Abstract
Set1-dependent H3K4 di- and tri-methylation (H3K4me2/3) have been associated with active transcription. Recent data indicate that the H3K4me2/3 also plays a poorly characterized RNA-dependent repressive role. Here, we show that GAL1 promoter is attenuated by the H3K4me2/3 deposited by cryptic transcription. The H3K4me2/3 delay the recruitment of RNA polymerase II (RNAPII) and TBP on GAL1 promoter. Inactivation of RNA decay components revealed the existence of the RNAPII-dependent unstable RNAs, initiating upstream of GAL1 (GAL1ucut). GAL1ucut RNAs are synthesized in glucose and require the Reb1 transcription factor. Consistent with a regulatory function of the cryptic transcription, Reb1 depletion leads to a decrease of H3K4me3 on GAL10-GAL1 locus in glucose and to an acceleration of GAL1 induction. A candidate approach shows that the RPD3 histone deacetylase attenuates GAL1 induction and is tethered at the GAL10-GAL1 locus by H3K4me2/3 upon repression. Strikingly, Set1-dependent Rpd3 recruitment represses also the usage of a hidden promoter within SUC2, suggesting a general function for H3K4me2/3 in promoter fidelity. Our data support a model wherein certain promoters are embedded in a repressive chromatin controlled by cryptic transcription.
Keywords: chromatin, CUT, regulatory RNA, Set1, transcription
Introduction
The condensation of eukaryotic DNA in arrays of nucleosomes, folded into higher-order chromatin fibres influences several aspects of DNA metabolism such as gene expression, DNA repair and recombination (Kornberg and Lorch, 1999). The fundamental unit of chromatin, the nucleosome, consists of 146 base pairs (bp) of DNA wrapped around a histone octamer containing two H2A/H2B heterodimers and two H3 and H4 dimers (Luger et al, 1997). Histone-modifying enzymes regulate nucleosome functions by adding or removing a large variety of covalent modifications, mainly on the N-terminal regions of histones (Peterson and Laniel, 2004). Chromatin structure has been proposed to be regulated by histone tail recognition modules, binding acetylated lysine such as bromodomains, or methylated lysine such as plant homeobox domain (PHD) and chromodomains (CHD) (Dhalluin et al, 1999; Sims and Reinberg, 2006). Histone methylation has received a lot of interest, as it marks active and inactive chromatin. In Saccharomyces cerevisiae, the histone methyl transferases Set1, Set2 and Dot1, catalysing H3K4, H3K36 and H3K79 methylation (H3K4me, H3K36me and H3K79me), respectively, have been associated with active transcription and euchromatin (Santos-Rosa et al, 2002; Strahl et al, 2002; Krogan et al, 2003). Remarkably, these histone methylations are conserved throughout evolution. In contrast to Dot1, Set1- and Set2-dependent histone methylations are surprisingly dynamic on gene activation and have been shown to be involved in early and late transcription processes (Santos-Rosa et al, 2003; Morillon et al, 2005). Set2 associates with RNA polymerase-II (RNAPII) carboxy-terminal domain (CTD) and marks the 3′ region of active genes (Li et al, 2003; Xiao et al, 2003). The main function of Set2 is to control transcription fidelity by preventing spurious transcription from the hidden promoter. Indeed, H3K36me3 is recognized by the histone deacetylase complex (HDAC) RPD3S, through Eaf3 and Rco1 modules (Joshi and Struhl, 2005; Keogh et al, 2005; Li et al, 2007). Subsequently, histone H3 and H4 de-acetylation, catalyzed by Rpd3, prevents inappropriate nucleosome displacement at the 3′ end of genes and maintains the dormancy of cryptic promoters embedded in coding regions (Carrozza et al, 2005).
Similar to Set2, Set1 associates with the RNAPII-CTD and its activity is controlled by factors found in the SET1C complex (Dehe and Geli, 2006). The seven subunits of SET1C are important either for its integrity or for controlling, specifically, H3K4 tri-, di- and mono-methylation (H3K4me3, H3K4me2 and H3K4me1). Among them Spp1 has an important function in maintaining high levels of H3K4me3, whereas Sdc1 is necessary for H3K4me3 and H3K4me2. In addition, trans-tail communication regulates H3K4me. H2B ubiquitylation and H3R2 methylation are required to control H3K4me3 (Briggs et al, 2002; Kirmizis et al, 2007; Lee et al, 2007; Vitaliano-Prunier et al, 2008). H3K4me3 is mainly found at the 5′ end of genes, consistent with high levels of Set1 at the promoter proximal regions of active genes (Ng et al, 2003). The H3K4me3 function is still poorly characterized, but has been linked with transcription initiation, elongation and RNA processing (Santos-Rosa et al, 2003; Sims et al, 2007; Vermeulen et al, 2007). Surprisingly, H3K4me2 is found throughout the coding region on transcribed gene, whereas H3K4me1 is excluded from the 5′ end of active genes to accumulate at the 3′ end (Morillon et al, 2005; Shahbazian et al, 2005). In agreement with its positive role in transcription, several active transcription-related factors have been shown to interact with H3K4me3 in higher eukaryotes such as CHD1, TAF1 and NURF (Flanagan et al, 2005; Wysocka et al, 2006; Vermeulen et al, 2007). Despite the general idea that Set1 activity is associated with transcription, some evidence suggested that Set1-dependent histone methylation might be involved in gene silencing. First, a genetic screen in Caenorhabditis elegans identified homologs of Set1 that are involved in ncRNA-mediated gene silencing (Ketting and Plasterk, 2000). Second, reports showed that H3K4me3 is recognized by ING2, subunit of a HDAC, to repress Cyclin D gene in mammalian cells (Pena et al, 2006; Shi et al, 2006). Finally, our results showed that trans-acting regulatory ncRNAs silence gene expression through Set1-dependent histone H3K4me2/3 in S. cerevisiae (Berretta et al, 2008). In addition to RNA-mediated gene silencing, Set1 plays a repressive role on PHO5, PHO84 and GAL1 expression in yeast, suggesting that H3K4me could create a repressive chromatin configuration (Carvin and Kladde, 2004).
In this work, we investigated the role of Set1 on gene repression in S. cerevisiae. Using mutations disabling the SET1C activity and H3K4 point mutation, we showed that H3K4me2/3 are repressive marks for GAL1 induction, inhibiting TBP and RNAPII recruitment to the GAL1 promoter. Furthermore, we showed the existence of long cryptic unstable RNAPII transcripts initiating upstream of GAL1, which are produced upon GAL1 repression and controlled by the Reb1 transcription factor. Inactivating the GAL1ucut cryptic transcription led to a large reduction of H3K4me3 at the GAL10-GAL1 locus. A candidate screen for PHD domain-containing factors, showed that Rco1, a subunit of the HDAC RPD3S (Li et al, 2007), attenuates GAL1 induction. Consistent with this result, we showed that ‘cryptic' H3K4me2/3 at the GAL10-GAL1 locus tethers Rpd3 to delay gene induction. In an attempt to generalize our observation to inducible genes known to contain cryptic transcription, we found that the Rpd3 occupancy is mediated by H3K4me2/3 at the PHO5, IMD2 and SUC2 promoter proximal regions. Importantly, Set1 and Rpd3 impaired the usage of a hidden SUC2 promoter, suggesting a general role for H3K4me2/3 in transcription fidelity. Altogether, our data support a model in which, cryptic transcription can generate a repressive chromatin configuration on RNAPII promoters to control transcription initiation.
Results
Set1-dependent H3K4me2/3 attenuate GAL1 mRNA accumulation
To study the repressive role of Set1 in transcription, we generated mutations affecting the Set1-dependent H3K4me2/3 and analyzed the kinetics of GAL1 induction. The GAL1 gene is located in the GAL10-GAL1 locus, where the two genes share the same promoter (Campbell et al, 2008). The GAL10 and GAL1 genes are controlled by carbon sources in the growth media, being repressed by glucose, poised by raffinose and transcriptionally activated by galactose. The different states of the GAL10-GAL1 transcriptional activity are the result of the interplay between the Gal80, Gal3 and Gal4 factors. Among the mutations affecting Set1, partial N-terminal truncations have been shown to reduce the levels of H3K4me2 and H3K4me3 when expressed from a multicopy plasmid (Schlichter and Cairns, 2005). To get robust results, we designed a strain labelled set1(463–1080), expressing the N-terminal truncated version of Set1 from its endogenous promoter. Western blot and chromatin immunoprecipitation (ChIP) experiments confirmed that the N-terminal part of Set1 was required for H3K4me2 and H3K4me3 (Supplementary Figure 1A and B). Then, we extracted total RNA from WT and set1(463–1080) strains. After probing with the GAL1 sense riboprobe P1 (Figure 2A), we observed that accumulation of GAL1 RNA in set1(463–1080) at 60 min of induction, was 3.5-fold higher than in WT (Figure 1A and D). This observation suggests that H3K4me3 and/or H3K4me2 attenuated the GAL1 induction.
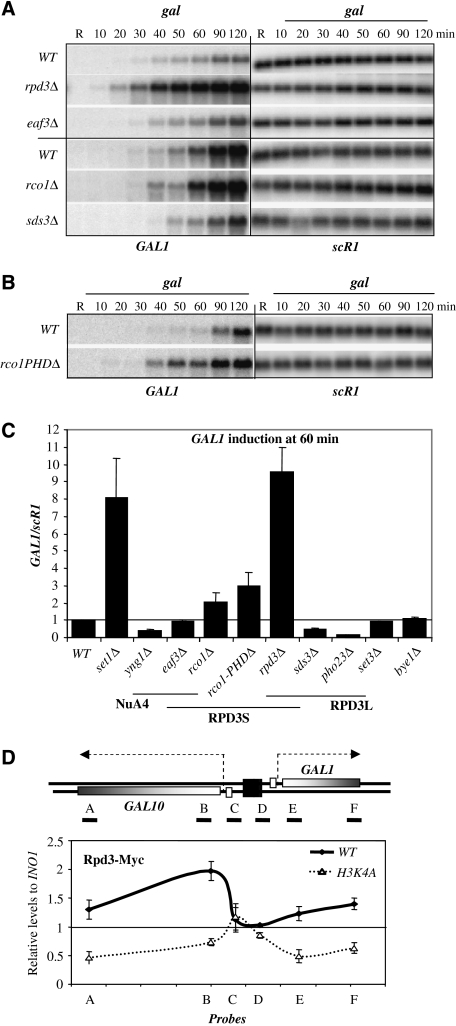
Figure 2.
Set1-dependent H3K4me2/3 control GAL1 transcription initiation. (A) Schematic view of the GAL10-GAL1 locus with positions of probes (P1, P2 and P3) and amplicons (A, B, C, D, E and F) for northern blot and real-time PCR, respectively. (B) Set1 and Sdc1 affect RNAPII occupancy on GAL1 gene. Chromatin immunoprecipitation (ChIP) experiments were carried out with an anti-RNAPII antibody in WT (YAM1242), set1Δ (YAM1243) and sdc1Δ (YAM1237) strains after transfer in galactose media (times in minutes). Amplicons correspond to E and F probes. Results are presented as percentages of input normalized with the 3′ end of RPO21 gene. (C) Set1 and Sdc1 regulate TBP-HA occupancy on GAL1 promoter. Same experiment as in (B), but with an anti-HA antibody. Amplicon corresponds to the D probe. Percentages of input were normalized with a tRNA region (tf(GAA)P2 in ChrXVI).
Figure 1.
Set1-dependent histone H3K4me2/3 attenuate GAL1 activation. Northern blot experiments with total RNA extracted from (A). WT (YAM908), set1Δ (YAM249), set1(463–1080) (YAM912), (B) WT (YAM92), set1Δ (YAM249), spp1Δ (YAM804), sdc1Δ (YAM800) and set2Δ (YAM678) and set1Δset2Δ (YAM623) and (C) with WT (YAM212), H3K4A (YAM216) and H3K36A (YAM215) strains. scR1 RNA is a loading control and GAL1 has been probed with P1 probe (see Figure 2A). Time of induction is indicated in minutes after a shift from glucose (G) to raffinose (R) and galactose (gal) containing media. (D) Quantification of GAL1/scR1 levels at 60 min of induction. GAL1/scR1 levels of WT have been arbitrary set to 1. Error bars represent the standard deviations of at least three independent experiments.
To distinguish between H3K4me2 and H3K4me3 and determine whether the regulation was restricted to Set1-dependent methylation, we performed similar experiments with WT, spp1Δ, sdc1Δ, set1Δ, set2Δ and set1Δset2Δ strains. The sdc1Δ and set1Δ strains showed 6- to 8-fold increase of GAL1 levels, respectively, when compared with the WT, whereas the spp1Δ strain had a milder effect with a fourfold increase (Figure 1B and D). In contrast, the set2Δ strain showed no change of GAL1 RNA accumulation at 60 min of induction (Figure 1B and D), and the kinetics of induction of set1Δset2Δ strain was similar to the set1Δ strain, showing that H3K36me has no function in GAL1 regulation. The induction folds of spp1Δ and sdc1Δ strains, affecting H3K4me3 only and both H3K4me2 and H3K4me3, respectively, point out that the two states of H3K4 methylation are important for the GAL1 regulation. As it is difficult to distinguish between the effects of H3K4me2 and H3K4me3, we will refer to both states of methylation as H3K4me2/3 in the remainder of the paper.
To confirm that histone methylation was indeed involved in the GAL1 regulation, we generated strains with H3K4A and H3K36A mutations, lacking methylation at the respective lysines. In the H3K4A mutant, the accumulation of GAL1 RNA was 2.5-fold higher than in WT at 60 min of induction (Figure 1C and D). In contrast, GAL1 levels were similar in H3K36A and WT strains, confirming that H3K36me was not involved in GAL1 regulation. Analyses of the 120-min-late time point of induction showed that the strains defective for H3K4me2/3 finally accumulated equivalent levels of GAL1 as the WT (Supplementary Figure 2), suggesting that Set1-dependent regulation controls only the rate of activation. Finally, we observed similar profiles for GAL10 induction in the different mutants (data not shown), suggesting that the Set1-dependent regulation exerts an effect in the same manner on GAL10 and GAL1, putatively through their shared promoter. From these data, we concluded that H3K4me2/3 marks can attenuate directly or indirectly the GAL10 and GAL1 RNA accumulation.
Set1-dependent H3K4me2/3 attenuate GAL1 transcription initiation
As the GAL1 RNA accumulation is the outcome of RNA degradation and synthesis, we asked whether the Set1-dependent histone methylation affects the GAL1 RNA stability or the GAL1 transcription. To discriminate between these two possibilities, our first strategy was to induce GAL1 transcription and then to arrest RNAPII activity to measure the GAL1 RNA stability in WT and set1Δ strains (Supplementary Figure 3A). Inactivating the main subunit of RNAPII by the use of rpb1-1 strain (Nonet et al, 1987) showed similar half-lives of the GAL1 RNA in set1Δ (27 min) and WT strains (34 min) (Supplementary Figure 3B and D), indicating that the GAL1 RNA was not stabilized in set1Δ strain but even slightly more unstable.
To test whether GAL1 transcription is regulated by Set1, we carried out ChIP experiments using RNAPII antibody and primers in the GAL1 gene (Figure 2A). The WT strain showed a twofold increase of RNAPII occupancy at 60 min in galactose, when compared with the non-induced condition (Figure 2B). In contrast, set1Δ and sdc1Δ strains showed a 10-fold increase of RNAPII levels at 60 min of induction. The levels of RNAPII occupancy remained high in sdc1Δ and set1Δ strains after 120 min of induction, but the difference with the WT levels decreased. This observation is consistent with the kinetics of GAL1 RNA and confirms that Set1 activity delayed the GAL1 transcription. Similar results were obtained with the H3K4A mutant but not with the H3K36A strain (Supplementary Figure 4A), showing that only H3K4me2/3 marks attenuate transcription of the GAL1 gene.
To determine whether GAL1 transcription was controlled at the initiation level, we measured by ChIP the levels of TBP-HA at the GAL10-GAL1 promoter (probe D). At 60 min, we observed a twofold increase of TBP-HA levels in WT, whereas set1Δ and sdc1Δ strains showed a 13- and 10-fold increase, respectively (Figure 2C). After 120 min of induction, the TBP–HA occupancy reached the same levels in WT, sdc1Δ and set1Δ strains, suggesting that the Set1 activity delayed the TBP occupancy. Furthermore, set1Δ and WT strains maintained identical levels of Gal4 during the kinetics of induction (Supplementary Figure 4B), ruling out an indirect effect of H3K4me2/3 on Gal4 binding. We propose that H3K4me2/3 control the pre-initiation complex (PIC) formation at the GAL1 promoter.
The cytoplasmic 5′–3′ RNA decay pathway degrades cryptic long unstable RNAs initiating upstream of GAL1
The repressive activity of Set1 is largely uncharacterized. Our recent data linked Set1-dependent H3K4me2/3 with RNA-mediated gene silencing (Berretta et al, 2008). To determine whether Set1 could control GAL1 transcription through an RNA-dependent mechanism, we tested the existence of cryptic unstable transcripts, which could regulate the GAL1 transcription. As this work was in progress, Houseley et al. (2008) showed the existence of unstable antisense GAL10 ncRNAs, reinforcing a model linking Set1 activity and ncRNA-mediated regulation of the GAL10-GAL1 locus. To facilitate the detection of putative regulatory RNAs, we probed total RNA extracted from xrn1Δ strain defective for the cytoplasmic 5′–3′ decay (Long and McNally, 2003) and from a trf4Δ strain defective for the nuclear surveillance 3′–5′ pathway (Wyers et al, 2005). Interestingly, with a GAL10-GAL1 double-stranded DNA probe (probe P2 in Figure 2A), we observed a significant stabilization of three species (A, B and C) of unstable RNAs ranging from 2, 4 and 6 kilobases (kb) long in the xrn1Δ strain upon repression (Figure 3A). Characterization of these unstable RNAs with different probes spanning the GAL10-GAL1 locus showed that they are transcribed through GAL10-GAL1 promoters in antisense orientation to the GAL10 gene. These RNAs were subsequently called GAL1 upstream cryptic unstable transcripts (GAL1ucut). 5′RACE experiments established that the main start site of the GAL1ucut is at +572 nucleotides from the GAL10 stop codon (Figure 3B), corresponding to one of the initiation sites determined for the GAL10 antisense ncRNA (Houseley et al, 2008). As shown earlier, the 3′ end of GAL10 contains a perfect consensus motif (TTACCCG) for the binding of Reb1 transcription factor (Morrow et al, 1989) at +380, and we identified two perfect TATA boxes (+159 and +538) in this region (Figure 3B). To test whether Reb1 could control GAL1ucut transcription, we used a reb1-1 thermosensitive degron strain (Ben-Aroya et al, 2008) in which XRN1 has been deleted and measured the presence of the GAL1ucut RNAs. After three hours at 37°C, we observed a two- to fourfold decrease of the three forms of GAL1ucut in the reb1-1xrn1Δ strain when compared with the xrn1Δ strain (Figure 3C). Remarkably, the Ty1 antisense regulatory ncRNA RTL, also sensitive to Xrn1 (Berretta et al, 2008), did not show any decrease on Reb1 depletion, showing that Reb1 is required for the synthesis of the three GAL1ucut RNAs specifically.
Figure 3.
Reb1-dependent cryptic transcription generates GAL1 upstream transcripts mainly destabilized by Xrn1. (A) Xrn1 exoribonuclease destabilizes GAL1ucut RNAs. Northern blot experiments with total RNA extracted from WT (YAM92), xrn1Δ (YAM97) and trf4Δ (YAM456) grown in glucose or galactose (gal) for 3 h. scR1 RNA is a loading control and GAL1ucut, GAL1and GAL10 have been probed with P2 probe (see Figure 2A). The three species of GAL1ucut (A, B and C) are labelled with arrows. (B) Schematic representation of the different RNAs detected at the GAL10-GAL1 locus and determination of the main starting site of the GAL1ucut (+572 from GAL10 stop codon). Classic and putative TATA boxes are represented by white boxes and the Reb1 binding site (Reb1BS) is indicated. (C) GAL1ucut A, B and C are controlled by Reb1. Same as in (A), but with WT (YAM1), xrn1Δ (YAM6), reb1-1 (YAM1591) and xrn1Δreb1-1 (YAM1650), grown at 30°C in glucose and transferred to 37°C during 3 h. Ratios of the different GAL1ucut and RTL ncRNA are indicated (Berretta et al, 2008). (D) GAL1ucut RNA transcription attenuates GAL1 and GAL10 induction. Same as in Figure 1A, but RNA extracted from WT (YAM1) and reb1-1 (YAM1591) strains. Cells were grown at 30°C in glucose, diluted in raffinose containing media for 2 h at 37°C then transferred in galactose containing media at 37°C and RNA analyzed at the indicated time (min). The GAL1 and GAL10 RNAs were detected with the P2 probe.
To test whether GAL1ucut transcription could attenuate GAL1 induction, we carried out kinetics analyses of GAL1 induction in the absence of Reb1. Northern-blot experiments and GAL1/ScR1 quantifications (Figure 3D and E) showed that GAL1 and GAL10 inductions were accelerated when Reb1 was depleted, supporting the idea that Reb1 transcription factor plays a repressive role on GAL10 and GAL1 expression.
Altogether these data reveal the existence of several cryptic transcripts, starting upstream of GAL1 gene, that are mainly degraded by the 5′–3′ cytoplasmic degradation pathway. Importantly, we show that Reb1 is required for the GAL10-GAL1 cryptic transcription and controls GAL1 and GAL10 induction, either by a transcription interference mechanism or in an RNA-dependent manner.
Accumulation of the GAL1ucut does not control GAL1 induction
To determine whether GAL1 induction is attenuated by the GAL1ucut RNA accumulation, we carried out kinetics studies in xrn1Δ and xrn1Δtrf4Δ strains mutated for the 5′–3′ and/or the 3′–5′ RNA decay pathways (Figure 4A). Using a GAL1 sense riboprobe detecting both GAL1 and GAL1ucut RNAs (probe P1), we made two key observations. First, we confirmed that the GAL1ucut B is the most abundant form of GAL1ucut RNA detected in xrn1Δ strain (Figure 4A). Furthermore, the xrn1Δtrf4Δ strain showed accumulation of the GAL1ucut C, suggesting that GAL1ucut C is sensitive to both RNA decay pathways. Importantly, GAL1ucut B and C species were detectable mainly in repressive or non-induced conditions and their levels decreased on induction, suggesting that the accumulation of GAL1ucut and GAL1 RNAs is antagonistic. Second, the GAL1 levels were similar in WT and xrn1Δ strains and only 1.5 fold reduced in xrn1Δtrf4Δ strain at 60 min of induction (Figure 4A and B). These results strongly suggest that stabilizing the GAL1ucut B and C RNAs does not attenuate the GAL1 induction.
Figure 4.
GAL1 upstream transcripts are not Set1 dependent. (A) The GAL1ucut RNAs do not control GAL1 induction. Same as in Figure 1A, but with WT (YAM92), xrn1Δ (YAM97) and xrn1Δtrf4Δ (YAM458). GAL1ucut are labelled with an arrow and the GAL1 read through RNA (GAL1RT) with an asterisk. (B) Quantification of GAL1/scR1 levels at 60 min of induction. Same as in Figure 1C. (C) GAL1ucut RNAs are synthesised upon repressive conditions, destabilized by Xrn1 but not controlled by Set1. Reverse transcriptions were performed with the GAL1UAS primer P3 (position in Figure 2A) and amplified by PCR with amplicon D of GAL1 UAS region. Data were normalized with scR1 RNA. WT (YAM92), xrn1Δ (YAM97), set1Δ (YAM249) and xrn1Δset1Δ (YAM448) strains were grown in glucose, transferred during 2 h in raffinose and shifted to galactose containing media. The samples were extracted at the indicated time (min).
From these data, we concluded that the GAL10-GAL1 locus is transcribed on repressive conditions, but that the produced GAL1ucut RNAs do not interfere with GAL1 induction. An attractive hypothesis is that the GAL1ucut transcription per se could regulate the GAL1 promoter through the Set1-dependent activity. Alternatively, Set1 could control the GAL1ucut transcription to repress GAL1 promoter through a transcriptional interference mechanism.
Set1 is not involved in GAL1ucut transcription
To distinguish between these two hypotheses, we first aimed to determine whether Set1 plays a role in the GAL1ucut transcription. We carried out high sensitive reverse-transcription experiments coupled with quantitative-PCR analyses on total RNAs extracted from WT, xrn1Δ, set1Δ and set1Δxrn1Δ strains. Using amplicons in the GAL1 UAS (probe P3), we detected low but significant levels of GAL1ucut in WT cells grown in glucose and raffinose containing media (Figure 4C). Consistent with our data in Figure 4A, GAL1ucut levels showed a threefold decrease when cells were transferred in the inducible galactose-containing media, confirming that GAL1 is fully active when GAL1ucut are not produced anymore. More importantly, the levels of GAL1ucut were 10-fold higher in the xrn1Δ strain than in the WT strain in glucose, confirming a key role of the 5′ end cytoplasmic RNA decay in GAL1ucut stability. Furthermore, deletion of SET1 did not affect the levels of GAL1ucut RNA when compared with the WT and xrn1Δ strains in glucose-containing media, suggesting that Set1 was not required for the synthesis of the GAL1ucut. In addition, we observed that GAL1ucut levels rapidly decreased upon induction in the set1Δxrn1Δ strain when compared with the xrn1Δ strain. This observation is consistent with a rapid induction of GAL1 when Set1 is inactivated. We concluded that Set1 controls GAL1 induction but not the GAL1ucut transcription. Instead, an interesting idea is that the cryptic transcription would deposit H3K4me2/3 marks to attenuate GAL1 induction.
Cryptic RNAPII transcription and Reb1 transcription factor are required for H3K4me2/3 deposition on GAL10-GAL1 region on repressive conditions
To test whether cryptic transcription was required for the deposition of H3K4me2/3 at the GAL10-GAL1 locus, we carried out ChIP with chromatin extracted from WT, set1Δ and rpb1-1 strains grown in glucose or galactose media. Using antiH3K4me3 antibody, we observed that, after 1 h at 37°C in glucose conditions, H3K4me3/H3 ratios in WT were 14-fold higher than those in the set1Δ strain and peaked at the 3′ end of GAL10 (probe A, Figure 5A and B). Interestingly, along the rest of the locus, H3K4me3 showed a two- to threefold difference with the threshold defined by the set1Δ strain (probes B, C, D, E and F), suggesting that Set1 maintained high levels of H3K4me3 in repressive conditions mainly at the 3′ end of GAL10. Importantly, the peak of H3K4me3 at the 3′ end of GAL10 is consistent with the start site of the GAL1ucut in this region. Strikingly, the rpb1-1 mutant showed almost no signal of H3K4me3/H3 from the beginning to the end of the GAL10-GAL1 locus (Figure 5B), suggesting that cryptic active transcription was required to maintain detectable levels of H3K4me3 at the GAL10-GAL1 locus in glucose. To test whether the peak of H3K4me3 at the 3′ end of GAL10 resulted from a leakage of the GAL10 promoter, we induced GAL1-GAL10 transcription and measured the H3K4me3 occupancy. As anticipated, the H3K4me3 re-localized to the 5′ end of GAL10 and GAL1 genes and dramatically decreased from the 3′ end region of GAL10, strongly suggesting that the peak of H3K4me3 at the 3′ end of GAL10 in glucose was not the consequence of a GAL10 promoter leakage but instead the result of an alternative transcription process.
Figure 5.
RNAPII- and Reb1-dependent cryptic transcription control residual H3K4me3 and H3K4me2 at the GAL10-GAL1 locus. (A) Schematic view of the GAL10-GAL1 locus with positions of amplicons for real-time PCR. (B) Cryptic transcription deposits H3K4me3 on GAL10-GAL1 locus in repressive condition. Chromatin immunoprecipitation (ChIP) experiments were performed with an anti-H3K4me3 antibody in WT (YAM15) and rpb1-1 (YAM268) and set1Δ (YAM1494) strains grown in glucose-containing media at 28°C, then shifted to 37°C in 1 h. H3K4me3 signals were normalized with H3 signals performed with an anti-H3 antibody on the same chromatin. Amplicons correspond to A, B, C, D, E and F shown in (A). Results are presented as relative levels of H3K4me3/H3 to those measured in set1Δ strain (WT/set1Δ and rpb1-1/set1Δ). (C) Cryptic transcription deposits H3K4me2 on GAL10-GAL1 locus in repressive condition. Same as in (B), but with an anti-H3K4me2 antibody. (D) Reb1 controls H3K4me3 on the GAL10-GAL1 locus. Same as in (B), but with WT (YAM1) and reb1-1 (YAM1591) strains. Cells were grown in glucose containing media at 28°C then shifted to 37°C in 3 h.
We next performed the same experiment with anti-H3K4me2 antibody (Figure 5C). First, similarly to H3K4me3/H3, H3K4me2/H3 peaked at the 3′ end of GAL10 (probe A) but remained detectable along the rest of the locus with signals up to 20-fold above the baseline defined by those measured in the set1Δ strain (probes B, C, D, E and F). Second, H3K4me2 levels showed a 10- to 2-fold decrease in the GAL10 coding region in the rpb1-1 strain (probes A and B, respectively), suggesting that RNAPII-dependent transcription was also required for H3K4me2 deposition at GAL10. Surprisingly, the levels of H3K4me2 remained unchanged over the rest of the locus in the rpb1-1 strain (probe C, D, E and F) when compared with the WT. This observation is in contrast to the decrease of H3K4me3 levels in this region, suggesting that the H3K4me2 demethylation process might be less efficient than H3K4me3 demethylation after transcription arrest. In addition, galactose induction led to a four fold decrease of H3K4me2 over the 3′ end region of GAL10 and showed constant but high levels over the GAL1-GAL10 locus, again reinforcing the idea that the peak of H3K4me2/3 at the 3′ end of GAL10 was not because of GAL10 promoter leakage in glucose.
To determine whether the H3K4me3 at the 3′ end of GAL10 in glucose was the result of alternative transcription, we depleted Reb1 in vivo, abolishing GAL1ucut synthesis, and measured the H3K4me3/H3 levels. ChIP experiments (Figure 5D) showed that inactivation of Reb1 resulted in a threefold decrease of H3K4me3 levels at the 3′ end of GAL10, showing that Reb1 indeed controls the GAL1ucut transcription mediating the H3K4me3 deposition onto GAL10 chromatin in glucose conditions.
Altogether these data argue that cryptic transcription controls the presence of H3K4me2/3 marks at the GAL10-GAL1 locus in repressive conditions.
The HDAC RPD3S attenuates GAL1 induction and is tethered on GAL10-GAL1 locus by H3K4me2/3
To understand how H3K4me2/3 regulate GAL1 transcription, we systematically screened for factors containing PHD domains, which could bind H3K4me2/3 in vitro (Shi et al, 2007). One of them is implicated in transcriptional elongation (Bye1); the others are part of histone acetylation (Yng1) and histone deacetylation (Rco1, Set3 and Pho23) complexes (Wang et al, 2002; Wu et al, 2003; Martin et al, 2006). Interestingly, among these factors, Pho23 and Rco1 belong to the distinct HDAC RPD3L and RPD3S complexes, respectively (Carrozza et al, 2005).
Our approach was to follow the GAL1 induction in strains mutated for these different PHD-factors and to discriminate between the RPD3S and RPD3L complexes. Thus, total RNAs were extracted from rpd3Δ, eaf3Δ, rco1Δ, sds3Δ and pho23Δ strains lacking subunits of the RPD3S and RPD3L complexes, and from bye1Δ, yng1Δ and set3Δ strains. The rpd3Δ and rco1Δ strains showed an acceleration of the GAL1 kinetics by eight- and two-fold at 60 min of induction, respectively, when compared with WT (Figure 6A and C). In contrast, strains deleted for genes encoding subunits of the RPD3L, such as pho23Δ and sds3Δ strains, but also set3Δ and yng1Δ strains showed a delay in GAL1 accumulation, suggesting that RPD3L is required for GAL1 activation, as shown earlier (Wang et al, 2002). This indicates that the RPD3S complex specifically attenuates GAL1 induction. Interestingly, the eaf3Δ strain showed a similar induction as the WT despite the fact that Eaf3 belongs to the RPD3S complex. One explanation is that Eaf3, in addition to being part of the RPD3S complex, is a subunit of the histone acetyl transferase (HAT) NuA4 complex (Eisen et al, 2001) and EAF3 deletion might impair both activities and would result in no change in gene induction (Figure 6C). Finally, the late time point at 120 min of induction showed that the rpd3Δ strain had very high levels of GAL1 RNA when compared with the WT or set1Δ strains, suggesting that Rpd3 has an additional repressive activity on the GAL1 promoter efficiency (Supplementary Figure 5).
Figure 6.
RPD3S complex attenuates GAL1 induction. (A) RPD3S attenuates the GAL1 induction. Same experiment as in Figure 1A, but with WT (YAM1), rpd3Δ (YAM4) and eaf3Δ (YAM5) and WT (YAM1483), rco1Δ (YAM1489) and sds3Δ (YAM1488) strains. (B) Rco1-plant homeobox domain (PHD) is required to attenuate GAL1 induction. Same as in Figure 1A, but with WT (YAM118) and rco1PHDΔ (YAM1465). (C) Quantification of GAL1/scR1 at 60 min of induction for strains used in (A) and (B) but, also for pho23Δ (YAM1370), bye1Δ (YAM1371), yng1Δ (YAM1369) and set3Δ (YAM1302) strains. (D) H3K4me2/3 control Rpd3 occupancy at the GAL10-GAL1 locus upon repressive conditions. Chromatin immunoprecipitant (ChIP) experiments were performed as in (A), but with an anti-Myc antibody in WT (YAM1477), H3K4A (YAM1481) strains grown in glucose. Results are presented as percentage of input normalized with the promoter of INO1.
To determine more precisely the role of Rco1 in histone recognition, we analyzed the effect of the PHD domain truncation of Rco1 subunit. We observed that the rate of induction of rco1PHDΔ strain was threefold higher than the WT strain in galactose-containing media (Figure 6B and C). It is interesting to note that rco1PHDΔ strain had a stronger effect with respect to GAL1 induction than rco1Δ strain, suggesting that Rco1 could be partially complemented to attenuate GAL1 transcription. Altogether, these results suggest that the PHD domain of Rco1 was required for the Rpd3 repressive activity at the GAL10-GAL1 locus. An interesting idea is that H3K4-methylated histone tethers Rco1 in order to recruit the RPD3S complex and to repress the transcription of GAL genes.
To test this hypothesis, we measured Rpd3-Myc occupancy on the GAL10-GAL1 region in WT and H3K4A strains. Consistent with earlier genome-wide analyses of Rpd3 occupancy (Wang et al, 2002), we observed that Rpd3 was detectable at the 5′ end of GAL10 and at the 3′ end of GAL1 genes in glucose (Figure 6D). Remarkably, the H3K4A strain showed a fourfold decrease of Rpd3 levels on the GAL10-GAL1 coding regions. In contrast, Rpd3-Myc levels were similar at the GAL10-GAL1 UAS region in WT and H3K4A strains, suggesting that H3K4 methylation controls only a sub-population of Rpd3. The remaining Rpd3 might represent the RPD3L complex, which was earlier shown to play a positive role on GAL1 induction (our results in Figure 6C and Wang et al, 2002). Finally, ChIP experiments with WT and set1Δ strains confirmed that Rpd3-Myc was tethered directly by Set1 activity (Supplementary Figure 6B). To test whether Set1 controls the RPD3S specifically, we attempted to measure the occupancy of the Rco1 specific subunit over the GAL10-GAL1 locus. Unfortunately, Rco1-Myc failed to produce consistent data by ChIP experiments (data not shown). Nevertheless, our results suggest that Set1 controls GAL1 induction by tethering the repressive RPD3S complex at the GAL10-GAL1 locus.
Set1-dependent binding of Rpd3 to SUC2 promoter represses the usage of a SUC2 hidden promoter
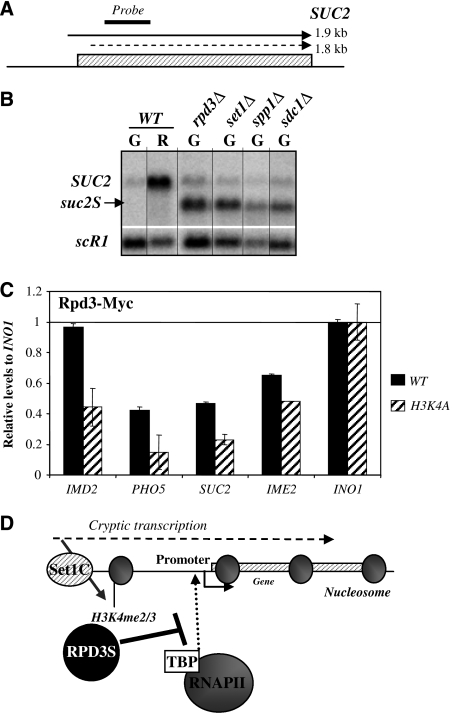
To generalize our observation to other loci in the genome, we selected a few genes of which the promoters have been shown to be recognized by Rpd3. Among them, we analyzed SUC2 RNAs levels in mutants defective for H3K4me2/3 and Rpd3 activity. The SUC2 gene encodes an invertase enzyme playing a role in carbon source metabolism (Carlson and Botstein, 1982). SUC2 is transcribed in 1.8 and 1.9 kb RNA species (schematic view in Figure 7A). The constitutive 1.8 kb SUC2 RNA is lowly expressed and results in a protein lacking the secretion signal. In contrast, the 1.9 kb SUC2 RNA is induced on glucose depletion and is translated into an active secreted protein (Carlson and Botstein, 1982). Consistently, our northern-blot experiments showed low levels of the 1.8 kb SUC2 RNA in glucose (repressed) and high levels of the 1.9 kb SUC2 RNA in raffinose (derepressed) for the WT strain (Figure 7B). We noted that the two SUC2 RNAs species co-migrated in the gel system used in this study. Strikingly, we observed a shorter SUC2 RNA species (suc2S that will be described elsewhere) in spp1Δ, sdc1Δ, set1Δ and rpd3Δ strains in repressed conditions. This suggests that Set1-dependent H3K4me2/3 inhibit the usage of a hidden promoter within the SUC2 gene, probably in a similar manner as for the GAL1 promoter, by targeting Rpd3 to the SUC2 promoter proximal region.
Figure 7.
H3K4me2/3 recruit Rpd3 to control SUC2 promoter fidelity. (A) Schematic view of the SUC2 transcripts with positions of the probes for northern blot and qPCR. (B) Set1-dependent H3K4me2/3 repress a SUC2 spurious transcript (suc2S). Northern blot with total RNAs extracted from WT (YAM1 and 92), rpd3Δ (YAM4), set1Δ (YAM249) spp1Δ (YAM804) and sdc1Δ (YAM800) strains. WT cells were grown in glucose (G) and transferred in raffinose (R) containing media. Mutants were grown in glucose containing media (G) only. scR1 is a loading control and SUC2 has been probed according to Figure 7A. (C) Rpd3-Myc is targeted to SUC2, IMD2 and PHO5 promoters and is dependent on H3K4me. Chromatin immunoprecipitant (ChIP) experiments were performed with an anti-Myc antibody in WT (YAM1477) and H3K4A (YAM1481) strains in glucose. Amplicons correspond to SUC2, PHO5, IMD2, IME2 and INO1 promoters. Results are presented as percentage of input normalized with the promoter of INO1. (D) Model of promoter attenuation by H3K4me2/3. Cryptic transcription activates the deposition of H3K4me2/3 on promoter proximal regions and triggers the recruitment of the RPD3S complex inhibiting the pre-initiation complex (PIC) formation and promoter activity.
To test our hypothesis, we measured the Rpd3-Myc occupancy on the SUC2 promoter proximal region upon repressed conditions (glucose) in WT and H3K4A strains. As controls, we probed other promoters such as IME2 and INO1 known to target the RPD3L complex through a Ume6-dependent mechanism (Kadosh and Struhl, 1997), and the PHO5 and IMD2 promoters described to be regulated by cryptic transcription (Uhler et al, 2007; Kuehner and Brow, 2008). We observed a consistent twofold reduction of Rpd3-Myc levels at SUC2, PHO5 and IMD2 promoter proximal regions in the H3K4A strain (Figure 7C) on non-induced conditions. In contrast, the IME2 promoter showed a mild 1.2-fold decrease of Rpd3-Myc in the H3K4A strain. These results suggest that Set1-dependent RPD3S recruitment could be extended to a specific class of promoters containing a cryptic transcription unit.
Discussion
With this work, we addressed the possibility of a repressive role of Set1 on gene expression. We showed that Set1-dependent H3K4me2/3 marks directly contribute to the repression mechanism of the GAL10-GAL1 promoter by tethering the RPD3S complex. H3K4me2/3 inhibit PIC formation and attenuate GAL1 induction. Importantly, we showed that Set1-dependent RPD3S recruitment might be widely generalized to regions containing cryptic transcription units and could be an important regulation process for the transcription fidelity (The model in Figure 7D).
H3K4me2 as a signalling mark for histone deacetylase recognition?
A key point of this study is the unexpected finding that H3K4me2/3 might provide a signal for tethering the RPD3S complex to repress GAL1 induction. Our mutant analysis suggests that both H3K4me2 and H3K4me3 are required for Rpd3 recruitment and gene attenuation. However, we could not distinguish which of the two marks was the main recruiting signal as published in vitro data showed that the Rco1-PHD domain is able to recognize H3K4me1, H3K4me2 and H3K4me3 (Shi et al, 2007). It is important to note, however, that Rpd3-Myc peaked in the GAL10 region where H3K4me3 levels are low, whereas H3K4me2 levels remain high, suggesting that H3K4me2 is sufficient to target Rpd3. One model is that H3K4me3 would attract another complex competing with RPD3S. This is compatible with the recognition of H3K4me3 by the NuA3 HAT complex (Taverna et al, 2006) and displacing RPD3S from H3K4me2.
Moreover, H3K4me3 controls the mSIN3 histone deacetylase complex in higher eukaryotes through the recruitment of ING2 containing a PHD domain (Shi et al, 2006). ING proteins are also present in HAT complexes, resulting in a surprising regulation where, a single histone mark tethers complexes with opposing activities (Mellor, 2006). It is tempting to speculate that, in yeast, these functions could be separated, with H3K4me2 tethering the HDAC RPD3S complexes and with H3K4me3 activity recruiting the NuA3 HAT complex to compete with RPD3S (Taverna et al, 2006).
RPD3S recognizes H3K36me or H3K4me?
Recent publications have shown that RPD3S recognizes H3K36me3 through Eaf3 and Rco1 modules (Joshi and Struhl, 2005; Keogh et al, 2005; Li et al, 2007). Subsequently, histone H3 and H4 de-acetylation, catalyzed by Rpd3, prevents inappropriate nucleosome displacement at the 3′ end of genes and maintains the dormancy of hidden promoters embedded in coding regions such as FLO8 and STE11 (Carrozza et al, 2005). Interestingly, none of those characterized spurious transcripts are produced in set1Δ, sdc1Δ or spp1Δ strains (data not shown), suggesting that Set1-dependent H3K4me2/3 are not the marks for tethering RPD3S at these regions. Houseley et al. proposed a model in which, H3K36me would be responsible for the GAL10-GAL1 promoter regulation through a similar mechanism, but we could not determine a negative role for Set2-dependent methylation for the GAL10-GAL1 induction. Instead, H3K4me2/3 marks play a crucial role in targeting RPD3S at the cryptic transcribed regions and are crucial for GAL10-GAL1 attenuation, suggesting that Set1 is important for targeting the RPD3S complex to control the hidden promoter on a specific transcriptional unit. An interesting hypothesis is that an RNA quality control process or a transcription termination-related mechanism, would exert an effect by way of distinguish between Set1- or Set2-dependent RPD3S recruitment. This idea is reinforced by the striking concomitant association to Ser5 phosphorylated RNAPII of Set1 and Nrd1/Nab3 complex, the latter triggering the degradation of unstable transcripts (Ng et al, 2003; Vasiljeva et al, 2008). Further work is necessary to understand the recognition mechanism of RPD3S in different transcriptional mode contexts.
Cryptic transcription marks an inactive promoter?
As cryptic ncRNAs produced within the genome are unstable, it has been suggested that the RNA molecules are not the regulatory aspect of this pathway (Struhl, 2007). Consistent with this model, recent works on S. cerevisiae showed that cryptic transcription could control gene expression by interfering with promoters of IMD2, PHO5, URA2 and SRG1 (Martens et al, 2004; Uhler et al, 2007; Kuehner and Brow, 2008; Thiebaut et al, 2008). In our work, we propose that Set1-dependent H3K4me2/3 deposited by the cryptic transcription is one of the signals that downregulates the promoter activity of the downstream gene. In the case of IMD2, the cryptic transcription machinery has been shown to have an important function in attenuating IMD2 expression through sensing intracellular nucleotide pools and the choice of initiation starting site (Kuehner and Brow, 2008). Here, we showed that, upon repression, the IMD2 promoter proximal region is targeted by RPD3S mediated by H3K4me2/3, reinforcing the link between cryptic transcription and Set1 activity. However, in the case of PHO5, even though Set1 represses the gene activity probably through Rpd3 recruitment, the characterized cryptic transcription has a positive role on PHO5 induction (Carvin and Kladde, 2004; Uhler et al, 2007). One explanation to this discrepancy is that the Set1-dependent regulation of PHO5 gene would be independent of the cryptic transcription. Alternatively, it remains possible that another uncharacterized cryptic transcription that we predict to be embedded in the PHO5 promoter could deposit the H3K4me2/3 repressive marks. Interestingly, H3K4me2/3 marks also repress a SUC2 spurious transcription and control Rpd3 recruitment on the SUC2 promoter. Our model proposes that a SUC2 cryptic transcription, that needs to be properly characterized, might deposit the H3K4me2/3 marks at the proximity of the SUC2 promoter to prevent inappropriate usage of a hidden promoter. Further investigation will determine the features of these transcripts.
Materials and methods
Yeast strains
The strains used in this study are described in Supplementary Table 1.
Media and culture conditions
Typically, all inductions have been performed with cultures grown overnight in YPA (Gibco) containing 2% glucose, diluted at OD 0.5 in 2% raffinose media. After 2 h, cells were transferred in 2% galactose-containing media.
RNA analyses
RNAs were analyzed following the described procedure (Berretta et al, 2008) and the mean of at least three independent experiments. The 5′RACE experiments were carried out as described earlier (Berretta et al, 2008) and following the manufacturer's instructions (kit Ambion).
Chromatin immunoprecipitation
ChIPs were carried out as described earlier (Berretta et al, 2008) using antibodies against H3K4me2, H3K4me3, H3, Gal4p (Abcam), the N-terminal domain of Rpb1 (Santacruz, Y80), HA (Santacruz, fp7) and Myc (Roche). Primer pairs are listed in Supplementary Table 2. Signals are expressed as % of input DNA relative to a specific region. Error bars correspond to standard deviations of 2–3 independent experiments.
Supplementary Material
Supplementary Text
Supplementary Information
Acknowledgments
We thank V Géli, J Workman, S Berger and D Stillman for providing strains; D Tollervey and F Stutz for communicating results before publications; L Kuras for fruitful suggestions; and the members of the Morillon's group for their critical reading of the paper; Special thanks are due to M Kwapisz for technical tips and comments. MP is a fellow of CNRS. Financial support has been provided by ANR blanc (REGULncRNA), ARC, FRM and HFSPO.
References
- Ben-Aroya S, Coombes C, Kwok T, O'Donnell KA, Boeke JD, Hieter P (2008) Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol Cell 30: 248–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta J, Pinskaya M, Morillon A (2008) A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev 22: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD (2002) Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Campbell RN, Leverentz MK, Ryan LA, Reece RJ (2008) Metabolic control of transcription: paradigms and lessons from Saccharomyces cerevisiae. Biochem J 414: 177–187 [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D (1982) Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28: 145–154 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Carvin CD, Kladde MP (2004) Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1-10. J Biol Chem 279: 33057–33062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehe PM, Geli V (2006) The multiple faces of Set1. Biochem Cell Biol 84: 536–548 [DOI] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491–496 [DOI] [PubMed] [Google Scholar]
- Eisen A, Utley RT, Nourani A, Allard S, Schmidt P, Lane WS, Lucchesi JC, Cote J (2001) The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J Biol Chem 276: 3484–3491 [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S (2005) Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438: 1181–1185 [DOI] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M (2008) A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell 32: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell 20: 971–978 [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K (1997) Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371 [DOI] [PubMed] [Google Scholar]
- Ahn SH, Boone C, Buratowski S, Chin K, Collins SR, Emili A, Greenblatt JF, Grunstein M, Hughes TR, Keogh MC, Krogan NJ, Kurdistani SK, Morris SA, Podolny V, Punna T, Schuldiner M, Strahl BD, Thompson NJ, Weissman JS (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Plasterk RH (2000) A genetic link between co-suppression and RNA interference in C. elegans. Nature 404: 296–298 [DOI] [PubMed] [Google Scholar]
- Bahler J, Green RD, Kirmizis A, Kouzarides T, Mann M, Penkett CJ, Santos-Rosa H, Singer MA, Vermeulen M (2007) Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449: 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y (1999) Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome [In Process Citation]. Cell 98: 285–294 [DOI] [PubMed] [Google Scholar]
- Boateng MA, Dean K, Dover J, Golshani A, Greenblatt JF, Heidt J, Johnston M, Krogan NJ, Ryan OW, Schneider J, Shilatifard A, Wood A (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Brow DA, Kuehner JN (2008) Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell 31: 201–211 [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Florens L, Lee JS, Schneider J, Shilatifard A, Shukla A, Swanson SK, Washburn MP (2007) Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL (2007) Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Howe L, Anderson S, Yates JR III, Workman JL (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 278: 8897–8903 [DOI] [PubMed] [Google Scholar]
- Long RM, McNally MT (2003) mRNA decay: x (XRN1) marks the spot. Mol Cell 11: 1126–1128 [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260 [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F (2004) Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L (2006) The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol 26: 7871–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J (2006) It takes a PHD to read the histone code. Cell 126: 22–24 [DOI] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, Nair A, Mellor J (2005) Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol Cell 18: 723–734 [DOI] [PubMed] [Google Scholar]
- Morrow BE, Johnson SP, Warner JR (1989) Proteins that bind to the yeast rDNA enhancer. J Biol Chem 264: 9061–9068 [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R (1987) Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol 7: 1602–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG (2006) Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442: 100–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA (2004) Histones and histone modifications. Curr Biol 14: R546–R551 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bernstein BE, Karabetsou N, Morillon A, Weise C, Schreiber SL, Mellor J, Kouzarides T (2003) Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol Cell 12: 1325–1332 [DOI] [PubMed] [Google Scholar]
- Schlichter A, Cairns BR (2005) Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. Embo J 24: 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M, Zhang K, Grunstein M (2005) Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Molecular Cell 19: 271–277 [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF et al. (2006) ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O (2007) Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem 282: 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ III, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D (2007) Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ III, Reinberg D (2006) Histone H3 Lys 4 methylation: caught in a bind? Genes Dev 20: 2779–2786 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, Allis CD (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22: 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K (2007) Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 14: 103–105 [DOI] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell 24: 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut M, Colin J, Neil H, Jacquier A, Seraphin B, Lacroute F, Libri D (2008) Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell 31: 671–682 [DOI] [PubMed] [Google Scholar]
- Uhler JP, Hertel C, Svejstrup JQ (2007) A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci USA 104: 8011–8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A (2008) The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 15: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Menant A, Hobeika M, Geli V, Gwizdek C, Dargemont C (2008) Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol. 10: 1365–1371 [DOI] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M (2002) Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298: 1412–1414 [DOI] [PubMed] [Google Scholar]
- Wu X, Rossettini A, Hanes SD (2003) The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA pol II to inhibit transcription elongation in Saccharomyces cerevisiae. Genetics 165: 1687–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A (2005) Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev 17: 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Supplementary Information