). Anesthetized mice were transcardially perfused with ice-cold PBS (pH 7.4) for 2 min, followed by cold zinc-containing buffered formalin (Z-Fix; Anatech Ltd., Battle Creek, MI) for 5 to 10 min. After perfusion, tissues were immediately removed, postfixed in Z-fix, and embedded in paraffin. Dewaxed tissue sections (4.0–5.0 μm) were immunostained as reported previously (Krajewski et al. 1999) using rabbit polyclonal antibodies to cleaved caspase-3 (c-caspase-3; Cell Signaling Technology, Inc., Beverly, MA), cleaved lamin A (c-lamin A; Cell Signaling Technology), and γH2AX (Upstate Biotechnology, Inc.; Lake Placid, NY), as well as a mouse monoclonal anti-cleaved PARP (c-PARP) antibody (Cell Signaling Technology). The antibody to cleaved caspase-3 specifically recognizes the large fragment (∼17 kDa) that results from cleavage after Asp175. The anti-PARP antibody detects the large (89 kDa) fragment of human PARP-1 that results from cleavage at aspartic acid 214 and does not recognize full-length PARP-1 or other PARP isoforms. The lamin A antibody detects the large (∼45 kDa) fragment of lamin A, and the site of proteolysis is mapped to a conserved aspartic acid residue at position 230 (Rao et al. 1996). Application of the primary antibody was followed by incubation with goat anti-mouse or goat anti-rabbit polymer-based EnVision-HRP-enzyme conjugate (DakoCytomation; Carpinteria, CA). DAB (DakoCytomation) and SG-Vector (Vector Lab, Inc.; Burlingame, CA) chromogens were applied, yielding brown (c-caspase-3, c-lamin A, γH2AX) and black (c-PARP) colors, respectively.
Terminal Deoxyribonucleotidyl Transferase–mediated dUTP Nick End Labeling (TUNEL) Assay
The detection of nuclei with fragmented DNA by TUNEL was accomplished using the ApopTag Peroxidase In situ Apoptosis Detection Kit (Chemicon, Temecula, CA) according to the instructions of the manufacturer. Nuclear Red (DakoCytomation) was used as a counterstain.
Human Tumor Specimens
Patient Cohort Characteristics
Tissue microarrays containing specimens from 78 patients diagnosed with American Joint Committee on Cancer stage II colorectal cancer (CRC) during the period from 1990 to 1999 were obtained from PathWest at the Sir Charles Gairdner Hospital, Western Australia. Information on patient demographics and tumor features was obtained from pathology records. Tumor site was classified as proximal to and including, or distal to, the splenic flexure. Information on histological grade, anatomical site, and microsatellite instability (MSI) determined using the BAT-26 marker was available for 99%, 90%, and 94% of cases, respectively. Information on disease-specific survival was obtained from the Cancer Registry of Western Australia. The median follow-up time was 81.7 months for this patient cohort. At the end of the study period, 14% of patients had died of disease recurrence and 26% had died from other causes. Information on the use of adjuvant chemotherapy with fluorouracil/leucovorin-based regimens was obtained from hospital medical records for 45% (n=35) of patients. Thirty-one percent of patients (11 of 35) received chemotherapy. Ethics approval for the project was obtained from the Sir Charles Gairdner Hospital Research Ethics Committee.
Bcl-2 IHC
Using mouse monoclonal anti-Bcl-2 antibody clone 124 (DakoCytomation), immunostaining was performed by Clarient, Inc. (Aliso Viejo, CA) according to the manufacturer's instructions.
Mouse Controlled Cortical Impact Model
Experimental Design
The controlled cortical impact (CCI) model was used as described elsewhere (Bermpohl et al. 2006). The initial CCI tests aimed to measure the dimensions of neuronal cell death in the brain lesion after CCI elicited at 5 m/sec velocity and a time course of 6, 24, and 48 hr. Two control transgenic lines representing Cre-lox system (lines 1 and 2) were studied in a cohort of eight animals per time point.
Assessment of Histopathological Consequences of CCI
The aim of this pilot study was to obtain quantitative characteristics of brain tissue damage such as: (1) the size (area) of brain lesion that can be used to perform volume calculation; (2) the percentage of hemorrhagia; and (3) the dimension of acute/chronic neuronal cell death using Masson's trichrome stain (American Master*Tech Scientific, Inc.; Lodi, CA). After manual annotation of the impact foci, image analysis tools were tuned and employed to perform morphometry of this histochemical staining.
To calculate brain lesion volume, the brains were cut into five equally spaced 2 mm coronal slabs with three cross sections placed on the rostral and caudal edges and in the middle of the cortical lesion. The hematoxylin and eosin–stained and Masson's trichrome–stained sections were digitalized using the Aperio (Aperio Technologies; Vista, CA) scanning system. Using the pen tool, virtual slides were annotated by encircling the lesion edges (penumbra) between intact and pathologically changed brain tissue, and the lesion area (mm2) was reported in the annotation window. The rostral–caudal dimensions of the foci were determined through the use of a stereotaxic atlas for the mouse brain (Paxinos and Franklin 2001), which was viewed electronically side by side with the coronal section images. Distance (mm) between coronal coordinates for bregma +1.54 (rostral position) and −3.88 (caudal position) determined the distribution of the lesion volume and was multiplied by the sum of lesion areas (mm2) from all cross sections, yielding the total lesion volume (mm3). The proposed calculation is based on Cavalieri's method (Rosen and Harry 1990), which was modified by us for the purpose of volumetric analysis performed on digital slides.
Cardiac Fibrosis Model
Induction of Cardiac Fibrosis via Angiotensin II Administration
Male wild-type C57Bl/6J mice at 8 weeks of age were administered saline or angiotensin II (1.1 μg/kg/day) via a subcutaneously implanted mini-osmotic pump (Alzet # 1004; Alzet Osmotic Pumps, Cupertino, CA) for 4 weeks to induce cardiac fibrosis (n=6/group).
Histology
Animals were euthanized under isoflurane anesthesia, and the hearts were collected and fixed in 10% zinc-buffered formalin (Z-Fix). Formalin-fixed tissue was serially dehydrated and embedded in paraffin, and 5-μm-thick cross sections were assessed for total cardiac fibrosis (including interstitial, perivascular, and coronary arterial fibrosis) by Masson's trichrome staining. Image analysis was performed by a single blinded investigator, as described below. For each heart, the total fibrosis was calculated from three to six serial sections. For the purpose of evaluating the algorithms, 72 analyses were performed. Statistical analysis was performed using Prism 5 (GraphPad; San Diego, CA).
Quantitative Real-time RT-PCR
RNA was isolated from four 10-μm-thick formalin-fixed, paraffin-embedded sections according to the RecoverAll™ total nucleic acid isolation kit (Ambion, Inc.; Austin, TX). cDNA synthesis was performed using SuperScript III (Invitrogen, Carlsbad, CA). Quantitative real-time PCRs were performed on the Realplex4 Real-time PCR Detection System (Eppendorf; Westbury, NY) using TaqMan probe sets. Quantitative PCR assays (Assays on Demand) were from Applied Biosystems (Palo Alto, CA). Target gene probe sets included TGF-β (Mm01178820_m1), collagen1a1 (Mm01302043_g1), fibronectin (Mm01256744_m1), and β-actin (Mn00607939_s1). Experimental cycle threshold (Ct) values were normalized to β-actin measured on the same plate, and fold differences in gene expression were determined using the 2−ΔΔ Ct method (Livak and Schmittgen 2001).
Quantitative Analysis of Immunostaining and Histochemical Staining
All slides were scanned at an absolute magnification of 400× [resolution of 0.25 μm/pixel (100,000 pix/in.)] using the Aperio ScanScope CS and XT systems (Aperio Technologies), typically scanning from 5 to up to 120 slides per session, respectively. The background illumination levels were calibrated using a prescan procedure. The acquired digital images representing whole-tissue sections were evaluated for image quality. Images for 2 of the 126 slides were not captured, owing to a failure of the “tissue finding” function to detect weak immunostaining. These slides had to be rescanned using the “faint” mode. All acquired images were subsequently labeled, placed in dedicated project folders, and stored in designated local servers. Slides were viewed and analyzed remotely using desktop personal computers employing the web-based ImageScope viewer. The Spectrum Analysis algorithm package and ImageScope analysis software (version 9; Aperio Technologies, Inc.) were applied to quantify IHC and histochemical stainings. These algorithms make use of a color deconvolution method (Ruifrok and Johnston, 2001) to separate stains, so that quantification of individual stains avoided cross contamination. Using the software, each stain was individually calibrated by analyzing single-stained sections and recording the average red, green, and blue (RGB) optical density (OD) vectors. Other algorithm parameters were set to achieve concordance with manual scoring on a number of high-power fields, including intensity thresholds for positivity and parameters that control cell segmentation using the nuclear algorithm. The algorithms calculate the area of positive staining, the average positive intensity (API; optical density), as well as the percentage of weak (1+), medium (2+), and strong (3+) positive staining. A colocalization algorithm uses the deconvolution method to separate the stains and classifies each pixel according to the number of stains present. For colocalization, the user specifies a detection threshold for each stain, and the algorithm reports the percentage of area for which each stain combination is detected: 1, 2, 3, 1+2, 1+3, 2+3, 1+2+3, none (up to 3 stains are supported). The algorithm also provides an eight-color mark-up image for visualization of the colocalized states.
Manual Quantification of Immunostaining and TUNEL Results
To obtain a reference value, the number of nuclei positive for cleaved-caspase-3 and the total number of nuclei were counted in 10 random high-power fields. These counts were expressed as a percentage of immunopositive nuclei and used as references for evaluating the accuracy of the automated nuclear quantification.
The Bcl-2 immunostaining results were arbitrarily scored according to intensity as 0, negative; 1+, weak; 2+, moderate; or 3+, strong. The scoring was based on the percentage of immunopositive cells (0 to 100) multiplied by staining intensity score (0/1/2/3), yielding scores of 0 to 300.
Statistical Analysis
Data were analyzed using the STATISTICA software package (StatSoft; Tulsa, OK). The agreement between manual and automated results was tested by Spearman's correlation coefficient. Differences in the distribution of variables were tested using the ANOVA test for continuous variables. Median Bcl-2 immunopercentages and immunoscores (IS) obtained from manual assessment were applied as cutoffs for Kaplan-Meier survival analysis. Similarly, continuous automated assessment of Bcl-2 expression was dichotomized using median autoscore or median OD × percentage of immunopositive cells. Tissue microarray (TMA) cores with non-malignant tissue were excluded from statistical analysis.
Results
Automated Assessment of Apoptosis Markers and DNA Damage
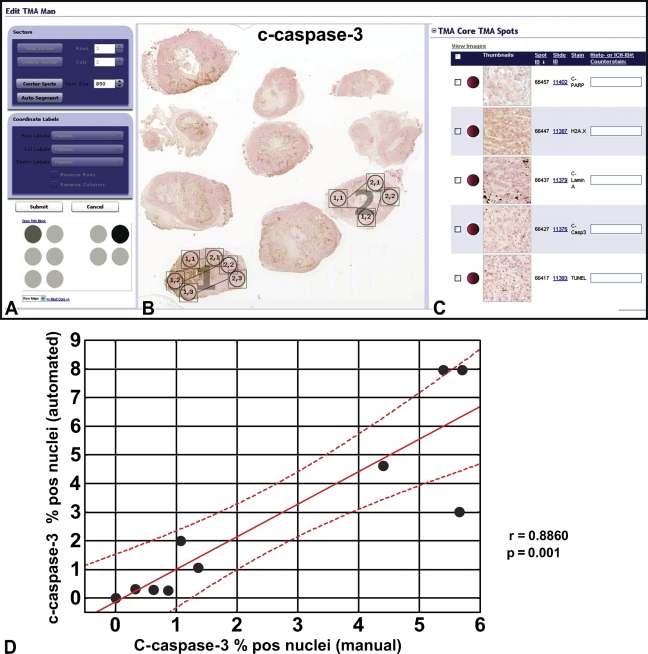
We developed automated procedures for quantifying markers of apoptosis representing cleaved caspases and their substrates as well as TUNEL-stained nuclei using a tumor xenograft model. Because pilot studies indicated that positive signals for all applied markers could be detected in nuclei, we selected the nuclear algorithm to perform the measurements. The algorithm counts the number of immunopositive and immunonegative nuclei, the number of pixels, and the average intensity of staining. Before applying the algorithm to the whole slide, we compared the automated and manual scoring on selected tumor areas, adjusting the algorithm parameters. For this purpose, a virtual TMA was assembled (Figure 1A) using TMA Lab software (Aperio Technologies, Inc.). The virtual TMA included images of tissue specimens stained for cleaved PARP-1, cleaved lamin A, cleaved caspase-3, and TUNEL. In addition, automated scoring for γH2AX, a marker of DNA damage, was also developed. Two representative tumor slices (Figure 1B) were assigned to sectors 1 and 2 (Figure 1A) with 10 TMA cores representing randomly selected areas of the tumor images. Each TMA core was viewed at a size corresponding to a square viewing field of 215 μm × 215 μm = 0.046 mm2 at 40× zoom magnification. TMA spot image thumbnails for all applied markers (Figure 1C) correspond to row 1, column 2 of sector 2 (Figure 1A) and were displayed in the Spectrum-Image database program, which allows data entries for manual and electronic measurements and image file management.
Figure 1.
Comparison of automated and manual scorings of apoptosis and DNA damage markers in tumor xenografts. Automated and manual scorings for cleaved caspase-3, cleaved lamin A, cleaved poly(ADP-ribose)-polymerase1 (PARP-1), γH2AX, and terminal deoxyribonucleotidyl transferase–mediated dUTP nick end labeling (TUNEL) were compared on the selected breast cancer areas using tissue microarray (TMA) Lab software (A–C). Correlation analysis compares automated and manual assessment of c-caspase-3–positive nuclei on 10 virtual TMA cores (D). r = 0.8860; p=0.001.
We adjusted the algorithm parameters, including averaging radius (0.8–1.5μm), curvature threshold (0.8–1.25), segmentation type (1) intensity for PARP-1; (2) cytoplasmic rejection for the other markers, threshold type (1) edge threshold method for TUNEL, cleaved caspase-3, and cleaved lamin A; (2) manual threshold method for H2AX, and PARP-1, and minimal nuclear size (15 μm2). Reducing the curvature threshold separated coalescing nuclei. In addition, we manually set RGB color components, and, if necessary, adjusted the OD values for particular channels using our single-stain procedure for each applied chromogen and counterstain. Weak, moderate, and strong positive thresholds were determined that conformed to intensity ranges on a scale of 0 to 255 (black to white, respectively). These adjustments were made for each marker to achieve optimal discrimination of tumor nuclei from cytosolic compartments. Mark-up images were reviewed to control the algorithm adjustment and were compared with the virtual images until optimal concordance was attained. Comparison of the percentage of cleaved caspase-3–positive nuclei obtained on 10 TMA cores by manual scoring and by the image analysis program showed a high degree of concordance, as confirmed by the correlation coefficient (r = 0.8860; p=0.001) (Figure 1D).
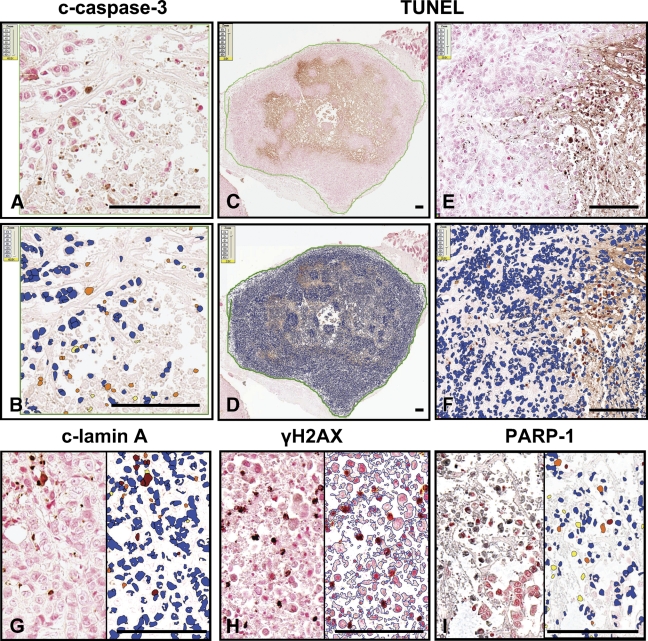
Once the algorithm had been fine-tuned on TMA cores, saved in a macro-file, and validated against manual scoring, full tumor sections were selected with a pen tool for quantitative analyses of cleaved caspase-3, cleaved PARP-1, cleaved lamin A, and H2AX immunostainings, and the TUNEL assay (Figure 2C) in tumor xenografts recovered from mice treated with vehicle (control) or a combination of anticancer agents (synthetic triterpenoid CDDO-Im and TRAIL). The adjusted algorithms were then reapplied to the full selected tumor areas. All results were saved in annotation format. Figure 2 presents digital images of slides stained for cleaved caspase-3, TUNEL, cleaved lamin A, γH2AX, and PARP-1. These are displayed side by side with pseudocolor mark-up images superimposed on the original images, which provides a visual representation of the numerical results. For histone H2AX, the analysis was done with nuclear edges as a mark-up image type (Figure 2H). Quantitative results obtained by application of the nuclear algorithm for all employed markers are provided in Table 1. Note that all markers of apoptosis significantly increased in the treated tumors compared with untreated. The marker of DNA damage was also slightly increased.
Figure 2.
Application of the nuclear algorithm to immunostained cancer xenograft specimens. The nuclear algorithm was applied to breast tumor specimens immunostained for cleaved caspase-3 (A,B), cleaved lamin A (G), γH2AX (H), PARP-1 (I), and labeled by TUNEL assay (C–F). On the digital slide images (A,C,E, and left side of G–I), brown and black colors denote immunopositive staining (brown, DAB chromogen; black, SG chromogen). Specimens were counterstained with Nuclear Red. In the mark-up images (B,D,F, and right side of G–I), red, orange, and yellow pixels visualize immunopositive nuclei (strong, moderate, and weak intensity, respectively), whereas blue pixels depict immunonegative nuclei. Bar = ∼100 μm.
Table 1.
Results of the nuclear algorithm application in tumor xenograft specimens
| Percentage of positive nuclei/cells
|
|||||
|---|---|---|---|---|---|
| Treatment | c-Caspase-3 | PARP-1 | c-Lamin A | γH2AX | TUNEL |
| Untreated: vehicle | 5.09 + 0.16 | 7.1 + 0.24 | 8.89 + 0.28 | 1.77 + 0.13 | 4.92 + 0.21 |
| Treated: CDDO-Im+TRAIL | 16.25 + 0.22 | 15.0 + 0.16 | 17.8 + 0.25 | 3.67 + 0.24 | 11.18 + 0.32 |
Mean percentage and SEM values are recorded for nuclei positive for cleaved caspase-3, cleaved lamin A, cleaved PARP-1, γH2AX, or terminal deoxyribonucleotidyl transferase–mediated dUTP nick end labeling (TUNEL). CDDO-Im, 1-(2-cyano-3,12-dioxooleana-1,9-dien-28-oyl) imidazole; TRAIL, tumor necrosis factor–related apoptosis-inducing ligand; PARP, poly(ADP-ribose)-polymerase.
Correlation of Manual and Automated Immunoscoring Methods for Apoptosis Biomarker Bcl-2 in Human Tumors
The apoptosis-suppressive protein Bcl-2 has previously been assessed by immunohistochemistry in human tumor specimens as a powerful prognostic or predictive biomarker (see, for example, Krajewska et al. 2005; Tewari et al. 2008). We sought to develop an automated immunoscoring method for Bcl-2 applied to patient tumor materials. For this purpose, we evaluated a collection of CRCs. Tumor areas were selected for computer-assisted analysis. Cytosolic Bcl-2 expression [3,3′-diaminobenzidine (brown)] was quantitatively determined by applying a color deconvolution algorithm. To perform separation of the Bcl-2 immunostain and nuclear counterstain, a color deconvolution plug-in was applied, utilizing the following vectors for the RGB channels: for color (1) hematoxylin, 0.65, 0.70, and 0.29; for color (3) DAB, 0.27, 0.57, and 0.78 (Ruifrok and Johnston, 2001). Thus, in this example of a single antigen and chromogen, the unmixing of the chromogen from the counterstain generates a partition of DAB and hematoxylin signals.
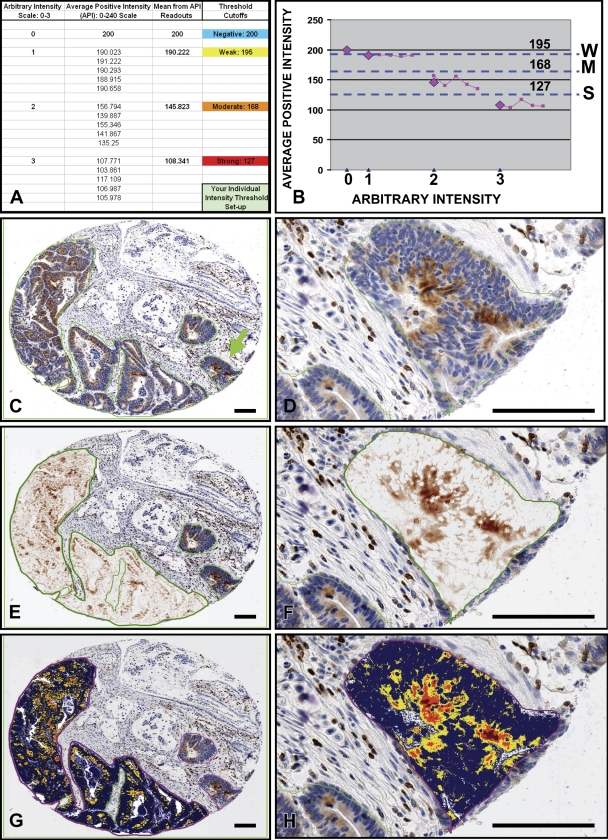
To fine-tune the algorithm, we sought to exclude the background staining (image noise) from the analysis. For this purpose, the algorithm was run on a selected region containing only the background DAB staining; by gradually reducing the initial 240 weak positive threshold value, the yellow (“weak positive”) color in the mark-up image was increasingly eliminated and replaced by blue (“negative”). The 200 API corresponding to the entirely blue annotated region was recorded (Figures 3A and 3B) as “0” positive intensity and tentatively entered as the weak positive threshold in the algorithm. Next, the algorithm was run on five regions representing weak positive intensity, and the corresponding API values were recorded as “1” intensity (Figure 3A). The same procedure was repeated for the medium (2) and subsequently for the strong (3) intensity regions (lymphocytes) (Figure 3A). The API values (Figure 3A) were plotted against arbitrary intensity (0–3), yielding means from the API readouts depicted as magenta diamonds on the presented graph (Figure 3B). Dotted lines drawn through the middle points between the API mean values (magenta diamonds) determined weak, medium, and strong positive thresholds (Figure 3B). After fine-tuning of the algorithm, the actual analysis was performed without human supervision; the TMA Lab program successively analyzed images based on the algorithm attributes established during the parameter adjustment. Only manually selected tumor areas were analyzed, thus eliminating stromal and non-malignant epithelial colon tissue (Figures 3C and 3D). All tissue cores could be viewed simultaneously for comparison or separately at different zoom levels. The color deconvolution algorithm separated the chromogen color from the counterstain, revealing undisguised expression of the Bcl-2 protein characterized by the brown DAB color (Figures 3E and 3F). Pseudocolor “mark-up” images visualized the Bcl-2 protein intensity range and distribution in tumors (Figures 3G and 3H). The obtained data were exported from the TMA Lab program to an Excel spreadsheet.
Figure 3.
Quantitative assessment of Bcl-2 immunostaining in tumors assembled on a TMA slide. TMAs containing tumor cores were stained for Bcl-2 using the diaminobenzidine-based detection method and hematoxylin counterstaining. The color deconvolution algorithm was fine-tuned and determined weak, medium, and strong positive thresholds (A,B). Tumor areas were selected (C) for analysis by the TMA Lab program. The selected regions (C,D) were visualized as deconvolved images (E,F) (DAB, brown color) and mark-up intensity range images (G,H). Bar = ∼100 μm.
To assess the level of concordance between results of visual and computer-assisted analyses, immunopercentages of Bcl-2–positive tumor cells were compared. In addition, manual IS, as defined in Materials and Methods, was compared with: (1) the “autoscore” [(1 × percentage weak positive + 2 × percentage medium positive + 3 × percentage strong positive): 3]; (2) OD × percentage total positive; and (3) OD × positive area (mm2). It is assumed that the signal on the slide is quantitatively related to the abundance of the antigens in the tissue section. According to the Lambert-Beer law, the amount of stain deposited determines the OD at stain-specific wavelengths, with the OD proportional to the stain concentration (Ruifrok and Johnston 2001). Thus, the amount of dye is linearly related to the OD (absorbance). The conversion to OD is performed by taking the negative (base 10) log of the intensity of light detected after passing the specimen (In) divided by the intensity of light that would have been received by the detector, if no object were in the light path (I0) (Jonker et al. 1997; Ruifrok and Johnston 2001). Thus, OD = −log (In/I0), whereby I0 is determined by recording a blank image (Jonker et al. 1997) and equals 240 in the ScanScope. OD units are dimensionless and logarithmic: zero absorbance means all photons transmitted; an OD of 1.0 absorbs 90% of all photons, and an OD of 2.0 absorbs 99% of all potentially detected photons; IHC stains can individually generate signals of 1 OD (Taylor and Levenson 2006).
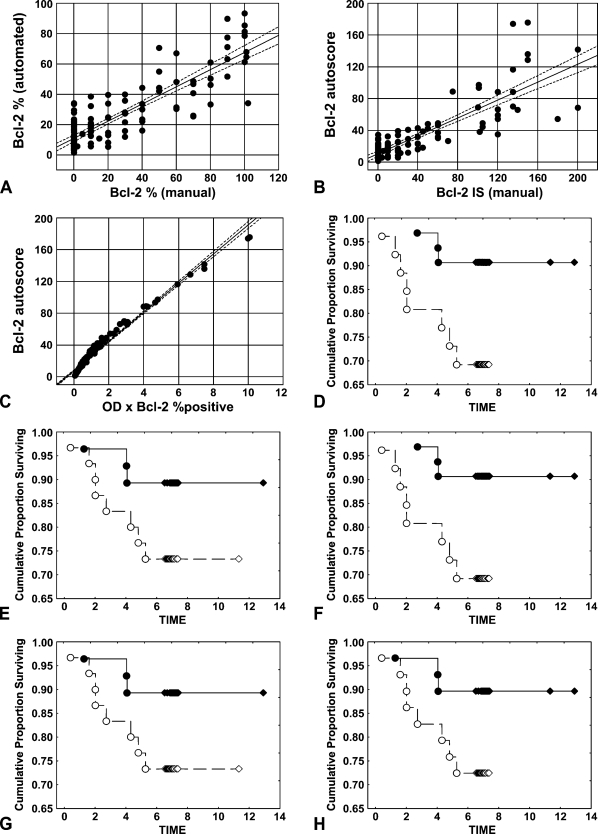
Strong positive correlations between automatically and manually evaluated Bcl-2 expression levels were determined with 95% concordance level for immunopercentage of Bcl-2–positive cells (Figure 4A; r = 0.8665; p<0.0001). Correlations were also observed for the manual Bcl-2 and automated IS for Bcl-2 (Figure 4B; r = 0.8453; p<0.0001) and for the manual IS and the “OD × percentage total positive” parameter (r = 0.8076; p<0.0001; not shown). Thus, both the autoscore and the “OD × percentage total positive” factor can be applied as automatically determined equivalents to the manual immunoscore. An excellent concordance between these two parameters (Figure 4C; r = 0.9877; p<0.0001) confirms this observation. Surprisingly, the “OD × positive area (mm2)” parameter showed only a low degree of agreement with the manual IS assessment (r = 0.5247; p=0.01).
Figure 4.
Assessment of concordance between visual and computer-assisted analyses of Bcl-2 immunostaining. TMAs containing tumor cores were stained for Bcl-2 using the diaminobenzidine-based detection method and counterstained with hematoxylin. Correlations were determined with 95% concordance level between (A) automatically vs manually determined immunopercentage of Bcl-2–positive cells (r = 0.8665; p<0.0001), (B) Bcl-2 autoscore vs Bcl-2 manual immunoscores (IS) (r = 0.8453; p<0.0001), and (C) Bcl-2 autoscore vs optical density (OD) × percentage of Bcl-2–positive cells as determined by algorithm (r = 0.9877; p<0.0001). The Kaplan-Meier method was applied to generate the survival curves, and univariate survival distributions were compared by the log-rank test (D–H). Manual (D) (p=0.04) and automated (E) (p=0.1) Bcl-2 immunopercentage, Bcl-2 manual IS (F) (p=0.04), and autoscore (G) (p=0.1), as well as OD × automated Bcl-2 immunopercentage (H) (p=0.1) were compared with overall survival for colorectal cancer patients. Continuous line depicts high and dashed line low Bcl-2 protein levels.
To further validate the algorithm, agreement levels between both automated and manual assessments of Bcl-2 protein expression and demographic characteristics or clinicopathological parameters were analyzed. Regardless of the employed assessment method, Bcl-2 expression did not correlate with MSI status, anatomic location of tumors, patient gender, or age. To correlate Bcl-2 protein expression in tumors with patient survival, we empirically dichotomized all of the above-mentioned parameters at the median, comparing the survival of patients whose scores were above the median with that of those below the median. For univariate survival analysis, the Kaplan-Meier method was applied, and the differences between survival curves were assessed by the log-rank test. In the investigated cohort, significant correlations were observed between shorter overall survival and visually determined immunopercentages and IS indicative of lower Bcl-2 expression (p=0.04; Figures 4D and 4F). The same tendency was maintained when the automatically established parameters, such as percentage of Bcl-2-immunopositive cells, Bcl-2 autoscore, and “OD × percentage total positive” were employed (Figures 4E, 4G, and 4H, respectively), but the results did not reach statistical significance (p=0.1).
Interpretation of Masson's Trichrome Stain in the Context of Cell Death in the Mouse CCI Model
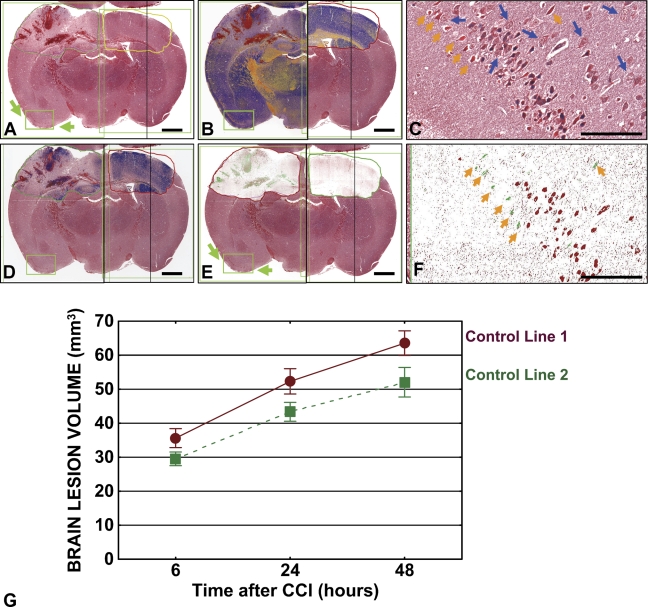
Measurements of IHC colorimetric intensity constitute a primary application of image analysis algorithms. We applied these measurements to a histochemical stain, Masson's trichrome, to qualitatively and quantitatively characterize cell death events in nervous tissue. Examples of this application are provided in Figure 5, which presents Masson's trichrome–stained images of a representative brain coronal section derived from a mouse 6 hr after CCI (Figure 5A). Color deconvolution (Figure 5B), nuclear (Figure 5D), and colocalization (Figure 5E) algorithms were employed to obtain quantitative characteristics of brain tissue damage, such as percentage of hemorrhagic area, as well as percentage and total number (not shown) of apoptotic and necrotic neurons in the annotated area of interest (Table 2). These measurements have been compared with those from the control contralateral cerebral hemisphere (Figure 5; Table 2). Using this approach, we performed a semiautomated analysis of brain lesion volumes in mice subjected to CCI. A comparison was performed of two control transgenic lines representing Cre-lox system (lines 1 and 2). Comparison of average brain lesion volumes in the control lines did not reveal significant differences (Figure 5G).
Figure 5.
Algorithm-assisted analysis of brain lesions. Tissue sections from mice subjected to controlled cortical impact were stained with Masson's trichrome stain. The digital slide image (A) presents the core lesion in the left hemisphere and intact contralateral right hemisphere with selected regions of interest. Application of color deconvolution (B), nuclear (D), and color colocalization (E) algorithms resulted in generation of mark-up images. (B) Micro-hemorrhage areas are indicated by red color, early ischemic changes in the gray matter by orange, normal brain tissue by blue. (C) Necrotic (bright red; orange arrowheads), apoptotic (pyknotic; dark-blue nuclei), and normal (blue arrows) neurons in the contrecoup focus in the digital slide image (A,E) (green arrows) are visualized as green (necrotic) or red (apoptotic) cells in the corresponding mark-up image (F). Bars in macroimages (A,B,D,E) = 1 mm, in high-zoom images (C,F) = ∼100 μm. (G) Graphical presentation of average volumes of brain lesions in control lines 1 (brown) and 2 (green). Markers depict mean volume; error bars indicate SEM. p values for 6 hr, 24 hr, and 48 hr time points equal 0.1, 0.07, and 0.06, respectively.
Table 2.
Algorithm-assisted measurements corresponding to Figure 5
| Abnormal neurons (%)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotated area
|
Hemorrhage (%)
|
Intact gray matter (%)
|
Apoptotic
|
Necrotic
|
|||||||
| Figure 5 Panel | Algorithm | Control (mm2) | CCI (mm2) | Control | CCI | Control | CCI | Control | CCI | Control | CCI |
| B | Deconvolution | 3.763 | 4.027 | 1.472 | 20.79 | 59.342 | 42.082 | 1.46 | 8.24 | 0.456 | 2.678 |
| D | Nuclear | 6.487 | 8.963 | 2.621 | 14.399 | 93.709 | 85.6 | 5.9 | 12.89 | 1.345 | 2.341 |
| E | Colocalization | 6.503 | 9.036 | 1.379 | 29.093 | NR | NR | 0.95 | 14.387 | 0.782 | 7.726 |
CCI, controlled cortical impact; NR, not revealed.
Morphologic criteria identified with Masson's trichrome stain assist in discerning apoptotic neurons (the chromatin clumps are basophilic, and the rest of the cell is eosinophilic) from shrunken necrotic neuronal cells characterized by brightly eosinophilic cytoplasm; different color and size/shape characteristics permit algorithm application. For example, the color deconvolution algorithm separates the image into three channels, allowing measurement of the area for each stain separately. As in the prior application, algorithms were tuned and tested on a variety of images to obtain the most accurate results on this particular tissue type and for this specific histochemical stain. Multiple images containing the same selected region were synchronized and viewed simultaneously, facilitating analysis of the results. Magnified (40×) views of areas of entorhinal cortex, which are indicated by green arrows on the digital slide images (Figures 5A and 5E) are presented in panels C (original image) and F (mark-up; colocalization algorithm), respectively. Note that the apoptotic nuclei are colored red and the necrotic nuclei are colored green.
Quantitative Assessment of Cardiac Fibrosis
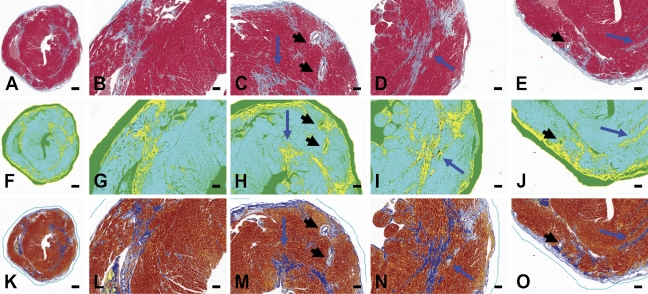
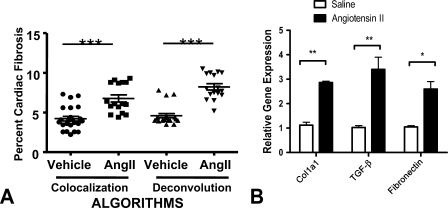
Total cardiac fibrosis was determined using two distinct yet complementary color analysis algorithms, employing tissues from a mouse model that had been stained with Masson's trichrome. First, color colocalization was performed (Figures 6F–6J). Aniline blue staining of collagen was identified and coded yellow (“pseudocolored” yellow). Cardiac myocytes and other cells were coded turquoise. Regions of overlap were coded black. The pixel area coded yellow or black was summed and expressed as a percent of total area (turquoise myocardium). White space surrounding the section or in the ventricular lumen was excluded from the analysis. Second, color deconvolution was performed (Figures 6K–6O). Collagen was identified and coded blue, whereas myocardial tissue was coded red. In the algorithm window, we input the following values for the RGB channels: for color (1) 0.65, 0.70, and 0.29; for color (2) 0.99, 0.105, and 0.268, for color (3) 0.27, 0.57, and 0.78. Percent fibrotic area was calculated automatically by the ScanScope software and corresponded to the blue color parameters. The colocalization analysis revealed significantly more fibrosis in the form of collagen deposition in hearts from animals treated with angiotensin II (6.8 ± 0.5%) compared with hearts from vehicle-treated animals (4.2 ± 0.3%; p<0.001, by Student's t-test) (Figure 7A). Analysis performed using the color deconvolution algorithm revealed similar results. In animals receiving angiotensin II, the percent area identified as collagen was significantly greater than in animals receiving saline vehicle (8.3 ± 0.4% angiotensin II vs 4.6 ± 0.3% saline; p<0.001 by Student's t-test, Figure 7A). The percent area of collagen observed in both treatment groups was consistent with prior literature (Billet et al. 2007). Both algorithms demonstrated a significant amount of collagen in the hearts from angiotensin II–treated animals. This finding is consistent with numerous reports of angiotensin II–induced cardiac fibrosis (see, for example, Ramires et al. 1998). The intensity of Masson's trichrome blue color staining depended on the content of collagen fibers in the investigated tissue. Fresh scar, containing a lower number of loose collagen fibers, showed a lower aniline blue signal that was identified by a more-sensitive colocalization algorithm. The extent of cardiac fibrosis as assessed by the automated algorithm was consistent with molecular markers of fibrosis (Figure 7B). Quantitative real-time PCR revealed a significant increase in the expression in angiotensin II–treated mice of Col1a1 (p<0.01), other profibrotic genes (TGF-β; p<0.01), and fibronectin (p<0.05) by Student's t-test.
Figure 6.
Analysis of tissue fibrosis. Tissues were obtained from mice treated with vehicle control or angiotensin II. Representative digital micrograph of a single cardiac section (5 μm) from a mouse administered angiotensin II (1.1 μg/kg/day) for 4 weeks. Tissue sections were stained with Masson's trichrome to identify collagen, as indicated by aniline blue staining (A–E). Collagen content was determined by two complementary algorithms: color colocalization (F–J) and color deconvolution (K–O). In both analyses, collagen deposition was separated from non-fibrotic tissue or empty space based on red, green, and blue pixel intensity thresholding. Panels A, F, K show the entire section, whereas the remaining panels show the section in quarters at higher magnification. Both analyses identified similar regions containing collagen. Blue arrows indicate areas of significant interstitial fibrosis. Note the perivascular collagen surrounding coronary arteries (black arrowheads).
Figure 7.
Quantitative assessment of tissue fibrosis. (A) The relative ratio of total fibrosis area to total area as determined by color colocalization and deconvolution. Data are presented as a scatter plot with mean ± SEM indicated by error bars. ***p<0.001. (B) Quantitative real-time PCR revealed significant increases in the expression of collagen1a1 and other profibrotic genes TGF-β and fibronectin. ** p>0.01; *p>0.05 by Student's t-test.
Discussion
To overcome limitations of laborious manual semiquantitative scoring of immuno- and histochemical stainings of tissue sections, more-objective, reproducible, and rapid computer-assisted quantification methods have been developed and applied to digital images of tissues (reviewed in Rojo et al. 2006; Mulrane et al. 2008). Examples of algorithm applications for quantitative analysis of histological images have previously been provided by us (Krajewska et al. 2007,2008) and others (Rubin et al. 2004; Brennan et al. 2008; Fine et al. 2008; Rexhepaj et al. 2008). In the present study, we illustrate specific applications of algorithms using biomarkers that reflect events commonly associated with cell death mechanisms. Unlike semiquantitative manual scoring, which is subjective, due to its inherent intra- and inter-observer variability, computer-based image analysis provides a means of objectively measuring areas and intensities of histological stains in tissue sections. Moreover, whereas tedious traditional morphometric analysis by manual inspection allows cell counting in only selected areas, which often do not reflect the cellular heterogeneity of specimens, virtual slide technology circumvents this problem by permitting the rapid automated analysis of entire tissue sections. By enabling researchers to view and analyze digital images via the Internet, this technology fosters collaboration, potentially increasing the accuracy of interpretation of experimental results.
Evaluation by the human eye presents difficulties in distinguishing subtle differences in staining intensity, especially at the extremes of the continuous color scale. TMAs provide almost ideal material for automated image analysis because of the concurrent application of identical staining conditions to all cores on a single slide. The rate of manual scoring is insufficient to keep pace with the high-throughput nature of TMA immunostainings. TMA Lab software enables rapid automated image analysis, thus overcoming many of the bottlenecks associated with traditional microscope-based TMA evaluations.
Due to variations in stainings, resulting from different fixation procedures, antibody properties, and retrieval methods, the establishment of individual cutoff values is an important step in algorithm application. Because the investigator can calibrate the algorithms, they can be adapted to the individual experiment. Our efforts focused, to a large degree, on establishing a method to optimize threshold values for scoring, as exemplified by the Bcl-2 immunostainings. OD of the marker, corrected for the intensity of the background, provides a more accurate assessment of the staining and allows comparisons between slides stained in different batches or in different laboratories. Although it is more time-consuming, the specific selection of tumor tissue within the tissue core increases the specificity of the analysis.
It is noteworthy that the proposed solutions do not necessarily depend on the availability of the Aperio system. Once the images are scanned, they can be analyzed in Image J, which, along with its Java source code, is freely available in the National Institutes of Health Image public domain and runs on Linux, Mac, Windows, and the Sharp Zaurus personal digital assistant. More than 300 macros and 500 plug-ins are available on the Image J Web site to facilitate analysis of scanned images and development of algorithms. For example, the color deconvolution algorithm that has been applied in this study is based on the National Institutes of Health Image macro provided and described by A. C. Ruifrok (Ruifrok and Johnston 2001). This algorithm is based on orthonormal transformation of the original RGB image and can be used for separation of two or three stains that are sufficiently different in their red, green, or blue absorption characteristics (Ruifrok and Johnston 2001). Depending on the applied chromogens and counterstaining reagents, new vectors can be determined by performing repeated measurements on specimens with single stains (stained only with a selected dye). The use of algorithms with capabilities for measurement of subcellular localization of proteins and colocalization of specific protein expression with cell-specific markers (Krajewska et al. 2007,2008) provides the potential for significant improvement in assessment of biomarker expression.
This study is the first to use automated quantitative algorithms to analyze immunohistochemical and histological parameters that characterize processes associated with cell death. In addition, we have applied two novel automated scoring models (i.e., autoscore and “OD × percentage total positive”) in our study of Bcl-2 protein expression in human tumor specimens. Both parameters significantly correlate with the conventional manual immunoscore. OD (absorbance) values are an intrinsic property of the sample, and both parameters significantly correlate with the conventional manual immunoscore. Furthermore, we demonstrate the utility of the color colocalization, deconvolution, and nuclear algorithms to qualitatively and quantitatively characterize cell death events in nervous tissue when applied to a routine histochemical stain, Masson's trichrome. This approach may provide a valuable tool for quantitative assessment of brain tissue damage, for example in genetically engineered mouse models (Krajewski et al., unpublished data). Moreover, the color colocalization and deconvolution algorithms presented are suitable for quantification of tissue fibrosis as reflected in Masson's trichrome staining. The tool is easily applied to cardiac fibrosis, but it may also be useful in determining the area of infarct after myocardial ischemia and perivascular fibrosis, as well as in the quantification of hepatic, renal, or pulmonary fibrosis. These examples illustrate that wider application of automated histological analysis tools could lead to better standardization and more-quantitative interpretation of stained tissue sections for experimental medicine and diagnostic pathology.
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grant NS-036821 (to SK), the California Breast Cancer Research Program (grant 10WB-0194) and NIH Grant U01-CA113318 (to JCR) and Florida Department of Health (grant 06-NIR-09 to LS). The TMA project was supported by National Cancer Institute Grant R44CA-117001 (to SPL).
We thank Lisa Spalding for high-quality TMA preparation and Jessica Groos and Judy Wade for excellent animal care assistance.
References
- Berger NA (1985) Poly(ADP-ribose) in the cellular response to DNA damage. Radiat Res 101:4–15 [PubMed] [Google Scholar]
- Bermpohl D, You Z, Korsmeyer SJ, Moskowitz MA, Whalen MJ (2006) Traumatic brain injury in mice deficient in Bid: effects on histopathology and functional outcome. J Cereb Blood Flow Metab 26:625–633 [DOI] [PubMed] [Google Scholar]
- Billet S, Bardin S, Verp S, Baudrie V, Michaud A, Conchon S, Muffat-Joly M, et al. (2007) Gain-of-function mutant of angiotensin II receptor, type 1A, causes hypertension and cardiovascular fibrosis in mice. J Clin Invest 117:1914–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredesen DE, Rao RV, Mehlen P (2006) Cell death in the nervous system. Nature 443:796–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan DJ, Rexhepaj E, O'Brien SL, McSherry E, O'Connor DP, Fagan A, Culhane AC, et al. (2008) Altered cytoplasmic-to-nuclear ratio of survivin is a prognostic indicator in breast cancer. Clin Cancer Res 14:2681–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine JL, Grzybicki DM, Silowash R, Ho J, Gilbertson JR, Anthony L, Wilson R, et al. (2008) Evaluation of whole slide image immunohistochemistry interpretation in challenging prostate needle biopsies. Hum Pathol 39:564–572 [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Maiuri MC, Vitale I, Zischka H, Castedo M, Zitvogel L, Kroemer G (2007) Cell death modalities: classification and pathophysiological implications. Cell Death Differ 14:1237–1243 [DOI] [PubMed] [Google Scholar]
- Ha HC, Snyder SH (1999) Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci USA 96:13978–13982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyer ML, Croxton R, Krajewska M, Krajewski S, Kress CL, Lu M, Suh N, et al. (2005) Synthetic triterpenoids cooperate with tumor necrosis factor-related apoptosis-inducing ligand to induce apoptosis of breast cancer cells. Cancer Res 65:4799–4808 [DOI] [PubMed] [Google Scholar]
- Jonker A, Geerts WJ, Chieco P, Moorman AF, Lamers WH, Van Noorden CJ (1997) Basic strategies for valid cytometry using image analysis. Histochem J 29:347–364 [DOI] [PubMed] [Google Scholar]
- Kivinen K, Kallajoki M, Taimen P (2005) Caspase-3 is required in the apoptotic disintegration of the nuclear matrix. Exp Cell Res 311:62–73 [DOI] [PubMed] [Google Scholar]
- Krajewska M, Kim H, Kim C, Kang H, Welsh K, Matsuzawa S, Tsukamoto M, et al. (2005) Analysis of apoptosis protein expression in early-stage colorectal cancer suggests opportunities for new prognostic biomarkers. Clin Cancer Res 11:5451–5461 [DOI] [PubMed] [Google Scholar]
- Krajewska M, Kitada S, Winter JN, Variakojis D, Lichtenstein A, Zhai D, Cuddy M, et al (2008) Bcl-B expression in human epithelial and nonepithelial malignancies. Clin Cancer Res 14:3011–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewska M, Olson AH, Mercola D, Reed JC, Krajewski S (2007) Claudin-1 immunohistochemistry for distinguishing malignant from benign epithelial lesions of prostate. Prostate 67:907–910 [DOI] [PubMed] [Google Scholar]
- Krajewski S, Krajewska M, Ellerby LM, Welsch K, Xie Z, Deveraux QL, Salvesen GS, et al. (1999) Release of caspase-9 from mitochondria during neuronal apoptosis and cerebral ischemia. Proc Natl Acad Sci USA 96:5752–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. (2009) Classification of cell death: recommendations of the nomenclature committee on cell death 2009. Cell Death Differ 16:3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S (1997) The apoptotic cysteine protease CPP32. Int J Biochem Cell Biol 29:393–396 [DOI] [PubMed] [Google Scholar]
- Lazebnik YA, Kaufman SH, Desnoyers S, Poirier GG, Earnshaw WC (1994) Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ice. Nature 371:346–347 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Meier P, Vousden KH (2007) Lucifer's labyrinth: ten years of path finding in cell death. Mol Cell 28:746–754 [DOI] [PubMed] [Google Scholar]
- Mulrane L, Rexhepaj E, Penney S, Callanan JJ, Gallagher WM (2008) Automated image analysis in histopathology: a valuable tool in medical diagnostics. Expert Rev Mol Diagn 8:707–725 [DOI] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10:886–895 [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin BJ (2001) The Mouse Brain in Stereotaxic Coordinates. San Diego, Academic Press
- Plesca D, Mazumder S, Almasan A (2008) DNA damage response and apoptosis. Methods Enzymol 446:107–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramires FJ, Sun Y, Weber KT (1998) Myocardial fibrosis associated with aldosterone or angiotensin II administration: attenuation by calcium channel blockade. J Mol Cell Cardiol 30:475–483 [DOI] [PubMed] [Google Scholar]
- Rao L, Perez D, White E (1996) Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol 135:1441–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JC (2004) Apoptosis mechanisms: implications for cancer drug discovery. Oncology 18:11–20 [PubMed] [Google Scholar]
- Reed JC (2006) Drug insight: cancer therapy strategies based on restoration of endogenous cell death mechanisms. Nat Clin Pract Oncol 3:388–398 [DOI] [PubMed] [Google Scholar]
- Rexhepaj E, Brennan DJ, Holloway P, Kay EW, McCann AH, Landberg G, Duffy MJ, et al. (2008) Novel image analysis approach for quantifying expression of nuclear proteins assessed by immunohistochemistry: application to measurement of oestrogen and progesterone receptor levels in breast cancer. Breast Cancer Res 10:R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273:5858–5868 [DOI] [PubMed] [Google Scholar]
- Rojo MG, Garcia GB, Mateos CP, Garcia JG, Vicente MC (2006) Critical comparison of 31 commercially available digital slide systems in pathology. Int J Surg Pathol 14:285–305 [DOI] [PubMed] [Google Scholar]
- Rosen GD, Harry JD (1990) Brain volume estimation from serial section measurements: a comparison of methodologies. J Neurosci Methods 35:115–124 [DOI] [PubMed] [Google Scholar]
- Rubin MA, Zerkowski MP, Camp RL, Kuefer R, Hofer MD, Chinnaiyan AM, Rimm DL (2004) Quantitative determination of expression of the prostate cancer protein alpha-methylacyl-CoA racemase using automated quantitative analysis (AQUA): a novel paradigm for automated and continuous biomarker measurements. Am J Pathol 164:831–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruifrok AC, Johnston DA (2001) Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol 23:291–299 [PubMed] [Google Scholar]
- Takahashi A, Alnemri ES, Lazebnik YA, Fernandes-Alnemri T, Litwack G, Moir RD, Goldman RD, et al. (1996) Cleavage of lamin A by Mch2 alpha but not CPP32: multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Natl Acad Sci USA 93:8395–8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura G, Fujiwara H (2004) Role of apoptosis in remodeling after myocardial infarction. Pharmacol Ther 104:1–16 [DOI] [PubMed] [Google Scholar]
- Taylor CR, Levenson RM (2006) Quantification of immunohistochemistry: issues concerning methods, utility and semiquantitative assessment II. Histopathology 49:411–424 [DOI] [PubMed] [Google Scholar]
- Tewari M, Krishnamurthy A, Shukla HS (2008) Predictive markers of response to neoadjuvant chemotherapy in breast cancer. Surg Oncol 17:301–311 [DOI] [PubMed] [Google Scholar]
- Vazquez A, Bond EE, Levine AJ, Bond GL (2008) The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov 7:979–987 [DOI] [PubMed] [Google Scholar]
- Weber KT (2004) From inflammation to fibrosis: a stiff stretch of highway. Hypertension 43:716–719 [DOI] [PubMed] [Google Scholar]
- Yip KW, Reed JC (2008) Bcl-2 family proteins and cancer. Oncogene 27:6398–6406 [DOI] [PubMed] [Google Scholar]